Abstract
Plasmodium vivax is a complex protozoan parasite with over 6,500 genes and stage-specific differential expression. Much of the unique biology of this pathogen remains unknown, including how it modifies and restructures the host reticulocyte. Using a recently published P. vivax reference genome, we report the proteome from two biological replicates of infected Saimiri boliviensis host reticulocytes undergoing transition from the late trophozoite to early schizont stages. Using five database search engines, we identified a total of 2000 P. vivax and 3487 S. boliviensis proteins, making this the most comprehensive P. vivax proteome to date. PlasmoDB GO-term enrichment analysis of proteins identified at least twice by a search engine highlighted core metabolic processes and molecular functions such as glycolysis, translation and protein folding, cell components such as ribosomes, proteasomes and the Golgi apparatus, and a number of vesicle and trafficking related clusters. Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 enriched functional annotation clusters of S. boliviensis proteins highlighted vesicle and trafficking-related clusters, elements of the cytoskeleton, oxidative processes and response to oxidative stress, macromolecular complexes such as the proteasome and ribosome, metabolism, translation, and cell death. Host and parasite proteins potentially involved in cell adhesion were also identified. Over 25% of the P. vivax proteins have no functional annotation; this group includes 45 VIR members of the large PIR family. A number of host and pathogen proteins contained highly oxidized or nitrated residues, extending prior trophozoite-enriched stage observations from S. boliviensis infections, and supporting the possibility of oxidative stress in relation to the disease. This proteome significantly expands the size and complexity of the known P. vivax and Saimiri host iRBC proteomes, and provides in-depth data that will be valuable for ongoing research on this parasite’s biology and pathogenesis.
Introduction
Malaria caused by Plasmodium vivax is a serious neglected disease that can result in extreme morbidity and possible death [1–3]. It is characterized by approximate 48-hour cycles of reticulocyte invasion, growth, development, and release of new invasive merozoite progeny. The clinical outcome is cyclical high fever and chills, paroxysms, violent headaches, vomiting, diarrhea, and muscle aches; and the parasite can pose a particular threat to pregnant women [4]. Additional clinical observations for both P. vivax and the closely related simian malaria model species P. cynomolgi [5–7], can include an enlarged spleen, thrombocytopenia and severe anemia. This pathogen clearly poses a great public health threat, and examination of its biology, biochemistry, and pathogenesis at the molecular level is important for the development of diagnostics, therapeutics, and vaccines that can reduce disease burden [1–9].
Plasmodium vivax is phylogenetically distant from the pathogen causing the more lethal form of malaria, Plasmodium falciparum [5], thus it is important to study both species in parallel to derive the species-specific interventions important for global efforts aiming to control, eliminate and ultimately eradicate malaria [10]. About 4% of estimated global malaria cases are due to P. vivax, but outside the African continent the proportion of P. vivax infections has been reported as 41% [11]. The problem is even greater when considering this species’ dormant stage reservoir in the liver that can account for relapsing infections and asymptomatic cases that may be a source of parasites for ongoing transmission by mosquitoes, as suggested from macaque infections [6].
Plasmodium vivax merozoites invade reticulocytes [12–13], requiring special receptor-ligand interactions for invasion [14] with a strong predilection for invading CD71 high cells [15], which represent the youngest form of reticulocytes [16–17]. This host cell preference differs significantly from the biology of P. falciparum, which has some preference for reticulocytes but invades RBCs (red blood cells) of all ages. These differences are largely unexplored at the molecular level, and are intriguing since under normal circumstances most CD71 high reticulocytes are known to reside in the bone marrow [16]. After invasion, unlike P. falciparum trophozoite and schizont infected RBC (iRBC) that display knob-like surface structures linked to variant adhesive proteins associated with cytoadhesion and virulence [17–18], P. vivax and P. cynomolgi synthesize elaborate caveolae-vesicle complexes (CVCs), which have been observed all along the host cell membrane, with external caveolae cup-like structures and associated cytoplasmic vesicular and tubular structures [19–20] containing a variety of uncharacterized parasite-encoded membrane proteins [21]. The CVCs have been examined by electron- and immunoelectron-tomography and shown in particular to contain a highly abundant protein, PHIST/CVC-8195, associated with the caveolae and the cytoplasmic face of the tubules [21–22]. Plasmodium falciparum iRBC strongly adhere to endothelial cells deep within the venular vasculature, a process linked to severe disease [23]; endothelial cell adhesion of P. vivax iRBCs, in particular relating to the schizont stage, has only recently been proposed [24–26] and remains to be understood in detail.
Another intriguing difference across the species is that P. vivax and P. cynomolgi express members of the multigene Plasmodium interspersed repeat (pir) family, encoding PIR proteins with multiple predicted localizations (ca. 1200 in P. vivax, including the variant antigen multigene subset known as vir) [27–31], but lack the high molecular weight variant antigen families shared by P. falciparum and Plasmodium knowlesi, which are encoded by the var and SICAvar multigene families, respectively [32–35]. The encoded P. falciparum erythrocyte membrane protein 1 (PfEMP1) [33] and schizont infected cell agglutination (SICA) proteins are expressed at the iRBC surface and undergo switching during an immune response (reviewed in [36]). Whether P. vivax has such mechanisms, involving VIR or other proteins expressed at the surface, remains to be more deeply explored.
For both P. vivax and P. falciparum, the expressed proteome during the pathogen's life cycle in both mosquito vectors and primate hosts would be expected to differ between stages [37–39]. Each of these parasites develops in the blood over a 48-hour time period, from the ring stage of development, through a trophozoite stage in which the pathogen undergoes morphological changes, grows in size, and remodels the host iRBC, to a schizont stage in which 16–24 daughter merozoites are produced and then released into the bloodstream [40]. Experimentally derived proteomes from specific stages can be complicated by low parasitemias (typically less than 1% iRBC) in patient-isolated samples, ethical or logistical barriers drawing blood from patients, an asynchronous life cycle stage composition, and the potential for multiple strains to be present in individual patient samples, which is also likely when samples from multiple patients are pooled.
Many of these challenges can be overcome by use of nonhuman primate (NHP) models, such as the Bolivian squirrel monkey, S. boliviensis [8, 41–43], which allows optimized blood-stage P. vivax infections with blood draws timed to enrich individual life cycle stages, allowing increased association of identified proteins with individual developmental stages and disease processes. Using this animal model and 2D lc/ms/ms, and searches based on the P. vivax (Salvador-I strain; Sal-I) genome with 5459 genes [37], we have previously reported 1375 P. vivax and 3209 S. boliviensis monkey host proteins in 71–91% enriched trophozoite-stage preparations [44]. Another earlier proteomics study based on an Aotus monkey-adapted Colombian isolate reported 238 P. vivax trophozoite-stage proteins and 485 Aotus monkey host proteins in a pooled sample enriched for 70% trophozoites, and 727 P. vivax schizont-stage and 310 Aotus monkey host proteins in a pooled sample enriched for 91% schizonts [45]. Other clinical studies have captured fewer proteins. These include 16 P. vivax proteins from a single patient [46], 154 from a multi-patient pool of isolates [47], and 314 P. vivax proteins from cultured schizont-stage enriched iRBCs from a multi-patient pool also containing gametocytes [48].
With the goal of continuing to expand knowledge of the P. vivax blood-stage cycle, including modeling potential, we have developed two biological replicate trophozoite-schizont transition-stage proteomes from P. vivax (Sal-I strain) iRBCs purified from infections of two S. boliviensis monkeys. These are the first proteomes focusing on the actively growing transition period between the P. vivax trophozoite and schizont stages, including large late-stage trophozoites and early multi-nucleated (2–4) schizonts. Using methods based on four 2D lc/ms/ms runs per proteome and analysis with five database search engines [44, 49] we identified 2000 P. vivax and 3487 S. boliviensis host RBC proteins at a ~2% false discovery rate (FDR) for each search engine. We identified a number of post-translational modifications including oxidized host and parasite proteins suggestive of stress responses, as also reported in the trophozoite-enriched proteome [44], and a number of proteins that may be involved in iRBC cytoadhesion with possible survival benefits to the parasite and pathology consequences for the host. In addition, GO-term enrichment analysis of P. vivax proteins and DAVID functional annotation clustering identified host and/or pathogen enriched protein clusters that are predicted to be important, and perhaps essential, for a variety of iRBC growth and development biological and pathogenic processes.
Materials and methods
Animals and pathogen isolation
Saimiri boliviensis monkeys were acquired by the Yerkes National Primate Research Center (YNPRC), an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) internationally-certified institution, from the Keeling Center for Comparative Medicine and Research, UT MD Anderson Cancer Center, supported by the National Institutes of Allergy and Infectious Diseases.
The animals were socially housed in pairs at the YNPRC, and all housing was in compliance with Animal Welfare Act regulations as well as the Guide for the Care and Use of Laboratory Animals. Standard procedures for splenectomy, monitoring the clinical conditions of the animals, collecting biological samples, treatment, and euthanasia were approved by Emory University’s Institutional Animal Care and Use Committee (IACUC) with the approval # YER-2003225. All nonhuman primates used in this study were provided regular environmental enrichment opportunities consisting of daily feeding enrichment, provision of manipulanda, and physical enrichment. Subjects were regularly monitored for any behavioral signs of distress by the YNPRC behavior management personnel. Animals were trained using positive reinforcement to allow blood collections from the ear without sedation.
Anesthesia was performed with Ketamine (5–10 mg/kg IM) or Telazol (3–5 mg/kg, IM). Euthanasia was performed per the recommendations of the "AVMA Guidelines for the Euthanasia of Animals: 2013 edition". Intravenous (iv) injection of barbiturates is an acceptable method of euthanasia for nonhuman primates, per these recommendations, and was approved by Emory’s IACUC. Animals are first anesthetized with either ketamine or telazol and when possible a catheter is placed in a peripheral vessel to ensure delivery. Pentobarbital 100mg/kg is injected intravenously and the animal is monitored and auscultated for cessation of heart beat and breathing.
Two independent P. vivax Sal-I blood-stage infections were initiated in S. boliviensis monkeys using procedures discussed previously [44]. Briefly, donor S. boliviensis monkeys were inoculated with cryopreserved and reconstituted P. vivax (Sal-I) iRBC monkey-adapted stocks acquired from the Centers for Disease Control and Prevention (CDC) and monitored daily; 0.5–1.0 ml of blood with a parasitemia of 0.5–1% was transferred from donor to recipient monkeys. These iRBCs had been passaged previously at the CDC in splenectomized S. boliviensis to ensure adequate peak parasitemias (typically 1–2%) and high iRBC yields; thus, splenectomies were performed in this study prior to infection to ensure comparable yields. The spleen modulates iRBC parasite variant antigen surface expression in P. knowlesi and P. falciparum infections [50–53]; but, as of yet, there is no known effect of splenectomy on the expression of any particular P. vivax protein.
The parasite density was estimated from microscopy analyses of thin blood smears, counting 2000 RBCs. Blood with respectively 0.5% and 0.9% parasitemia was collected from SB3256 and SB5115 monkeys into sodium heparin tubes and processed through ADP-glass beads and a Plasmodipur filter using standard procedures to remove platelets and white blood cells, respectively. The infected blood was then layered onto a 52% Percoll gradient to concentrate and further purify the iRBCs; platelets were less than 0.1% of the original platelet count after purification. The late-stage trophozoite/early-stage, 2–4 nucleated schizont/gametocyte differential microscopy readings for Proteomes 1 and 2 respectively were 44%/56%/0% and 40%/60%/<1%. These iRBC samples were frozen at -80 C and thawed at a later date for generation of tryptic peptides and subsequent proteomic analysis.
Proteome analysis
Two biological replicate proteomes were analyzed. Plasmodium vivax (Sal-I) iRBC proteins and peptides were prepared for analysis using the FASP-II protocol [54] and analyzed using 2D SCX (strong cation exchange)/C18 RP (reversed phase) lc/ms/ms on a Thermo Scientific (San Jose, CA) LTQ-XL ETD Orbitrap mass spectrometer [44]. Four 2D lc/ms/ms runs were analyzed separately (except as noted) and identified proteins concatenated for each proteome. For Proteome 1 ca. 42 μg peptides were analyzed per run, with 12–15 salt step elutions in each run; four test C18 column runs were also included in run 1. For Proteome 2, ca. 94 μg peptides were analyzed in each of four runs, using 20–27 salt step elutions due to the larger peptide load of the SCX column. All runs used internal lock masses, selected by the mass spectrometer depending on presence of individual polysiloxane or bis(2-ethylhexyl)phthalate lock mass ions, to increase precursor ion mass accuracy, including ions at 371.101233, 391.284286, 445.120024, 519.138815 and 593.157607 m/z [55].
Database search engines used a PvP01 P. vivax reference genome [31] derived database combined with the NCBI S. boliviensis fasta protein database (both obtained from the University of Georgia Informatics Core as part of the Malaria Host-Pathogen Interaction Center, Emory University), combined with common contaminants such as the trypsin used for protein digestion, human cytokeratins, etc. [44]. Although the recently released PvP01 genome is based on a P. vivax Indonesian clinical isolate [31], it was used for analysis in preference to the Sal-I P. vivax reference genome [40], due to improved sequencing reducing fragmentation from over 2500 to 226 scaffolds, improved curation increasing the number of genes with functional attributes from 38% (Sal-I reference genome) to 58%, and improved subtelomere assembly resulting in identification of over 1200 pir genes vs. 346 in the Sal-1 genome [31].
Data analysis utilized five search engines. Andromeda (v. 1.2.0.14, embedded in Maxquant software v. 1.2.0.18) [56] used precursor and fragment ion uncertainties of 13 ppm and 0.8 Da respectively; for each proteome all four 2D lc/ms/ms runs were included in a single analysis with a maximum peptide FDR (false discovery rate) of 0.2 and maximum protein FDR of 0.1; identified proteins were then selected to a maximum protein PEP (posterior error probability) of 0.02. Mascot [57] v. 2.3.02 with Mascot Distiller v. 2.4.2.0 included proteins up to a protein FDR of 2.17%, and used Percolator peptide spectrum match scoring [58]. SEQUEST [59] utilized Percolator peptide scoring embedded in Thermo Proteome Discoverer v. 1.3.0.339 software, with protein PEP maximally 2% calculated using custom Excel macros based on the Protein Prophet algorithm without the mixture model [60, 44]. Two sets of runs were analyzed, one using only carbamidomethyl-cys and met sulfoxide as variable modifications, the second using a more extensive set of oxidative modifications (below). The fourth search engine utilized was Crux with Percolator scoring [61]; proteins were accepted to a maximal protein q value of 0.02. The fifth search engine was MSGF Plus [62], using Mascot to convert Thermo.raw files to mgf format, and Proteowizard [63] to convert output.mzid files to.pepXML files for analysis by IDPicker 3 and IDPAssemble [64]. The maximum MSGF Plus protein FDR was 2%. For protein identification, searches were conducted with a precursor ion tolerance of 13 ppm and product ion tolerance of 0.8 Da, required a single unique peptide for identification [65] and full tryptic specificity, and used a maximum of two missed tryptic cleavage sites.
Identification at least twice by a search engine (similar to [66]) was required for consideration of the protein's function when assessing P. vivax or S. boliviensis biology. Different search engines used different algorithms for protein grouping; proteins are thus presented as individual proteins independent of groups, with information on individual search engine results presented in S1 Table, S2 Table, S3 Table and S4 Table. Pseudogenes were not listed as identified proteins.
To streamline the modification identification protocol used previously for trophozoite-enriched proteomes [41], post-translational modifications (mainly oxidized) were identified in a single SEQUEST database search with Percolator scoring. Variable modifications included optional monoisotopic mass additions of 15.995 Da (oxidation) or 31.990 Da (dioxidation) for C, F, H, M, W and Y, 44.985 Da (nitration) or 60.980 (nitrohydroxylation) for F, H, W and Y, 47.985 Da (trioxidation) for C, W and Y, and 57.021 (carboxamidomethylation) for C, with a maximum of 3 identical modifications per peptide. Only b and y ions were considered for identification; peptides accepted as oxidatively modified had a Percolator [58]—calculated posterior error probability of 0.01 or less, a delta score of 1.00 (i.e. the modified peptide was the only identification for the peptide-spectral match found in the search), and a search engine rank of 1. For oxidative modifications, both trophozoite-schizont transition proteomes were analyzed, and the trophozoite proteomes [44] re-analyzed, using a recently released PvP01 genome database [31] to allow direct comparisons based on the best annotated P. vivax genome to date. Identifications by SEQUEST from separate searches for oxidative modifications and basic modifications (M oxidation, C carbamidomethylation) were combined.
Functional annotation
Functional annotation for piecharts, of proteins identified at least twice by a search engine, was from PlasmoDB [67], Uniprot [68], NCBI Entrez [69], CDD [70], PubMed or the primary literature, KEGG [71], or InterPro [72]. When proteins were associated with multiple functions, the apparent most prominent functional category was listed. The function of human homologs of S. boliviensis proteins is generally listed when available. The source of the functional annotation, when not obvious from the protein description, is listed in a separate annotation column. The "transcription" functional category includes transcription factors, RNA polymerase complex proteins, and proteins involved in RNA polyadenylation, capping and splicing, and RNA transport to the cytoplasm. The "translation" category includes ribosome assembly proteins, ribosomal proteins, and proteins involved in elongation on tRNA and termination.
GO term enrichment (relative to the whole genome) for P. vivax proteins used PlasmoDB [67] release 31, included proteins identified at least twice by a search engine, included GO biological process, molecular function, and cell compartment, and included the default P-value cutoff of 0.05. The Benjamini-Hochberg FDR [73] is listed for each enriched cluster.
For analysis of S. boliviensis identified proteins, we used Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 Bioinformatics Resources [74–75]. Saimiri boliviensis proteins were first blasted against the human genome using BlastP [76] with the top-ranked human protein saved for analysis. A list of human protein accession numbers was then submitted to DAVID for medium stringency classification, against a background of the Homo sapiens genome, using the annotation terms GOTERM_MF_ALL, GOTERM_BP_ALL, GOTERM_CC_ALL, KEGG_PATHWAY, BIOCARTA, BBID, COG ontology, UP_KEYWORDS, and OMIM disease. Individual annotation clusters were generally named by the top term or a consensus term. Clusters are listed with Enrichment Score, and the Benjamini-Hochberg FDR for the top term of the cluster.
Results
Protein identifications
Fig 1 illustrates the representative iRBC stages isolated from biological replicate 1, which contained 44% trophozoites, 56% schizonts, and no apparent gametocytes. The morphology of individual iRBCs illustrates the nature of the trophozoite/schizont transition stages used to derive Proteome 1, as iRBCs 1, 4 and 5 are early nucleated schizonts, while iRBCs 2 and 3 represent large late-stage trophozoites. Biological replicate 2, used to obtain Proteome 2, contained 40% trophozoites, 60% schizonts, and <1% gametocytes.
Fig 1. Giemsa-stained P. vivax iRBC isolated from biological replicate 1.
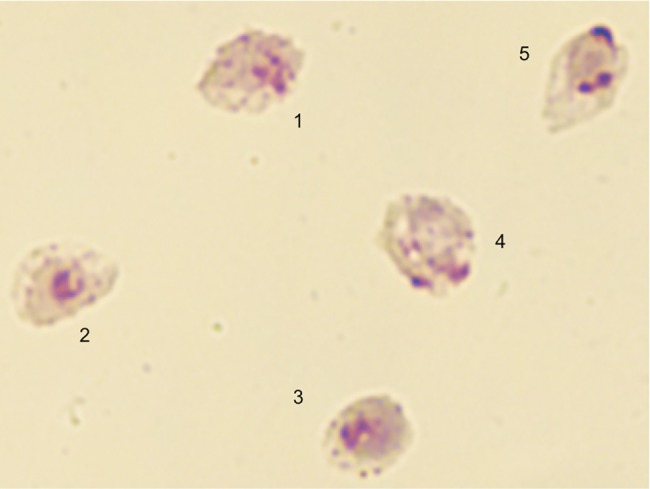
Thin smears were prepared from Percoll-gradient enriched iRBC, and show early nucleated schizonts (iRBC 1, 4 and 5) and large late-stage trophozoites (iRBC 2 and 3).
A summary of proteins identified in Proteomes 1 and 2 is listed in S1 Table (Proteome 1) and S2 Table (Proteome 2). S3 Table lists S. boliviensis and P. vivax proteins combined from both proteomes; in total, 2000 P. vivax and 3487 S. boliviensis proteins were identified by at least one search engine.
The relative abundances of P. vivax and S. boliviensis proteins are listed in S4 Table, calculated using all 8 2D lc/ms/ms runs from both proteomes, as the exponentially multiplied protein abundance index emPAI [77] by Mascot [57]. The most abundant P. vivax protein, glyceraldehyde-3-phosphate dehydrogenase (GPDH), is ca. 15-fold more abundant than the same (but distinct) S. boliviensis protein. Other abundant P. vivax proteins include a number of additional glycolytic enzymes (enolase, phosphoglycerate kinase, lactate dehydrogenase, fructose 1,6-bisphosphate aldolase, pyruvate kinase, triose phosphate isomerase, phosphoglycerate mutase), the four core histones comprising a nucleosome, two heat shock proteins (HSP), ribosomal protein P2, elongation factor 1 alpha, the CVC protein PHIST/CVC-8195 (PVP01_0119200) [22], and an endoplasmic reticulum (ER) calcium binding protein proposed as a target of anti-malarial endoperoxides in P. falciparum [78]. The most abundant S. boliviensis proteins include five hemoglobin chains, ribosomal and ubiquitin-related proteins, the Ig superfamily hypothetical protein C17orf99 homolog, cytoplasmic actin, flavin reductase, two carbonic anhydrase enzymes, and the proinflammatory cytokine IL-36 beta. Nine different complement proteins are identified at least twice by a search engine; of these three (C9, factor I, C1Q) have a relative abundance in the range of 0.15–0.21, and six (factor B, C4-A, C3, C5, C7, C4b binding protein alpha) have a relative abundance in the range of 0.02–0.07).
Protein functional categories
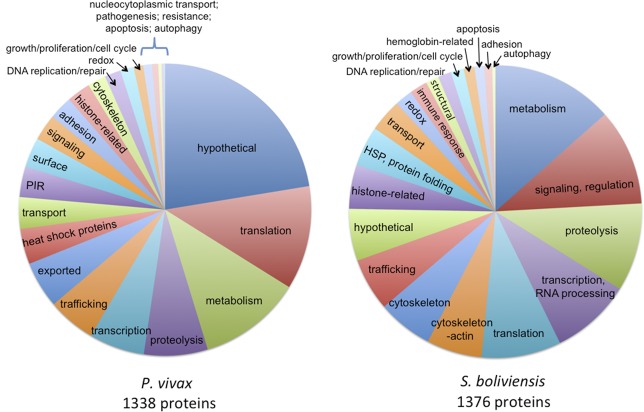
Fig 2 shows pie charts for one overview of functional categories of proteins identified at least twice by a search engine in the analysis of the two trophozoite-schizont transition proteomes; this includes a subset totaling 1338 P. vivax and 1376 S. boliviensis proteins. Individual protein functional categories are listed in S5 Table, along with 15 P. vivax and 64 S. boliviensis proteins annotated as mitochondrial proteins. The largest P. vivax category, including ca. 22% of this proteome, consists of hypothetical proteins without functional annotation. Other major categories include translation, metabolism, proteolysis, transcription and trafficking. Two additional significant categories include Plasmodium exported proteins and PIR/VIR proteins. Saimiri boliviensis iRBC proteins were more evenly divided among a number of categories, the largest being metabolism, signaling, proteolysis, transcription and translation. About half of the identified cytoskeletal proteins were linked to actin. For both organisms, the functional pie charts are similar to those from our previously reported trophozoite-enriched stage [44]; use of the new PvP01 reference genome for database searches here may have decreased the fraction of P. vivax hypothetical proteins. Identification of PIR/VIR proteins was also substantially increased (to 45) compared to analysis with the Sal-I database, which could reflect stage or isolate specific differences though also likely reflects the improved assembly and annotation of the telomeres in this reference sequence
Fig 2. Functional categories of P. vivax and S. boliviensis trophozoite-schizont transition iRBC proteins.
Major categories for both organisms include metabolism, translation, transcription, proteolysis and trafficking. Over 25% of P. vivax proteins (hypothetical and PIR proteins) have no annotated function. Numerous cytoskeletal proteins, particularly actin-related, are identified for S. boliviensis. Details of individual proteins, each identified more than once by a database search engine in the two combined biological replicate proteomes, are in S5 Table. These pie charts are similar to those of trophozoite-stage iRBC proteins [44], however the P. vivax hypothetical protein fraction has decreased. New P. vivax categories include PIR/VIR proteins and Plasmodium exported proteins.
Protein functional clustering using PlasmoDB and DAVID
To further characterize overall function of proteins expressed in Proteomes 1 and 2, we used GO term enrichment analysis, implemented in PlasmoDB [66], to analyze P. vivax proteins (Table 1), and DAVID v. 6.8 [74–75] functional annotation clustering to analyze S. boliviensis proteins (Tables 2 and 3).
Table 1. GO-term enrichment analysis of P. vivax proteins in proteomes 1 and 2.
| GO ID | GO Term, Proteome 1 | Fold enrichment | Benjamini FDR | GO ID | GO Term, Proteome 2 | Fold enrichment | Benjamini FDR |
|---|---|---|---|---|---|---|---|
| Biological Process | Biological Process | ||||||
| GO:0006096 | glycolytic process | 3.96 | 0.010 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER | 3.79 | 0.038 |
| GO:0006090 | pyruvate metabolic process | 3.6 | 0.013 | GO:0009064 | glutamine family amino acid metabolic process | 3.79 | 0.043 |
| GO:0006414 | translational elongation | 3.05 | 0.019 | GO:0006096 | glycolytic process | 3.03 | 0.035 |
| GO:0051169 | nuclear transport | 2.86 | 0.010 | GO:0006090 | pyruvate metabolic process | 2.76 | 0.040 |
| GO:0015991 | ATP hydrolysis coupled proton transport | 2.83 | 0.020 | GO:0006414 | translational elongation | 2.62 | 0.040 |
| GO:0009166 | nucleotide catabolic process | 2.64 | 0.017 | GO:0015991 | ATP hydrolysis coupled proton transport | 2.44 | 0.041 |
| GO:0006184 | GTP catabolic process | 2.55 | 0.019 | GO:0016052 | carbohydrate catabolic process | 2.44 | 0.041 |
| GO:0006457 | protein folding | 2.45 | 0.001 | GO:0006511 | ubiquitin-dependent protein catabolic process | 2.27 | 0.009 |
| GO:0045454 | cell redox homeostasis | 2.38 | 0.019 | GO:0006413 | translational initiation | 2.17 | 0.040 |
| GO:0006511 | ubiquitin-dependent protein catabolic process | 2.14 | 0.017 | ||||
| GO:0006413 | translational initiation | 2.17 | 0.040 | ||||
| Molecular Function | Molecular Function | ||||||
| GO:0070003 | threonine-type peptidase activity | 3.81 | 0.005 | GO:0070003 | threonine-type peptidase activity | 3.28 | 0.009 |
| GO:0002161 | aminoacyl-tRNA editing activity | 3.67 | 0.046 | GO:0051082 | unfolded protein binding | 2.22 | 0.017 |
| GO:0016209 | antioxidant activity | 3.42 | 0.040 | GO:0004812 | aminoacyl-tRNA ligase activity | 2.15 | 0.013 |
| GO:0051020 | GTPase binding | 3.3 | 0.043 | GO:0003743 | translation initiation factor activity | 2.11 | 0.024 |
| GO:0019829 | cation-transporting ATPase activity | 2.75 | 0.036 | ||||
| GO:0051082 | unfolded protein binding | 2.73 | 0.007 | ||||
| GO:0003743 | translation initiation factor activity | 2.28 | 0.036 | ||||
| Cell Component | Cell Component | ||||||
| GO:0005852 | eukaryotic translation initiation factor 3 complex | 4 | 0.009 | GO:0044433 | cytoplasmic vesicle | 3.79 | 0.006 |
| GO:0000785 | chromatin | 3.85 | 0.023 | GO:0005798 | Golgi-associated vesicle | 3.79 | 0.017 |
| GO:0005839 | proteasome core complex | 3.81 | 0.003 | GO:0030137 | COPI-coated vesicle | 3.79 | 0.024 |
| GO:0005773 | vacuole | 3.67 | 0.039 | GO:0000785 | chromatin | 3.32 | 0.027 |
| GO:0015935 | small ribosomal subunit | 3.3 | 0.023 | GO:0005839 | proteasome core complex | 3.28 | 0.008 |
| GO:0030135 | coated vesicle | 3.05 | 0.023 | GO:0033176 | proton-transporting V-type ATPase complex | 2.76 | 0.033 |
| GO:0005798 | Golgi-associated vesicle | 2.93 | 0.044 | GO:0005794 | Golgi apparatus | 2.59 | 0.022 |
| GO:0033176 | proton-transporting V-type ATPase complex | 2.8 | 0.039 | GO:0015935 | small ribosomal subunit | 2.53 | 0.043 |
| GO:0005794 | Golgi apparatus | 2.32 | 0.038 | GO:0031410 | cytoplasmic vesicle | 2.41 | 0.022 |
| GO:0031410 | cytoplasmic vesicle | 2.8 | 0.011 |
Table 2. DAVID-derived S. boliviensis functional annotation clusters in proteomes 1 and 2.
| Enrichment Score | Proteome 1 | Benjamini top term | Enrichment Score | Proteome 2 | Benjamini top term |
|---|---|---|---|---|---|
| vesicles, trafficking | vesicles, trafficking | ||||
| 154.35 | extracellular exosome | 9.10E-178 | 80.19 | extracellular exosome | 5.20E-92 |
| 52.94 | intracellular organelle | 1.50E-58 | 14.81 | membrane-bounded organelle | 1.80E-29 |
| 25.72 | organelle lumen | 6.80E-37 | |||
| 3.71 | cytoplasmic vesicle | 3.10E-06 | |||
| 3.01 | endocytic vesicle lumen | 2.50E-04 | |||
| 2.36 | nucleocytoplasmic transport | 1.40E-05 | |||
| 2.09 | exocytosis | 5.60E-04 | |||
| 2.08 | autophagy | 3.00E-02 | |||
| 2.04 | RNA export from nucleus | 5.60E-05 | |||
| cytoskeleton | cytoskeleton | ||||
| 50.2 | non-membrane-bounded organelle/cytoskeleton | 5.40E-60 | 11.07 | non-membrane-bounded organelle/cytoskeleton | 2.60E-17 |
| 29.67 | adherens junction | 9.50E-53 | 17.49 | adherens junction | 1.10E-25 |
| 19.14 | cytoskeleton | 1.80E-27 | 7.91 | cortical cytoskeleton | 7.00E-10 |
| 11.06 | cortical cytoskeleton | 1.80E-13 | 6.14 | spectrin-associated cytoskeleton | 1.30E-07 |
| 7.32 | actin cytoskeleton | 2.80E-18 | 5.89 | contractile fiber | 5.30E-09 |
| 6.16 | Intermediate filament | 4.90E-13 | 3.89 | actin cytoskeleton | 6.20E-10 |
| 5.91 | microtubules | 4.40E-07 | 2.95 | microtubule cytoskeleton | 1.80E-05 |
| 5.58 | contractile fiber | 4.40E-10 | 2.08 | dynein complex | 2.20E-04 |
| 5.27 | actin filament bundle | 5.90E-06 | |||
| 4.43 | spectrin-associated cytoskeleton | 9.90E-07 | |||
| 2.83 | myosin complex | 4.10E-05 | |||
| 2.01 | actin-myosin filament sliding | 5.50E-06 | |||
| oxidation | oxidation | ||||
| 7.67 | oxidation-reduction process | 5.30E-12 | 5.14 | oxidation-reduction process | 1.20E-06 |
| 5.85 | antioxidant activity | 2.70E-08 | 4.78 | response to oxidative stress | 3.00E-07 |
| 3.28 | cell redox homeostasis | 5.70E-05 | 3.77 | Oxidoreductase | 6.10E-04 |
| 2.99 | hydrogen peroxide metabolic process | 1.10E-06 | 2.94 | glutathione metabolic process | 5.90E-03 |
| 2.85 | Oxidation | 2.40E-07 | |||
| 2.83 | glutathione metabolic process | 1.10E-03 | |||
| 2.4 | response to oxidative stress | 3.00E-07 | |||
| pathogen | pathogen | ||||
| 31.5 | symbiosis, encompassing mutualism through parasitism | 2.80E-34 | 9.7 | symbiosis, encompassing mutualism through parasitism | 4.20E-11 |
| 12.2 | Viral nucleoprotein | 3.00E-15 | 2.62 | inclusion body assembly | 3.00E-03 |
| 7.84 | Pathogenic Escherichia coli infection | 1.80E-11 | |||
| 2.16 | establishment of viral latency | 3.10E-02 | |||
| 3.22 | viral genome replication | 1.40E-04 | |||
| 3.25 | inclusion body assembly | 2.00E-03 | |||
| homeostasis | homeostasis | ||||
| 4.81 | erythrocyte homeostasis | 9.10E-09 | 2.88 | tissue homeostasis | 1.60E-04 |
Table 3. DAVID-derived S. boliviensis functional annotation clusters in proteomes 1 and 2.
| Enrichment Score | Proteome 1 | Benjamini top term | Enrichment Score | Proteome 2 | Benjamini top term |
|---|---|---|---|---|---|
| protein folding | protein folding | ||||
| 14.5 | protein folding | 3.80E-15 | 5.32 | protein folding | 6.80E-16 |
| 3.58 | protein stabilization | 4.90E-06 | 2.71 | response to unfolded protein | 3.40E-06 |
| 3.26 | chaperonin-containing T-complex | 2.80E-06 | 2.62 | inclusion body assembly | 3.00E-03 |
| 3.25 | inclusion body assembly | 2.00E-03 | |||
| 2.31 | unfolded protein response | 1.50E-03 | |||
| 2.19 | protein folding in endoplasmic reticulum | 1.10E-04 | |||
| 2.14 | chaperone mediated protein folding | 4.10E-02 | |||
| macromolecular complexes | macromolecular complexes | ||||
| 25.4 | cellular macromolecular complex assembly | 1.20E-32 | 11.1 | macromolecular complex assembly | 4.40E-13 |
| 25.1 | ribonucleoprotein complex | 1.10E-69 | 11.7 | Proteasome | 3.30E-26 |
| 13.2 | spliceosome | 6.60E-28 | 4.31 | proteasome regulatory particle | 5.50E-11 |
| 9.97 | nucleosome | 1.80E-15 | 3.66 | regulation of proteasomal catabolic process | 1.30E-09 |
| 6.5 | ribosome assembly | 7.80E-10 | 3.02 | Protease | 1.10E-03 |
| 4.48 | proteasome accessory complex | 1.10E-08 | 2.04 | chromatin | 2.00E-04 |
| 2.64 | protein localization to chromatin | 3.10E-04 | |||
| 2.3 | lysosome | 2.00E-03 | metabolism | ||
| metabolism | 9.00 | small molecule metabolic process | 5.90E-19 | ||
| 11.99 | ATP metabolic process | 1.40E-17 | 6.93 | phosphorus metabolic process | 2.20E-09 |
| 11.09 | energy derivation by oxidation of organic compounds | 3.30E-17 | 6.85 | cofactor metabolic process | 5.10E-12 |
| 9.42 | small molecule metabolic process | 2.20E-16 | 4.86 | Protein biosynthesis | 6.20E-10 |
| 5.27 | tricarboxylic acid cycle | 3.20E-06 | 3.6 | aldehyde/NADP metabolic process | 3.10E-07 |
| 4.54 | NADP metabolic process | 1.70E-06 | 2.6 | Porphyrin biosynthesis | 2.80E-02 |
| 2.62 | ribonucleotide metabolic process | 1.50E-10 | 2.47 | regulation of transmembrane transport | 1.40E-04 |
| 2.43 | hemoglobin complex | 2.40E-05 | 2.43 | Purine biosynthesis | 7.90E-04 |
| 2.29 | glycogen metabolism | 5.50E-03 | 2.41 | hemoglobin complex | 1.60E-04 |
| 2.25 | pyruvate metabolism | 8.40E-04 | 2.39 | cysteine metabolic process | 9.90E-04 |
| 2.21 | DNA metabolic process | 5.30E-03 | 2.19 | L-ascorbic acid metabolic process | 2.20E-03 |
| RNA | RNA | ||||
| 46.96 | poly(A) RNA binding | 1.10E-69 | 7.89 | poly(A) RNA binding | 1.80E-10 |
| 9.08 | regulation of RNA stability | 1.30E-23 | 3.33 | translation elongation | 1.00E-04 |
| 5.48 | regulation of translation | 2.10E-08 | 2.56 | Aminoacyl-tRNA synthetase | 3.20E-03 |
| 2.99 | regulation of RNA splicing | 1.20E-05 | 2.22 | translational initiation | 1.10E-02 |
| 2.59 | translation elongation factor activity | 4.10E-03 | |||
| 2.09 | aminoacyl-tRNA ligase activity | 4.20E-07 | |||
| cell death | cell death | ||||
| 4.19 | negative regulation of programmed cell death | 9.20E-06 | 3.44 | regulation of cell death | 1.50E-04 |
| 3.42 | mitochondrion | 2.90E-08 | 3.32 | regulation of neuron apoptotic process | 1.90E-03 |
| 3.42 | apoptotic mitochondrial changes | 2.40E-06 | 2.44 | apoptotic mitochondrial changes | 1.70E-03 |
| 3.31 | regulation of apoptotic signaling pathway | 3.40E-05 | |||
| 3.25 | regulation of neuron apoptotic process | 2.30E-03 |
Table 1 lists some representative P. vivax non-redundant GO-term enrichment clusters for both proteomes, with two-fold or higher enrichment. The entire set of clusters is listed in S7 Table (Proteome 1) and S8 Table (Proteome 2). A number of enriched clusters are common to both biological replicate proteomes, as expected, including glycolysis and pyruvate metabolism; translation initiation, elongation, the small ribosomal subunit and aminoacyl tRNA ligase activity; the ubiquitin-proteasome system; unfolded protein binding, and chromatin. Another set of related clusters common to both proteomes includes intracellular vesicle systems (e.g. cytoplasmic vesicles, Golgi-associated vesicles and the Golgi apparatus, coated vesicles), and related processes such as V-type ATPase activity. Proteome 1 uniquely also included clusters for antioxidant activity and cell redox homeostasis.
Table 2 presents results from DAVID-based protein annotation clustering of S. boliviensis proteins. Each individual entry represents a cluster that is at least two-fold enriched, with an enrichment score (relative to the human genome) and the Benjamini-Hochberg FDR listed for the top term of each cluster. The clusters are grouped into sets of similar clusters, representing vesicles and trafficking, the cytoskeleton, oxidation, pathogen processes, and homeostasis. The most highly enriched single cluster includes extracellular exosomes for both Proteomes 1 and 2, with an 80-154-fold enrichment. The cytoskeleton set of clusters consists of 8–12 individual clusters for Proteomes 1 and 2, including enrichment of e.g. the actin cytoskeleton, microtubules, spectrin-associated cytoskeleton, and adherens junctions. Analysis of both proteomes highlights a set of oxidation-related clusters, which includes response to oxidative stress and glutathione metabolic processes, and in the more oxidized Proteome 1, antioxidant activity, cell redox homeostasis, hydrogen peroxide metabolic processes and oxidation clusters were prominently identified. Both proteomes contain the ~10–30 fold enriched cluster "symbiosis encompassing mutualism through parasitism"; Proteome 1 also contains clusters for pathogenic E. coli infection and three viral infection-related clusters. Details of the clusters are presented in S9 Table.
Table 3 presents additional S. boliviensis enriched protein annotation clusters related to protein folding and the response to unfolded proteins, macromolecular complexes (e.g. the proteasome (both proteomes), and ribosome, spliceosome, and nucleosome in Proteome 1), metabolism, RNA-related clusters (particularly translation and splicing), and a set of cell death clusters including apoptotic mitochondrial changes, regulation of neuron apoptotic processes, and regulation or negative regulation of (programmed) cell death. Additional clusters in Proteome 1 include regulation of apoptotic signaling pathway and the mitochondrion. Details of each individual cluster are presented in S9 Table.
Cytoadhesion proteins
S10 Table lists P. vivax proteins from the trophozoite-schizont transition proteomes that are annotated as, or predicted to be potentially involved in cytoadhesion, and identified at least twice by a search engine. These include four merozoite rhoptry proteins [45, 79–80] among five proteins associated with host-cell binding or invasion, including an erythrocyte binding protein and a Duffy receptor-binding protein [14]. An additional 29 MAAP-predicted adhesins [81] were identified, as were 45 PIR/VIR proteins, some of which may have functions other than adhesion, e.g. antigenic variation.
Sixteen S. boliviensis proteins are also annotated as involved in cytoadhesion, including the transferrin receptor (CD71 antigen, here identified as transferrin receptor protein 1), shown to be an important marker (when at high levels) of reticulocytes preferentially invaded by P. vivax [15] and P. yoelli [16]. Other proteins may directly mediate cell-cell adhesion, such as thrombospondin-3, fermitin family homolog 3, or protocadherin 10, or may be less directly involved in cell adhesion, such as CECAM 18, ankyrin-3 or talin-2.
Protein oxidation and nitration
We previously reported significant oxidation and nitration of both P. vivax and S. boliviensis trophozoite-stage enriched proteins in two biological replicates [44], including as examples S. boliviensis hemoglobin, actin, and the P. vivax CVC protein PHIST/CVC-81-95 [22]. Table 4 compares these observations to the same proteins in the transition Proteomes 1 and 2 identified here. Datasets for all four proteomes were analyzed, using the newly released P. vivax reference genome sequence [31] for residue oxidation and nitration in a single Sequest database search, as discussed in the methods section. Both of the hemoglobin alpha and beta chains are significantly oxidized in all four proteomes. Oxidation of cytoplasmic actin and the PHIST/CVC-81-95 protein are more variable, particularly in the trophozoite proteomes; details of high-confidence trophozoite-schizont transition oxidized peptides, and their cognate proteins, are contained in S6 Table. With two exceptions, all four proteins have a significant number of oxidized residues in both the trophozoite and trophozoite-schizont transition proteomes.
Table 4. Hemoglobin, actin, PHIST/ CVC-8195 protein oxidized/nitrated residues.
| Proteome: | Trophozoite 1 | 2 | Trophozoite-Schizont Transition 1 | 2 |
|---|---|---|---|---|
| Protein | # oxidized residues1,2 | |||
| S. boliviensis Hb β chain | 17 | 20 | 36 | 24 |
| S. boliviensis Hb α chain | 18 | 14 | 37 | 28 |
| S. boliviensis actin, cytoplasmic1 | 20 | 0 | 10 | 6 |
| P. vivax PHIST/CVC-8195 (PVP01_0119200) | 6 | 0 | 8 | 6 |
1 oxidized and nitrated residues include met, cys, his, trp, tyr and phe (see methods section)
but exclude met sulfoxide as met can be oxidized in solutions exposed to air.
2 peptide PEP ≤ 0.01
Table 5 examines the oxidized amino acids present in high-confidence tryptic peptides from both Proteomes 1 and 2, comparing the fraction of oxidized residues to residues in a control Mycobacterium smegmatis proteome prepared using the same FASP-II protocol, electrosprayed under identical conditions [44], and analyzed with the identical Sequest/Percolator workflow as the two trophozoite-schizont proteomes. Proteome 1 generally contains a higher fraction of oxidized residues than Proteome 2. With two exceptions (singly oxidized met- which is readily produced by dissolved oxygen in solution [82], and trp), oxidized met, tyr, trp, cys, phe and his occur at higher levels in Proteome 1 than in the control proteome. Proteomes 1 and 2 have higher levels of nitrated or nitrohydroxylated residues than the control proteome, in which neither modification is observed under these analytical conditions. Proteome 2 has several modifications at lower levels than the control proteome (e.g. singly oxidized met, tyr, and trp), but most highly oxidized residues (his, phe, cys, tyr, met) are more prevalent in Proteome 2 than in the control proteome. Excluding proteins with only methionine oxidized to met sulfoxide, and now analyzed identically to the transition proteomes, trophozoite Proteome 1 had the largest number of oxidized host (274) and parasite (72) proteins, compared to trophozoite Proteome 2 (18 host and 12 parasite highly oxidized proteins). Combined, transition trophozoite-schizont Proteomes 1 and 2 have 74 oxidized S. boliviensis proteins or subunits with at least one high confidence oxidized residue and 144 oxidized P. vivax proteins or subunits.
Table 5. Trophozoite-schizont transition proteome oxidative modifications1.
| Proteome 1 | Proteome 2 | Control | ||||
|---|---|---|---|---|---|---|
| peptides: | 5028 | 4377 | 3043 | |||
| aa, mod | # | Fraction | # | Fraction | # | Fraction |
| met | 1255 | 1449 | 604 | |||
| unmodified | 133 | 0.1060 | 899 | 0.6204 | 48 | 0.0795 |
| O | 994 | 0.7920 | 491 | 0.3389 | 522 | 0.8642 |
| O2 | 128 | 0.1020 | 109 | 0.0752 | 34 | 0.0563 |
| tyr | 2162 | 2282 | 1137 | |||
| unmodified | 1990 | 0.9204 | 2240 | 0.9816 | 1121 | 0.9859 |
| O | 81 | 0.0375 | 13 | 0.0057 | 14 | 0.0123 |
| O2 | 12 | 0.0056 | 7 | 0.0031 | 2 | 0.0018 |
| O3 | 13 | 0.0060 | 15 | 0.0066 | 0 | 0.0000 |
| NO2 | 60 | 0.0278 | 6 | 0.0026 | 0 | 0.0000 |
| NO2OH | 6 | 0.0028 | 1 | 0.0004 | 0 | 0.0000 |
| trp | 180 | 295 | 401 | |||
| unmodified | 122 | 0.6778 | 254 | 0.8610 | 233 | 0.5810 |
| O | 25 | 0.1389 | 25 | 0.0847 | 136 | 0.3392 |
| O2 | 17 | 0.0944 | 16 | 0.0542 | 26 | 0.0648 |
| O3 | 11 | 0.0611 | 0 | 0.0000 | 6 | 0.0150 |
| NO2 | 2 | 0.0111 | 0 | 0.0000 | 0 | 0.0000 |
| NO2OH | 7 | 0.0389 | 0 | 0.0000 | 0 | 0.0000 |
| cys | 457 | 796 | 149 | |||
| unmodified | 20 | 0.0438 | 1 | 0.0013 | 0 | 0.0000 |
| CAM2 | 403 | 0.8818 | 784 | 0.9849 | 149 | 1.0000 |
| O | 16 | 0.0350 | 0 | 0.0000 | 0 | 0.0000 |
| O2 | 8 | 0.0175 | 2 | 0.0025 | 0 | 0.0000 |
| O3 | 10 | 0.0219 | 9 | 0.0113 | 0 | 0.0000 |
| phe | 2639 | 2659 | 1870 | |||
| unmodified | 2513 | 0.9523 | 2611 | 0.9819 | 1846 | 0.9872 |
| O | 68 | 0.0258 | 30 | 0.0113 | 21 | 0.0112 |
| O2 | 32 | 0.0121 | 16 | 0.0060 | 3 | 0.0016 |
| NO2 | 13 | 0.0049 | 1 | 0.0004 | 0 | 0.0000 |
| NO2OH | 13 | 0.0049 | 1 | 0.0004 | 0 | 0.0000 |
| his | 1922 | 2600 | 1259 | |||
| unmodified | 1813 | 0.9433 | 2539 | 0.9765 | 1246 | 0.9897 |
| O | 42 | 0.0219 | 26 | 0.0100 | 10 | 0.0079 |
| O2 | 41 | 0.0213 | 33 | 0.0127 | 3 | 0.0024 |
| NO2 | 16 | 0.0083 | 2 | 0.0008 | 0 | 0.0000 |
| NO2OH | 10 | 0.0052 | 0 | 0.0000 | 0 | 0.0000 |
1 All peptides have an observed precursor ion mass less than 5 ppm from the theoretical mass;
2 carboxamidomethyl
Discussion
We have identified 2000 P. vivax and 3487 S. boliviensis trophozoite-schizont transition proteins by at least one of five search engines, using two biological replicates, expanding identifications from previous P. vivax trophozoite and schizont proteomes, and enhancing the modeling potential of this parasite’s biology by the broad research community. All data has been deposited in PlasmoDB. In addition to analysis of each biological replicate with four separate 2D lc/ms/ms runs, and use of higher amounts of tryptic peptides than our P. vivax trophozoite proteome published in 2015 with 1375 P. vivax and 3209 S. boliviensis proteins [44], analysis here using five search engines [83–84, 66, 44] has aided this process. The current analysis also benefits from use of a database derived from a more comprehensive reference genome sequence [31], which may reduce the percentage of unannotated proteins from 33% in [44] to 22% here.
Interestingly, the S. boliviensis host transferrin receptor protein 1 (transferrin receptor, or CD71 antigen) was identified in the trophozoite-enriched as well as the current proteomes, a point worth noting given P. vivax’s known preference of invading CD71 high reticulocytes and open questions surrounding the parasite-host receptor-ligand and invasion requirements [15, 16, 85]. The late trophozoite represents the growing parasite with dramatic host cell modifications, including the development of CVCs; and the early, 2–4 nucleated schizont stage represents the beginning of parasite division and the development of new progeny, the infectious merozoite forms of the parasite. In the future, a comparable proteome of the matured P. vivax schizont stage–with up to 16 merozoites–would be valuable for the characterization of late-stage schizont proteins including a predominance of merozoite proteins that are important for egress from their host RBCs and invasion of reticulocytes to propagate the blood-stage infection as well as development of new sexual stage forms for infection of the Anopheles mosquito vector and continued transmission. While genome and transcriptome studies (37–39, 31) can predict this parasite species and stage-specific proteomes and help generate hypotheses, proteomic studies provide the evidence of protein expression. Future time course analyses with integrated omics (e.g. transcriptome, proteome, lipidome, and metabolome) will help develop a deeper appreciation of the parasite’s unique biology, over ~48 hours between the invasion, takeover, and destruction of the reticulocyte with the release of new merozoite progeny.
The most abundant P. vivax trophozoite-schizont stage protein is GPDH, ca. 15-fold more abundant than the same (but distinct) S. boliviensis protein, and ca. 6–10 fold more abundant than additional P. vivax glycolytic enzymes. Elevated levels of the P. vivax glycolytic enzymes are consistent with 50–100 fold increased glucose consumption in P. falciparum-infected erythrocytes [86], as glycolysis is this pathogen's sole source of energy [87]. Enzymes such as GPDH can have additional roles beyond metabolism, including functions in oxidative stress and apoptosis [87]. Other abundant P. vivax proteins include heat shock proteins (HSPs), histones H4 and H2B, and elongation factor-1-alpha, as well as the P. vivax caveolae vesicle complex PHIST protein CVC-81-95 reported originally to be a main protein of the CVCs [21–22]. The P. falciparum schizont-stage proteome also includes as highly abundant proteins GPDH and other glycolytic enzymes, histones H4 and H2B, three HSPs and elongation factor-1-alpha [88].
As expected, the most abundant S. boliviensis protein identified is hemoglobin, whose alpha subunit is ca. 160-fold above levels of the next most abundant non-hemoglobin protein, GPDH. Two host carbonic anhydrases are identified here at relatively high levels. Carbon dioxide is essential for iRBC growth, pyrimidine biosynthesis and control of intracellular pH [89]; carbonic anhydrase facilitates CO2 transport across the RBC plasma membrane. This parasite enzyme has been noted as a potential malaria drug target [89–90]; the presence of significant levels of host iRBC carbonic anhydrases may be a consideration for development of such inhibitors. Identification of nine complement components may be relevant given observations of complement fixation in P. vivax malaria [91] and complement activation by the surface of P. falciparum iRBC [92–93] and P. knowlesi [94].
Protein functional clusters
Protein function is presented using several methods. Piecharts give a broad overview of the proteome and highlight some significant areas such as hypothetical proteins, exported proteins, and PIR proteins less readily observed by P. vivax GO-term enrichment. DAVID functional annotation clustering and PlasmoDB-based GO-term enrichment focus on gene sets enriched for particular molecular functions, biological processes or cellular locations.
Little is known about the restructuring of the host reticulocyte and the development of numerous caveolae vesicle complexes (CVCs), which include parasite-encoded proteins positioned all along the P. vivax infected host cell membrane [20–22]. In addition to the P. vivax PHIST protein CVC-8195 [22], other CVC proteins [21] may well be represented among the hypothetical proteins identified and further research will be required to confirm their presence, locations and functions. Pertinent to these distinctive biological features, with much yet to be discovered on the functions of the CVCs with their complex array of tubules and vesicles [22], results from both PlasmoDB and DAVID highlight intracellular vesicles and processes, and organelle and macromolecule trafficking. These data suggest that these processes are part of both the host and parasite encoded iRBC biology. The highest DAVID enrichment score is obtained for extracellular exosomes, perhaps reflecting exosome-mediated loss of cell surface molecules in the transition from reticulocytes to mature RBCs (or iRBCs) [15, 95–96]; P. yoelii exosomes have in fact been shown to contain parasite proteins, and these may be involved in cell-cell communication and immune modulation [97]. Proteins important for vesicle trafficking have also been identified in an uninfected murine reticulocyte proteome [98]. Retrograde vesicle transport from the Golgi apparatus to the ER, identified as a P. vivax cluster from Proteome 2, is important for secreted proteins; 69 exported proteins, related proteins, or Plasmodium exported proteins were also identified. This retrograde transport can involve COPI-coated vesicles, the Golgi apparatus, and Golgi-associated vesicles [99], all of which are enriched P. vivax GO-term clusters, as well as protein folding in the ER. Gautier et al. [100] identified 33 Golgi proteins that partitioned 20% or more into cultured human reticulocytes after orthochromatic cell enucleation. Organelle trafficking can involve autophagy, important for elimination of mitochondria during reticulocyte development [101–103], apicoplast maintenance and protein/organelle trafficking [104], and apicoplast-mediated [105–106] iRBC cell death by atypical autophagy [107]. ROS and RNS can be linked to autophagy [108] and may connect autophagy and apoptosis [109].
A second area of protein annotation cluster enrichment in Proteomes 1 and 2 includes DAVID-derived host cytoskeleton-related clusters, e.g. the actin cytoskeleton, microtubules, the spectrin-associated cytoskeleton, contractile fibers, and adherens junctions. When orthochromatic erythroblasts enucleate to form reticulocytes, cytoskeletal proteins, including the spectrin cytoskeleton, microtubules, myosin and actin [110] are retained in the reticulocyte. The cytoskeleton significantly rearranges during reticulocyte maturation, which includes proteasome degradation of tubulin and actin [111]; both DAVID and PlasmoDB also identify enriched proteasome or ubiquitin-dependent protein catabolism clusters here. Although not enriched at least twofold in GO-term clusters, a number of P. vivax cytoskeletal proteins (e.g. alpha and beta tubulin, dynein, myosin A and E, several actins and actin-like proteins) were also identified.
A third set of enriched clusters includes host protein DAVID-identified cell death-related clusters in both proteomes, e.g. regulation of (programmed) cell death, apoptotic mitochondrial changes, and regulation of neuron apoptotic processes. Reticulocytes are enucleated; however anucleate cells can undergo forms of apoptosis. Anucleate platelet apoptosis has some features in common with nucleated cell apoptosis, e.g. expression of pro- and anti-apoptotic proteins, depolarization of the mitochondrial inner membrane and cytochrome c release, surface exposure of phosphatidyl serine, activation of caspase 3, platelet shrinkage and fragmentation [112]. Red blood cells, without a nucleus or mitochondria, undergo programmed cell death (eryptosis) similar to apoptosis, in which the cells shrink, undergo membrane blebbing, with phosphatidylserine exposed at the surface to enable phagocytosis by macrophages [113–114]. Eryptosis can be induced by a variety of xenobiotics, Ca2+ entry, ceramide formation, caspase stimulation, energy depletion, and oxidative stress [114]. Plasmodium falciparum infection of erythrocytes can induce oxidative stress and trigger eryptosis [115]; premature eryptosis of iRBC before Plasmodium release may protect against malaria [115]. Reticulocytes contain mitochondria, which are eliminated by the time they mature to normocytes [98, 101, 116]. We identified ca. 15 host proteins annotated as involved in apoptosis including phospholipid scramblase 1, which may expose phosphatidyl serine on the surface of iRBC, allowing iRBC phagocytosis by macrophages [117]. We also identified the P. vivax anti-apoptotic protein bax inhibitor 1, which could inhibit bax-mediated apoptosis signaling and premature iRBC death, as well as 64 host and 15 parasite mitochondrial proteins. It is possible that some of these proteins are contaminants from platelets not completely removed during iRBC purification; however in a proteome of human reticulocytes isolated from cell culture, 6 apoptosis proteins partition after orthochromatic erythroblast enucleation into the reticulocyte at a level of 5% or higher, as do 105 mitochondrial proteins and 198 nuclear or nucleolar proteins [100]. It is thus possible that a form of apoptosis or eryptosis may be relevant to P. vivax iRBC biology and the normal host response to parasitism.
Protein oxidation and nitration
We evaluated oxidized P. vivax and S. boliviensis proteins in the two transition proteomes, as before for a trophozoite proteome [44], as they are indicative of possible stress reactions. To control for protein oxidation due to sample preparation and handling, we compared the levels of oxidized residues to those in a proteome from cultured Mycobacterium smegmatis [44], which does not contain food vacuoles; generally, higher levels of oxidized residues were observed in both transition proteomes when compared to the control proteome. The same FASP-II sample preparation protocol, and electrospray conditions, were used for all three proteomes. Peptides from each proteome were electrosprayed from silica capillary columns at or below 2.1 kV, avoiding higher (3.5 kV) spray voltages associated with corona discharge-associated oxidation from stainless steel capillary columns [118], which were not used here. Although the concentration of all oxidizing species will be significantly diluted upon addition of 4% SDS for cell lysis, it is possible that some oxidative modifications could occur after cell lysis if oxidizing species continue to be produced.
Proteome 1 appears to be more extensively oxidized than Proteome 2 (Table 5), which in turn appears to be mostly more oxidized than the control proteome. The identification by DAVID of host S. boliviensis functional annotation clusters for response to oxidative stress, antioxidant activity, cell redox homeostasis, oxidation, hydrogen peroxide metabolic processes, glutathione metabolic processes, and P. vivax cell redox homeostasis and antioxidant enriched GO-term clusters in the more heavily oxidized Proteome 1 (Table 5), suggests the presence of iRBC proteins to counteract oxidative stress in these iRBC. The observation of a variety of oxidized protein residues, compared to the control proteome, is consistent with observations of an internal oxidizing environment in P. falciparum iRBC [40, 119]. As such, these are worth further consideration on the path to understanding P. vivax pathogenesis.
Since protein nitration or oxidation in this context is a chemical process, the rate of reaction may follow second order kinetics, thus the rate, and extent of reaction at a fixed time, will depend on the concentration of both the protein side chains accessible to a reaction, and the concentration and reactivity of oxidizing/nitrating species; this may bias identifications to abundant proteins (e.g. hemoglobin or actin) or those close to the site of ROS (reactive oxygen species) or RNS (reactive nitrogen species) generation. Highly reactive hydroxyl radicals with a short lifetime (ca. 2 ns) will derivatize proteins only within ca. 20 Å of their source [120]. Our observations provide a list of hydroxylated proteins in this category and may suggest their presence close to or within food vacuoles or other cellular sources (Fig 3). The presence of antioxidants or enzymes degrading ROS (e.g. peroxiredoxins [121], superoxide dismutases, glutathione and thioredoxin systems [122]) will also affect the process of protein oxidation/nitration.
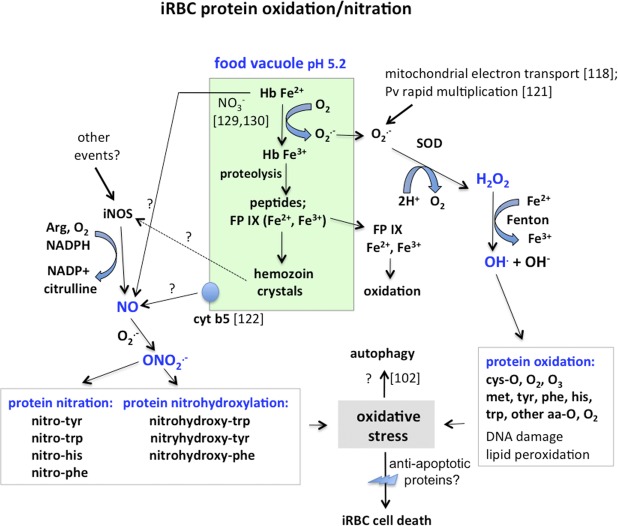
Fig 3. Summary of P. vivax iRBC oxidative reactions [40, 119–122].
Hemoglobin (Hb) oxidation in food vacuoles may be central to generation of superoxide and production of hydrogen peroxide (H2O2), leading to production of hydroxyl radicals and observed protein sidechain oxidations. Food vacuoles may also be central to production of nitric oxide (NO) [123–124], which could be produced by additional pathways [125–128], leading to reaction with superoxide, generation of peroxynitrite (ONO2), and nitration and nitrohydroxylation of additional protein sidechains. This protein oxidation and nitration may lead to or be part of iRBC oxidative stress. FP IX (ferri- or ferroprotoporphyrin IX), iNOS, inducible nitric oxide synthase; SOD, superoxide dismutase; Cyt b5, Cytochrome b5.
ROS can be generated by degradation of hemoglobin in food vacuoles [121], the high metabolic rate of the multiplying parasite and increased mitochondrial respiratory chain production of superoxide [122], and the host immune response [119]. The low food vacuole pH of ca. 5.2 [119] is associated with oxidation of oxyhemoglobin ferroprotoporphyrin IX to ferriprotoporphyrin IX and production of superoxide, which can spontaneously dismutate in food vacuoles to H2O2 and O2 [119]; in the cytosol superoxide is degraded to H2O2 and O2 by superoxide dismutase. The Fenton reaction, utilizing Fe2+, produces OH. radicals from H2O2 [125], which can oxidize protein residues. Oxidation of some proteins, such as S. boliviensis actin at Y240, may in fact result in altered function, as noted earlier [44].
The significant protein nitration we observe could result from induction of the pathophysiological NO/ONOO cycle, some elements of which are observed here [125]. However the source(s) of nitric oxide (NO) and peroxynitrite are unclear; due to the high reactivity of peroxynitrite, NO generation should be local [129]. Nitric oxide and RNS are present in P. falciparum trophozoite food vacuoles [123–124]; their production could be due, in P. falciparum, to a food vacuole-associated cytochrome b5 [123–124]. Plasmodium vivax trophozoite and schizont iRBC also contain food vacuoles; thus, the S. boliviensis cytochrome b5 identified in the transition proteome could potentially contribute to NO generation. Nitric oxide can also be generated by reaction of deoxyhemoglobin with the erythrocyte cytosolic metabolite nitrite [130–131], which is present at ca. 300 nM levels in uninfected erythrocytes [123].
It is also possible that NO production is initiated in P. vivax iRBC by other events. The hemoglobin catabolite hemozoin, produced in iRBC food vacuoles, increases iNOS (inducible nitric oxide synthase) in macrophages [126]. Induction of iNOS [127] in iRBC could result in high levels of NO, and reaction of the NO with superoxide to produce peroxynitrite (Fig 3), which in turn can nitrate and nitrohydroxylate [120, 132] proteins; and, in some cells iNOS can induce apoptosis [133].
Cytoadhesion
Virulence of P. falciparum is due in part to strong iRBC adhesion to the endothelium in the deep venular vasculature [134]. Plasmodium vivax iRBC may adhere to endothelial and other cell types under static and flow conditions [24], adhere to ICAM-1 under flow conditions via specific VIR proteins [28]; or undergo (cytoadhesive) rosetting with uninfected RBC [135]. Lower levels of P. vivax schizont iRBC observed in the blood may be due to enhanced schizont adhesion compared to other stages of asexual development [26]. As iRBC cytoadherence could utilize both host and pathogen proteins, we have examined the P. vivax trophozoite-schizont transition proteome for proteins that might be involved in adhesion. Enhanced sequestration of P. falciparum iRBC may involve altered expression of endothelial cell adhesion receptors linked to oxidative stress [119]. In view of our observation of highly oxidized host and parasite proteins, observation of a number of host protein enriched functional clusters involved in oxidative stress or the response to oxidation, and parasite protein GO term clusters enriched in proteins involved in antioxidant activity or cell redox homeostasis, it is possible that oxidative stress and cytoadhesion are linked for P. vivax as well.
The proteome identified here contains 45 PIR/VIR proteins identified at least twice by a search engine; we recognize that details of the expressed VIR proteins (e.g. sequences outside of identified tryptic peptides) could differ between the PvP01 isolate and Sal-I strain, and that a Sal-I genome with improved subtelomere sequencing may enable the identification of additional VIR proteins. Individual VIR proteins could have multiple functions, including immune evasion by adherence to spleen barrier cells to escape clearance by spleen macrophages [27, 136], adhesive rosette formation and/or invasion [137], and antigenic variation [50–52, 27–28, 84, 136]. A definitive role in adhesion will require further tests of individual VIR proteins. Other cytoadhesive iRBC parasite proteins include a number of erythrocyte binding or reticulocyte invasion proteins, or P. vivax analogs of P. falciparum erythrocyte binding or adhesion proteins, as well as 29 additional MAAP-predicted adhesins [81]. Surface proteins with roles in immune evasion and/or cytoadhesion, are of interest also as possible vaccine candidates [136]. A number of identified S. boliviensis proteins are implicated in cell adhesion processes, including thrombospondin-3, fermitin family homolog 3, protocadherin 10-like, vitronectin, talin-2, neuroplastin, retinoschisin and PDZ domain-containing protein 2. Thus both host and/or parasite proteins may potentially be involved in adhesion.
Conclusions
In this paper, we have examined the P. vivax trophozoite-schizont transition iRBC proteome using a new P. vivax reference genome, identifying 2000 P. vivax and 3487 host S. boliviensis proteins. Over 22% of P. vivax proteins still have no functional annotation, highlighting a large gap in the basic molecular understanding of P. vivax biology. Oxidation and nitration observed in the trophozoite stage-enriched proteome appear also to be present at the transition to the schizont stage and could reflect iRBC oxidative stress. GO-term enrichment analysis highlighted a number of parasite metabolic processes and molecular functions including glycolysis, translation and protein folding, cell components such as ribosomes, proteasomes and the Golgi apparatus, and a number of vesicle- and macromolecule/organelle trafficking-related clusters. DAVID analysis of S. boliviensis proteins included cytoskeletal actin-, tubulin- and spectrin-related clusters, oxidative processes and response to oxidative stress, vesicle and trafficking-related clusters including exosomes and exocytosis, macromolecular complexes such as the proteasome and ribosome, metabolism, translation, and cell death. Host and parasite proteins potentially involved in cell adhesion were also identified. These identifications may provide details for a deeper understanding of P. vivax biology as well as hypotheses for the functional involvement of individual proteins in iRBC biology, parasitism, immunity and pathogenesis.
Supporting information
This table shows Proteome 1 identifications using 5 different search engines. P. vivax proteins are indicated by accession numbers beginning with PvP01_ and S. boliviensis proteins are identified by accession numbers consisting of nine numbers. Crux did not include protein annotations with identifications. XID is the identification number of the protein; Xrep is the number of search engines identifying the protein. Proteins identified by all 5 engines are listed in the worksheet "5x of 5x"; proteins identified by only one of 5 engines are in the worksheet "1x of 5x" etc. Avg. Xrep is the average number of times a search engine identified a protein from within the four individual 2D lc/ms/ms runs for each proteome. For Maxquant, all four individual 2D lc/ms/ms runs were analyzed at once (giving more identifications than analyzing each run separately), thus Avg. Xrep is 1 for all proteins. For Sequest, all four 2D lc/ms/ms runs were analyzed twice (see the methods section), thus Avg. Xrep is maximally 8 for Sequest.
(XLS)
This table shows Proteome 2 identifications using 5 different search engines. P. vivax proteins are indicated by accession numbers beginning PvP01_, and S. boliviensis proteins are identified by accession numbers consisting of nine numbers. XID is the identification number of the protein; Xrep is the number of search engines identifying the protein. Proteins identified by all 5 engines are listed in the worksheet "5x of 5x"; proteins identified by only one of 5 engines are in the worksheet "1x of 5x" etc. Xrep is the number of search engines identifying the protein; Avg. Xrep (as for S1 Table) is the average number of times a search engine identified a protein from within the four individual 2D lc/ms/ms runs for each proteome. Crux did not include protein annotations with identifications.
(XLS)
This table contains all of the P. vivax and S. boliviensis proteins identified at least once by a search engine, obtained by combining S1 Table and S2 Table. For this table Xrep refers to identification in Proteome 1 and/or Proteome 2.
(XLS)
This table contains the relative abundance of P. vivax and S. boliviensis proteins, for all proteins identified by Mascot using the exponentially multiplied protein abundance index (emPAI) [65]. Values were averaged for identifications from both proteomes 1 and 2. Xrep is the number of times the protein was identified by Mascot in the total of 8 2D lc/ms/ms runs included from both proteomes.
(XLS)
This table shows the functional categorization of proteins identified in combined trophozoite-schizont transition Proteomes 1 and 2 for both P. vivax and S. boliviensis. Proteins included have been identified at least twice by a search engine in the combined proteomes. The functional categories are listed to the right of the proteins, which are identified by accession number. Xrep represents identification of the protein in one or both proteomes; Avg. Xrep is the average number of times a single search engine identified a protein from within the four individual 2D lc/ms/ms runs for each proteome, and must be at least 2. The protein descriptions from the database used for identification are also listed. Some details from additional annotation (e.g. from Uniprot) are listed in the Annotation column. Proteins are placed into broad functional categories of interest, e.g. cytoskeleton(-related) as indicated. More than one functional category may be listed when this is supported by available information.
(XLS)
This table lists all Sequest-identified proteins containing at least one high confidence (Percolator posterior error probability of 0.01 or less) oxidized peptide, in trophozoite-schizont transition Proteomes 1 and 2. Proteins from both P. vivax and S. boliviensis are listed in layer 1, and details of peptide Sequest scores, and modifications of individual peptides, are listed in layer 2 for each proteome. Proteins containing only methionine oxidized to met sulfoxide, which can occur in solutions exposed to air, are excluded. Proteome 1 contains 52 oxidized S. boliviensis and 83 oxidized P. vivax proteins or subunits, Proteome 2 has 38 oxidized S. boliviensis and 86 oxidized P. vivax proteins or subunits; the combined proteome contains 74 different oxidized S. boliviensis and 143 different oxidized P. vivax proteins or subunits.
(XLS)
Enrichments were obtained using PlasmoDB. The table lists all GO terms, including GO_biological process, GO_molecular function, and GO_cellular component, enriched for P. vivax proteins from proteome 1.
(XLS)
Enrichments were obtained using PlasmoDB. The table lists all GO terms, including GO_biological process, GO_molecular function, and GO_cellular component, enriched for P. vivax proteins from proteome 2.
(XLS)
This table presents results for all DAVID-derived annotation clusters of S. boliviensis proteins identified at least twice by a search engine. Each cluster is composed of a set of similar GO terms (e.g. biological process or BP, molecular function or MF, cell component or CC). The overall cluster is generally named for the top term or a consensus of individual terms; each GO term has a p-value and Benjamini-Hochberg false discovery rate calculated by DAVID 6.8. Annotation clusters for Proteomes 1 and 2 are in separate tabs.
(XLS)
This table lists P. vivax and S. boliviensis proteins identified in trophozoite-schizont transition Proteomes 1 and/or 2 at least twice by a search engine, that are either annotated as involved in cell adhesion, or predicted to be adhesins [78]. We also included 45 VIR proteins, which mainly have no known function but have been hypothesized to be involved in adhesion, and/or trafficking, signaling, or antigenic variation/immune evasion.
(XLS)
Acknowledgments
This project was funded in part by federal funds from the US National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under grant # R01-AI24710 and contract # HHSN272201200031C (PI, MRG), and supported in part by the Office of Research Infrastructure Programs/OD P51OD011132 (formerly National Center for Research Resources P51RR000165). The Squirrel Monkey Breeding and Research Resource at the Keeling Center for Comparative Medicine and Research, University of Texas MD Anderson Cancer Center, was supported by NIH Grant P40-OD010938. We also thank Drs. Walter Moos and Krishna Kodukula and SRI International for their support of this work, and Dr. Evelina Basenko (EupathDB) for a number of helpful comments and suggestions.
Data Availability
The raw data are deposited in PRIDE: https://www.ebi.ac.uk/pride/archive/projects/PXD005769.
Funding Statement
This project was funded in part by federal funds from the US National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under grant # R01-AI24710 and contract #HHSN272201200031C (PI, MRG), and supported in part by the Office of Research Infrastructure Programs/OD P51OD011132 (formerly National Center for Research Resources P51RR000165). The Squirrel Monkey Breeding and Research Resource at the Keeling Center for Comparative Medicine and Research, University of Texas MD Anderson Cancer Center, was supported by NIH Grant P40-OD010938. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rahimi B, Thakkinstian A, White N, Sirivichayakul C, Dondorp A, Chokejindachai S. Severe vivax malaria: a systematic review and meta-analysis of clinical studies since 1900. Malar J 2014; 13: 481 doi: 10.1186/1475-2875-13-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Price RN, Douglas NM, Anstey NM. New developments in Plasmodium vivax malaria: severe disease and the rise of chloroquine resistance. Curr Opin Infect Dis. 2009; 22:430–5. doi: 10.1097/QCO.0b013e32832f14c1 [DOI] [PubMed] [Google Scholar]
- 3.Mueller I, Galinski MR, Baird JK, Carlton JM., Kochar DK, Alonso PL, et al. Key gaps in the knowledge of Plasmodium vivax, a neglected human malaria parasite. Lancet Infect Dis. 2009; 9, 555–66. doi: 10.1016/S1473-3099(09)70177-X [DOI] [PubMed] [Google Scholar]
- 4.McGready R, Wongsaen K, Chu C, Tun N, Chotivanich K, White N, et al. Uncomplicated Plasmodium vivax malaria in pregnancy associated with mortality from acute respiratory distress syndrome. Malaria Journal 2014; 13: 191 doi: 10.1186/1475-2875-13-191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waters AP, Higgins DG, McCutchan TF. Evolutionary relatedness of some primate models of Plasmodium. Mol Biol Evol 1993; 10: 914–23. [DOI] [PubMed] [Google Scholar]
- 6.Joyner C, Moreno A, Meyer E, Cabrera-Mora M., The MaHPIC Consortium, Kissinger J et al. Plasmodium cynomolgi infections in rhesus macaques display clinical and parasitological features pertinent to modelling vivax malaria pathology and relapse infections. Malaria J. 2016; 15:451–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joyner C, the MaHPIC Consortium, Wood JS, Moreno A, Garcia A, Galinski MR. Severe cynomolgi malaria in a rhesus macaque resulted in similar histopathological changes as those seen in human malaria. Am. J. Tropical Medicine and Hygiene March 2017 in press. [DOI] [PMC free article] [PubMed]
- 8.Galinski MR, Barnwell JW. Plasmodium vivax: who cares? Malaria Journal 2008; 7, S9–27. doi: 10.1186/1475-2875-7-S1-S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galinski M, Meyer E, Barnwell JW. Plasmodium vivax: modern strategies to study a persistent parasite's life cycle. Adv Parasitol. 2013; 81, 1–26. doi: 10.1016/B978-0-12-407826-0.00001-1 [DOI] [PubMed] [Google Scholar]
- 10.Alonso P, Brown G, Arevalo-Herrera M, Binka F, Chitnis C, Collins F et al. A research agenda to underpin malaria eradication. PLoS Med 2011; 8: e1000406 doi: 10.1371/journal.pmed.1000406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization, World Malaria Report 2016, Table 6–1 and 6–2.
- 12.Kitchen SK. The infection of reticulocytes by Plasmodium vivax. Am J Trop Med Hyg 1938; 18, 347–53. [Google Scholar]
- 13.Prajapati SK, Singh OP. Insights into the invasion biology of Plasmodium vivax. Front Cell Infect Microbiol. 2013;3:8 doi: 10.3389/fcimb.2013.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galinski M, Medina C, Ingravallo P and Barnwell J. A reticulocyte-binding protein complex of Plasmodium vivax merozoites Cell 69, 1213–1226, 1992. [DOI] [PubMed] [Google Scholar]
- 15.Martín-Jaular L, Elizalde-Torrent A, Thomson-Luque R, Ferrer M, Segovia JC, Herreros-Aviles E, et al. Reticulocyte-prone malaria parasites predominantly invade CD71hi immature cells: implications for the development of an in vitro culture for Plasmodium vivax. Malar J. 2013;12: 434 doi: 10.1186/1475-2875-12-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malleret B, Li A, Zhang R, Tan KS, Suwanarusk R, Claser C, et al. Plasmodium vivax: restricted tropism and rapid remodeling of CD71-positive reticulocytes. Blood. 2015;125:1314–24. doi: 10.1182/blood-2014-08-596015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winograd E, Sherman I. Characterization of a modified red cell membrane protein expressed on erythrocytes infected with the human malaria parasite Plasmodium falciparum: possible role as a cytoadherent mediating protein. J Cell Biol 1989; 108, 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma YD. Knobs, knob proteins and cytoadherence in falciparum malaria. Int J Biochem. 1991;23: 775–89. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto Y, Matsuda S, Yoshida Y. Ultrastructure of human erythrocytes infected with Plasmodium ovale. Am J Trop Med Hyg 1986; 35, 697–703. [DOI] [PubMed] [Google Scholar]
- 20.Aikawa M, Miller LH, Rabbege J. Caveola—vesicle complexes in the plasmalemma of erythrocytes infected by Plasmodium vivax and P cynomolgi. Unique structures related to Schuffner's dots. Am J Pathol. 1975; 79, 285–300. [PMC free article] [PubMed] [Google Scholar]
- 21.Barnwell JW, Ingravallo P, Galinski MR, Matsumoto Y, Aikawa M. Plasmodium vivax: malarial proteins associated with the membrane-bound caveola-vesicle complexes and cytoplasmic cleft structures of infected erythrocytes. Exp Parasitol. 1990; 70:85–99. [DOI] [PubMed] [Google Scholar]
- 22.Akinyi S, Hanssen E, Meyer EV, Jiang J, Korir CC, Singh B, et al. A 95 kDa protein of Plasmodium vivax and P. cynomolgi visualized by three-dimensional tomography in the caveola-vesicle complexes (Schuffner's dots) of infected erythrocytes is a member of the PHIST family. Mol Microbiol. 2012; 84, 816–31. doi: 10.1111/j.1365-2958.2012.08060.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller LH, Baruch DI, Marsh K, Doumbo OK. The pathogenic basis of malaria. Nature 2002; 415:673–9. doi: 10.1038/415673a [DOI] [PubMed] [Google Scholar]
- 24.Carvalho B, Lopes S, Nogueira P, Orlandi P, Bargieri D, Blanco Y, et al. On the Cytoadhesion of Plasmodium vivax–Infected Erythrocytes. J Infectious Dis. 2010; 202, 638–47. [DOI] [PubMed] [Google Scholar]
- 25.Costa FT, Lopes SC, Ferrer M, Leite JA, Martin-Jaular L, Bernabeu M, et al. On cytoadhesion of Plasmodium vivax: raison d'être? Mem Inst Oswaldo Cruz. 2011;106 Suppl 1:79–84. [DOI] [PubMed] [Google Scholar]
- 26.Lopes SC, Albrecht L, Carvalho BO, Siqueira AM, Thomson-Luque R, Nogueira PA, et al. Paucity of Plasmodium vivax mature schizonts in peripheral blood is associated with their increased cytoadhesive potential. J Infect Dis. 2014; 209:1403–7. doi: 10.1093/infdis/jiu018 [DOI] [PubMed] [Google Scholar]
- 27.del Portillo HA, Fernandez-Becerra C, Bowman S, Oliver K, Preuss M, Sanchez CP, et al. A superfamily of variant genes encoded in the subtelomeric region of Plasmodium vivax. Nature. 2001;410, 839–42. doi: 10.1038/35071118 [DOI] [PubMed] [Google Scholar]
- 28.Bernabeu M, Lopez FJ, Ferrer M, Martin-Jaular L, Razaname A, Corradin G, et al. Functional analysis of Plasmodium vivax VIR proteins reveals different subcellular localizations and cytoadherence to the ICAM-1 endothelial receptor. Cell Microbiol. 2012;14, 386–400. doi: 10.1111/j.1462-5822.2011.01726.x [DOI] [PubMed] [Google Scholar]
- 29.Lopez FJ, Bernabeu M, Fernandez-Becerra C, del Portillo HA. A new computational approach redefines the subtelomeric vir superfamily of Plasmodium vivax. BMC Genomics. 2013;14:8 doi: 10.1186/1471-2164-14-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tachibana S, Sullivan SA, Kawai S, Nakamura S, Kim HR, Goto N, et al. Plasmodium cynomolgi genome sequences provide insight into Plasmodium vivax and the monkey malaria clade. Nat Genet. 2012;44,1051–5. doi: 10.1038/ng.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Auburn S, Bohme U, Steinbiss S, Trimarsanto H, Hostetler J, Sanders M, et al. A new Plasmodium vivax reference sequence with improved assembly of the subtelomeres reveals an abundance of pir genes. Wellcome Open Res. 2016. 1:4 doi: 10.12688/wellcomeopenres.9876.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.al-Khedery B, Barnwell JW, Galinski MR. Antigenic variation in malaria: a 3' genomic alteration associated with the expression of a P. knowlesi variant antigen. Mol Cell. 1999;3: 131–41. [DOI] [PubMed] [Google Scholar]
- 33.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 1995, 82: 77–87. [DOI] [PubMed] [Google Scholar]
- 34.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, et al. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 1995, 82:89–100. [DOI] [PubMed] [Google Scholar]
- 35.Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82: 101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnot DE, Jensen AT. Antigenic Variation and the Genetics and Epigenetics of the PfEMP1 Erythrocyte Surface Antigens in Plasmodium falciparum Malaria. Adv Appl Microbiol. 2011;74:77–96. doi: 10.1016/B978-0-12-387022-3.00007-0 [DOI] [PubMed] [Google Scholar]
- 37.Carlton J, Adams J, Silva J, Bidwell S, Lorenzi H, Caler E et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008; 455, 757–763. doi: 10.1038/nature07327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci USA. 2008; 105:16290–5. doi: 10.1073/pnas.0807404105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Westenberger SJ, McClean CM, Chattopadhyay R, Dharia NV, Carlton JM, Barnwell JW, et al. A systems-based analysis of Plasmodium vivax lifecycle transcription from human to mosquito. PLoS Negl Trop Dis 2010;4:e653 doi: 10.1371/journal.pntd.0000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bautista J, Marin-Garcia P, Diez A, Azcarate I, Puyet A. Malaria proteomics: Insights into the parasite-host interactions in the pathogenic space. J Proteomics 2014; 97: 107–25. doi: 10.1016/j.jprot.2013.10.011 [DOI] [PubMed] [Google Scholar]
- 41.Galinski MR, Barnwell JW. 2012. Non-human primate model for human malaria research. In: Nonhuman Primates in Biomedical Research: Diseases, 2012; 2e; 299–323. CR Abee, K Mansfield, SD Tardif, T Morris (Eds). Elsevier Inc.: Academic Press.
- 42.Collins WE, Sullivan JS, Galland GG, Williams A, Nace D, Williams T et al. Plasmodium simium and Saimiri boliviensis as a model system for testing candidate vaccines against Plasmodium vivax. Am J Trop Med Hyg 2005; 73, 644–648. [PubMed] [Google Scholar]
- 43.Langhorne J, Buffet P, Galinski M, Good M, Harty J, Leroy D, et al. The relevance of non-human primate and rodent malaria models for humans. Malaria J. 2011; 10, 23–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anderson DC, Lapp SA, Akinyi S, Meyer EV, Barnwell JW, Korir-Morrison C et al. 2015. Plasmodium vivax trophozoite-stage proteomes. J Proteomics. 115:157–76. doi: 10.1016/j.jprot.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno-Perez D, Degano R, Ibarrola N, Muro A, Patarroyo M. Determining the Plasmodium vivax VCG-1 strain blood stage proteome. J. Proteomics 2015; 113, 268–80. [DOI] [PubMed] [Google Scholar]
- 46.Acharya P, Pallavi R, Chandran S, Chakravarti H, Middha S, Acharya J, et al. A glimpse into the clinical proteome of human malaria parasites Plasmodium falciparum and Plasmodium vivax. Proteomics Clin Appl. 2009; 3, 1314–25. doi: 10.1002/prca.200900090 [DOI] [PubMed] [Google Scholar]
- 47.Acharya P, Pallavi R, Chandran S, Dandavate V, Sayeed S, Rochani A, et al. Clinical proteomics of the neglected human malarial parasite Plasmodium vivax. PLoS One 2011; 6, e26623 doi: 10.1371/journal.pone.0026623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roobsoong W, Roytrakul S, Sattabongkot J, Li J, Udomsangpetch R, Cui L. Determination of the Plasmodium vivax schizont stage proteome. J. Proteomics 2011;74, 1701–10. doi: 10.1016/j.jprot.2011.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chopra S., Ramkissoon K, Anderson DC. A Systematic Quantitative Proteomic Examination of Multidrug Resistance in Acinetobacter baumannii. Journal of Proteomics 2013; 84,17–39. doi: 10.1016/j.jprot.2013.03.008 [DOI] [PubMed] [Google Scholar]
- 50.Barnwell JW, Howard RJ, Coon HG, Miller LH. Splenic requirement for antigenic variation and expression of the variant antigen on the erythrocyte membrane in cloned Plasmodium knowlesi malaria. Infect Immun. 1983; 40: 985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hommel M, David PH, Oligino LD. Surface alterations of erythrocytes in Plasmodium falciparum malaria. Antigenic variation, antigenic diversity, and the role of the spleen. J Exp Med. 1983; 157: 1137–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lapp SA, Korir-Morrison C, Jiang J, Bai Y, Corredor V, Galinski MR. Spleen-Dependent Regulation of Antigenic Variation in Malaria Parasites: Plasmodium knowlesi SICAvar Expression Profiles in Splenic and Asplenic Hosts. PLoS One. 2013; 8: e78014 doi: 10.1371/journal.pone.0078014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho M, Bannister L, Looareesuwan S, Suntharasamai P. Cytoadherence and ultrastructure of Plasmodium falciparum-infected erythrocytes from a splenectomized patient. Infect. Immun. 1992; 60, 977 2225–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wisniewski J., Zougman A., Nararaj N., Mann M. Universal sample preparation method for proteome analysis. Nature Methods 2009; 6, 359–362. doi: 10.1038/nmeth.1322 [DOI] [PubMed] [Google Scholar]
- 55.Olsen J, de Godoy L, Li G, Macek B, Mortensen P, Pesch R, et al. Parts per million mass accuracy on an orbitrap mass spectrometer via lock mass injection into a C-trap. Molecular and Cellular Proteomics 2005; 4: 2010–25. doi: 10.1074/mcp.T500030-MCP200 [DOI] [PubMed] [Google Scholar]
- 56.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 2011;10, 1794–1805. doi: 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 57.Perkins D., Pappin D., Creasy D., Cottrell J. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999;20, 3551–67. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2 [DOI] [PubMed] [Google Scholar]
- 58.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss M. A semi-supervised machine learning technique for peptide identification from shotgun proteomics datasets. Nat. Methods 2007;4, 923–5. doi: 10.1038/nmeth1113 [DOI] [PubMed] [Google Scholar]
- 59.Eng J., McCormack A., Yates J. III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 1994;5, 976–989. doi: 10.1016/1044-0305(94)80016-2 [DOI] [PubMed] [Google Scholar]
- 60.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem 2003;75, 4646–58. [DOI] [PubMed] [Google Scholar]
- 61.Park C, Klammer A, Kall L, MacCoss M, Noble W. Rapid and accurate peptide identification from tandem mass spectra. J. Proteome Res. 2008, 7, 3022−7. doi: 10.1021/pr800127y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim S, Mischerikow N, Bandeira N, Navarro JD, Wich L, Mohammed S, et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol Cell Proteomics. 2010; 9: 2840–52. doi: 10.1074/mcp.M110.003731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kessner D, Chambers M, Burke R, Agus D, Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008; 24: 2534–6. doi: 10.1093/bioinformatics/btn323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang B, Chambers MC, Tabb DL. Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J Proteome Res. 2007;6:3549–57. doi: 10.1021/pr070230d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta N, Pevzner P. False discovery rates of protein identifications: a strike against the two-peptide rule. J. Proteome Res. 2009; 8: 4173–81. doi: 10.1021/pr9004794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu W, Taylor JA, Davis MT, Bonilla LE, Lee KA, Auger PL, et al. Maximizing the sensitivity and reliability of peptide identification in large-scale proteomic experiments by harnessing multiple search engines. Proteomics 2010;10, 1172–89. doi: 10.1002/pmic.200900074 [DOI] [PubMed] [Google Scholar]
- 67.Bahl A, Brunk B, Crabtree J, Fraunholz M, Gajria B, Grant G et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003; 31: 212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015; 43: D204-D212. [DOI] [PMC free article] [PubMed]
- 69.Ostell J. The Entrez Search and Retrieval System. In: McEntyre J, Ostell J, editors. The NCBI Handbook Internet. Bethesda (MD): National Center for Biotechnology Information (US); 2002. Chapter 15.
- 70.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 2011;39, D225–9. doi: 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, and Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014; 42, D199–D205. doi: 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Research 2012; 40: D306–D312. doi: 10.1093/nar/gkr948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, Series B, 1995; 57: 289–300. [Google Scholar]
- 74.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009. a; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 75.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009. b; 37:1–13. doi: 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. Journal of Molecular Biology. 1990; 215: 403–410. STOP doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 77.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics 2005; 4, 1265–72. doi: 10.1074/mcp.M500061-MCP200 [DOI] [PubMed] [Google Scholar]
- 78.Morita M, Sanai H, Hiramoto A, Sato A, Hiraoka O, Sakura T, et al. Plasmodium falciparum endoplasmic reticulum-resident calcium binding protein is a possible target of synthetic antimalarial endoperoxides, N-89 and N-251. J Proteome Res 2012; 11: 5704–11. doi: 10.1021/pr3005315 [DOI] [PubMed] [Google Scholar]
- 79.Arévalo-Pinzón G, Bermúdez M, Curtidor H, Patarroyo MA. The Plasmodium vivax rhoptry neck protein 5 is expressed in the apical pole of Plasmodium vivax VCG-1 strain schizonts and binds to human reticulocytes. Malar J. 2015; 14:106 doi: 10.1186/s12936-015-0619-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pinzón C, Curtidor H, Reyes C, Méndez D, Patarroyo ME. Identification of Plasmodium falciparum RhopH3 protein peptides that specifically bind to erythrocytes and inhibit merozoite invasion. Protein Sci. 2008; 17: 1719–1730. doi: 10.1110/ps.035923.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ansari FA, Kumar N, Bala Subramanyam M, Gnanamani M, Ramachandran S. MAAP: malarial adhesins and adhesin-like proteins predictor. Proteins. 2008; 70:659–66. doi: 10.1002/prot.21568 [DOI] [PubMed] [Google Scholar]
- 82.Fransson J, Florin-Robertsson E, Axelsson K, Nyhlén C. Oxidation of human insulin-like growth factor I in formulation studies: kinetics of methionine oxidation in aqueous solution and in solid state. Pharm Res. 1996; 13:1252–7. [DOI] [PubMed] [Google Scholar]
- 83.Alves G, Wu WW, Wang G, Shen RF, Yu YK. Enhancing peptide identification confidence by combining search methods. J Proteome Res. 2008; 7: 3102–13. doi: 10.1021/pr700798h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jones AR, Siepen JA, Hubbard SJ, Paton NW. Improving sensitivity in proteome studies by analysis of false discovery rates for multiple search engines. Proteomics. 2009; 9:1220–9. doi: 10.1002/pmic.200800473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Malleret B, Renia L, Russell B. The unhealthy attraction of Plasmodium vivax to reticulocytes expressing transferrin receptor 1 (CD71). Int J Parasitol. epub 13 April 2017. [DOI] [PubMed]
- 86.Roth EF Jr, Calvin MC, Max-Audit I, Rosa J, Rosa R. The enzymes of the glycolytic pathway in erythrocytes infected with Plasmodium falciparum malaria parasites. Blood. 1988; 72:1922–5. [PubMed] [Google Scholar]
- 87.Alam A, Neyaz M, Hasan S. Exploiting Unique Structural and Functional Properties of Malarial Glycolytic Enzymes for Antimalarial Drug Development. Malar Res Treat. 2014:451065 doi: 10.1155/2014/451065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.LeRoch K, Johnson J, Florens L, Zhou Y, Santrosyan A, Grainger M, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004; 14: 2308–18. doi: 10.1101/gr.2523904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sein K, Aikawa M. The pivotal role of carbonic anhydrase in malaria infection. Med Hypotheses. 1998; 50: 19–23. [DOI] [PubMed] [Google Scholar]
- 90.Reungprapavut S, Krungkrai SR, Krungkrai J. Plasmodium falciparum carbonic anhydrase is a possible target for malaria chemotherapy. J Enzyme Inhib Med Chem. 2004; 19: 249–56. doi: 10.1080/14756360410001689577 [DOI] [PubMed] [Google Scholar]
- 91.Dulaney A, Watson RB. Complement fixation in relapsing Plasmodium vivax malaria. Am J Trop Med Hyg. 1945; 25:473–80. [DOI] [PubMed] [Google Scholar]
- 92.Stanley HA, Mayes JT, Cooper NR, Reese RT. Complement activation by the surface of Plasmodium falciparum infected erythrocytes. Mol Immunol. 1984; 21:145–50. [DOI] [PubMed] [Google Scholar]
- 93.Biryukov S, Stoute J. Complement activation in malaria: friend or foe? Trends Molecular Medicine 20, 293–301, 2014. [DOI] [PubMed] [Google Scholar]
- 94.Eaton M, Coggeshall L. Complement fixation in human malaria with an antigen prepared from the monkey parasite Plasmodium knowlesi. J Exp Med 1939; 69: 379–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J Biol Chem. 1987; 262: 9412–20. [PubMed] [Google Scholar]
- 96.Griffiths RE, Kupzig S, Cogan N, Mankelow TJ, Betin VM, Trakarnsanga K, et al. : The ins and outs of human reticulocyte maturation: autophagy and the endosome/exosome pathway. Autophagy 2012, 8:1150–1151 doi: 10.4161/auto.20648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martin-Jaular L, Nakayasu ES, Ferrer M, Almeida IC, del Portillo HA. Exosomes from Plasmodium yoelii-Infected Reticulocytes Protect Mice from Lethal Infections. PLoS ONE 2011; 6: e26588 doi: 10.1371/journal.pone.0026588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Prenni J, Vidal M, Olver C. Preliminary characterization of the murine membrane reticulocyte proteome. Blood Cells, Molecules, and Diseases 2012; 49: 74–82. doi: 10.1016/j.bcmd.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 99.Spang A. Retrograde Traffic from the Golgi to the Endoplasmic Reticulum. Cold Spring Harb Perspect Biol 2013;5: a013391 doi: 10.1101/cshperspect.a013391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gautier E, Ducamp S, Leduc M, Sainot V, Guillonneau F, Dussiot M, et al. Comprehensive proteomic analysis of human erythropoiesis. Cell Rep 2016; 16: 1470–84. doi: 10.1016/j.celrep.2016.06.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gronowicz G, Swift H, Steck T. Maturation of the reticulocyte in vitro. J. Cell Sci. 1984; 71, 177–97. [DOI] [PubMed] [Google Scholar]
- 102.Zhang J, Ney PA. Reticulocyte mitophagy. Monitoring mitochondrial clearance in a mammalian model. Autophagy 2010; 6: 405–8. [DOI] [PubMed] [Google Scholar]
- 103.Coppens I. Metamorphoses of malaria: the role of autophagy in parasite differentiation. Essays Biochem. 2011;51:127–36. doi: 10.1042/bse0510127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hain AU, Bosch J. Autophagy in Plasmodium, a multifunctional pathway? Comput Struct Biotechnol J. 2013; 8: e201308002 doi: 10.5936/csbj.201308002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jayabalasingham B, Voss C, Ehrenman K, Romano JD, Smith ME, Fidock DA, et al. Characterization of the ATG8-conjugation system in two Plasmodium species with special focus on the liver stage: possible linkage between the apicoplastic and autophagic systems? Autophagy. 2014;10: 269–84. doi: 10.4161/auto.27166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cervantes S, Bunnik EM, Saraf A, Conner CM, Escalante A, Sardiu ME, et al. The multifunctional autophagy pathway in the human malaria parasite, Plasmodium falciparum. Autophagy. 2014;10: 80–92. doi: 10.4161/auto.26743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Totino PR, Daniel- R. Plasmodium falciparum: erythrocytic stages die by autophagic-like cell death under drug pressure. Exp Parasitol. 2008;118:478–86. doi: 10.1016/j.exppara.2007.10.017 [DOI] [PubMed] [Google Scholar]
- 108.Dodson M, Darley-Usmar V, Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic Biol Med. 2013; 63: 207–21. doi: 10.1016/j.freeradbiomed.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kaminskyy V, Zhivotovsky B. Free radicals in cross talk between autophagy and apoptosis. Antioxid Redox Signal 2014; 21: 86–102. doi: 10.1089/ars.2013.5746 [DOI] [PubMed] [Google Scholar]
- 110.Bell A, Satchwell T, Heesom K, Hawley B, Kupzig S, Hazell M, et al. Protein distribution during human erythroblast enucleation In vitro. PLoS ONE 2013; 8: e60300 doi: 10.1371/journal.pone.0060300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu J, Guo X, Mohandas N, Chasis JA, An X. Membrane remodeling during reticulocyte maturation. Blood. 2010;115: 2021–7. doi: 10.1182/blood-2009-08-241182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gyulkhandanyan AV, Mutlu A, Freedman J, Leytin V. Markers of platelet apoptosis: methodology and applications. J Thromb Thrombolysis. 2012: 33, 397–411. doi: 10.1007/s11239-012-0688-8 [DOI] [PubMed] [Google Scholar]
- 113.Pretorius E, du Plooy JN, Bester J. A Comprehensive Review on Eryptosis. Cell Physiol Biochem. 2016;39:1977–2000. doi: 10.1159/000447895 [DOI] [PubMed] [Google Scholar]
- 114.Lang E, Lang F. Triggers, inhibitors, mechanisms, and significance of eryptosis: the suicidal erythrocyte death. Biomed Res Int. 2015: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Föller M, Bobbala D, Koka S, Huber SM, Gulbins E, Lang F. Suicide for survival—death of infected erythrocytes as a host mechanism to survive malaria. Cell Physiol Biochem. 2009; 24: 133–40. doi: 10.1159/000233238 [DOI] [PubMed] [Google Scholar]
- 116.Blanc L, Vidal M. Reticulocyte membrane remodeling: contribution of the exosome pathway. Curr Opin Hematol. 2010;17:177–83. doi: 10.1097/MOH.0b013e328337b4e3 [DOI] [PubMed] [Google Scholar]
- 117.Sims P, Wiedmer T. Unraveling the mysteries of phospholipid scrambling. Thromb Haemost. 2001; 86: 266–75. [PubMed] [Google Scholar]
- 118.Boys B, Kuprowski M, Noe J, Konermann L. Protein oxidative modifications during electrospray ionization: solution phase electrochemistry or corona discharge-induced radical attack? Anal Chem. 2009; 81:4027–34. doi: 10.1021/ac900243p [DOI] [PubMed] [Google Scholar]
- 119.Becker K, Tilley L, Vennerstrom JL, Roberts D, Rogerson S, Ginsburg H. Oxidative stress in malaria parasite-infected erythrocytes: host-parasite interactions. Int J Parasitol. 2004;34:163 doi: 10.1016/j.ijpara.2003.09.011 [DOI] [PubMed] [Google Scholar]
- 120.Hoshi T, Heinemann S. Regulation of cell function by methionine oxidation and reduction. J Physiol. 2001;531, 1–11. doi: 10.1111/j.1469-7793.2001.0001j.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hakimi H, Asada M, Angeles JM, Kawai S, Inoue N, Kawazu S. Plasmodium vivax and Plasmodium knowlesi: cloning, expression and functional analysis of 1-Cys peroxiredoxin. Exp Parasitol. 2013;133:101–5. doi: 10.1016/j.exppara.2012.10.018 [DOI] [PubMed] [Google Scholar]
- 122.Kehr S, Sturm N, Rahlfs S, Przyborski JM, Becker K. Compartmentation of redox metabolism in malaria parasites. PLoS Pathog. 2010; 6: e1001242 doi: 10.1371/journal.ppat.1001242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ostera G, Tokumasu F, Oliveira F, Sa J, Furuya T, Teixeira C, et al. Plasmodium falciparum: food vacuole localization of nitric oxide-derived species in intraerythrocytic stages of the malaria parasite. Exp Parasitol. 2008;120: 29–38. doi: 10.1016/j.exppara.2008.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ostera G, Tokumasu F, Teixeira C, Collin N, Sa J, Hume J, et al. Plasmodium falciparum: nitric oxide modulates heme speciation in isolated food vacuoles. Exp Parasitol. 2011; 127: 1–8. doi: 10.1016/j.exppara.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pall ML. The NO/ONOO-cycle as the central cause of heart failure. Int J Mol Sci. 2013; 14:2 2274–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Jaramillo M, Gowda DC, Radzioch D, Olivier M. Hemozoin increases IFN-gamma-inducible macrophage nitric oxide generation through extracellular signal-regulated kinase- and NF-kappa B-dependent pathways. J Immunol. 2003;171: 4243–53. [DOI] [PubMed] [Google Scholar]
- 127.Eghbalzadeh K, Brixius K, Bloch W, Brinkmann C. Skeletal muscle nitric oxide (NO) synthases and NO-signaling in "diabesity"—what about the relevance of exercise training interventions? Nitric Oxide. 2014; 37:28–40. doi: 10.1016/j.niox.2013.12.009 [DOI] [PubMed] [Google Scholar]
- 128.Pino P, Vouldoukis I, Dugas N, Conti M, Nitcheu J, Traore B, et al. Induction of the CD23/nitric oxide pathway in endothelial cells downregulates ICAM-1 expression and decreases cytoadherence of Plasmodium falciparum-infected erythrocytes. Cell Microbiol. 2004; 6: 839–48. doi: 10.1111/j.1462-5822.2004.00406.x [DOI] [PubMed] [Google Scholar]
- 129.Heijnen HF, van Donselaar E, Slot JW, Fries DM, Blachard-Fillion B, Hodara R, et al. Subcellular localization of tyrosine-nitrated proteins is dictated by reactive oxygen species generating enzymes and by proximity to nitric oxide synthase. Free Radic Biol Med. 2006; 40: 1903–13. doi: 10.1016/j.freeradbiomed.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 130.Reutov VP, Sorokina EG. NO-synthase and nitrite-reductase components of nitric oxide cycle. Biochemistry (Mosc). 1998; 63: 874–84. [PubMed] [Google Scholar]
- 131.Jensen FB. The role of nitrite in nitric oxide homeostasis: a comparative perspective. Biochim Biophys Acta. 2009; 1787: 841–8. doi: 10.1016/j.bbabio.2009.02.010 [DOI] [PubMed] [Google Scholar]
- 132.Uzasci L, Bianchet M, Cotter R, Nath A. Identification of Nitrated Immunoglobulin Variable Regions in the HIV-Infected Human Brain: Implications in HIV Infection and Immune Response. J Proteome Res. 2014, 13, 1614–23. doi: 10.1021/pr401117m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chowdhury AA, Chaudhuri J, Biswas N, Manna A, Chatterjee S, Mahato SK, et al. Synergistic apoptosis of CML cells by buthionine sulfoximine and hydroxychavicol correlates with activation of AIF and GSH-ROS-JNK-ERK-iNOS pathway. PLoS One. 2013;8: e73672 doi: 10.1371/journal.pone.0073672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ocampo M, Rodríguez LE, Curtidor H, Puentes A, Vera R, Valbuena JJ, et al. Identifying Plasmodium falciparum cytoadherence-linked asexual protein 3 (CLAG 3) sequences that specifically bind to C32 cells and erythrocytes. Protein Sci. 2005; 14: 504–13. doi: 10.1110/ps.04883905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Marín-Menéndez A, Bardají A, Martínez-Espinosa FE, Bôtto-Menezes C, Lacerda MV, Ortiz J, et al. Rosetting in Plasmodium vivax: a cytoadhesion phenotype associated with anaemia. PLoS Negl Trop Dis. 2013; 7:e2155 doi: 10.1371/journal.pntd.0002155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fernandez-Becerra C, Yamamoto M, Vencio R, Lacerda M, Rosanas-Urgell A, del Portillo H. Plasmodium vivax and the importance of the subtelomeric multigene vir superfamily. Trends Parasitol. 2009; 25, 44–51. doi: 10.1016/j.pt.2008.09.012 [DOI] [PubMed] [Google Scholar]
- 137.Yam X, Brugat T, Siau A, Lawton J, Wong D, Farah A, et al. Characterization of the Plasmodium interspersed repeats (PIR) proteins of Plasmodium chabaudi indicates functional diversity. Sci Rep. 2016; 6: 23449 doi: 10.1038/srep23449 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table shows Proteome 1 identifications using 5 different search engines. P. vivax proteins are indicated by accession numbers beginning with PvP01_ and S. boliviensis proteins are identified by accession numbers consisting of nine numbers. Crux did not include protein annotations with identifications. XID is the identification number of the protein; Xrep is the number of search engines identifying the protein. Proteins identified by all 5 engines are listed in the worksheet "5x of 5x"; proteins identified by only one of 5 engines are in the worksheet "1x of 5x" etc. Avg. Xrep is the average number of times a search engine identified a protein from within the four individual 2D lc/ms/ms runs for each proteome. For Maxquant, all four individual 2D lc/ms/ms runs were analyzed at once (giving more identifications than analyzing each run separately), thus Avg. Xrep is 1 for all proteins. For Sequest, all four 2D lc/ms/ms runs were analyzed twice (see the methods section), thus Avg. Xrep is maximally 8 for Sequest.
(XLS)
This table shows Proteome 2 identifications using 5 different search engines. P. vivax proteins are indicated by accession numbers beginning PvP01_, and S. boliviensis proteins are identified by accession numbers consisting of nine numbers. XID is the identification number of the protein; Xrep is the number of search engines identifying the protein. Proteins identified by all 5 engines are listed in the worksheet "5x of 5x"; proteins identified by only one of 5 engines are in the worksheet "1x of 5x" etc. Xrep is the number of search engines identifying the protein; Avg. Xrep (as for S1 Table) is the average number of times a search engine identified a protein from within the four individual 2D lc/ms/ms runs for each proteome. Crux did not include protein annotations with identifications.
(XLS)
This table contains all of the P. vivax and S. boliviensis proteins identified at least once by a search engine, obtained by combining S1 Table and S2 Table. For this table Xrep refers to identification in Proteome 1 and/or Proteome 2.
(XLS)
This table contains the relative abundance of P. vivax and S. boliviensis proteins, for all proteins identified by Mascot using the exponentially multiplied protein abundance index (emPAI) [65]. Values were averaged for identifications from both proteomes 1 and 2. Xrep is the number of times the protein was identified by Mascot in the total of 8 2D lc/ms/ms runs included from both proteomes.
(XLS)
This table shows the functional categorization of proteins identified in combined trophozoite-schizont transition Proteomes 1 and 2 for both P. vivax and S. boliviensis. Proteins included have been identified at least twice by a search engine in the combined proteomes. The functional categories are listed to the right of the proteins, which are identified by accession number. Xrep represents identification of the protein in one or both proteomes; Avg. Xrep is the average number of times a single search engine identified a protein from within the four individual 2D lc/ms/ms runs for each proteome, and must be at least 2. The protein descriptions from the database used for identification are also listed. Some details from additional annotation (e.g. from Uniprot) are listed in the Annotation column. Proteins are placed into broad functional categories of interest, e.g. cytoskeleton(-related) as indicated. More than one functional category may be listed when this is supported by available information.
(XLS)
This table lists all Sequest-identified proteins containing at least one high confidence (Percolator posterior error probability of 0.01 or less) oxidized peptide, in trophozoite-schizont transition Proteomes 1 and 2. Proteins from both P. vivax and S. boliviensis are listed in layer 1, and details of peptide Sequest scores, and modifications of individual peptides, are listed in layer 2 for each proteome. Proteins containing only methionine oxidized to met sulfoxide, which can occur in solutions exposed to air, are excluded. Proteome 1 contains 52 oxidized S. boliviensis and 83 oxidized P. vivax proteins or subunits, Proteome 2 has 38 oxidized S. boliviensis and 86 oxidized P. vivax proteins or subunits; the combined proteome contains 74 different oxidized S. boliviensis and 143 different oxidized P. vivax proteins or subunits.
(XLS)
Enrichments were obtained using PlasmoDB. The table lists all GO terms, including GO_biological process, GO_molecular function, and GO_cellular component, enriched for P. vivax proteins from proteome 1.
(XLS)
Enrichments were obtained using PlasmoDB. The table lists all GO terms, including GO_biological process, GO_molecular function, and GO_cellular component, enriched for P. vivax proteins from proteome 2.
(XLS)
This table presents results for all DAVID-derived annotation clusters of S. boliviensis proteins identified at least twice by a search engine. Each cluster is composed of a set of similar GO terms (e.g. biological process or BP, molecular function or MF, cell component or CC). The overall cluster is generally named for the top term or a consensus of individual terms; each GO term has a p-value and Benjamini-Hochberg false discovery rate calculated by DAVID 6.8. Annotation clusters for Proteomes 1 and 2 are in separate tabs.
(XLS)
This table lists P. vivax and S. boliviensis proteins identified in trophozoite-schizont transition Proteomes 1 and/or 2 at least twice by a search engine, that are either annotated as involved in cell adhesion, or predicted to be adhesins [78]. We also included 45 VIR proteins, which mainly have no known function but have been hypothesized to be involved in adhesion, and/or trafficking, signaling, or antigenic variation/immune evasion.
(XLS)
Data Availability Statement
The raw data are deposited in PRIDE: https://www.ebi.ac.uk/pride/archive/projects/PXD005769.