Abstract
We have a limited understanding of the genetic and molecular basis of evolutionary changes in the size and proportion of limbs. We studied wing and pectoral skeleton reduction leading to flightlessness in the Galapagos cormorant (Phalacrocorax harrisi). We sequenced and de novo assembled the genomes of four cormorant species and applied a predictive and comparative genomics approach to find candidate variants that may have contributed to the evolution of flightlessness. These analyses and cross-species experiments in C. elegans and in chondrogenic cell lines implicated variants in genes necessary for transcriptional regulation and function of the primary cilium. Cilia are essential for Hedgehog signaling, and humans affected by skeletal ciliopathies suffer from premature bone growth arrest, mirroring skeletal features associated with loss of flight.
Introduction
The evolution of loss of flight is one the most recurrent limb modifications encountered in nature (1). In fact, Darwin used the occurrence of flightless birds as an argument in favor of his theory of natural selection (2). He proposed that loss of flight could evolve selection in favor of larger bodies and relaxed selection due to the absence of predators. Loss of flight has evolved repeatedly, and is found among 26 families of birds in 17 different orders (1). Moreover, recent studies strongly suggest that the ratites (ostriches, emus, rheas, cassowaries and kiwis), long thought to derive from a single flightless ancestor, may constitute a polyphyletic group characterized by multiple independent instances of loss of flight and convergent evolution (3–5). However, despite the ubiquity and evolutionary importance of loss of flight (6), the underlying genetic and molecular mechanisms remain unknown.
The Galapagos cormorant (Phalacrocorax harrisi) is the only flightless cormorant among approximately 40 extant species (7). The entire population is distributed along the coastlines of Isabela and Fernandina Islands in the Galapagos archipelago. P. harrisi has a pair of short wings, which are smaller than those of any other cormorant (Fig. 1A); a deviation from the allometric relationship between wing length and body mass (7). The radius and ulna are disproportionally small compared to the humerus, but no digits have been fused or lost, unlike in some ratites (8). In addition, the Galapagos cormorant differs from its flighted relatives in a delay in the onset of several developmental landmarks after hatching (9), shortened remiges (flight feathers), underdeveloped pectoral muscles, a long and narrow skull and pelvis, a disproportionally long tibiotarsus, a 1.6-fold increase in body mass, and a highly reduced keel (7). The keel is an extension of the sternum that runs along its midline and provides an attachment surface for the flight muscles, the largest muscles in birds. Flightless taxa, such as ratites and Cretaceous Hesperornis have evolved flat sternums in which the keel has been largely reduced or lost (10).
Fig. 1. The Galapagos Cormorant, a model to study flightlessness evolution.
(A) The average wing length of an adult Galapagos cormorant male is 19 cm (3.6 kg. body mass), whereas the wing length of its closest relative, the double-crested cormorant, is 31.5 cm. (2.2 kg. body mass). Illustration by Katie Bertsche from specimens 134079 and 151575 from the Museum of Vertebrate Zoology at Berkeley. (B) CEGMA score is a good predictor of genome completeness from a gene-centric perspective. Blue line is the linear regression model (r2 = 0.75 P = 4.3×10−07). Genomes reported in this study are red triangles and other published avian genomes are black circles (table S2). (C) Bayesian phylogram reconstructed with fourfold degenerate sites from whole genome sequences. The orange bar illustrates the time span between the approximate origin of proto-Galapagos archipelago (9 MYA) and the origin of the oldest extant island, San Cristobal (4 MYA). Nodes represent median divergence ages. Blue bars indicate the 95% Highest Posterior Density (HPD) Interval. (D) Heterozygosity levels inferred from whole genome sequences. Birds are not drawn to scale.
In contrast to ratites and penguins, which became flightless over 50 million years ago (MYA) (5, 11), the Galapagos cormorant and its flighted relatives are estimated to share a common ancestor ~2 MYA (12). This recent and extreme modification of wing size and pectoral skeleton makes P. harrisi an attractive model to study loss of flight.
High-quality genome sequences of four cormorant species
To identify variants associated with loss of flight, we sequenced and de novo assembled the 1.2 Gb genomes of the Galapagos cormorant (Galapagos Islands, Ecuador) and three flighted cormorant species: the Double–crested cormorant (Phalacrocorax auritus; Minnesota, USA), the Neotropical cormorant (Phalacrocorax brasilianus; Valdivia, Chile), and the Pelagic cormorant (Phalacrocorax pelagicus; Alaska, USA). P. auritus and P. brasilianus are the closest relatives of P. harrisi (12–14), and P. pelagicus is part of a sister clade and served as an outgroup. Genomes were assembled from a combination of short insert and mate-pair Illumina libraries with SOAPdenovo2 (15) (table S1). Among these four genomes, the Galapagos cormorant’s assembly had the longest contig and scaffold N50 metrics (contig N50: 103 kb; scaffold N50: 4.6 Mb; table S1B). We evaluated the completeness of the cormorants’ genomes by estimating the total number of uniquely annotated proteins in each assembly and by using the CEGMA pipeline (16, 17). Overall, we found agreement between these two independent metrics in a dataset including the four cormorant genomes and 17 recently published bird genomes (r2 = 0.75 P = 4.3×10−07; Fig. 1B and table S2).
Interestingly, commonly used metrics of assembly quality, such as contig and scaffold N50, were very poor predictors of the total number of proteins present in each assembly (r2 = 0.13 P = 0.15 and r2 = 0.06 P = 0.81; fig. S1; table S2). Three of the four cormorant genomes (P. harrisi, P. auritus and P. pelagicus) obtained the highest CEGMA scores and number of uniquely annotated genes among all bird genomes (red triangles, Fig. 1B). The following CEGMA scores, a means to predict gene annotation, were obtained for the cormorants: P. harrisi, 90.3%; P. auritus, 91.3%; P. brasilianus, 72.6%; P. pelagicus, 87.1%. In contrast, Sanger and PacBio genomes had lower scores for other birds: Gallus gallus (Sanger assembly) (18), 80.7%; Taeniopygia guttata (Sanger assembly) (19), 71.4%; and Melopsittacus undulatus (PacBio assembly) (20), 79.0%. Thus, our cormorant genomes perform even better than genomes assembled from Sanger sequences and PacBio long reads (complete statistics in table S2).
Phylogeny and genetic diversity
We reconstructed the cormorant phylogeny using a Bayesian framework (17), confirming the phylogenetic relationship among the four sequenced species (Fig. 1C). Moreover, our results indicate that P. harrisi last shared a common ancestor with P. auritus and P. brasilianus ~2.37 MYA, in agreement with an estimate from mitochondrial DNA (12) (Fig. 1C). The oldest extant island in the Galapagos archipelago, Española, emerged at most 4 MYA, and proto-Galapagos islands existed at least 9 MYA (21). Our results are consistent with the view that P. harrisi lost the ability to fly while inhabiting the archipelago.
We calculated the proportion of single nucleotide polymorphism (SNP) heterozygous sites for each sequenced individual to estimate the levels of intra-specific genetic diversity (Fig. 1D). P. harrisi showed the lowest proportion of heterozygous SNPs among the sequenced cormorants (0.00685%; Fig. 1D). The heterozygosity of P. harrisi is even lower than that of the Crested Ibis, Nipponia nippon, highly endangered bird with a small effective population size and a known recent population bottleneck (22) (0.0172%; Fig. 1D). The low level of heterozygosity found in the Galapagos cormorant is most likely due to its small population size (~1,500 individuals) and multiple population bottlenecks (23).
Discovery and characterization of function-altering variants in P. harrisi
To investigate the genetics of flightlessness evolution, we developed a comparative and predictive genomics approach (24, 25) that uses the genome sequences of P. harrisi and its flighted relatives to identify genetic variants that likely contributed to the evolution of loss of flight. Both coding and cis-regulatory variants have been implicated in the evolution of morphological traits (26, 27). However, determining the impact of regulatory variants is not straightforward. To identify the contribution of regulatory variants to the evolution of loss of flight in P. harrisi, we searched for ultra-conserved noncoding sequences showing accelerated molecular evolution (28–30). We identified eleven ultra-conserved noncoding regions in tetrapods that show accelerated evolution in P. harrisi but not in the other cormorants (False Discovery Rate (FDR) < 5%). One of these regions was located in an intron of the gene FTO (fig. S2), which been associated with obesity in humans (31); however, none of these regions overlapped with experimentally validated or putative mouse limb enhancers (17, 32, 33) (table S7).
We thus focused on characterizing coding variants because we are better able to predict their molecular consequences. For our variant discovery approach to be comprehensive, it was imperative to interrogate most of the Galapagos cormorant’s genes. To increase our power to do so, we annotated genes using homology-based and transcriptome-based gene annotations. The latter was derived with mRNA expression data from the developing wing of a double-crested cormorant embryo (fig. S3C) (17). We then predicted all missense, deletion, and insertion variants in ortholog pairs between P. harrisi and each of its three flighted relatives (fig. S4) (17).
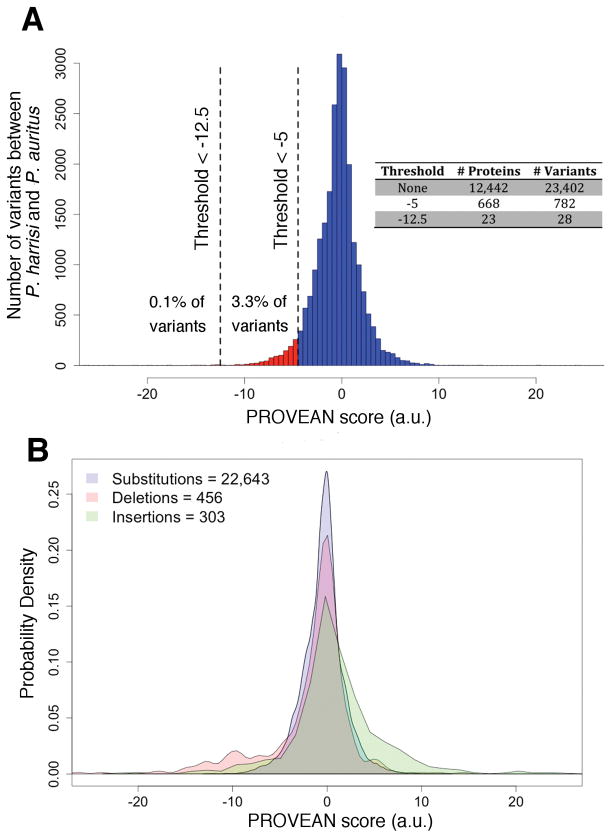
We used PROVEAN (34), a phylogeny-corrected variant effect predictor, to evaluate the impact on protein function of each of the Galapagos cormorant’s variants on a genome-wide scale. PROVEAN predictions have been validated in experimental evolution studies that mimic the process of gradual accumulation of mutations in nature (35). A PROVEAN score is calculated for each variant; the more negative the score, the more likely a given variant is to alter protein function. We examined the distribution of PROVEAN scores obtained when comparing 12,442 ortholog pairs between P. harrisi and P. auritus (Fig. 2A). Of these, 4,959 pairs (40%) did not contain coding variants; the remaining 7,483 pairs contained a total of 23,402 coding variants: 22,643 single amino acid substitutions, 456 deletions, and 303 insertions (Fig. 2B). Most variants were predicted to be neutral (the distribution is centered around zero). As expected, deletion and insertions were enriched in the tails of the distribution (Fig. 2B). Very similar numbers of variants and PROVEAN score distributions were obtained for the other homology-based (fig. S5) and transcriptome-based annotations (fig. S3).
Fig. 2. Distribution of the effect of variants between P. auritus and P. harrisi.
(A) We used PROVEAN to predict the effect on protein function of 23,402 variants contained in 12,449 orthologous pairs between P. auritus and P. harrisi. 4,966 pairs contained no variants. The more negative the score; the more likely the variant affects protein function. PROVEAN score thresholds used in this study are drawn as vertical dashed lines. Number of proteins and variants found for each threshold are presented in inset table. (B) Density of PROVEAN scores for each class of variant. The same variants presented in (A) were classified as single amino acid substitutions, deletions, and insertions. Number of variants in each class is indicated in the legend.
Enrichment for genes mutated in skeletal ciliopathies
To identify proteins carrying function-altering variants in the Galapagos cormorant, we applied a stringent threshold to our four prediction datasets: PROVEAN score < −5, two times the threshold for human disease variants discovery (17, 34) (Fig. 2A). In our dataset, variants with a PROVEAN score < −5 typically occur at residues that have been perfectly conserved at least since mammals and birds last shared a common ancestor (~300 MYA; fig. S6). Consequently, changes in these residues are likely to alter protein function or stability.
On the basis of theoretical and experimental considerations (36), we hypothesized that flightlessness is likely to have a polygenic basis and that the underlying variants would be enriched in certain biological pathways. Consistent with this hypothesis, gene enrichment analysis of function-altering variants in the Galapagos cormorant revealed that genes implicated in human developmental disorders were significantly overrepresented (17) (table S3A). Strikingly, 8 out of the 19 significantly enriched categories consisted of genes implicated in disorders affecting limb development, such as polydactyly, syndactyly, and duplication of limb bones. Control analyses showed no enrichment of these categories in the flighted cormorants (17) (table S3B, S3C).
Many of the genes underlying the enrichment for limb syndromes are those mutated in a family of human disorders known as ciliopathies. For instance, 17 out of 25 genes (65%) in the “duplication of hand bones” category and 12 out of 12 genes (100%) in the “preaxial hand polydactyly” category are mutated in human ciliopathies (table S4). Moreover, ciliopathy-associated genes were present in all of the enriched categories (table S4). Ciliopathies compromise a phenotypically diverse group of rare genetic disorders that result from defects in the formation or function of cilia (37).
Cilia are hair-like microtubule-based structures that are nucleated by the basal body (centriole and associated proteins) and project from the surface of cells. Primary cilia are essential for mediating Hedgehog (Hh) signaling in vertebrates, serving as antennae for morphogens during development (38). We confirmed by Sanger sequencing the presence of predicted function-altering variants in Ofd1, Evc, Talpid3, Dync2h1, Ift122, Wdr34, and Kif7, all of which are necessary for the assembly or functioning of the primary cilium and are mutated in human ciliopathies, particularly those affecting the skeleton (Table 1). We also found a likely function-altering variant in Gli2, a transcription factor necessary for Hh signaling (39) (Table 1). Humans affected by diverse skeletal ciliopathies have small limbs and rib cages (37), suggesting a parallel with the main features of the Galapagos cormorant: small wings and a flattened sternum. However, the consequences of ciliopathies in humans are often more severe and pleotropic, likely as a consequence of the overrepresentation of loss of function alleles in patients (40). Interestingly, although ciliopathies do not exclusively affect the forelimb in humans, differences between forelimbs and hindlimbs are commonly found among patients. For example, digital abnormalities affect the hands more often than the feet of Oral-facial-digital syndrome Type I (40, 41) and Ellis van Creveld (42) patients, suggesting that forelimbs and hindlimbs differ in their intrinsic sensitivity to ciliary dysfunction.
Table 1.
Function-altering variants in P. harrisi are enriched for genes that cause skeletal ciliopathies in humans.
| Gene | Pathway | Variant | PROVEAN score | Human Syndrome |
|---|---|---|---|---|
| Ofd1 | Cilia/Hh | R325C | −6.913 | Orofaciodigital and Joubert |
| K517T | −5.673 | |||
| E889G | −5.068 | |||
|
| ||||
| Talpid3 | Cilia/Hh | D759V | −7.805 | Joubert and Jeune |
|
| ||||
| Evc | Cilia/Hh | T341I | −5.546 | Ellias-Van Creveld |
|
| ||||
| Dync2h1 | Cilia/Hh | P2733S | −7.431 | Short-rib thoracic dysplasia |
|
| ||||
| Ift122 | Cilia/Hh | Q691L | −5.491 | Cranioectodermal dysplasia |
|
| ||||
| Wdr34 | Cilia/Hh | P188R | −6.337 | Short-rib thoracic dysplasia |
|
| ||||
| Kif7 | Cilia/Hh | R833W | −6.827 | Joubert and Acrocallosal |
|
| ||||
| Gli2 | Hh | P1086T | −5.117 | Culler-Jones |
|
| ||||
| Fat1 | PCP | S1717L | −5.858 | Facioscapulohumeral Dystrophy (*) |
| Y2462C | −8.592 | |||
|
| ||||
| Dchs1 | PCP | G2063D | −6.45 | Van Maldergem |
|
| ||||
| Dvl1 | PCP | P103L | −8.23 | Robinow |
Sanger validated examples of function-altering variants (PROVEAN score <−5) in P. harrisi. Cilia/Hh related genes were found based on functional enrichment for human syndromes. PCP (Planar cell polarity) genes were selected based on literature evidence linking cilia and PCP. These variant are fixed in the population.
(*) Based on phenotypic similarity to mutant mouse model.
The Galapagos cormorant ortholog of human OFD1 (mutated in Oral-facial-digital syndrome 1) contains three predicted function-altering variants with PROVEAN score < −5 (R325C −6.913, K517T −5.673 and E889G −5.068; fig. S6A and S6B). Ofd1 knockout mice display polydactyly and shortened long bones (43). Also, a function-altering missense variant (Q691L −5.491) was found in IFT122, a component of the IFT complex that controls ciliogenesis and the ciliary localization of Shh pathway regulators (44). Null Ift122 mutants show severe limb and skeletal phenotypes in mice (45), and mutations in Ift122 have been associated with Sensenbrenner Syndrome in humans, which is characterized by craniofacial, ectodermal, and skeletal abnormalities, including limb shortening (46). Strikingly, the mutated glutamine in IFT122 is virtually invariant among eukaryotes ranging from green algae, C. elegans, and Drosophila to vertebrates (fig. S6C). To directly test whether the non-synonymous substitution in IFT122 affects protein function, we generated a C. elegans knock-in strain using CRISPR/Cas9 homology directed genome editing (Fig 3A). The edited strain carries the Galapagos cormorant missense variant at the corresponding orthologous position in daf-10 (Q862L), the ortholog of IFT122 in C. elegans (47).
Fig. 3. The Galapagos cormorant variant IFT122 Q691L affects ciliary function in vivo.
(A) The daf-10 gene (IFT122 ortholog) was targeted with CRISPR/Cas9 homology mediated repair in C. elegans to introduce a non-synonymous substitution present exclusively in the Galapagos Cormorant (IFT122 Q691L). The resulting edited knock-in strain contains the daf-10 Q862L substitution and ten synonymous substitutions (17). Edited strains were sequenced with Sanger sequencing to confirm genotypes. Representative bordering behavior of (B) N2 wild type worms, (C) daf-10(e1387) containing a premature stop codon Q892X, and (D) daf-10 Q862L knock-in strain. (E) Quantification of bordering behavior in N2, daf-10(e1387) and two independently generated knock-in ) daf-10 Q862L strains (n=3, t-test, (*) P <0.05).
In invertebrates, cilia do not mediate Shh signaling but are necessary for detecting external sensory inputs (48). The only ciliated cells in C. elegans are sensory neurons and mutations in cilia components affect dispersal behavior, chemotaxis, and dauer formation (49). We tested the bordering behavior of worms -accumulation of animals on the thickest part of a bacterial lawn- which is known to be mediated by ciliated neurons (50). We found that daf-10(e1387) mutants carrying a premature stop (Q869X) displayed an increased bordering behavior in a dispersal assay compared to wild type (73% for daf-10(e1387) vs. 30% for N2; P = 0.019, t-test, Fig. 3B, C, E). Two independently generated daf-10 Q862L knock-in lines phenocopied the effect of the premature stop allele (70% for line 1 and 69% for line 2 vs. 30% for N2; P = 0.018 and P = 0.016 respectively; Fig. 3B, D, E; see movie S1). Daf-10(e1387) ciliary neurons fail to incorporate fluorescent dyes, like many other loss of function mutants in cilia components (49). In contrast, the daf-10 Q862L knock-in worms incorporated the DiO dye like wild type worms, suggesting that this allele is hypomorphic (fig. S7). Overall, these results indicate that the IFT122 Q691L missense variant present in the Galapagos cormorant can affect protein function in vivo in C. elegans.
The Planar Cell Polarity (PCP) pathway exhibits a genetic link to cilia (38, 51–53). We found function-altering variants in members of the PCP pathway in P. harrisi: Fat1 atypical cadherin (Fat1), Dachsous cadherin-related 1 (Dchs1) and Disheveled-1 (Dvl1) (Table 1). The Galapagos Cormorant FAT1 contains two function-altering variants (S1717L and Y2462C, Table 1). The mutated serine and tyrosine are conserved from zebrafish to humans (fig. S5D). Fat1 knockout mice show very selective defects in muscles of the upper body, but not in posterior muscles (54). In addition, Dvl1 is mutated in humans with Robinow syndrome, characterized by limb shortening (55, 56).
Sanger sequencing of 20 Galapagos cormorant individuals from two different populations (Cabo Hammond and Cañones Sur) (57) revealed only homozygous carriers for all of the variants in Table 1, indicating that these variants are most likely fixed in the Galapagos Cormorant. In summary, we found an overrepresentation of predicted function-altering variants in genes that, when mutated in humans and mice, cause skeletal ciliopathies and bone growth defects.
CUX1 is mutated in P. harrisi
To identify the most likely function-altering variants in P. harrisi, we applied a more stringent PROVEAN score threshold: −12.5 delta alignment score, five times the threshold used for human disease variants discovery (34). This strategy narrowed our search to 23 proteins (0.16% of annotated proteins in P. harrisi) (table S5). We manually curated these 23 proteins and performed additional Sanger sequencing, reducing the list of proteins with confirmed or putative variants to 12 (table S5) (17). These variants were exclusively small deletions. Among these 12 proteins, two stood out from their known role in development: LGALS-3 and CUX1. LGALS-3 is affected by a 7 amino acid deletion in P. harrisi (PROVEAN score −26.319). LGALS-3 (Galectin-3) is localized at the base of the primary cilium and is necessary for correct ciliogenesis in mice (58), but it has not been implicated in human ciliopathies. Moreover, LGALS-3 physically interacts with SUFU, an important regulator of mammalian Hh signaling (59), and knockout mice show pleiotropic defects in chondrocyte differentiation (60).
In addition, we found a 4 amino acid deletion (PROVEAN score −15.704) in CUX1. CUX1 (cut-like homeobox 1), also known as CDP, is a highly conserved transcription factor with diverse roles in development. CUX1 contains four DNA binding domains: three CUT domains (CR1–3) and one homeodomain (HD) (Fig. 4A) (61). The full-length isoform, which contains four DNA binding domains (CR1–3HD), acts exclusively as a transcriptional repressor and has rapid and unstable DNA binding dynamics. In contrast, smaller isoforms such as CR2–3HD and CR3HD can act as both repressors and activators of gene expression, and show slow and stable DNA binding dynamics in vitro (61). Although insect and bird wings evolved independently, it is interesting to note that cut, the Drosophila ortholog of Cux1, is necessary for the proper development of wings and flight muscles in flies (62). In chicken, Cux1 mRNA expression in the limb at embryonic stage 23 is restricted to the ectoderm bordering the Apical Ectodermal Ridge (AER) (63). The AER is one of the key signaling centers that drive limb development. At later stages, Cux1 is expressed in the developing joints of both chicken (64) and mice (fig. S8) and is also detected in chondrocytes in developing bones of mice (fig. S9). A function-altering variant in CUX1 is a strong candidate to contribute to loss of flight in P. harrisi because adenovirus-mediated overexpression in the developing chicken wing of a form of CUX1 missing the Cut2 DNA binding domain results in severe wing truncation (63, 65). These truncations most strongly affect distal skeletal elements (digits, radius and ulna). Interestingly, in P. harrisi the radius and ulna are disproportionately small compared to the humerus (7).
Fig. 4. The Galapagos Cormorant Cux1 is a transcriptional activation hypomorph.
(A) Western blot showing the expression of CUX1 isoforms in the developing wing of a mallard embryo (22 days). The most abundant band corresponds to the predicted size of the CR3HD CUX1 isoform. (B) Protein alignment showing the deleted –AGSQ-residues in the Galapagos Cormorant CUX1 and their high degree of conservation among vertebrates. (C) Differential up-regulation of genes by CUX1-Ancestral and CUX1-Δ4aa variants in ATDC5 cells. Pooled stable lines carrying CR3HD CUX1-Ancestral or CUX1-Δ4aa variants were generated by lentiviral transduction and puromycin selection. Controls cells were transduced with an empty vector. Gene expression levels were measured by reverse transcriptase quantitative PCR (n=5 biological replicates, each compromising 3 technical replicates). (D) Luciferase based assay to test the repression activity of CUX1 C-terminal domain lacking CR3 and HD domains. GAL4 DNA binding domain was fused to CUX1-Ancestral or CUX1-Δ4aa variants. Both constructs equally repressed a promoter containing UAS binding sites in COS-7 cells (n=3 biological replicates, each compromising 3 technical replicates). Gene expression levels were measured by RT-qPCR (n=5 biological replicates, each compromising 3 technical replicates). Error bars indicate standard errors. We used ANOVA and Tukey’s HSD to test for statistical significance. Black stars indicate a significant difference between a CUX1 variant and the control. Red starts indicate a significant difference between CUX1-Ancestral and CUX1-Δ4aa variants. Absence of stars indicates no significant difference. (*) P <0.05, (**) P<0.01, and (***) P<0.001
We Sanger-sequenced and confirmed the predicted Cux1 12 base pair (bp) deletion in P. harrisi. We also confirmed that this variant is fixed in the population and absent in the other cormorant species (fig. S10A). The 12 bp deletion in Cux1 removes four amino acids, AGSQ, immediately adjacent to the C-terminal end of the homeodomain (Fig. 4B). We will refer to this variant as CUX1-Δ4aa. Alignment of CUX1 orthologs from available vertebrate genomes revealed that the 4 missing residues are extremely conserved among tetrapods (Fig. 4B). The deleted serine is phosphorylated in human cells (66), but the consequences of this modification are unknown.
The Cux1 deletion does not include any of the predicted residues responsible for DNA contact and recognition (67), but given its close proximity to the homeodomain, we decided to test whether the DNA binding activity of CUX1 was affected. We chose to express the CR3HD isoform because western blot analysis revealed that this was the most abundant CUX1 isoform expressed in the developing wing of mallard embryos (~50 kDa; Fig. 4A). We performed Electrophoretic Mobility Shift Assay (EMSA) with purified CR3HD CUX1-Ancestral and CUX1-Δ4aa protein variants (fig. S10B) as previously described (68) and found that DNA binding was not abolished in the deletion variant (fig. S10C). CUX1 is able to both directly repress and activate gene expression through its C-terminal tail (69, 70). We performed a luciferase reporter assay (69, 71) and found that both variants were equally capable of repressing the expression of a UAS/tk luciferase reporter (Fig. 4D). Thus, the Galapagos cormorant CUX1-Δ4aa variant appears to not affect DNA binding in vitro or the C-terminal repression activity in COS-7 cells.
CUX1 regulates the expression of cilia and PCP genes
We hypothesized that the Cux1 deletion variant is mechanistically related to the enrichment of function-altering variants in ciliopathy-related genes. This inference came from the fact that transgenic mice overexpressing the CUX1-CR3HD isoform develop polycystic kidneys. Cilia in cystic epithelial cells from these animals were twice as long as the ones in control epithelial cells (72). Furthermore, the CUX1-CR2CR3HD isoform has been shown to directly upregulate the expression of RPGRIPL1, also known as FTM, a component of the cilia basal body that is involved in Shh signaling and mutated in human ciliopathies (73). Also, Cux1 knockout mice show deregulation of SHH expression in hair follicles (74).
To test whether Cux1 could globally regulate expression of cilia genes, we analyzed expression array data from human-derived Hs578t cells stably expressing a shRNA against Cux1, as well as cells overexpressing the human CUX1-CR2CR3HD isoform (75). In concordance with the role of Cux1 as a regulator of cell growth and proliferation (76), genes significantly up- or down-regulated in both conditions (p < 0.05 and > 1.1 fold change) were enriched for pathways such as “Cell cycle” and “Mitotic G1-G1/S phases” (P = 3.99×10−5 and 0.016, respectively; table S6). Importantly, we also found enrichment for cilia-related categories such as “Assembly of the primary cilium” and “Intraflagellar transport” (P = 1.2×10−4 and 5.7×10−3 respectively; table S6). These results suggest that cilia-related genes are enriched among Cux1 targets.
To further test whether Cux1 can regulate ciliary genes in an appropriate cellular context, we generated ATDC5 stable lines expressing N-terminal HIS-tagged versions of CR3HD CUX1-Ancestral and CUX1-Δ4aa variants. ATDC5 is a well-characterized mouse chondrogenic cell line that largely recapitulates in vitro the differentiation landmarks of chondrocytes (77). We performed quantitative reverse transcription PCR (RT-qPCR) on a selected number of genes containing predicted strong function-altering variants in P. harrisi (Table 1) and showing detectable levels of expression in ATDC5 cells. In addition, we measured the expression of Ptch1, the receptor of the Hh pathway. Our experiments indicate that the CUX1-Ancestral variant transcriptionally upregulated the expression of Ofd1 (1.7 fold, P = 1.2×10−6; Fig. 4C) and Fat1 (1.8 fold, P = 2.9×10−2; Fig. 4C) and downregulated the expression of Ift122 (0.77 fold, P = 2.5×10−3; Fig. 4C) and Ptch1 (0.53 fold, P = 1.4×10−2; Fig. 4C) compared to the control line. In contrast, neither Dync2h1 nor Wdr34 expression levels were changed by CUX1-Ancestral overexpression (Fig. 4C). These results suggest that cilia and Hh related genes are likely transcriptional targets of CUX1 in chondrocytes.
Impaired transcriptional activity of the Galapagos cormorant CUX1
The Galapagos cormorant CUX1 showed impaired transcriptional activity compared to the ancestral variant. Ofd1 was significantly upregulated in CUX1-Δ4aa cells compared to control cells (1.2 fold, P = 2.1×10−2, ANOVA and Tukey’s HSD test ; Fig. 4C); however, Ofd1 upregulation was significantly reduced in CUX1-Δ4aa cells compared to CUX1-Ancestral cells (1.2 vs. 1.7 fold, P = 6×10−5; Fig. 4C). Similarly, although Fat1 was significantly upregulated in CUX1-Ancestral cells compared to control (1.8 fold, P = 2.9×10−2), Fat1 expression levels in CUX1-Δ4aa cells were not significantly different from control lines (1.2 fold, P = 0.57; Fig. 4C). The difference between Fat1 upregulation in CUX1-Δ4aa and CUX1-Δ4aa cells was not significant (1.8 fold vs. 1.3 fold, P = 0.17; Fig. 4C). In contrast, CUX1-Δ4aa cells significantly repressed both Ift122 (0.78 fold, P = 2.6×10−3) and Ptch1 (0.56 fold, P = 2.4×10−2) and there were no significant differences between CUX1-Ancestral and CUX1-Δ4aa cells (0.78 fold vs. 0.78 fold for Ift122; P = 0.99) and 0.53 fold vs. 0.56 fold for Ptch1, P = 0.96; Fig. 4C). These results suggest that the four amino acid deletion in the Galapagos Cormorant CUX1 affects its ability to activate but not to repress gene expression, and are consistent with our luciferase reporter assays, which showed no effect on repression (Fig. 4D). It is notable that both the transcriptional activator (Cux1) and its target genes (Ofd1 and Fat1) exhibit function-altering variants in the Galapagos Cormorant.
CR3HD-CUX1 promotes chondrogenesis
Chondrocytes are the main engine of bone growth. The growth of skeletal elements depends on the precise regulation of chondrocyte proliferation and hypertrophy. Mutations that affect cilia result in premature arrest of bone growth due defects in Indian Hedgehog (IHH) signaling in chondrocytes (78). To test the role of CUX1-CR3HD in chondrogenesis, we differentiated control, CUX1-Ancestral and CUX1-Δ4aa ATDC5 cell lines and quantified the expression of Ihh and Sox9, two well-established markers of chondrocyte differentiation in vitro and in vivo (78). Overexpression of both CR3HD CUX1-Ancestral and CUX1-Δ4aa variants promoted chondrogenic differentiation of ATDC5 cells after 7 and 12 days of differentiation (Fig. 5A). However, the CUX1-Δ4aa variant was not as efficient as the ancestral variant, showing significant differences from CUX1-Ancestral in Ihh expression after 7 days of differentiation (~50% decrease, P = 5.9×10−4, ANOVA and Tukey’s HSD test; Fig. 5A) and in Sox9 expression after 12 days (~15% decrease, P = 1.6×10−2; Fig. 5A). These results suggest that the Galapagos cormorant CUX1 is probably not as effective as the ortholog from its flighted relatives in promoting chondrogenic differentiation, and that mutations in Cux1 may affect the dynamics of chondrogenesis. This observation is further supported by the fact that CUX1 is expressed in the hypertrophic chondrocytes of developing bones in mice, and that the bones of Cux1 mutant mice are thin and flaky (79)
Fig. 5. CR3HD CUX1 promotes chondrogenesis.
(A) ATDC5 control cells and cells carrying CR3HD CUX1-Ancestral or CUX1-Δ4aa variants were differentiated into chondrocytes. Gene expression levels were measured by RT-qPCR (n=4 biological replicates, each compromising 3 technical replicates) after 7 and 12 days. Error bars indicate standard errors. We used ANOVA and Tukey’s HSD to test for statistical significance. Black stars indicate a significant difference between a CUX1 variant and the control. Red starts indicate a significant difference between CUX1-Ancestral and CUX1-Δ4aa variants. Absence of stars indicates no significant difference. (*) P <0.05, (**) P<0.01, and (***) P<0.001. (B) Proposed mechanism for the reduction of wing size in P. harrisi. The left panel depicts the normal functioning of IHH signaling pathway in vertebrates. CUX1 regulates the expression of cilia related genes such as Ofd1 and promotes chondrogenesis. The right panel depicts the state of the IHH pathway in P. harrisi. Proteins in red have predicted function-altering variants in P. harrisi. We propose that these variants will affect both cilia formation and functioning leading to a reduction in IHH pathway activity. As a result, the pool of proliferating chondrocytes would decrease in wing bones and the number of hypertrophic chondrocytes would increase resulting in impaired bone growth.
Possible evolutionary scenarios
Loss of flight has traditionally been attributed to relaxed selection. In this scenario, the first cormorants that inhabited the Galapagos Islands found a unique environment that lacked predators and provided food year-round, drastically reducing the need to migrate. However, we found no evidence for pseudogenization of developmental genes in P. harrisi (17) (table S10 and S11). On the other hand, loss of flight in the Galapagos Cormorant is thought to confer an advantage for diving by decreasing buoyancy via shorter wings and by indirectly allowing an increase oxygen storage via larger body size (80). This advantage could make flightlessness a target of positive selection.
We evaluated whether any of our candidate genes (Table 1) showed signatures of positive selection in the Galapagos Cormorant lineage by estimating the ratio of non-synonymous to synonymous substitutions (ω = dN/dS). This is a very stringent test of selection because it assumes that all sites in a protein are evolving under the same selective pressure, a condition rarely met in highly conserved regulatory genes (81). We found that three out of eleven tested genes showed signs of positive selection (ω>1) in the Galapagos cormorant lineage compared to a background phylogeny of 35 taxa (Ofd1 ω =1.92, Evc ω=1.93, Gli2 ω=1.10; table S8).
One of these three genes, Gli2, showed a statistically significant difference (ω=1.10 (Galapagos branch) vs. ω=0.11 (Background branch), P = 2.4×10−3; table S8). In contrast, Gli2 showed no sign of selection in the sister group of P. harrisi (P. auritus and P. brasilianus) (ω=0.11 (Galapagos branch) vs. ω=1×10−4 (Background branch), P = 0.46). As a control, we also analyzed Gli3, the partially redundant paralog of Gli2, which also mediates Hh signaling but has no predicted function-altering variants in P. harrisi and found no evidence for positive selection (ω=0.04 (Galapagos branch) vs. ω=0.15 (Background branch), P = 0.11; table S8). These results suggest that selection towards flightlessness may be partially responsible for the phenotype of P. harrisi.
Discussion
The study of evolution of flightlessness in the Rallidae family led to the hypothesis that flightlessness could be a fast-evolving heterochronic condition (10, 82). Heterochrony, the relative change in the rate or timing of developmental events among species, is thought to be an important factor contributing to macroevolutionary change (83). Yet, virtually nothing is known about its genetic and molecular mechanisms.
Diverse myological, osteological, and developmental observations suggest that flightlessness in the Galapagos cormorant is caused by the retention into adulthood of juvenile characteristics affecting pectoral and forelimb development (a class of heterochrony known as paedomorphosis) (7). Here, we propose a genetic and molecular model that may explain this heterochronic condition, where the perturbations of cilia/Ihh signaling may be responsible for the reduction in growth of both wings and keel in the Galapagos cormorant. However, we cannot rule out a role of Cux1 also in the AER. Of special interest is the gene Fat1, a target of Cux1 (Fig. 4D), which contains two putative function-altering variants (Table 1). Fat1−/− mouse mutants are viable and show selective defects in face, pectoral and shoulder muscles but not in in hindlimb muscles (54). Thus, variants in Fat1 could explain the underdeveloped pectoral muscles of P. harrisi.
Although we have identified multiple variants that likely contribute to the flightless phenotype of P. harrisi, we cannot exclude that other genes and pathways may contribute to the phenotype, nor the contribution of non-coding regulatory variants (17). Further characterization of the individual and joint contributions of the variants found in this study will help us to reconstruct the chain of events leading to flightlessness and to genetically dissect macroevolutionary change. We hypothesize that mutations in cilia or functionally related genes could be responsible for limb and other skeletal heterochronic transformations in birds and diverse organisms, including humans.
Supplementary Material
Table S1. Sequenced species and genome assembly statistics.
Table S2. Genome completeness statistics of a diverse selection of avian genomes.
Table S3. Functional profiling of proteins with predicted function-altering variants (PROVEAN score < −5)
Table S4. List of genes in each of the enriched categories with predicted function-altering variants in P. harrisi.
Table S5. List of the proteins with the strongest function-altering variants in P. harrisi.
Table S6. Gene enrichment analysis of putative CUX1 target genes in Hs578t cells. P-values were corrected for multiple testing.
Table S7. Ultra-conserved noncoding elements in tetrapods showing accelerated molecular evolution in P. harrisi. Elements are annotated using human coordinates as a reference
Table S8. Test for positive selection in previously identified cilia related genes carrying function-altering mutations in P. harrisi.
Table S9. Sequenced libraries used for genome assembly.
Table S10. List of genes not annotated in P. harrisi but annotated in P. auritus, P. brasilianus and P.pelagicus.
Table S11. Curated list of proteins with putative premature stop codons in P. harrisi that are absent in in P. auritus, P. brasilianus and P.pelagicus.
Table S12. Primers used to verify predicted variants in the sequenced cormorants.
Table S13. Primers used for quantitative real time PCR in ATDC5 cell lines.
Acknowledgments
A.B. is funded by the Jane Coffin Childs Memorial Fund for Medical Research. L.K. is funded by the Howard Hughes Medical Institute (HHMI). A.R. is funded by the U.S. Geological Survey through the Wildlife Program of the Ecosystems Mission area. Any use of trade names is for descriptive purposes only and does not imply endorsement by the U.S. Government. E.B-D is supported by a Gruss-Lipper postdoctoral fellowship from the EGL foundation. C.V. is funded by FONDECYT (11130305). K.L is supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS/NIH). We thank C. Koo and Dr. M. Arbing at the UCLA-DOE Protein Expression Laboratory Core Facility for protein purification and Dr. S. Feng at the Broad Stem Cell Research Center High Throughput Sequencing Core for assistance. Dr. C. Cicero for granting access to cormorant specimens (134079 & 151575) at the Museum of Vertebrate Zoology at Berkeley. We also thank Dr. C. Valle (USFQ) for advice and Dr. K. Garrett (Natural History Museum of Los Angeles), Dr. K. Burns (San Diego Natural History Museum and SDSU Museum of Biodiversity), and Dr. R. Duerr (International Bird Rescue) for providing samples used in preliminary stages of this study. A.B. and L.K. conceived the study. A.B. coordinated the collection of samples, prepared libraries, assembled and annotated genomes, and performed analyses and experiments. E.B-D and A.B. performed the accelerated evolution analysis. P.W., A.R., C.V., and P.P. provided DNA or tissue samples. W.W. and A.B. carried out ATDC5 cell line experiments supervised by K.L. AB. and L.K. wrote the manuscript. All authors discussed and agreed on the final version of the manuscript. All sequencing data from this study is available through the NCBI Sequence Read Archive (SRA) under Bioproject accession number PRJNA327123. Alignments used for phylogenetic analysis and selection test are available at DRYAD doi:10.5061/dryad.8m2t5.
Footnotes
The authors declare no competing financial interests.
References and Notes
- 1.Roff DA. The evolution of flightlessness: Is history important? Evol Ecol. 1994;8:639–657. [Google Scholar]
- 2.Darwin C. On the Origin of Species by Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life. John Murray; London: 1859. [PMC free article] [PubMed] [Google Scholar]
- 3.Harshman J, et al. Phylogenomic evidence for multiple losses of flight in ratite birds. Proc Natl Acad Sci U S A. 2008;105:13462–13467. doi: 10.1073/pnas.0803242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker AJ, Haddrath O, Mcpherson JD, Cloutier A. Genomic support for a moa-tinamou clade and adaptive morphological convergence in flightless ratites. Mol Biol Evol. 2014;31:1686–1696. doi: 10.1093/molbev/msu153. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell KJ, et al. Ancient DNA reveals elephant birds and kiwi are sister taxa and clarifies ratite bird evolution. Science. 2014;344:898–900. doi: 10.1126/science.1251981. [DOI] [PubMed] [Google Scholar]
- 6.Wright NA, Steadman DW, Witt CC. Predictable evolution toward flightlessness in volant island birds. Proc Natl Acad Sci. 2016;113:201522931. doi: 10.1073/pnas.1522931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livezey BC. Flightlessness in the Galápagos Cormorant (Compsohalieus [Nannopterum] harrisi): heterochrony, gigantism, and specialization. Zool J Linn Soc. 1992;105:155–224. [Google Scholar]
- 8.de Bakker MaG, et al. Digit loss in archosaur evolution and the interplay between selection and constraints. Nature. 2013;500:445–8. doi: 10.1038/nature12336. [DOI] [PubMed] [Google Scholar]
- 9.The timing of several homologous developmental events such as the appearance of immature feathers, dropping of egg tooth, and independence from parental care, occur progressively later in the Galapagos cormorant chick compared to P. auritus.
- 10.Feduccia A. The Origin and Evolution of Birds. 2. Yale University Press; New Haven: 1999. [Google Scholar]
- 11.Slack KE, et al. Early penguin fossils, plus mitochondrial genomes, calibrate avian evolution. Mol Biol Evol. 2006;23:1144–1155. doi: 10.1093/molbev/msj124. [DOI] [PubMed] [Google Scholar]
- 12.Kennedy M, Valle Ca, Spencer HG. The phylogenetic position of the Galápagos Cormorant. Mol Phylogenet Evol. 2009;53:94–98. doi: 10.1016/j.ympev.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy M, Spencer HG. Classification of the cormorants of the world. Mol Phylogenet Evol. 2014;79:249–257. doi: 10.1016/j.ympev.2014.06.020. [DOI] [PubMed] [Google Scholar]
- 14.A recent phylogeny of the Phalacrocoracidae suggested classifying P. harrisi, P. auritus, and P. brasilianus as members of the monophyletic genera Nannopterum.
- 15.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parra G, Bradnam K, Ning Z, Keane T, Korf I. Assessing the gene space in draft genomes. Nucleic Acids Res. 2009;37:289–297. doi: 10.1093/nar/gkn916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.see Material and Methods.
- 18.I. C. G. S. Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 19.Warren WC, et al. The genome of a songbird. Nature. 2010;464:757–762. doi: 10.1038/nature08819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganapathy G, et al. High-coverage sequencing and annotated assemblies of the budgerigar genome. Gigascience. 2014;3:11. doi: 10.1186/2047-217X-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harpp KS, Mittelstaedt E, D’Ozouville N, Graham DW. Geophysical Monograph Series. John Wiley & Sons, Inc; Hoboken, New Jersey: 2014. The Galápagos: A Natural Laboratory for the Earth Sciences; pp. 145–166. [Google Scholar]
- 22.Wang G, Li X. Population Dynamics and Recovery of Endangered Crested Ibis (Nipponia nippon ) in Central China. Waterbirds. 2008;31:489–494. [Google Scholar]
- 23.Valle CA, Coulter MCM. Present status of the flightless cormorant, Galapagos penguin and greater flamingo populations in the Galapagos Islands, Ecuador, after the 1982–83 El Nino. Condor. 1987;89:276–281. [Google Scholar]
- 24.Alfoldi J, Lindblad-Toh K. Comparative genomics as a tool to understand evolution and disease. Genome Res. 2013;23:1063–1068. doi: 10.1101/gr.157503.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jelier R, Semple JI, Garcia-Verdugo R, Lehner B. Predicting phenotypic variation in yeast from individual genome sequences. Nat Genet. 2011;43:1270–1274. doi: 10.1038/ng.1007. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro MD, et al. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 27.Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution (N Y) 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pollard KS, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- 29.Booker BM, et al. Bat Accelerated Regions Identify a Bat Forelimb Specific Enhancer in the HoxD Locus. PLoS Genet. 2016;12:e1005738. doi: 10.1371/journal.pgen.1005738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prabhakar S, et al. Human-specific gain of function in a developmental enhancer. Science. 2008;321:1346–1350. doi: 10.1126/science.1159974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frayling TM, et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–8. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotney J, et al. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012;22:1069–1080. doi: 10.1101/gr.129817.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. Predicting the Functional Effect of Amino Acid Substitutions and Indels. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rockah-Shmuel L, Tóth-Petróczy Á, Tawfik DS. Systematic Mapping of Protein Mutational Space by Prolonged Drift Reveals the Deleterious Effects of Seemingly Neutral Mutations. PLOS Comput Biol. 2015;11:e1004421. doi: 10.1371/journal.pcbi.1004421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stern DL. Perspective: Evolutionary Developmental Biology and the Problem of Variation. Evolution (N Y) 2000;54:1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 37.Waters AM, Beales PL. Ciliopathies: An expanding disease spectrum. Pediatr Nephrol. 2011;26:1039–1056. doi: 10.1007/s00467-010-1731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haycraft CJ, et al. Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1:e53. doi: 10.1371/journal.pgen.0010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bisschoff IJ, et al. Novel Mutations Including Deletions of the Entire OFD1 Gene in 30 Families with Type 1 Orofaciodigital Syndrome: A Study of the Extensive Clinical Variability. Hum Mutat. 2013;34:237–247. doi: 10.1002/humu.22224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coene KLM, et al. OFD1 Is Mutated in X-Linked Joubert Syndrome and Interacts with LCA5-Encoded Lebercilin. Am J Hum Genet. 2009;85:465–481. doi: 10.1016/j.ajhg.2009.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baujat G, Le Merrer M. Ellis-van Creveld syndrome. Orphanet J Rare Dis. 2007;2:27. doi: 10.1186/1750-1172-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bimonte S, et al. Ofd1 is required in limb bud patterning and endochondral bone development. Dev Biol. 2011;349:179–191. doi: 10.1016/j.ydbio.2010.09.020. [DOI] [PubMed] [Google Scholar]
- 44.Ocbina PJR, Eggenschwiler JT, Moskowitz I, Anderson KV. Complex interactions between genes controlling trafficking in primary cilia. Nat Genet. 2011;43:547–553. doi: 10.1038/ng.832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin J, Lin Y, Norman RX, Ko HW, Eggenschwiler JT. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc Natl Acad Sci U S A. 2011;108:1456–61. doi: 10.1073/pnas.1011410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walczak-Sztulpa J, et al. Cranioectodermal Dysplasia, Sensenbrenner syndrome, is a ciliopathy caused by mutations in the IFT122 gene. Am J Hum Genet. 2010;86:949–956. doi: 10.1016/j.ajhg.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell LR. The Molecular Identities of the Caenorhabditis elegans Intraflagellar Transport Genes dyf-6, daf-10 and osm-1. Genetics. 2006;173:1275–1286. doi: 10.1534/genetics.106.056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singla V, Reiter JF. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 49.Perkins La, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 50.Macosko EZ, et al. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature. 2009;458:1171–5. doi: 10.1038/nature07886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park TJ, Haigo SL, Wallingford JB. Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet. 2006;38:303–311. doi: 10.1038/ng1753. [DOI] [PubMed] [Google Scholar]
- 52.Gray RS, et al. The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nat Cell Biol. 2009;11:1225–1232. doi: 10.1038/ncb1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng H, Hoover AN, Liu A. PCP effector gene Inturned is an important regulator of cilia formation and embryonic development in mammals. Dev Biol. 2010;339:418–28. doi: 10.1016/j.ydbio.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 54.Caruso N, et al. Deregulation of the Protocadherin Gene FAT1 Alters Muscle Shapes: Implications for the Pathogenesis of Facioscapulohumeral Dystrophy. PLoS Genet. 2013;9:e1003550. doi: 10.1371/journal.pgen.1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White J, et al. DVL1 Frameshift Mutations Clustering in the Penultimate Exon Cause Autosomal-Dominant Robinow Syndrome. Am J Hum Genet. 2015;96:612–622. doi: 10.1016/j.ajhg.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bunn KJ, et al. Mutations in DVL1 Cause an Osteosclerotic Form of Robinow Syndrome. Am J Hum Genet. 2015;96:623–630. doi: 10.1016/j.ajhg.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duffie CV, Glenn TC, Vargas FH, Parker PG. Genetic structure within and between island populations of the flightless cormorant (Phalacrocorax harrisi) Mol Ecol. 2009;18:2103–2111. doi: 10.1111/j.1365-294X.2009.04179.x. [DOI] [PubMed] [Google Scholar]
- 58.Koch A, Poirier F, Jacob R, Delacour D. Galectin-3, a novel centrosome-associated protein, required for epithelial morphogenesis. Mol Biol Cell. 2010;21:219–31. doi: 10.1091/mbc.E09-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paces-Fessy M, Boucher D, Petit E, Paute-Briand S, Blanchet-Tournier M-F. The negative regulator of Gli, Suppressor of fused (Sufu), interacts with SAP18, Galectin3 and other nuclear proteins. Biochem J. 2004;378:353–62. doi: 10.1042/BJ20030786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Colnot C, Sidhu SS, Balmain N, Poirier F. Uncoupling of Chondrocyte Death and Vascular Invasion in Mouse Galectin 3 Null Mutant Bones. Dev Biol. 2001;229:203–214. doi: 10.1006/dbio.2000.9933. [DOI] [PubMed] [Google Scholar]
- 61.Sansregret L, Nepveu A. The multiple roles of CUX1: Insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 62.Sudarsan V, Anant S, Guptan P, Vijayraghavan K, Skaer H. Myoblast Diversification and Ectodermal Signaling in Drosophila. Dev Cell. 2001;1:829–839. doi: 10.1016/s1534-5807(01)00089-2. [DOI] [PubMed] [Google Scholar]
- 63.Tavares AT, Tsukui T, Izpisúa Belmonte JC. Evidence that members of the Cut/Cux/CDP family may be involved in AER positioning and polarizing activity during chick limb development. Development. 2000;127:5133–5144. doi: 10.1242/dev.127.23.5133. [DOI] [PubMed] [Google Scholar]
- 64.Lizarraga G, Lichtler A, Upholt WB, Kosher Ra. Studies on the role of Cux1 in regulation of the onset of joint formation in the developing limb. Dev Biol. 2002;243:44–54. doi: 10.1006/dbio.2001.0559. [DOI] [PubMed] [Google Scholar]
- 65.The chicken CUX1-WT variant studied by Tavarez, et al. was inadvertently missing the Cut2 DNA binding domain, as revealed by the chicken genome generated subsequently to their study (Ensembl ID: ENSGALP00000002788). This isoform is likely hypomorphic, and its overexpression may exert a dominant negative effect.
- 66.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 67.Iyaguchi D, Yao M, Watanabe N, Nishihira J, Tanaka I. DNA Recognition Mechanism of the ONECUT Homeodomain of Transcription Factor HNF-6. Structure. 2007;15:75–83. doi: 10.1016/j.str.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 68.Moon NS, Bérubé G, Nepveu A. CCAAT displacement activity involves CUT repeats 1 and 2, not the CUT homeodomain. J Biol Chem. 2000;275:31325–31334. doi: 10.1074/jbc.M002912200. [DOI] [PubMed] [Google Scholar]
- 69.Mailly F, et al. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16:5346–5357. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Truscott M, et al. Carboxyl-terminal proteolytic processing of CUX1 by a caspase enables transcriptional activation in proliferating cells. J Biol Chem. 2007;282:30216–30226. doi: 10.1074/jbc.M702328200. [DOI] [PubMed] [Google Scholar]
- 71.Nishio H, Walsh MJ. CCAAT displacement protein/cut homolog recruits G9a histone lysine methyltransferase to repress transcription. Proc Natl Acad Sci U S A. 2004;101:11257–11262. doi: 10.1073/pnas.0401343101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cadieux C, et al. Polycystic kidneys caused by sustained expression of Cux1 isoform p75. J Biol Chem. 2008;283:13817–13824. doi: 10.1074/jbc.M709332200. [DOI] [PubMed] [Google Scholar]
- 73.Stratigopoulos G, LeDuc Ca, Cremona ML, Chung WK, Leibel RL. Cut-like Homeobox 1 (CUX1) Regulates Expression of the Fat Mass and Obesity-associated and Retinitis Pigmentosa GTPase Regulator-interacting Protein-1-like (RPGRIP1L) Genes and Coordinates Leptin Receptor Signaling. J Biol Chem. 2011;286:2155–2170. doi: 10.1074/jbc.M110.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ellis T, et al. The transcriptional repressor CDP (Cutl1) is essential for epithelial cell differentiation of the lung and the hair follicle. Genes Dev. 2001;15:2307–2319. doi: 10.1101/gad.200101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vadnais C, et al. Long-range transcriptional regulation by the p110 CUX1 homeodomain protein on the ENCODE array. BMC Genomics. 2013;14:258. doi: 10.1186/1471-2164-14-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ramdzan ZM, Nepveu A. CUX1, a haploinsufficient tumour suppressor gene overexpressed in advanced cancers. Nat Rev Cancer. 2014;14:673–682. doi: 10.1038/nrc3805. [DOI] [PubMed] [Google Scholar]
- 77.Shukunami C, et al. Chondrogenic differentiation of clonal mouse embryonic cell line ATDC5 in vitro: Differentiation-dependent gene expression of parathyroid hormone (PTH)/PTH-related peptide receptor. J Cell Biol. 1996;133:457–468. doi: 10.1083/jcb.133.2.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 79.Sinclair AM, et al. Lymphoid apoptosis and myeloid hyperplasia in CCAAT displacement protein mutant mice. Blood. 2001;98:3658–3667. doi: 10.1182/blood.v98.13.3658. [DOI] [PubMed] [Google Scholar]
- 80.Watanabe YY, Takahashi A, Sato K, Viviant M, Bost C-A. Poor flight performance in deep-diving cormorants. J Exp Biol. 2011;214:412–421. doi: 10.1242/jeb.050161. [DOI] [PubMed] [Google Scholar]
- 81.Yang Z. Likelihood Ratio Tests for Detecting Positive Selection and Application to Primate Lysozyme Evolution. 1997:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 82.Olson SL. Evolution of the rails of the South Atlantic islands (Aves: Rallidae) Smithson Contrib to Zool. 1973;152:1–53. [Google Scholar]
- 83.Gould SJ. Ontogeny and Phylogeny. The Belknap Press of Harvard Universiry Press; Cambridge, Massachusetts: 1977. [Google Scholar]
- 84.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leggett RM, Clavijo BJ, Clissold L, Clark MD, Caccamo M. Next clip: An analysis and read preparation tool for nextera long mate pair libraries. Bioinformatics. 2014;30:566–568. doi: 10.1093/bioinformatics/btt702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346:1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.She R, Chu JSC, Wang K, Pei J, Chen N. genBlastA: Enabling BLAST to identify homologous gene sequences. Genome Res. 2009;19:143–149. doi: 10.1101/gr.082081.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Powell ADC, Aulerich RJ, Balander RJ, Stromborg KL, Bursian SJ. A Photographic Guide to the Development of Double-Crested Cormorant Embryos. Colon Waterbirds. 1998;21:348–355. [Google Scholar]
- 90.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ronquist F, et al. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Green RE, et al. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science. 2014;346:1254449–1254449. doi: 10.1126/science.1254449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hubisz MJ, Pollard KS, Siepel A. Phastand Rphast: Phylogenetic analysis with space/time models. Brief Bioinform. 2011;12:41–51. doi: 10.1093/bib/bbq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 99.Reimand J, Arak T, Vilo J. g:Profiler--a web server for functional interpretation of gene lists (2011 update) Nucleic Acids Res. 2011;39:W307–W315. doi: 10.1093/nar/gkr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Paix A, Folkmann A, Rasoloson D, Seydoux G. High Efficiency, Homology-Directed Genome Editing in Caenorhabditis elegans Using CRISPR-Cas9 Ribonucleoprotein Complexes. Genetics. 2015;201:47–54. doi: 10.1534/genetics.115.179382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhai Z, Yao Y, Wang Y. Importance of Suitable Reference Gene Selection for Quantitative RT-PCR during ATDC5 Cells Chondrocyte Differentiation. PLoS One. 2013;8 doi: 10.1371/journal.pone.0064786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suyama M, Torrents D, Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:609–612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 104.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–5. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stratigopoulos G, et al. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J Clin Invest. 2016;1:1–14. doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Osborn DPS, et al. Loss of FTO antagonises wnt signaling and leads to developmental defects associated with ciliopathies. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Arts HH, et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat Genet. 2007;39:882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- 108.Bell GW, Yatskievych TA, Antin PB. GEISHA, a Whole-Mount in Situ Hybridization Gene Expression Screen in Chicken Embryos. Dev Dyn. 2004;229:677–687. doi: 10.1002/dvdy.10503. [DOI] [PubMed] [Google Scholar]
- 109.Diez-Roux G, et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Sequenced species and genome assembly statistics.
Table S2. Genome completeness statistics of a diverse selection of avian genomes.
Table S3. Functional profiling of proteins with predicted function-altering variants (PROVEAN score < −5)
Table S4. List of genes in each of the enriched categories with predicted function-altering variants in P. harrisi.
Table S5. List of the proteins with the strongest function-altering variants in P. harrisi.
Table S6. Gene enrichment analysis of putative CUX1 target genes in Hs578t cells. P-values were corrected for multiple testing.
Table S7. Ultra-conserved noncoding elements in tetrapods showing accelerated molecular evolution in P. harrisi. Elements are annotated using human coordinates as a reference
Table S8. Test for positive selection in previously identified cilia related genes carrying function-altering mutations in P. harrisi.
Table S9. Sequenced libraries used for genome assembly.
Table S10. List of genes not annotated in P. harrisi but annotated in P. auritus, P. brasilianus and P.pelagicus.
Table S11. Curated list of proteins with putative premature stop codons in P. harrisi that are absent in in P. auritus, P. brasilianus and P.pelagicus.
Table S12. Primers used to verify predicted variants in the sequenced cormorants.
Table S13. Primers used for quantitative real time PCR in ATDC5 cell lines.