Abstract
The fluids of the intestine serve as a physical barrier to pathogens, a medium for the diffusion of nutrients and metabolites, and an environment for commensal microbes. The rheological properties of intestinal mucus have therefore been the subject of many investigations, thus far limited to in vitro studies due to the difficulty of measurement in the natural context of the gut. This limitation especially hinders our understanding of how the gut microbiota interact with the intestinal space, since examination of this calls not only for in vivo measurement techniques, but for techniques that can be applied to model organisms in which the microbial state of the gut can be controlled. We have addressed this challenge with two complementary approaches. We performed passive microrheological measurements using thermally driven nanoparticles and active microrheology using micron-scale ellipsoidal magnetic microparticles, in both cases using light-sheet fluorescence microscopy to optically access the intestinal bulb of the larval zebrafish, a model vertebrate. We present viscosity measurements in germ-free animals (devoid of gut microbes), animals colonized by a single bacterial species, and conventionally reared animals, and find that in all cases, the mucin-rich intestinal liquid is well described as a Newtonian fluid. Surprisingly, despite known differences in the number of secretory cells in germ-free zebrafish and their conventional counterparts, the fluid viscosity for these two groups is very similar, as measured with either technique. Our study provides, to our knowledge, the first in vivo microrheological measurements of the intestinal space in living animals, and we comment on its implications for timescales of host-microbe interactions in the gut.
Introduction
The rheological properties of the materials that make up animals are important to physiology (1, 2), development (3), the transport of drugs and biomolecules (4, 5, 6), and the activities of commensal and pathogenic microorganisms (7). These properties can depend on the length, time, and shear scales at which they are probed and may furthermore vary in space, mature over time, or respond to biophysical or biochemical stimuli. Although many biological materials can be extracted and characterized in vitro via traditional rheological techniques, there are systems for which the complexity of the in vivo environment, the small sizes or volumes of biomaterials present, or the fragility of the substances with respect to changes in their context demand in vivo rheological assays. Such is the case with the intestinal fluid of a living animal host, a biomaterial dominated by mucus, a glycopolymer solution or gel whose rheological properties are known to span orders of magnitude depending on, for example, the health of the animal (4).
The rheology of the intestine also impacts its vast number of microbial occupants, which must colonize the gut, respond to flows induced by peristaltic motions, and interact with host cells and other microbial cells, all by propelling themselves through the gastrointestinal fluid. In fact, it has been shown in vitro that bacteria can alter the rheological properties of mucus directly by chemically modifying it, and it is theorized that this is done specifically to facilitate movement (7). It is also known from studies of larval zebrafish (Danio rerio) that the presence or absence of intestinal bacteria modulates the number of mucus-producing secretory cells that develop within the host (8). Despite these observations, the rheology of intestinal mucus in germ-free versus conventional animals has never been directly measured in any species.
Inferring the physical properties of the gastrointestinal environment is complicated by a variety of challenges. Especially at early stages in the development of many animals, when the intestine is being colonized by microbes, the volume of the gut is small and difficult to access noninvasively. Extrapolating from in vitro measurements to in vivo properties is hindered by changes in temperature, pH, hydration, and other factors that are unavoidable if mucus is extracted from its source (4, 9). We note also that rheological properties of mucus have been found in vitro to be highly length-scale dependent (4), a consequence of the polymeric nature of mucin glycoproteins, which implies a different effective viscosity at the scales occupied by microbes compared with the macroscopic scales that are typically measured. Passive microrheological techniques that rely on tracking the diffusive displacement of tracer particles have proven useful in a wide variety of studies (10, 11, 12, 13), including this one. Interpreting passive tracer response in living organisms, however, can easily be confounded by adhesion to particular anatomical structures and by fluid flows, e.g., related to circulation, peristalsis, or the active motion of agents such as bacteria. For all these reasons, there is a shortage of rheological studies capable of revealing gastrointestinal fluid properties in living, developing organisms at the length-scales relevant to commensal microbes. In fact, the only reported in vivo measurement of gastrointestinal viscosity is based on echo-planar magnetic resonance imaging, which reports viscosity at molecular length-scales and moreover requires highly specialized equipment and large volumes of probe material (“model food”) (14).
To address this, we implemented both passive microrheology of spherical tracer particles and active microrheology of magnetically driven ellipsoids, visualizing both passive and driven motion using light-sheet fluorescence microscopy (15, 16, 17, 18, 19), which enables rapid, spatially resolved imaging well inside a living organism. Notably, to the best of our knowledge, ours is the first integration of active microrheological methods with light-sheet fluorescence microscopy. We applied these techniques to measure the intestinal rheology of larval zebrafish, a powerful model organism for studying the gut microbiota and their impact on host physiology and development (8, 20, 21, 22, 23, 24) due to the zebrafish’s biological commonalities with other vertebrates, its optical transparency at young ages, its genetic tractability, and its amenability to gnotobiotic techniques by which larval fish can be kept germ-free (devoid of microbes) or exposed to particular microbial constituents (25, 26). Using the methods detailed below, we are able to measure the viscosity of the intestinal interior in larvae that are germ-free, that are colonized by a single bacterial species, and that are conventionally colonized by many microbial species. To our knowledge, these represent the first such measurements to be performed in any living animals. We note that because traditional rheological tools would require sample sizes of over 0.10 mL, or the contents of more than larval zebrafish intestines, our approach will prove useful in overcoming similar limitations of sample volume, spatial heterogeneity, and developmental maturation in other systems that remain resistant to conventional techniques.
Unlike humans or mice, whose intestines contain dense, mucus-rich layers adhered to the epithelial wall along with a more liquid lumenal space, there is no observable adherent layer in the larval zebrafish gut. There is, however, mucus within the gut, as indicated by multiple lines of evidence. First, the lumenal space of the gut is highly autofluorescent, indicating the presence of some sort of organic material. Second, similar but brighter autofluorescence is seen in the mucus-secretory goblet cells of the gut (8). Third, and most importantly, recent studies have specifically characterized larval zebrafish intestinal mucus, identifying mucin gene types, labeling intestinal mucins, and perturbing mucus secretion (27, 28). Given all this, it is reasonable to characterize the larval zebrafish gut interior as a mucin-rich fluid. We caution against drawing extrapolations from our findings in larval zebrafish to human gastrointestinal mucus properties, given the inherent structural differences. Rather, we consider larval zebrafish intestinal rheology as being important in itself. Zebrafish are a valuable model organism for studies of vertebrate physiology, development, and host-microbe interactions, and a quantitative understanding of such studies requires a quantitative understanding of the zebrafish environment.
Materials and Methods
Zebrafish handling and preparation
All zebrafish experiments were performed using protocols approved by the University of Oregon Institutional Animal Care and Use Committee. Fish husbandry and handling followed standard procedures. Larvae were sustained on yolk-derived nutrients and were not fed during experiments. Zebrafish embryos were derived germ-free as previously described (8, 26). In earlier work, bacteria of the genus Vibrio, strain ZWU0020, were isolated from the zebrafish intestinal tract and engineered to express the fluorescent protein dTomato. In experiments involving Vibrio mono-association, bacterial strains were grown overnight in Luria Broth at 30°C; an inoculum of colony-forming units per milliliter was added directly to 15 mL flasks housing 15 germ-free larvae, and fish were examined 24 h postinoculation, as in prior work (24). From prior and current studies, the Vibrio abundance in the intestine is known to be bacteria per gut, corresponding to a concentration of bacteria/mL (24). Conventional zebrafish were raised by bypassing the germ-free derivation, or by adding water from aquarium tanks to larval flasks for at least 24 h prior to experiments. From prior studies (e.g., (8)) and observations, the total bacterial abundance in the intestine of conventional fish is bacteria per gut.
Light-sheet fluorescence microscopy
Light-sheet imaging was performed using a home-built, light-sheet fluorescence microscope, based on the design of Keller et al. (15) and described in detail elsewhere (21, 29). In brief, a laser beam is rapidly scanned and demagnified to provide a thin sheet of fluorescence excitation light. An objective lens mounted perpendicular to the sheet captures fluorescence emission from the optical slice, detected with an sCMOS scientific camera (Pco.edge, PCO AG). The excitation laser power was 5 mW, as measured before the excitation objective, and the wavelength was 488 nm for all experiments in this study.
Larval zebrafish were mounted for imaging as described in (21). In brief, specimens were anesthetized using 120 μg/mL tricaine methanesulfonate (Western Chemical, Ferndale, WA), briefly immersed in 0.5% agar (maximum temperature: 42°C) and drawn into a glass capillary that was then mounted onto a sample holder. The agar-embedded specimens were partially extruded from the capillary so that the excitation and emission paths did not intersect glass interfaces. The specimen holder was maintained at 28°C, with tricaine present as an anesthetic. All specimens were 5 days postfertilization when imaged.
Passive microrheology: experimental methods
Passive microrheological studies used nominally 28 nm diameter fluorescent carboxylate-modified polystyrene spheres (Thermo Fisher cat. #F8787), with peak excitation and emission wavelengths 505 and 515 nm, respectively. For studies of the intestine, larval zebrafish ingested nanospheres added to their aqueous environment. Imaging via light-sheet fluorescence microscopy ∼2 h after delivery of particles to the medium revealed mobile particles within the gut. At earlier times, particle numbers were low; at later times, nanospheres aggregated into dense boluses. For each fish examined, a time-series of a single optical section was captured at ∼75 frames per second for a duration of ∼15 s. Movies of passive tracers in vivo are provided as Movies S1 and S2.
Passive microrheology: analysis
Time-series images of single optical sections were analyzed to yield particle locations and trajectories using a symmetry-based localization algorithm (30) with a localization uncertainty in individual images estimated at 2.5 nm, based on the measured signal-to-noise ratio. Diffusion coefficients were calculated from particle trajectories in two ways. First, we used the conventional approach of fitting for each trajectory the mean-squared-displacement (msd, ) versus time-delay (τ) via to yield the diffusion coefficient D, or to yield the scaling exponent α. Second, we used an optimal, covariance-based estimator of D that provides greater accuracy (31), and that also yields a goodness-of-fit of trajectories to Brownian diffusion as well as an independent estimate of the localization error. The latter was found to be 16 nm, larger than the single-frame estimate, as expected, as it accounts for both uncertainty within individual images and vibration across image frames.
To relate D to the fluid viscosity, η, we used the Stokes-Einstein-Sutherland relation: , where is Boltzmann’s constant, T is temperature, and a is the particle radius. Because the effective a may differ from the nominal radius due, for example, to particle aggregation, we examined Brownian motion of the nanospheres in pure water, imaged with both epifluorescence microscopy (Nikon TE2000 inverted fluorescence microscope, Hamamatsu ORCA II CCD camera) and light-sheet fluorescence microscopy, described above. Light-sheet imaging of particles in water was performed with the same frame rates, magnification, and exposure times as used in live-imaging studies. In both cases, using the known viscosity of water (0.9 cP at 25°C) gives an effective particle radius of 75 nm, indicative of aggregation. For the in vivo studies, we use the measured rather than nominal a to relate D to η.
To determine whether the effective particle radius is intrinsic or dependent on the environment, we compared the fluorescence intensities of the nanospheres in 1/10× phosphate-buffered saline and in the larval zebrafish gut, in both cases imaging with the same light-sheet fluorescence microscope with the same excitation power, camera exposure time, and other experimental parameters. In both of these dissimilar fluids, the particle intensity distributions are similar, with a mean integrated pixel intensity roughly 100× that of the background (Fig. S1), suggesting that the effective radius can be considered an intrinsic property of the particles.
Like the intestines of other vertebrate, the larval zebrafish gut contains narrow crypt-like invaginations. Particles in these spaces are highly confined and effectively stuck, and so their motion is not representative of lumenal fluid properties. A representative movie with these “crypts” labeled is provided as Movie S2. Regions containing these confined particles are manually identified and excluded from analysis.
The peristaltic motility of the zebrafish gut leads to large-scale intestinal flows, typically occurring over a few seconds approximately twice per minute, in between which the gut is fairly quiescent. A representative movie of peristalsis-driven flow of probe particles is provided as Movie S3. Images taken during these events were manually identified and excluded from analysis.
Magnetic probe particles
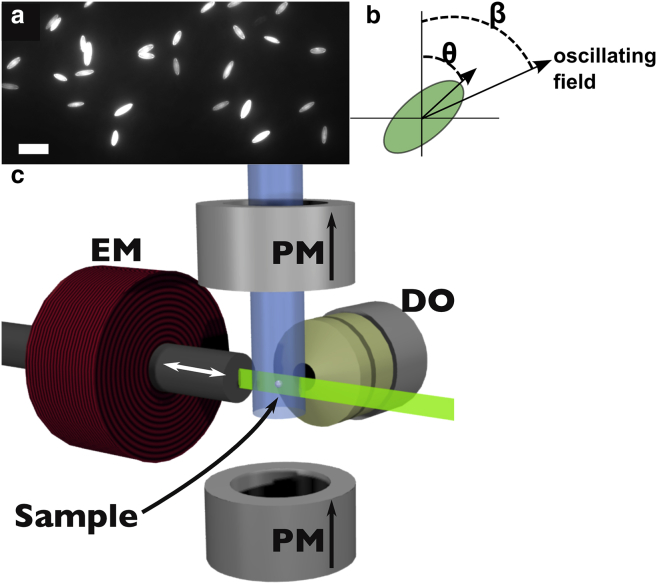
In order to visualize the rotational movement of a particle subject to an oscillating magnetic field, fluorescent, super-paramagnetic polystyrene microspheres (4.88 μm diameter; Spherotech, Lake Forest, IL) were elongated into prolate spheroids (32, 33). Briefly, particles are first embedded in a thin vinyl film, which is fitted onto a mechanical stretcher. This film is then immersed in toluene, which dissolves the polystyrene. Next, the mechanical stretcher is used to elongate the film, deforming the particles along the axis of pull. After removing the toluene, the particles solidify in their new shape and can be recovered by dissolving the vinyl and centrifuging out the particles, which are now shaped as shown in Fig. 1 a.
Figure 1.
Magnetic probe particles and experimental setup. (a) Fluorescent, super-paramagnetic microspheres are stretched into prolate spheroids (Scale bars, ). (b) The resulting shape allows visualization of the angular orientation θ as the spheroid is driven by an external field with orientation angle β. (c) Schematic of the arrangement of magnetic elements and imaging optics, which create the necessary field in the sample volume of a light-sheet fluorescence microscope. In this schematic, which excludes the water-filled sample chamber, a vertical field is created by a pair of permanent magnets (PM) while a perpendicularly oriented electromagnet (EM) is used to tilt the direction of the field within the focal plane, imaged through the detection optics (DO). Arrows indicate the direction of magnetic field due to each element, the excitation laser (shown in green) enters from the right, and the sample is held by a capillary that extends through the top magnet into the sample volume. To see this figure in color, go online.
Active microrheology: theoretical background
A particle with a magnetic moment m and angular orientation θ, located in an external magnetic field of amplitude B and direction β (as in Fig. 1 b), will experience a torque of magnitude , for small values of . This torque tends to align the particle’s magnetic moment with the external field, and the particle must perturb its surrounding environment to do so. For the low Reynolds number system of a colloidal particle in a fluid, a field oscillating as will result in a particle orientation of . If the surrounding fluid is characterized by the constitutive law for the stress σ, viscosity η, and strain rate (i.e., it is a Newtonian fluid), Eq. 1 relates the amplitude of angular displacement of the particle to the driving frequency of the field:
| (1) |
Here, and ξ is a constant that contains the geometrical factors necessary to make the correspondence between stress/torque and strain/angle (34). Using this expression, the frequency dependence of a particle’s response to an oscillating field can yield a measure of the surrounding fluid’s viscosity. More generally, a material may exhibit non-Newtonian behavior, more fully characterized by the complex shear modulus, as in (34). For our present discussion, we restrict ourselves to Newtonian fluids described by Eq. 1, as this will be shown to sufficiently describe the fluids in question.
We also note that in principle, the phase angle ϕ can be used to identify signatures of non-Newtonian rheology, and that more generally one can reconstruct the storage and loss moduli from the in- and out-of-phase components of the particle response. Determination of ϕ is very susceptible to systematic error and noise from asynchrony between the imaging camera and the function generator that controls the field oscillation. In our attempts to reconstruct the phase signal, its uncertainty at low frequencies is small, but approaches 100% above roughly 30 Hz, although the camera is triggered by the function generator. As noted also in the Discussion, precision studies at higher frequencies would be valuable for identifying signatures of elasticity, beyond the scope of this work.
Microrheology apparatus and measurement procedure
Our experimental setup includes a pair of neodymium magnets located above and below the sample, such that they create a permanent uniform magnetic field directed vertically (Fig. 1 c). Additionally, a ferrite core electromagnet is mounted to one side of the sample such that, when supplied with a current, the orientation of the field is tilted to an angle β with respect to vertical. This is contained within the sample chamber of a light-sheet fluorescence microscope with the excitation laser entering from the side opposite the electromagnet, allowing the generation of the magnetic field described above, whose vector orientation oscillates within the imaging focal plane.
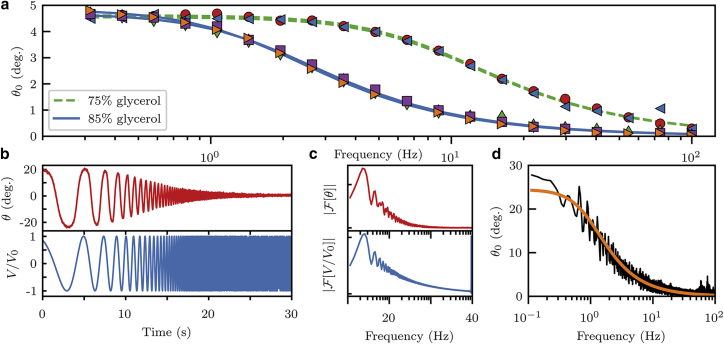
In Eq. 1, m, B, and ξ are unknown system parameters; however, the quantity does not vary appreciably for different particles driven by the same field, as seen in Fig. 2 a. We therefore use a fluid of known viscosity to determine this quantity and calibrate the system for subsequent measurements on fluids of unknown viscosity. For this calibration, we independently measured the viscosity of a water/glycerol mixture by tracking the diffusion of 200 nm diameter, spherical, colloidal particles using an inverted wide-field fluorescence microscope and applied the Stokes-Einstein-Sutherland relation to determine the mixture’s viscosity. Measurements of for actively driven ellipsoids in the same glycerol solution, fit to Eq. 1, provided the necessary information to calibrate the method.
Figure 2.
Frequency response in a water/glycerol calibration fluid. (a) is measured at discrete frequencies in two different water/glycerol preparations. The curves are fits to Eq. 1. The consistency between different probe particles demonstrates the uniformity in their shape and magnetization, allowing calibration using the known viscosity of these solutions. (b–d) Particles are driven with a continuously increasing frequency, allowing a rapid measurement. First, the orientation of the particle is measured (b, top) in response to the externally supplied electromagnet voltage (b, bottom). Next, Fourier transforms are computed for the two signals (c). Finally, taking the absolute value of the ratio of these computed signals yields (d), which can be fit to Eq. 1 to obtain a measure of viscosity η. To see this figure in color, go online.
To further ensure the properties of the calibration fluid, we also measured the viscosity of aqueous glycerol solutions using the method of falling sphere viscometry. In brief, we recorded videos of glass spheres of diameter 2.25 0.05 mm and mass 0.136 0.002 grams falling in the liquid of interest. Analysis of the steady-state velocities using Oseen’s correction to Stokes’ formula for the hydrodynamic drag on a sphere, taken to first order in the Reynolds number, yields the liquid viscosity. For 75 and 85% glycerol solutions, we measured viscosities of 0.034 0.013 and 0.139 0.015 Pa s, respectively, in accord with literature values of 0.046 and 0.131 Pa s, respectively (35). We conclude, therefore, that the glycerol solutions provide a sensible calibration of our particle properties.
The most obvious procedure to measure as a function of diving frequency ω would be to incrementally drive the particle through a series of discrete frequencies while recording high-speed video of the resulting motion. Once the orientation is extracted from the video, the amplitude and phase of the signal can be computed by using the known driving signal and the orthogonality of sine waves over an integral number of periods. This is the procedure followed in collecting the data in Fig. 2 a. When performing a measurement in a system such as the intestinal bulb of a larval zebrafish, however, this procedure is too time consuming, as other sources of fluid flow (e.g., peristaltic motion in the gut) that may occur during data collection corrupt the particle motion and invalidate the measurement. We therefore drive the magnetic field with an exponentially chirped signal of the form with determined by the initial frequency , final frequency , and the time (T) taken to sweep from the former to the later.
Fluorescence images were captured with an sCMOS scientific camera (Pco.edge, PCO AG) at frames per second and the particle orientation was tracked using custom code written using OpenCV (36). A triggering signal from the camera ensured that the driving signal begins its frequency modulation in sync with image acquisition and a time stamp encoded in each image enabled us to account for occasionally dropped frames. The spectra of the input and output signals are related by the system transfer function , as
| (2) |
| (3) |
| (4) |
where denotes the Fourier transform and is the voltage applied to the electromagnet, to which β is proportional. Using this relationship, is calculated from the measured orientation of the particle and the known driving voltage on the electromagnet and can be fitted to Eq. 1 in order to extract and, therefore, the viscosity η, as summarized in Fig. 2, b–d.
To estimate the uncertainty in , we simulated signals for various values of , adding various levels of Gaussian distributed noise. These signals were then used to compute the transfer function from Eq. 4, which was fit to Eq. 1. The inferred value of was compared with its actual value, giving a mapping from noise level and value to expected error. Data from experiments were assigned uncertainty values based on their individual noise levels and value of .
Microgavage of zebrafish larvae
In order to perform our active microrheological measurements, we must first introduce magnetic tracer particles into the intestinal bulb of zebrafish larvae. In principle, this could be done by passive ingestion (as in the passive microrheology experiments) or doping a food source with the magnetic particles prior to feeding the zebrafish larvae. The former strategy, while barely feasible for sub-100 nm particles, fails for micron-scale particles; uptake rates are extremely small. The latter strategy would likely change the gut environment substantially by the introduction of food matter. (At these ages, larvae can subsist on yolk material, and the fish in this study are unfed.) We therefore adopted a recently developed technique of orally gavaging zebrafish larvae (37), in which fluid is delivered to the intestinal bulb by inserting a micropipette tip into the esophagus of the fish and injecting a small volume of material. Using this method, we are able to introduce a volume of fluid small enough that it does not visually displace mucus from the gut (as seen by mucus autofluorescence), but carries with it the desired tracer particles. In order to control for the fluid that accompanies the tracer particles, we estimated the time required for the gut environment to return to its preinjection composition by gavaging zebrafish larvae with an aqueous solution of a fluorescent dye (fluorescein-conjugated dextran, molecular mass 150 kDa; Sigma-Aldrich #46946) and measuring its brightness in the gut over time. As seen in Fig. 4 a, such a solution is not detectably present in the gut 2 h after the initial injection. We therefore restrict our data collection to times that are at least 2 h after gavaging tracer particles into the fish. Notably, in many cases, the particles do not remain in the fish for such long periods, and measurement is therefore impossible. All zebrafish larvae were microgavaged and imaged at 5 days postfertilization.
Figure 4.
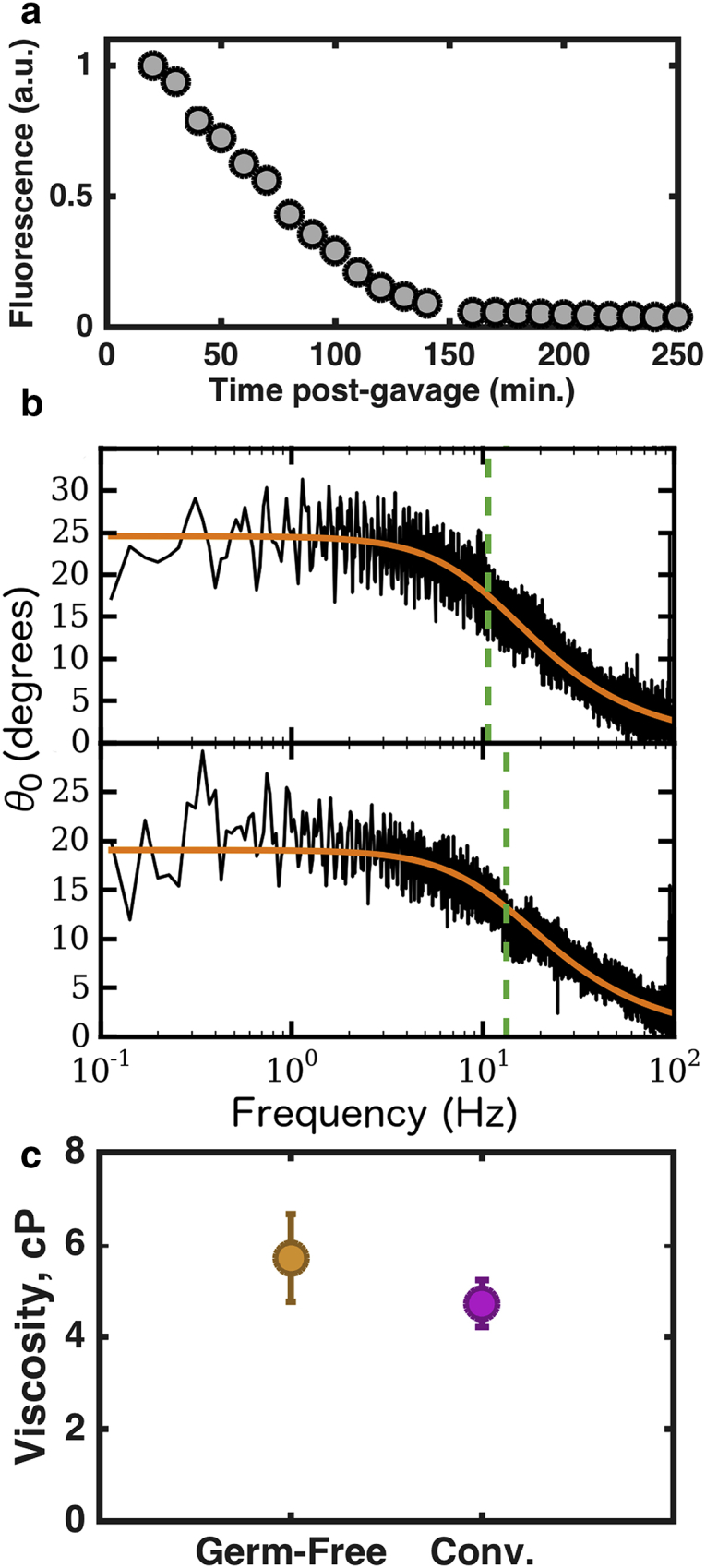
Active microrheology. (a) Fluorescence intensity of a gut gavaged with 150 kDa fluorescein-conjugated dextran shows that fluid is cleared from the gut within 2 h. (b) Typical response curves from driven magnetic microparticles in the intestinal mucus of germ-free (upper panel) and conventional (lower panel) zebrafish larvae. In each case, the smooth curve shows the fit of the experimental data to Eq. 1, with the fit parameter indicated by the vertical dashed line. (c) Measured viscosities of intestinal mucus in conventional and germ-free zebrafish larvae. Circles indicate the weighted mean values, and bars show SE of the weighted mean (with calibration error included). To see this figure in color, go online.
Results
Passive microrheology
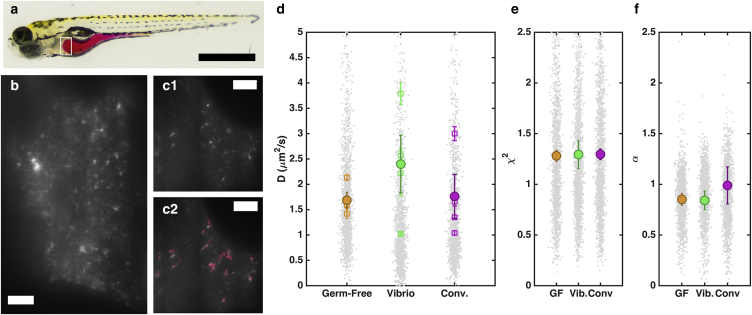
For passive rheological studies, fluorescent polystyrene nanospheres in the intestinal bulb of larval zebrafish (Fig. 3 a) were imaged and their dynamics analyzed as detailed in Materials and Methods (Fig. 3, b and c; Movie S1). We obtained an average of ∼400 particle trajectories per fish with an average duration within the light-sheet plane of 1.6 s (120 frames). For each trajectory, we calculated the diffusion coefficient using both mean-square-displacement analysis and a covariance-based estimator . We performed passive rheological measurements in three types of fish, described in Materials and Methods: germ-free animals, fish mono-associated with a commensal Vibrio species, and conventionally colonized fish, with specimens examined in each case. The Vibrio species was chosen because it is known to colonize with high abundance (/gut) and because the individual bacteria are highly motile (24), suggesting the possibility of bacterially enhanced particle motion. Fluorescently labeled Vibrio, as in (24), sample the entire intestinal volume, and there is no apparent segregation of particles and bacteria (Fig. S2; Movie S2).
Figure 3.
Passive microrheology. (a) A larval zebrafish at five days postfertilization (dpf). The intestine is highlighted for illustration by orally gavaged phenol red dye. The voluminous intestinal bulb is evident at the anterior (left). Bar, 0.5 mm. (b) A subset of an optical section, corresponding roughly to the white rectangle region in (a), showing fluorescent tracer particles at the anterior of the gut. Bar, . (c) Another subset of a light-sheet field of view, showing particles (c1) and their trajectories (c2). Bar, . (d) Diffusion coefficients of nanospheres in the larval intestinal bulb. All data points are shown in gray; fewer than 3% are outside the plotted range. Average values within individual fish are shown as open squares, with error bars indicating mean ± SE. Averages across fish for each type, germ-free, monoassociated with a commensal Vibrio species, and conventionally colonized by microbes, are shown as solid circles, with error bars indicating mean ± SE ( fish each). (e) Reduced for the fit of the covariance-based estimator of D to the measured trajectories, for each fish type. (f) The scaling exponent, α, for the measured mean-squared-displacement as a function of time, for each fish type. For both (d) and (e), all data points are shown in gray, and solid symbols and error bars indicate the mean and SD of the mean, respectively, across fish of each type. The SD of α within each individual fish is ∼0.2 in all cases. For both (d) and (e), a value of one for or α is consistent with Brownian motion in a Newtonian fluid. To see this figure in color, go online.
Both diffusion measures, and , were in agreement with the average values for each fish from the two methods being within 10% of each other (Fig. S3). The values from all trajectories, their averages within individual fish, and their averages across fish type are plotted in Fig. 3 d. Across type, the data show no notable difference, with , and for germ-free fish, fish mono-associated with Vibrio, and conventional fish, respectively, where the uncertainty indicates the mean ± SE in each case. This gives viscosity values of , , and cP, slightly higher than that of water (0.9 cP). Passive measurements of from particles in the same zebrafish gut imaged over a 60-min interval show a roughly constant diffusion coefficient over time (Fig. S4 a). The fluorescence intensity of the particles is also unchanging over this interval (Fig. S4 b), implying that neither local particle aggregation nor large changes in the intestinal viscosity occur on this timescale. Notably, significant aggregation does occur, but does not take the form of gradual clustering of discrete, colliding particles but rather is manifested as the sweeping up of all particles into a dense bolus, described further below. Until this large-scale aggregation, we observe discrete particles with roughly constant dynamical properties.
For the covariance-based estimator of D, the goodness-of-fit (reduced ) to Brownian diffusion was close to one for all fish (Fig. 3 e). From mean-square-displacement analysis, the scaling exponent α (msd ; see Materials and Methods) was close to one (Fig. 3 f). Both of these measures are consistent with diffusion in a Newtonian fluid, and indicate neither a significant elastic response nor super-diffusive dynamics.
Combining all the in vivo measurements, regardless of the host’s bacterial state, we find a value of cP, or ∼1.9 times the viscosity of water, for the viscosity of the intestinal bulb at the nm length-scale probed by the tracer particles.
As noted earlier, nanospheres eventually aggregate into dense boluses along the central axis of the gut. Such aggregation is a consequence of peristaltic activity, which is well known to condense and transport material through the digestive tract. These boluses are highly autofluorescent and therefore presumably especially mucin-dense. Notably, these regions are not adhered to the intestinal wall, and are a small fraction of the intestinal space. Rarely, individual tracer particles are discernible in these boluses; these particles show little motion (Movie S4). Their apparent diffusion coefficient ( ; mean SD for particles in the fish shown in Movie S4) sets an upper-bound on the minimal D we can resolve with our instrumentation, which is two orders of magnitude lower than D for free particles in the gut. Equivalently, these values set a lower-bound for the viscosity of the bolus material, cP.
Active microrheology
For active microrheological studies, we gavaged ellipsoidal microparticles into the intestinal bulb of 5-day-old conventionally reared and germ-free zebrafish larvae and performed the active microrheology procedure described in Materials and Methods, using the calibration illustrated in Fig. 2. We did not perform active microrheological measurements in fish mono-associated with Vibrio, focusing on possible generic effects of bacterial species rather than specific effects from one species. Magnetically driven oscillation of microparticles, with an oscillation amplitude that decreases with frequency, is readily evident in the intestinal bulb (Movie S5). The measured response curves are well fit by the form expected of a Newtonian fluid (Eq. 1), as shown in Fig. 4 b. An elastic component of the fluid response would manifest as a nonzero lower limit to the oscillation amplitude at high frequencies (34), which is not seen in any of the data collected (e.g., Fig. 4 b). We note, however, that we cannot directly probe frequency scales above 100 Hz, and so cannot rule out the possibility of an upturn in oscillation amplitude above our accessible scale. Also, as noted in Materials and Methods, our techniques at present cannot accurately determine the phase angle between the magnetic field oscillations and particle response at the upper end of our accessible frequency range, which would provide another potential signature of elastic response.
Repeating this measurement over several specimens, we compute the weighted mean of the viscosity to be and cP for the intestinal fluid of germ-free and conventional intestines, respectively (as shown in Fig. 4 c; and 10 specimens, respectively), i.e., values of mucus viscosity that are ∼5 times larger than that of water, with a slight enhancement in germ-free relative to conventional fish. Uncertainties are the SE of the weighted mean, as described in Materials and Methods.
Discussion
From both passive and active in vivo microrheological measurements of the larval zebrafish gut, we find responses that are well described by viscous fluid models, with a negligible elastic response from intestinal mucus. Though extensions of the technique to higher frequencies and greater phase sensitivity are clearly called for to uncover potential elasticity, it is notable that in vitro macroscopic studies of mucus generally show strong elastic signatures at frequencies well within the range examined here (4). In vitro microrheological studies, however, are more varied. Georgiades et al. (38), for example, find strongly pH-dependent viscoelasticity from particle-tracking assays in porcine gastrointestinal mucus extracts, with purely viscous behavior at neutral pH and elasticity developing under acidic conditions. In vitro microrheology of human cervicovaginal mucus shows signatures of non-Newtonian behavior at length-scales above a few 100 nm (39, 40). Together, these and other data suggest that animal species, developmental stage, mucus type, and possible in vitro preparation artifacts have major impacts on mucus rheology, a more thorough understanding of which calls for considerable effort.
Our own work is motivated especially by the utility of larval zebrafish for studies of the vertebrate microbiome, and past work has shown that bacterial motility in the intestinal space can be a major determinant of community composition (24). The length-scale probed by our magnetic microparticles is similar to the size of typical bacteria, implying that microbes occupying the larval intestine will experience a predominantly viscous fluid environment, and that models of bacterial motility in viscoelastic fluids (41) will not be necessary in this context.
The similarity of viscosity values between germ-free and bacterially colonized intestines, measured with either technique, is perhaps surprising, as zebrafish which have been derived germ-free possess ∼50% fewer mucin-producing secretory cells than conventional fish (8). Our observation may reflect the developmental stage of the animal, which at the time of experiment is still sustained by its yolk and has yet to ingest solid food, which would likely stimulate secretory cell activity.
Regardless of its origin, our finding of a fluid environment with a viscosity a few times that of water provides useful data for understanding mechanisms of host-microbe interactions. In many contexts, bacterial communication involves secreted factors, whose dispersal timescales depend on their diffusion coefficients. For example, a recently discovered 29 kDa protein secreted by a member of the zebrafish intestinal microbiota stimulates the proliferation of insulin-producing β cells in the pancreas, through mechanisms that are unclear (23). Low viscosity implies a short time, on the order of minutes, for diffusion of a secreted protein through the entire few 100 micron intestinal space or through the duct that connects the intestine and the pancreas, suggesting that specific localization of the bacteria to the duct site or to targeted cells is unlikely to be necessary. We further expect that information about intestinal viscosity will set constraints on strategies for chemotaxis and other bacterial behaviors in the gut.
Our observation of a larger apparent viscosity for the few-micron, actively driven particles than the sub-100 nm, passive particles suggests a size-dependent viscosity, known from in vitro studies to be possible even in nonelastic polymer solutions (42). The size and spacing of mucin polymers in the intestinal fluid, even in the absence of entanglement, will set a length-scale that impacts probe dynamics; at very small probe sizes, the polymer network is invisible. It would, of course, be useful to examine passive diffusion of larger, m, particles. The Brownian motion of these objects would be small, however, and may be difficult to accurately assess given the short measurement window allowed by peristaltic motility. Nonetheless, this would be a worthwhile goal for future work, as it also may enable the observation of super-diffusive motion for passive particles in the presence of gut microbes, expected to be more pronounced for tracers of similar sizes to the surrounding bacteria (43).
Mucus provides a rich set of biophysical mysteries that are of both fundamental and practical importance. Our understanding of this fluid is still in its infancy, and is based largely on in vitro assays that may poorly mirror in vivo environments, and that are prone to artifacts. For example, in contrast to measurements of reconstituted mucus that report a lowering of viscosity by Helicobacter pylori (7), other studies report an increase in mucus viscosity following H. pylori infection, ascribed to possible differences in extraction and handling methods (9).
We suggest that the methodology we describe here offers a path to in vivo measurements that will be crucial for making sense of mucus biophysics. This opens the doors to examinations of mucin response in relation to host or microbial physiology, and comparisons to the rheologies of other complex fluids. Notably, this path is so far rather low-throughput and technically challenging, and would be well served by further methodological improvement. Of course, other approaches to in vivo microrheology also exist, such as optical tweezers (44), magnetic tweezers (45), and magnetic droplets (46). All of these, as well as our approach, have advantages and disadvantages. The optical inhomogeneity of tissues in a larval zebrafish can complicate force calibration, and the fairly high laser powers required for trapping can damage the animal, at least locally (44). Driving spherical magnetic particles requires large fields that may be challenging to create in a setup that is densely occupied by optical equipment. In contrast, oscillation of nonspherical magnetic particles, as in our study, requires a large magnetic field gradient, but not a large field. Magnetic droplets are an intriguing approach for determining rheological properties, and could be delivered to the gut. Use of these oil-based droplets might be confounded by concerns of toxicity, or by the droplets being digested by the fish. Despite these potential issues, all these techniques are powerful, and we hope that our work encourages further quantitative studies of intestinal rheology.
The experimental combination we introduce of passive microrheology, active microrheology, and light-sheet fluorescence microscopy should be applicable to a wide variety of small, structurally complex biophysical environments. For example, many marine invertebrates such as salps and the larvae of sea urchins, starfish, and other animals are quite transparent and possess digestive tracts that make use of mucus-rich organs, particle filtration, and other structures and phenomena whose biophysics could be investigated using these approaches. In addition to being interesting in themselves, processes associated with these organisms are important for ecological food webs and oceanic carbon capture. The techniques we have described could also be applied to studies of the gut environment in Caenorhabditis elegans, another transparent model organism, as well as recently developed ex vivo mimics of intestinal environments (47). Techniques such as two-photon light-sheet excitation (17) could further enable quantitative investigation of more opaque specimens.
Author Contributions
M.J.T., E. A. H., and R.P. designed the research. M.J.T. and E.A.H. performed the research and analyzed data. M.J.T., E.A.H, and R.P. wrote the article.
Acknowledgments
We thank Rose Sockol and the University of Oregon Zebrafish Facility staff for fish husbandry; Vince Thoms for assistance with particle tracking; and Karen Guillemin, Eric Corwin, Judith Eisen, Kyle Welch, and Ryan Baker for useful discussions and comments. Research reported in this publication was supported by the National Science Foundation (NSF) under awards 0922951 and 1507115, the M. J. Murdock Charitable Trust, the University of Oregon through a faculty research award, and by the National Institutes of Health (NIH) as follows: by the National Institute of General Medical Sciences under award number P50GM098911 and by the National Institute of Child Health and Human Development under award P01HD22486, which provided support for the University of Oregon Zebrafish Facility. The content is solely the responsibility of the authors and does not represent the official views of the NIH, NSF, or other funding agencies.
Editor: Alexander Dunn.
Footnotes
Michael J. Taormina and Edouard A. Hay contributed equally to this work.
Four figures and five movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30797-X.
Supporting Material
Fig. S2 shows a subset of a still frame of this movie, together with an image of bacteria in the same region.
These contractions occur roughly twice per minute.
Note that the bolus is not adhered to the epithelial wall. The embedded particles show little motion, in contrast to free particles elsewhere in the gut.
References
- 1.Vogel S. Oxford University Press; UK: 1993. Vital Circuits. On Pumps, Pipes and the Workings of Circulatory Systems. [Google Scholar]
- 2.Gambini C., Abou B., Cornelissen A.J. Micro- and macrorheology of jellyfish extracellular matrix. Biophys. J. 2012;102:1–9. doi: 10.1016/j.bpj.2011.11.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freund J.B., Goetz J.G., Vermot J. Fluid flows and forces in development: functions, features and biophysical principles. Development. 2012;139:1229–1245. doi: 10.1242/dev.073593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai S.K., Wang Y.-Y., Hanes J. Micro- and macrorheology of mucus. Adv. Drug Deliv. Rev. 2009;61:86–100. doi: 10.1016/j.addr.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ensign L.M., Cone R., Hanes J. Oral drug delivery with polymeric nanoparticles: the gastrointestinal mucus barriers. Adv. Drug Deliv. Rev. 2012;64:557–570. doi: 10.1016/j.addr.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saltzman W.M., Radomsky M.L., Cone R.A. Antibody diffusion in human cervical mucus. Biophys. J. 1994;66:508–515. doi: 10.1016/s0006-3495(94)80802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celli J.P., Turner B.S., Bansil R. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc. Natl. Acad. Sci. USA. 2009;106:14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates J.M., Mittge E., Guillemin K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 2006;297:374–386. doi: 10.1016/j.ydbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 9.Markesich D.C., Anand B.S., Graham D.Y. Helicobacter pylori infection does not reduce the viscosity of human gastric mucus gel. Gut. 1995;36:327–329. doi: 10.1136/gut.36.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardel M., Valentine M., Weitz D. Microrheology. In: Breuer K., editor. Microscale Diagnostic Techniques. Springer-Verlag; Berlin: 2005. pp. 1–49. [Google Scholar]
- 11.Weihs D., Mason T.G., Teitell M.A. Bio-microrheology: a frontier in microrheology. Biophys. J. 2006;91:4296–4305. doi: 10.1529/biophysj.106.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Squires T.M., Mason T.G. Fluid mechanics of microrheology. Annu. Rev. Fluid Mech. 2010;42:413–438. [Google Scholar]
- 13.Bennett J.S., Gibson L.J., Rubinsztein-Dunlop H. Spatially-resolved rotational microrheology with an optically-trapped sphere. Sci. Rep. 2013;3:1759. [Google Scholar]
- 14.Marciani L., Gowland P.A., Fillery-Travis A.J. Gastric response to increased meal viscosity assessed by echo-planar magnetic resonance imaging in humans. J. Nutr. 2000;130:122–127. doi: 10.1093/jn/130.1.122. [DOI] [PubMed] [Google Scholar]
- 15.Keller P.J., Schmidt A.D., Stelzer E.H.K. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 16.Huisken J., Stainier D.Y.R. Selective plane illumination microscopy techniques in developmental biology. Development. 2009;136:1963–1975. doi: 10.1242/dev.022426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truong T.V., Supatto W., Fraser S.E. Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods. 2011;8:757–760. doi: 10.1038/nmeth.1652. [DOI] [PubMed] [Google Scholar]
- 18.Planchon T.A., Gao L., Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santi P.A. Light sheet fluorescence microscopy: a review. J. Histochem. Cytochem. 2011;59:129–138. doi: 10.1369/0022155410394857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roeselers G., Mittge E.K., Rawls J.F. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011;5:1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jemielita M., Taormina M.J., Parthasarathy R. Spatial and temporal features of the growth of a bacterial species colonizing the zebrafish gut. MBio. 2014;5 doi: 10.1128/mBio.01751-14. e01751–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rolig A.S., Parthasarathy R., Guillemin K. Individual members of the microbiota disproportionately modulate host innate immune responses. Cell Host Microbe. 2015;18:613–620. doi: 10.1016/j.chom.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill J.H., Franzosa E.A., Guillemin K. A conserved bacterial protein induces pancreatic beta cell expansion during zebrafish development. eLife. 2016;5:e20145. doi: 10.7554/eLife.20145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiles T.J., Jemielita M., Parthasarathy R. Host gut motility promotes competitive exclusion within a model intestinal microbiota. PLoS Biol. 2016;14:e1002517. doi: 10.1371/journal.pbio.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milligan-Myhre K., Charette J.R., Kim C.H. Study of host-microbe interactions in zebrafish. Methods Cell Biol. 2011;105:87–116. doi: 10.1016/B978-0-12-381320-6.00004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melancon E., Gomez De La Torre Canny S., Guillemin K. Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods Cell Biol. 2017;138:61–100. doi: 10.1016/bs.mcb.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jevtov I., Samuelsson T., Ribbeck K. Zebrafish as a model to study live mucus physiology. Sci. Rep. 2014;4:6653. doi: 10.1038/srep06653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oehlers S.H., Flores M.V., Crosier P.S. Retinoic acid suppresses intestinal mucus production and exacerbates experimental enterocolitis. Dis. Model. Mech. 2012;5:457–467. doi: 10.1242/dmm.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taormina M.J., Jemielita M., Guillemin K. Investigating bacterial-animal symbioses with light sheet microscopy. Biol. Bull. 2012;223:7–20. doi: 10.1086/BBLv223n1p7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parthasarathy R. Rapid, accurate particle tracking by calculation of radial symmetry centers. Nat. Methods. 2012;9:724–726. doi: 10.1038/nmeth.2071. [DOI] [PubMed] [Google Scholar]
- 31.Vestergaard C.L., Blainey P.C., Flyvbjerg H. Optimal estimation of diffusion coefficients from single-particle trajectories. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2014;89:022726. doi: 10.1103/PhysRevE.89.022726. [DOI] [PubMed] [Google Scholar]
- 32.Champion J.A., Katare Y.K., Mitragotri S. Making polymeric micro- and nanoparticles of complex shapes. Proc. Natl. Acad. Sci. USA. 2007;104:11901–11904. doi: 10.1073/pnas.0705326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Güell O., Sagués F., Tierno P. Magnetically driven Janus micro-ellipsoids realized via asymmetric gathering of the magnetic charge. Adv. Mater. 2011;23:3674–3679. doi: 10.1002/adma.201100902. [DOI] [PubMed] [Google Scholar]
- 34.Wilhelm C., Browaeys J., Bacri J.C. Rotational magnetic particles microrheology: the Maxwellian case. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2003;67:011504. doi: 10.1103/PhysRevE.67.011504. [DOI] [PubMed] [Google Scholar]
- 35.Cheng N.-S. Formula for the viscosity of a glycerol-water mixture. Ind. Eng. Chem. Res. 2008;47:3285–3288. [Google Scholar]
- 36.Itseez. 2015. Open Source Computer Vision Library. https://github.com/itseez/opencv.
- 37.Cocchiaro J.L., Rawls J.F. Microgavage of zebrafish larvae. J. Vis. Exp. 2013;72:e4434. doi: 10.3791/4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgiades P., Pudney P.D.A., Waigh T.A. Particle tracking microrheology of purified gastrointestinal mucins. Biopolymers. 2014;101:366–377. doi: 10.1002/bip.22372. [DOI] [PubMed] [Google Scholar]
- 39.Lai S.K., O’Hanlon D.E., Hanes J. Rapid transport of large polymeric nanoparticles in fresh undiluted human mucus. Proc. Natl. Acad. Sci. USA. 2007;104:1482–1487. doi: 10.1073/pnas.0608611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olmsted S.S., Padgett J.L., Cone R.A. Diffusion of macromolecules and virus-like particles in human cervical mucus. Biophys. J. 2001;81:1930–1937. doi: 10.1016/S0006-3495(01)75844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spagnolie S.E., Liu B., Powers T.R. Locomotion of helical bodies in viscoelastic fluids: enhanced swimming at large helical amplitudes. Phys. Rev. Lett. 2013;111:068101. doi: 10.1103/PhysRevLett.111.068101. [DOI] [PubMed] [Google Scholar]
- 42.Poling-Skutvik R., Krishnamoorti R., Conrad J.C. Size-dependent dynamics of nanoparticles in unentangled polyelectrolyte solutions. ACS Macro Lett. 2015;4:1169–1173. doi: 10.1021/acsmacrolett.5b00616. [DOI] [PubMed] [Google Scholar]
- 43.Patteson A.E., Gopinath A., Arratia P.E. Particle diffusion in active fluids is non-monotonic in size. Soft Matter. 2016;12:2365–2372. doi: 10.1039/c5sm02800k. [DOI] [PubMed] [Google Scholar]
- 44.Johansen P.L., Fenaroli F., Koster G. Optical micromanipulation of nanoparticles and cells inside living zebrafish. Nat. Commun. 2016;7:10974. doi: 10.1038/ncomms10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desprat N., Supatto W., Farge E. Tissue deformation modulates twist expression to determine anterior midgut differentiation in Drosophila embryos. Dev. Cell. 2008;15:470–477. doi: 10.1016/j.devcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 46.Serwane F., Mongera A., Campàs O. In vivo quantification of spatially varying mechanical properties in developing tissues. Nat. Methods. 2017;14:181–186. doi: 10.1038/nmeth.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim H.J., Huh D., Ingber D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S2 shows a subset of a still frame of this movie, together with an image of bacteria in the same region.
These contractions occur roughly twice per minute.
Note that the bolus is not adhered to the epithelial wall. The embedded particles show little motion, in contrast to free particles elsewhere in the gut.