Abstract
Leiomodins (Lmods) are a family of actin filament nucleators related to tropomodulins (Tmods), which are pointed end-capping proteins. Whereas Tmods have alternating tropomyosin- and actin-binding sites (TMBS1, ABS1, TMBS2, ABS2), Lmods lack TMBS2 and half of ABS1, and present a C-terminal extension containing a proline-rich domain and an actin-binding Wiskott-Aldrich syndrome protein homology 2 (WH2) domain that is absent in Tmods. Most of the nucleation activity of Lmods resides within a fragment encompassing ABS2 and the C-terminal extension. This fragment recruits actin monomers into a polymerization nucleus. Here, we revise a recently reported structure of this region of Lmod2 in complex with actin and provide biochemical validation for the newly revised structure. We find that instead of two actin subunits connected by a single Lmod2 polypeptide, as reported in the original structure, the P1 unit cell contains two nearly identical copies of actin monomers, each bound to Lmod2’s ABS2 and WH2 domain, with no electron density connecting these two domains. Moreover, we show that the two actin molecules in the unit cell are related to each other by a local twofold noncrystallographic symmetry axis, a conformation clearly distinct from that of actin subunits in the helical filament. We further find that a proposed actin-binding site within the missing connecting region of Lmod2, termed helix h1, does not bind actin in vitro and that the electron density assigned to it in the original structure corresponds instead to a WH2 domain with opposite backbone directionality. Polymerization assays using Lmod2 mutants of helix h1 and the WH2 domain support this conclusion. Finally, we find that deleting the C-terminal extension of Lmod1 and Lmod2 results in an approximately threefold decrease in the nucleation activity, which is only partially accounted for by the lack of the WH2 domain.
Introduction
Actin nucleators are proteins that accelerate the rate of actin filament formation, playing essential roles in cells by controlling the spatiotemporal transition between the monomeric and filamentous forms of actin (1, 2). Different nucleators regulate the assembly of distinct actin-based cytoskeletal networks and in connection with different cellular functions, including organelle trafficking, membrane fusion and fission events, endo/exocytosis, and cell motility. Leiomodins (Lmods) are a family of actin filament nucleators, consisting of three isoforms in humans, which are expressed primarily in muscle cells (3, 4). Lmods are related in their domain organization to tropomodulins (Tmods), which comprise a family of filament pointed end capping proteins in muscle sarcomeres. Tmods contain alternating tropomyosin- and actin-binding sites (TMBS1, ABS1, TMBS2, and ABS2). Lmods lack TMBS2 and the C-terminal half of ABS1, but retain TMBS1 and ABS2, with these two domains being separated in Lmods by long, low-complexity sequences, substituting for TMBS2 in Tmods. More notably, Lmods are distinguished from Tmods by the presence of a C-terminal extension bearing a proline-rich domain (PRD) and an actin-binding Wiskott-Aldrich syndrome protein homology 2 (WH2) domain. In this way, ABS2, which is mostly folded as a leucine-rich repeat domain (LRR) (5, 6), is found toward the middle of the sequence in Lmods, whereas it is C-terminal in Tmods. Together, the central ABS2 and C-terminal extension account for most of the nucleation activity of Lmods, as Lmod1 and Lmod2 constructs comprising these two domains (Lmod1ABS2-C and Lmod2ABS2-C) have nucleation activities approaching those of the corresponding full-length proteins (7).
A recently reported crystal structure of Lmod2ABS2-C bound to actin (8) is of considerable interest for its potential to provide mechanistic insights into actin nucleation by Lmods. The authors of this study conclude that their structure corresponds to that of an actin-Lmod2 polymerization nucleus (or seed), and that it supports a mechanism of nucleation whereby one molecule of Lmod2 organizes two actin subunits into a nonfilamentous-like conformation to initiate polymerization (8). However, the authors’ interpretation of the structure as a polymerization nucleus hinges entirely on the construction of a 78-amino-acid region linking the two known actin-binding sites of Lmod2, i.e., ABS2 (which they refer to as the “LRR”) and the C-terminal WH2 domain (which they refer to as “W”), such that a single Lmod2ABS2-C molecule connects two adjacent actin molecules in the crystal lattice. The connecting region consists of Lmod2 residues P441–Q518, and in the reported structure it was modeled as a PRD (which they refer to as “polyP”), two helical segments (h1 and h2), and intervening loops. Here, we reexamine this structure and associated crystallographic data (deposited in the PDB: 4RWT). We find no electron density for any of the elements of the reported structure connecting ABS2 to the WH2 domain. We further find that the region of the structure modeled as helix h1, and described as a novel actin-binding site in Lmod2, corresponds instead to the WH2 domain of a second Lmod2 molecule present in the unit cell, and binds actin with opposite directionality of the polypeptide chain than that of helix h1 in the original structure. We present a revised structure that consists of two nearly identical actin monomers, each bound to Lmod2’s ABS2 and WH2 domain, with no connecting density in between these two domains. Furthermore, the two actin molecules in the P1 unit cell are related to each other by a noncrystallographic twofold symmetry axis, a conformation distinct from that of actin subunits in the filament. Thus, the structure cannot be interpreted as that of Lmod2’s polymerization nucleus, as suggested by Chen et al. (8). Consistent with our revised structure, biochemical studies confirm a role for the WH2 domain, but not helix h1, in actin binding and filament nucleation. Polymerization assays with Lmod2 and Lmod1 constructs show that the C-terminal extension is responsible for a two- to threefold increase in the nucleation activity compared to constructs consisting only of ABS2. This activity is distributed throughout the entire C-terminal extension, because a mutation that incapacitates the WH2 domain has only a minor effect on actin nucleation.
Materials and Methods
Structure revision and refinement
The previously deposited atomic coordinates and structure factor amplitudes of the actin-Lmod2 structure (PDB: 4RWT) were used to begin the refinement, and the same Rfree-flagged reflection subset was maintained to monitor the progress of the refinement. Crystal packing was examined and the diffraction data were analyzed using the Xtriage program of the PHENIX suite (9), allowing us to confirm that the space group was correctly assigned as P1. However, rather than two independent actin molecules connected by a single Lmod2 polypeptide, we found that the P1 unit cell contains two nearly identical actin-Lmod2 complexes related by a noncrystallographic twofold symmetry axis (Fig. 1). A SIGMAA-weighted 2Fo-Fc electron density map calculated based on the deposited model and data did not show interpretable density for Lmod2 residues P441–Q518, which in the original model comprised the PRD, helices h1 and h2, and intervening loops (Fig. 1). Thus, these residues were removed from the model at the beginning of the refinement. The structure was refined with the program PHENIX (https://www.phenix-online.org/) (9), with rounds of manual model rebuilding and inspection with the program Coot (10). First, two rounds of positional refinement were performed, using individual B-factors, x-ray/stereochemistry weight optimization, and riding hydrogen atoms (i.e., with hydrogen atom positions calculated from the positions of the main atoms to which they are bound). We note that the inclusion of riding hydrogen atoms improves substantially the stereochemistry of the model, without adding additional refinement parameters (as explained in the PHENIX documentation). After this initial refinement, the electron density for the second WH2 domain was clearly defined and this region was added to the model. Superimposition of the two actin-Lmod2ABS2-C complexes in the P1 unit cell revealed that they were nearly identical, even when refined without noncrystallographic symmetry restraints (root mean squared deviation = 0.22 Å for 525 equivalent Cα atoms). Therefore, from this point on, torsion-angle noncrystallographic symmetry restraints and secondary structure restraints were maintained. Additional PHENIX refinement and Coot inspection cycles allowed corrections of the Ramachandran plot and rotamer outliers, and six amino acids were newly added; one at the N-terminus of each of the actin chains and two at the N-terminus of each of the Lmod2 chains. We note that TLS and Simulated Annealing were not used during refinement. Electron density maps of the structure published by Chen et al. (8) were calculated from the deposited coordinates and structure factors using the “Create Maps” module in the program PHENIX. Illustrations of the structures and electron density maps were prepared with the program PyMOL (Schrödinger, New York, NY). The data and refinement statistics reported in Table 1 were calculated with the program PHENIX, using the diffraction data between 3.0 and 45.0 Å resolution.
Figure 1.
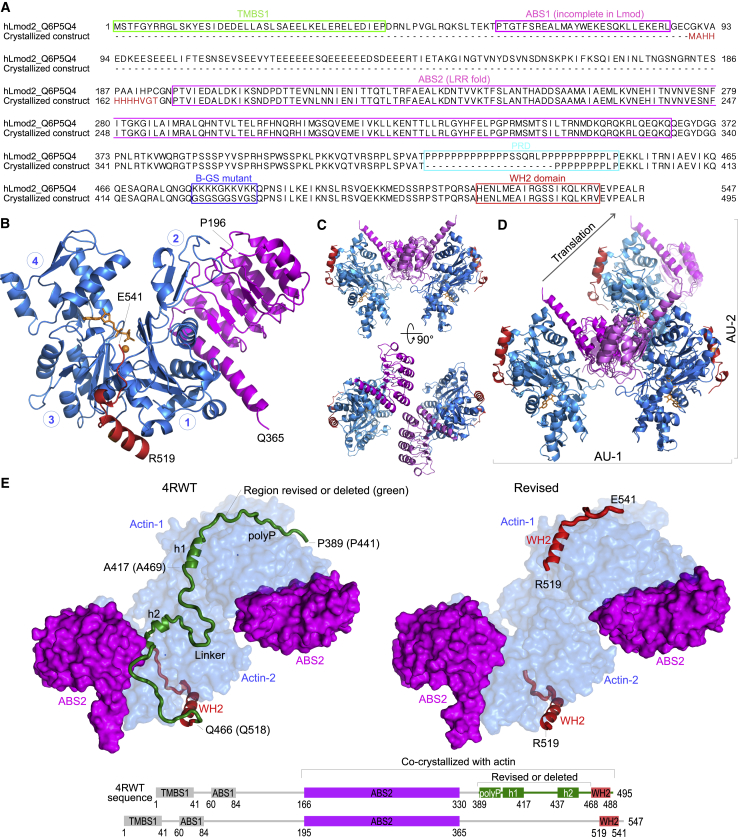
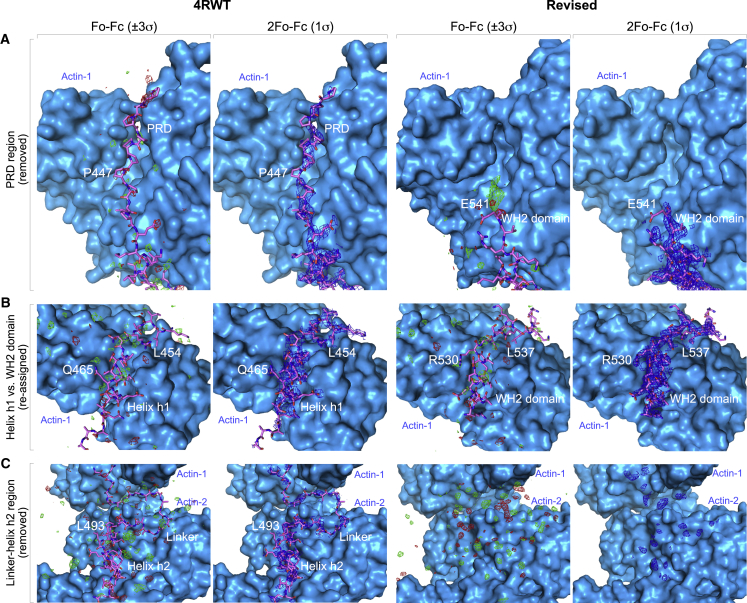
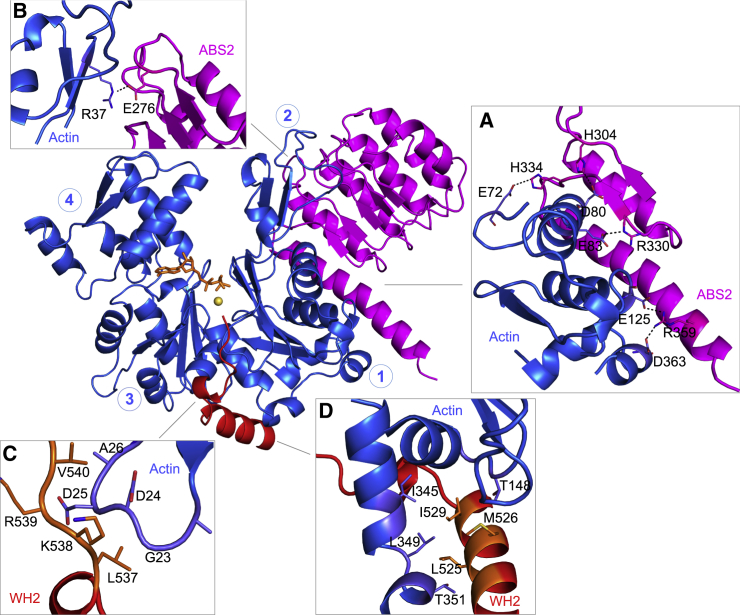
Revised structure of actin-Lmod2ABS2-C. (A) Shown here is an alignment of the correct sequence of human Lmod2 (UniProt ID: Q6P5Q4) and the Lmod2 construct cocrystallized with actin (8). The purification tag present in the crystallized Lmod2 construct is also shown at the N-terminus. Boxed regions correspond to the major Lmod2 subdomains implicated in interactions with actin (ABS1, ABS2, WH2 domain) and tropomyosin (TMBS1). The boxed region labeled “B-GS mutant” corresponds to a mutation introduced by Chen et al. (8) into the crystallized fragment to replace a basic patch prone to degradation by a Gly-Ser linker. (B) Given here is a ribbon representation of one of the two nearly identical actin-Lmod2 complexes present in the P1 unit cell. Circled numbers indicate the four subdomains of actin. The ABS2 and WH2 domain of Lmod2 correspond to the boxed regions shown in (A). ABS2 binds subdomains 1 and 2 of actin, whereas the WH2 domain binds at the barbed end of the same actin monomer, occupying the cleft between subdomains 1 and 3, known as the target-binding or hydrophobic cleft (20). The ATP analog AMP-PNP and associated Mg2+ ion in the nucleotide-binding cleft of actin are also shown. Terminal residues in ABS2 and the WH2 domain are labeled. (C) The P1 unit cell contains two copies of the actin-Lmod2ABS2-C complex, related by a local twofold symmetry axis, distinct from the helical symmetry of the actin filament. Two orthogonal views are shown. (D) Given here are two alternate but equivalent definitions of the contents of the asymmetric unit, AU, corresponding to the unit cell in the P1 space group. The AU-2 definition corresponds to that used by Chen et al. (8). AU-2 is the same as the twofold related complex (AU-1), but translated to an adjacent unit cell, as the arrow indicates. (E) Shown here is a comparison of the original (left) and revised (right) structures of actin-Lmod2ABS2-C according to the AU-2 definition. The region of the Lmod2 C-terminal extension present only in the original structure (residues P441–Q518, using the correct sequence numbering) is shown in the model and accompanying diagram and labeled as “revised or deleted”. The domains that are not present in the crystallized Lmod2 fragment or not visualized in the structures are shown in light gray in the diagram at the bottom, where both the original (top) and the correct (bottom) numbering schemes are shown for reference. The revised structure has two extra amino acids at the N-terminus of ABS2 and 83 fewer amino acids in the region between ABS2 and the WH2 domain, and also lacks the last six amino acids after the WH2 domain. Note also that there are two WH2 domains in the revised structure, whereas in the structure of Chen et al. (8), the position of the WH2 domains bound to one of actin molecules (labeled Actin-1) is occupied by a portion of the PRD (polyP) and helix h1, with reverse polypeptide directionality. To see this figure in color, go online.
Table 1.
Crystallographic Data and Refinement Statistics
| Crystallographic Data | |
|---|---|
| Resolution range (Å) | 44.24–3.0 (3.107–3.0) |
| Space group | P1 |
| Unit cell | |
| a, b, c (Å) | 65.35, 65.65, 81.92 |
| α, β, γ (°) | 101.29, 90.94, 107.97 |
| Total reflections | 24,696 (2487) |
| Completeness (%) | 97.39 (97.30) |
| Wilson B-factor | 53.71 |
| Reflections used in refinement | 24,696 (2487) |
| Reflections used for R-free | 1261 (104) |
| Refinement | PDB: 4RWT | Revised (PDB: 5WFN) |
|---|---|---|
| R-work | 0.2480 (0.2982) | 0.2081 (0.2670) |
| R-free | 0.2570 (0.3006) | 0.2463 (0.3374) |
| Number of nonhydrogen atoms | 9317 | 8908 |
| Protein atoms | 9253 | 8844 |
| Ligand atoms (AMPPNP, Mg2+) | 64 | 64 |
| Number of amino acids | 1186 | 1126 |
| Root mean square bonds (Å) | 0.011 | 0.009 |
| Root mean square angles (°) | 1.30 | 0.89 |
| Ramachandran plot | ||
| Favored (%) | 87.93 | 93.00 |
| Allowed (%) | 8.33 | 6.46 |
| Outliers (%) | 3.74 | 0.54 |
| Rotamer outliers (%) | 11.74 | 3.09 |
| Clashscore | 25.47 | 5.06 |
| Average B-factor (Å2) | 50.35 | 56.74 |
| Protein atoms | 50.36 | 56.68 |
| Ligand atoms | 48.08 | 64.69 |
Statistics for the highest-resolution shell are shown in parentheses.
Proteins
The cDNA encoding for human Lmod1 (UniProt: P29536-1) was purchased from Open Biosystems (now part of GE Healthcare, Little Chalfont, UK), and that of human Lmod2 (UniProt: Q6P5Q4-1) was reconstructed from an incomplete clone purchased previously from Open Biosystems as described in Boczkowska et al. (7). All the Lmod constructs (depicted in Figs. 5 A and 6 A) were cloned between the NdeI and SapI sites of vector pTYB1 (New England Biolabs, Ipswich, MA), which combines a chitin-binding domain for affinity purification and an intein domain for self-cleavage of the affinity tag after purification. Mutations of residues 451EKKL454 and 537LKRV540 to AAAA within helix h1 and the WH2 domain of Lmod2 were generated using the QuikChange Mutagenesis Kit (Stratagene, San Diego, CA). The hybrid construct Lmod1ABS2Lmod2C (depicted in Fig. 6 A) was obtained by introducing silent mutations at the junction between the two proteins using the forward and reverse primers to generate a new RsaI restriction site that was then used for ligation. The ligation product was cloned between the NdeI and SapI sites of vector pTYB1. Peptides (depicted in Fig. 4 A) corresponding to Lmod2 residues P445–Q470 (helix h1) and R519–E544 (WH2 domain) were synthesized by Biomatik (Wilmington, DE).
Figure 5.
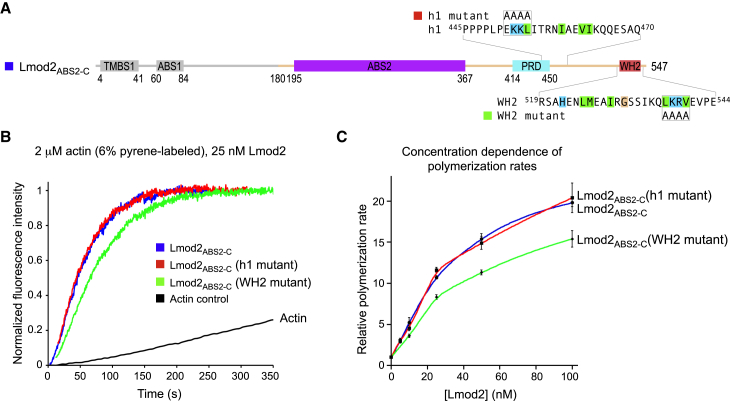
Mutating the WH2 domain but not helix h1 affects the nucleation activity. (A) Domain diagram of Lmod2, showing the sequences and mutations of helix h1 and the WH2 domain. The canonical residues of the WH2 domain implicated in interactions with actin and their proposed equivalents in the inverted helix h1 are background-highlighted according to their chemical character. The two mutants tested here target the conserved LKKV motif of the WH2 domain (537LKRV540) and the corresponding sequence in helix h1 (451EKKL454), which were both mutated to AAAA within construct Lmod2ABS2-C (residues N180–R547). The region N-terminal to ABS2 (light gray) was not included in the constructs tested here. (B) Shown here is nucleation activity of Lmod2ABS2-C compared to those of the helix h1 and WH2 domain mutants. The figure shows the time course of polymerization of 2 μM Mg-ATP-actin (6% pyrene-labeled) in the presence of 25 nM Lmod2 constructs depicted in (A) or the actin control (as indicated). Note that in these experiments, actin alone (control) polymerized at a rate of 2 nM s−1. (C) Shown here is the dependence of the polymerization rates on the concentration of Lmod2 constructs, displayed as the mean of three independent experiments ± SE. To see this figure in color, go online.
Figure 6.
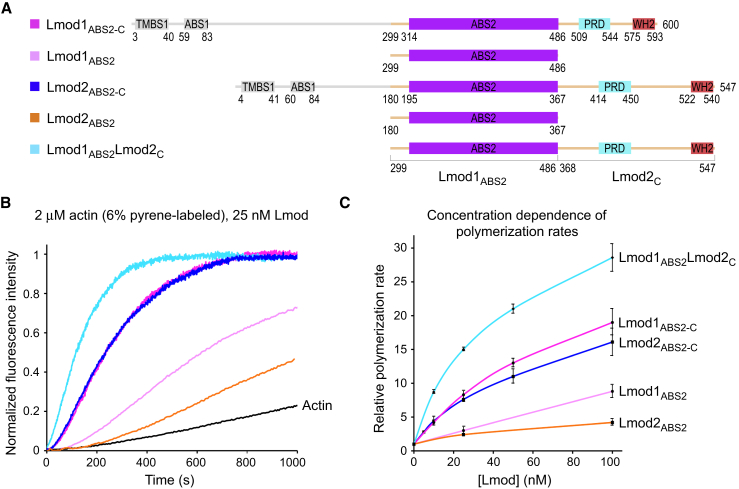
Contribution of the C-terminal extension to the nucleation activity. (A) Shown is the domain diagram of the Lmod1, Lmod2, and Lmod1-Lmod2 hybrid constructs analyzed here. (B) Given here is the time course of polymerization of 2 μM Mg-ATP-actin (6% pyrene-labeled) in the presence of 25 nM of the constructs depicted in (A) or the actin control (as indicated). Note that in these experiments actin alone (control) polymerized at a rate of 0.6 nM s−1. (C) Given here is the concentration dependence of the polymerization rates, displayed as the mean of three independent experiments ± SE. To see this figure in color, go online.
Figure 4.
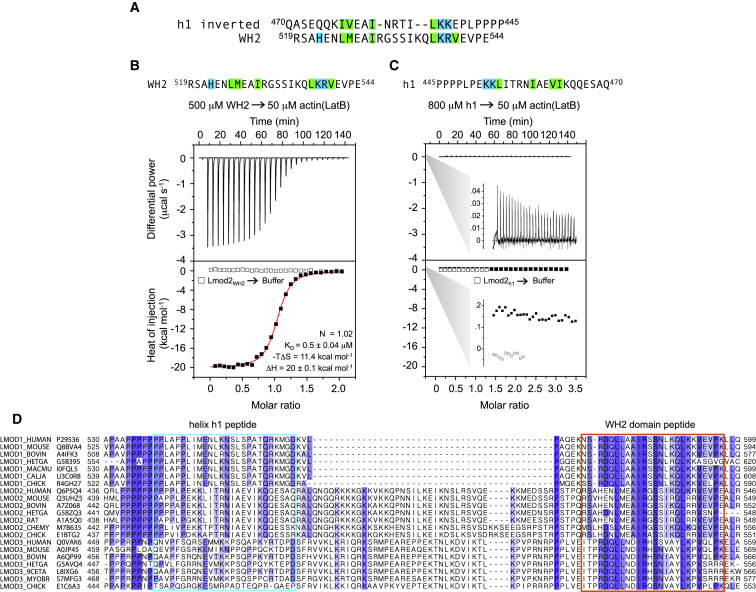
The WH2 domain binds actin whereas helix h1 does not. (A) Shown here is the alignment of the WH2 domain and the inverted helix h1 of Lmod2 proposed by Chen et al. (8) to explain their reported binding of these two sequences to the same site on the actin monomer. The canonical residues of the WH2 domain implicated in interactions with actin and their proposed equivalents in the inverted helix h1 are background-highlighted according to their chemical character. (B and C) Shown here are ITC titrations of the WH2 domain and helix h1 into LatB-actin. The titration of helix h1 is shown using the same scale as in (B), with zooms showing that the heat changes are ∼100-fold weaker for helix h1 than for the WH2 domain. The experiments were performed at 25°C and at the indicated protein concentrations. Open symbols correspond to control titrations of peptides into buffer. Only the titration of the WH2 domain could be fitted to a one-site binding isotherm (solid curve, parameters of the fit shown inside the graph), whereas helix h1 did not bind. Errors correspond to the SD of the fits. (D) Shown here is an alignment of a representative group of Lmod sequences for the region of the C-terminal extension encompassing from the PRD to the WH2 domain. The name of each sequence reflects the specific isoform, organism, and UniProt accession code. For each column, the amino acid conservation decreases from a dark to white background. The boxed regions span the length of the synthetic peptides used in the ITC experiments for the proposed helix h1 actin-binding site and the WH2 domain. Note that whereas the WH2 domain is highly conserved among Lmod isoforms and across different species, helix h1 is not. To see this figure in color, go online.
All the proteins were expressed in BL21(DE3) cells (Invitrogen, Carlsbad, CA), grown in Terrific Broth medium at 37°C until the OD600 reached a value of 1.5–2, followed by 16 h at 20°C in the presence of 0.5 mM IPTG. Cells were harvested by centrifugation, resuspended in 20 mM HEPES pH 7.5, 500 mM NaCl, 1 mM EDTA, and 100 μM PMSF protease inhibitor and lysed using a Microfluidizer apparatus (Microfluidics, Newton, MA). Proteins were purified on a chitin affinity column according to the manufacturer’s protocol (New England Biolabs), followed by gel filtration on a SD200HL 26/600 column (GE Healthcare) in 20 mM HEPES pH 7.5 and 200 mM NaCl. Actin was extracted from actomyosin acetone powder with G-buffer (2 mM Tris pH 8.0, 0.2 mM CaCl2, 0.2 mM ATP, 0.5 mM DTT, and 0.01% NaN3), centrifuged at 20,000 × g for 30 min and polymerized with the addition of 50 mM NaCl and 2 mM MgCl2. The actin pellet was homogenized in G-buffer with the addition of 10 mM DTT. After 1 h, actin was dialyzed exhaustively against G-buffer to remove the DTT and then centrifuged for 45 min at 277,000 × g to pellet any filamentous actin that did not depolymerize, as well as any denatured actin.
Actin polymerization assay
Unlabeled actin and actin labeled at residue C374 with pyrene-iodoacetamide (pyrene-actin) were mixed in G-buffer to produce a stock of 6% pyrene-labeled actin. Actin polymerization was measured as the time-course of the fluorescence increase (20-fold) resulting from the incorporation of pyrene-actin (excitation: 365 nm; emission: 407 nm) into filaments using a Cary Eclipse fluorescence spectrophotometer (Varian Medical Systems, Palo Alto, CA). Before data acquisition, 200 μL Mg-ATP-actin at 2 μM concentration (6% pyrene-labeled) was mixed with 5 μL Lmod constructs in 10 mM Tris pH 8.0, 1 mM MgCl2, 50 mM KCl, 1 mM EGTA, 0.2 mM ATP, 0.5 mM DTT, 0.1 mM NaN3. The concentration of Lmod in the polymerization reaction varied from 5 to 100 nM (as indicated in Figs. 5 and 6). Data acquisition started 10 s after mixing. All the measurements were done at 25°C and were repeated three times. The polymerization of actin alone was carried out with the addition of 5 μL buffer. Relative polymerization rates were calculated as the maximal slope of a polymerization curve (between 0.1 and 0.4 of the maximum fluorescence) divided by the maximal slope of the curve corresponding to actin alone. We note that the polymerization rate of actin alone varies considerably from preparation to preparation. Therefore, here we only compare the polymerization rates of Lmod fragments calculated using the same actin control (i.e., the same actin preparation).
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) measurements were performed on a VP-ITC apparatus (MicroCal, Northampton, MA). Actin was dialyzed for two days against 20 mM HEPES pH 7.5, 100 mM NaCl, 0.2 mM ATP, 1 mM DTT, and 55 μM LatB (ITC buffer). Peptides corresponding to helix h1 and the WH2 domain (Fig. 4 A) were resuspended in ITC buffer, followed by three cycles of lyophilization/resolubilization in 50% (v/v) methanol to remove any trifluoroacetic acid remaining after reverse-phase purification. The peptides were then resuspended in ITC buffer and titrated at a concentration 10- to 15-fold higher (500–800 μM) than that of actin (50 μM) in the ITC cell of total volume 1.44 mL. The experiments were carried out at 25°C. Titrations consisted of 10 μL injections, lasting for 10 s, with an interval of 5 min between injections. The heat of binding was corrected for the heat of injection, determined by injecting the peptides into buffer (open symbols in Fig. 4, B and C). Data were analyzed using the program Origin (OriginLab, Northampton, MA).
Results and Discussion
Revised structure of Lmod2ABS2-C in complex with actin
In studying the published structure of an actin complex with a fragment of Lmod2 comprising from ABS2 to the end of the WH2 domain-containing C-terminal extension (Lmod2ABS2-C; Fig. 1 A) (8), we found that several regions of the modeled Lmod2 polypeptide lacked supporting electron density or did not fit the observed density. Thus, we revised and rerefined the structure using the diffraction data deposited by the authors in the Protein Data Bank (PDB: 4RWT) (Table 1). The revised structure reveals two essentially identical copies of the actin-Lmod2ABS2-C complex in the P1 unit cell (Fig. 1, B–D). For each complex, clear density is observed for actin and the ABS2 (residues P196–Q365) and WH2 domain (residues R519–E541) of Lmod2, but we find no density for any of the residues between these two domains (Fig. 2). Note that we use the corrected sequence and numbering scheme for human Lmod2 (UniProt ID: Q6P5Q4), whereas the deposited structure used an older version of the sequence that is 52-aa shorter overall (Fig. 1 A). Specifically, the crystallized fragment of Lmod2 is 20-aa shorter than the correct sequence, lacking a portion of the PRD, and contains a substitution of a 10 residue stretch of basic amino acids prone to degradation by a Gly-Ser linker of equal length (Fig. 1 A).
Figure 2.
Representative regions of the Fo-Fc and 2Fo-Fc electron density maps in the original (PDB: 4RWT) and revised structures. From top to bottom, the figure shows three consecutive regions of the Lmod2 polypeptide in the original (PDB: 4RWT) and revised (PDB: 5WFN) structures: (A) the PRD, (B) helix h1 (or WH2 domain), and (C) a portion of helix h2 and the connecting linker between helices h1 and h2. For each structure, the first column shows the Fo-Fc electron density map contoured at ±3σ (according to convention, negative densities are red and positive densities are green) and the second column shows the 2Fo-Fc map contoured at 1σ around the Lmod2 atoms in view. The maps of the original and revised structures were calculated using the “Create Maps” module in the program PHENIX (9), with phases derived from the original and revised coordinates, respectively, and using the same structure factor amplitudes deposited with the PDB. Actin, which is mostly unchanged between the two structures, is shown in surface representation. Note that helix h1, immediately after the PRD, was reinterpreted as a WH2 domain with opposite polypeptide directionality in the revised structure. The other regions shown (PRD, helix h2, and the linker between helices h1 and h2) are not present in the revised structure. To see this figure in color, go online.
The two actin molecules in the asymmetric unit are related by a local noncrystallographic twofold symmetry axis
The ABS2 and WH2 domain bound to a given actin molecule in the asymmetric unit (equivalent to the unit cell in the P1 space group) could belong to the same Lmod2 polypeptide, as there is ample space in the crystal lattice to accommodate the missing portions of Lmod2, but it is also possible that they arise from neighboring Lmod2 polypeptides in the crystal lattice. Independent of this consideration, the two actin molecules in the asymmetric unit are related by a local noncrystallographic twofold symmetry axis (Fig. 1 C), which is evidently distinct from the helical symmetry of the actin filament, where successive actin subunits along the left-handed short-pitch helix are rotated by −166.6° (11). Thus, we do not ascribe any biological significance to the dimer formed by this noncrystallographic twofold symmetry, or to those formed by other crystallographically equivalent definitions of the contents of the unit cell (Fig. 1 D).
Differences between the original and revised structures of actin-Lmod2ABS2-C
The two actin molecules in the P1 unit cell and the ABS2 fragments of Lmod2 bound to them are essentially the same in the original and revised structures (Fig. 1 E). However, Chen et al. (8) modeled a long, continuous segment of Lmod2 (P389–R495, corresponding to P441–R547 in the correct sequence, Fig. 1 A) in a manner that spans the two actin molecules of the unit cell (Fig. 1 E, left, green, and red), according to the alternative unit cell definition used in their study (Fig. 1 D). Within this segment, the WH2 domain bound to the second actin molecule is essentially the same in the original and revised structures (residues R519–E541, Fig. 1 E, red). In contrast, we find that the electron density (Fig. 2) does not support the other 78 residues of the Lmod2 C-terminal extension modeled in the original structure (residues P441–Q518, Fig. 1 E, left, green). In particular, the original structure lacks the WH2 domain associated with the first actin molecule. In its place, Chen et al. (8) built a portion of the Lmod2 polypeptide that includes parts of the PRD (which they called “polyP”) and a helix that they called “h1”, and with opposite N- to C-terminal directionality of the polypeptide chain compared to the WH2 domain (Figs. 1 E and 2, A and B). Our reexamination of this region of the structure shows that the electron density clearly corresponds to a WH2 domain (Fig. 2 B). We also do not observe electron density to support modeling of other elements of the Lmod2 sequence between ABS2 and the WH2 domain present in the original structure; this includes a second helical segment (helix h2) and the long loop connecting helices h1 and h2, as well as the last six amino acids after the WH2 domain (Figs. 1 E and 2 C).
Interactions between Lmod2ABS2-C and actin
The specific interactions of ABS2 and the WH2 domain of Lmod2 with actin have been previously delineated, but for completeness are described here briefly. Most of ABS2 consists of an LRR domain, and associates with actin subdomains 1 and 2, burying 11% of the actin surface area, essentially as previously described for the ABS2s of Lmod1 (7) and Tmod1 (6). Key interactions with actin subdomain 1 include conserved salt bridges and electrostatic contacts, and an interaction between Lmod2 R359 and residues E125 and D363 in actin subdomain 1 (Fig. 3 A). Another conserved salt bridge involves R37 in actin subdomain 2 and Lmod2 E276 (Fig. 3 B). The WH2 domain exhibits the typical bipartite binding mode, with its LKKV motif (537LKRV540 in Lmod2) rising along the front face of the actin monomer, roughly following the rift between actin subdomains 1 and 3 (Fig. 3 C) and the helical portion (residues R519–R530) binding in the mostly hydrophobic groove between actin subdomains 1 and 3 (Fig. 3 D). These interactions closely resemble those of other WH2 domains (12, 13, 14).
Figure 3.
Interactions between actin and Lmod2. Residues involved in interactions are labeled and their side chains are shown for actin and Lmod2’s ABS2 (A and B) and the WH2 domain (C and D). (A) Shown here are salt bridges between actin subdomain 1 and Lmod2 ABS2. (B) R37 in actin subdomain 2 interacts with E276 in Lmod2 ABS2. (C) The 537LKRV540 motif of the WH2 domain makes backbone interactions with a loop in actin subdomain 1, and V540 inserts into a hydrophobic pocket in actin. (D) The helix of the WH2 domain makes extensive hydrophobic contacts in the cleft formed between actin subdomains 1 and 3. To see this figure in color, go online.
The WH2 domain binds actin whereas helix h1 does not
Although the PRD and helix h1 are not visualized in the revised structure (Figs. 1 E and 2), we sought to determine whether this poorly conserved region of the Lmod sequence was nonetheless important for Lmod2 function by measuring its ability to bind actin as well as its potential role in nucleation. We used ITC to compare the actin binding affinities of synthetic peptides corresponding to helix h1 and the WH2 domain. Chen et al. (8) noted that the sequence of helix h1 when inverted resembles that of the WH2 domain (Fig. 4 A). Therefore, peptides corresponding to helix h1 and the WH2 domain were designed such that they have the same length (26 amino acids), and correspond to the same actin-binding interface in the structure of Chen et al. (8). In the case of helix h1, the peptide also includes several proline residues of the PRD domain that appeared to interact with actin in their structure (Fig. 1 E). The titration of the WH2 domain peptide (Lmod2 residues R519–E544) at 500 μM into nonpolymerizable LatB-actin at 50 μM produced an exothermic binding reaction at 25°C best described by a single-site binding model with KD = 0.5 μM (Fig. 4 B). Under these conditions, the titration of helix h1 (Lmod2 residues P445–Q470) into LatB-actin did not produce any significant heat release compared to the buffer control, and further increasing the concentration of the peptide to 800 μM did not change this outcome (Fig. 4 C). We therefore conclude that the sequence similarity between the WH2 domain and the inverted helix h1 is purely circumstantial, and does not translate into an ability of helix h1 to bind actin. This conclusion is also consistent with helix h1 being disordered in the crystal structure (i.e., we do not know whether this sequence is actually folded as a helix), whereas the WH2 domain occupies the hydrophobic cleft of both actin subunits in the unit cell (Fig. 1 E). Finally, the inability of helix h1 to bind actin is consistent with the absence of sequence conservation for this region of Lmod among different isoforms and species, in contrast to the WH2 domain that is highly conserved (Fig. 4 D).
The WH2 domain contributes to the nucleation activity whereas helix h1 does not
We then tested the effect of mutations in helix h1 on the nucleation activity of Lmod2. A key structural and functional determinant of the WH2 domain is the so-called LKKV (or LKKT) motif (2), corresponding in human Lmod2 to the sequence 537LKRV540. The proposed equivalent motif in the inverted helix h1 is 451EKKL454 (8) (Fig. 4 A). To assess the effect of mutations that disable either the WH2 domain or helix h1 on the nucleation activity of Lmod2, we mutated these two motifs independently to AAAA within construct Lmod2ABS2-C (Lmod2 residues N180–R547, Fig. 5 A). Using the pyrene-actin polymerization assay, we found that the rate of polymerization of 2 μM Mg-ATP-actin (6% pyrene-labeled) with 25 nM Lmod2ABS2-C (WH2 mutant) was ∼25% lower than that of wild-type Lmod2ABS2-C (Fig. 5 B). This effect was reproducible over a range of concentrations (Fig. 5 C). In contrast, the polymerization activity of Lmod2ABS2-C (h1 mutant) was unaffected compared to that of the wild-type control (Fig. 5, B and C). Collectively, our structural findings and biochemical results argue against a role for helix h1 in actin binding and Lmod2-mediated nucleation.
The C-terminal extension of Lmod contributes to the nucleation activity other than through the WH2 domain
Although we found that helix h1 is not specifically involved in actin binding (Figs. 1 E and 4) or nucleation (Fig. 5), actin binding through the WH2 domain cannot fully account for the contributions of the C-terminal extension to the overall nucleation activity of Lmod2, because completely deleting this extension (construct Lmod2ABS2) results in a ∼75% drop in the polymerization rate over a range of concentrations (Fig. 6). Moreover, we had previously established that the polymerization rate of full-length Lmod2 was slightly higher than that of Lmod1, whereas the isolated ABS2 of Lmod1 had approximately two- to threefold stronger activity than that of Lmod2 (7), as also reproduced here (Fig. 6). Because most of the nucleation activity is contained within the fragment encompassing from ABS2 to the C terminus, this finding suggested that there must be something unique about the C-terminal extension of Lmod2 that makes it an overall stronger nucleator than Lmod1. Therefore, to further explore the contributions of the C-terminal extension to the polymerization activity, we made a hybrid construct consisting of Lmod1’s ABS2 and Lmod2’s C-terminal extension (Lmod1ABS2Lmod2C), i.e., combining the superior nucleation activities of Lmod1’s ABS2 and Lmod2’s C-terminal extension (Fig. 6 A). As anticipated, this construct had approximately twofold stronger nucleation activity than either Lmod1ABS2-C or Lmod2ABS2-C (Fig. 6 B), and this effect was reproducible over a range of concentrations (Fig. 6 C). We thus conclude that the similar nucleation activities of Lmod1ABS2-C and Lmod2ABS2-C result from different contributions of their ABS2s and C-terminal extensions. Specifically, the ABS2 of Lmod1 is a better nucleator compared to that of Lmod2, whereas the C-terminal extension of Lmod2 contributes more to the nucleation activity than that of Lmod1. Because the sequence of the C-terminal extension varies substantially among Lmod isoforms (Fig. 4 D), the actual source of these differences is not precisely understood. However, from the results presented here, we can conclude that the WH2 domain accounts only partially for the contributions of the C-terminal extension to nucleation.
Conclusions
Although the revised structure of actin-Lmod2 presented here cannot be used to support a specific nucleation (or elongation) model, as reported by Chen et al. (8), the results are nonetheless consistent with a previously proposed model of Lmod-driven actin nucleation (3, 4, 7). Biochemical studies have shown that Lmod binds up to three actin subunits to form a polymerization nucleus (3), and that the ABS2 of Lmod displays significant nucleation activity on its own (Fig. 6), contrary to that of Tmod, which has none (7). Structurally, the ABS2 of Lmod is expected to bind at the interface between three actin subunits in a filament-like arrangement to form a polymerization nucleus (4, 7). In support of this model, ABS2 can be readily docked onto the actin filament, as its binding surface on the primary actin subunit (i.e., the one engaged by ABS2 in this structure) is fully exposed in the actin filament. Furthermore, the docked ABS2 closely approaches two additional actin subunits in the filament, one on the short-pitch helix and another one on the long-pitch helix, and the Lmod residues that make contacts with all three actin subunits are well conserved (4, 7). Thus, alone, ABS2 can, in principle, stabilize a polymerization nucleus consisting of three actin subunits disposed in a filament-like arrangement (3, 4, 7), which may explain the ability of this isolated domain to nucleate polymerization. The flexible C-terminal region of Lmod could wrap around the primary (ABS2-bound) actin subunit of the nucleus to position the WH2 domain in the cleft between its subdomains 1 and 3. Yet, considering the length of the C-terminal extension, which varies significantly among Lmod isoforms, it is also possible that the WH2 domain engages an adjacent subunit. Because the WH2 domain-binding cleft on actin participates in intersubunit contacts in the filament (15), the WH2 domain must dissociate to allow the primary polymerization nucleus to elongate; i.e., to allow the addition of actin subunits to the barbed end of the polymerization nucleus. Analogous to Tmods (6), the flexible N-terminal region of Lmod could wrap around the first actin subunit at the pointed end of the nucleus, recruiting one tropomyosin coiled-coil through TMBS1. However, Tmod could ultimately outcompete these interactions of Lmod to bind and cap the pointed end, because Lmod lacks the second tropomyosin binding site of Tmod (i.e., TMBS2) and half of ABS1 (4). Interestingly, Lmod also lacks the D-loop-binding site found immediately N-terminal to ABS2 in Tmod, which is critical for pointed end capping (4, 7). Furthermore, because of the absence of the D-loop-binding site, Lmod binding is not restricted to the pointed end, likely explaining why Lmod can decorate thin filaments along their length in cells (16, 17, 18, 19) and in vitro (16), whereas Tmod cannot (4). The structural and biochemical analysis of Lmod2 and its interaction with actin described here should facilitate further analysis and understanding of Lmod function in sarcomere formation and maintenance.
Author Contributions
Z.Y., M.J.E., and R.D. revised and refined the structure. M.B. and G.R. collected the biochemical data on actin binding and nucleation. M.J.E., Z.Y., M.B., and R.D. wrote the paper and prepared the figures.
Acknowledgments
This work was funded by National Institutes of Health (NIH) grants R01 GM073791 to R.D. and GM110352 to M.J.E.
Editor: David Thomas.
Footnotes
Malgorzata Boczkowska and Zeynep Yurtsever contributed equally to this work.
Contributor Information
Michael J. Eck, Email: eck@crystal.harvard.edu.
Roberto Dominguez, Email: droberto@mail.med.upenn.edu.
References
- 1.Pollard T.D. Actin and actin-binding proteins. Cold Spring Harb. Perspect. Biol. 2016;8:a018226. doi: 10.1101/cshperspect.a018226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominguez R. The WH2 domain and actin nucleation: necessary but insufficient. Trends Biochem. Sci. 2016;41:478–490. doi: 10.1016/j.tibs.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chereau D., Boczkowska M., Dominguez R. Leiomodin is an actin filament nucleator in muscle cells. Science. 2008;320:239–243. doi: 10.1126/science.1155313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fowler V.M., Dominguez R. Tropomodulins and leiomodins: actin pointed end caps and nucleators in muscles. Biophys. J. 2017;112:1742–1760. doi: 10.1016/j.bpj.2017.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krieger I., Kostyukova A., Maéda Y. Crystal structure of the C-terminal half of tropomodulin and structural basis of actin filament pointed-end capping. Biophys. J. 2002;83:2716–2725. doi: 10.1016/S0006-3495(02)75281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rao J.N., Madasu Y., Dominguez R. Mechanism of actin filament pointed-end capping by tropomodulin. Science. 2014;345:463–467. doi: 10.1126/science.1256159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boczkowska M., Rebowski G., Dominguez R. How leiomodin and tropomodulin use a common fold for different actin assembly functions. Nat. Commun. 2015;6:8314. doi: 10.1038/ncomms9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X., Ni F., Wang Q. Mechanisms of leiomodin 2-mediated regulation of actin filament in muscle cells. Proc. Natl. Acad. Sci. USA. 2015;112:12687–12692. doi: 10.1073/pnas.1512464112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adams P.D., Baker D., Terwilliger T.C. Advances, interactions, and future developments in the CNS, Phenix, and Rosetta structural biology software systems. Annu. Rev. Biophys. 2013;42:265–287. doi: 10.1146/annurev-biophys-083012-130253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emsley P., Lohkamp B., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominguez R., Holmes K.C. Actin structure and function. Annu. Rev. Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park E., Graziano B.R., Eck M.J. Structure of a Bud6/Actin complex reveals a novel WH2-like actin monomer recruitment motif. Structure. 2015;23:1492–1499. doi: 10.1016/j.str.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chereau D., Kerff F., Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rebowski G., Namgoong S., Dominguez R. Structure of a longitudinal actin dimer assembled by tandem W domains: implications for actin filament nucleation. J. Mol. Biol. 2010;403:11–23. doi: 10.1016/j.jmb.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von der Ecken J., Müller M., Raunser S. Structure of the F-actin-tropomyosin complex. Nature. 2015;519:114–117. doi: 10.1038/nature14033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skwarek-Maruszewska A., Boczkowska M., Lappalainen P. Different localizations and cellular behaviors of leiomodin and tropomodulin in mature cardiomyocyte sarcomeres. Mol. Biol. Cell. 2010;21:3352–3361. doi: 10.1091/mbc.E10-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsukada T., Pappas C.T., Gregorio C.C. Leiomodin-2 is an antagonist of tropomodulin-1 at the pointed end of the thin filaments in cardiac muscle. J. Cell Sci. 2010;123:3136–3145. doi: 10.1242/jcs.071837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuen M., Sandaradura S.A., Clarke N.F. Leiomodin-3 dysfunction results in thin filament disorganization and nemaline myopathy. J. Clin. Invest. 2014;124:4693–4708. doi: 10.1172/JCI75199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gokhin D.S., Ochala J., Fowler V.M. Tropomodulin 1 directly controls thin filament length in both wild-type and tropomodulin 4-deficient skeletal muscle. Development. 2015;142:4351–4362. doi: 10.1242/dev.129171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez R. Actin-binding proteins—a unifying hypothesis. Trends Biochem. Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]