Abstract
Capsanthin/capsorubin synthase (Ccs) gene is a key gene that regulates the synthesis of capsanthin and the development of red coloration in pepper fruits. There are three tandem repeat units in the promoter region of Ccs, but the potential effects of the number of repetitive units on the transcriptional regulation of Ccs has been unclear. In the present study, expression vectors carrying different numbers of repeat units of the Ccs promoter were constructed, and the transient expression of the β-glucuronidase (GUS) gene was used to detect differences in expression levels associated with the promoter fragments. These repeat fragments and the plant expression vector PBI121 containing the 35s CaMV promoter were ligated to form recombinant vectors that were transfected into Agrobacterium tumefaciens GV3101. A fluorescence spectrophotometer was used to analyze the expression associated with the various repeat units. It was concluded that the constructs containing at least one repeat were associated with GUS expression, though they did not differ from one another. This repeating unit likely plays a role in transcription and regulation of Ccs expression.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0437-8) contains supplementary material, which is available to authorized users.
Keywords: Ccs, GUS, Promoter, Tandem repeat units, Transient expression
Introduction
Transcriptional promoters are regions of DNA that initiate transcription of specific downstream genes; generally, they are located quite near the transcription start site of the genes they regulate (Smale and Kadonaga 2003). As an important cis-regulatory element, promoters are involved in the regulation of gene transcription and expression (Segal et al. 2002), and a promoter can activate RNA polymerase to interact with the DNA template and begin transcription (Smale and Kadonaga 2003).
Capsanthin/capsorubin synthase (Ccs) gene is a key gene that regulates the synthesis of capsanthin in pepper fruits, which are unable to develop red coloration if Ccs has been deleted or contains a loss of function mutation. The promoter of Ccs gene is distinctive; it contains three tandem repeat units of 176 bp in Ccs promoter region. These repeat units start 165 bp upstream of the Ccs gene transcription start site, which is the main area in which promoters occur (Li et al. 2013). In plants, it is very rare that a tandem repeat appears within 165 bp upstream of a promoter. The existence of these tandem repeats may have important significance for capsanthin synthesis in pepper fruits.
In this study, ‘the ‘Yiduhong’ pepper cultivar was used as the experimental organism to examine the effect of the repeat units within the Ccs promoter; the main research objective was to elucidate the function of different numbers of repetitive units of the promoter. Three main experiments were conducted to fulfil this objective: (1) construction of expression vectors of different repeat unit lengths, (2) GUS reporter gene expression levels were assayed for transient expression, and (3) the expression activity associated with promoters of different repeat units was analyzed using a fluorescence spectrophotometer to detect differences among Ccs promoter repeat unit lengths in pepper fruits.
Materials and methods
Plant and expression vector materials
Seeds for the ‘Yiduhong’ pepper cultivar (Capsicum annuum) and the ‘Micro-Tom’ tomato (Solanum lycopersicum) cultivar were provided by the Capsicum Research Group in the College of Horticulture at Northwest A&F University, Shaanxi Province, People’s Republic of China. The PBI121 expression vector was also provided by the Capsicum Research Group at Northwest A&F University.
Construction of the cloning vector
PCR amplification bands (L1, L2, L2-1, and L3) were cut and purified with a Wizard PCR purification kit (TianGen Co., Ltd., Beijing, China). The purified PCR products (i.e., L1, L2, L2-1, and L3) were cloned into a pMD19-T vector and transformed into E. coli DH5α. The positive clones were screened on LB media plates with ampicillin, and detected by colony PCR. The recombinant plasmids (pMD-L0, pMD-L1, pMD-L2, pMD-L2-1, and pMD-L3, respectively) were digested with both BamHI and HindIII. The fragments were identified and sent to Sangon, Shanghai Co., Ltd. for sequencing.
Construction of the expression vector
Extraction of genomic DNA and detection in pepper fruits
Seeds were removed from pepper fruits, and five fruit pericarps from the same cultivar were mixed together and triturated in liquid nitrogen. A cetyltrimethylammonium bromide (CTAB) extraction solution was added to the resulting powder. DNA was extracted from samples following the procedure described by Tian et al. (2015). The DNA samples were dissolved in Tris–EDTA (TE) buffer solution and stored at −20 °C before gene identification. DNA primers were designed using Primer 5 software based on pepper gene sequences obtained from GenBank (Ccs: GenBank No. X77289.1).
Cloning Ccs promoters with various numbers of repeat units
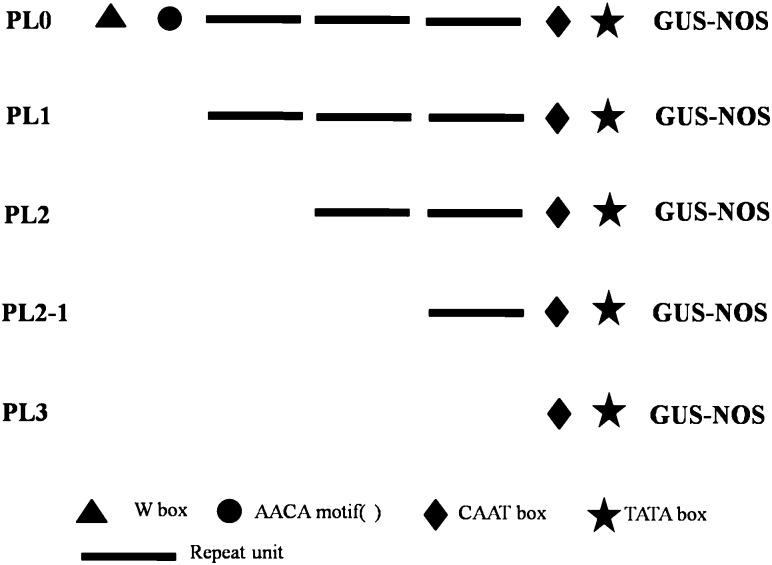
A primer was designed based upon sequences (GenBank) from the Ccs gene promoter in Capsicum (Supplementary Table 1). The repeat units of the promoters of various lengths were amplified using PCR. The PCR products were detected using agarose gel electrophoresis, purified with a wizard PCR purification kit (Promega, USA) and then cloned into a pMD19-T vector using T4 DNA ligase. The cloned vector was transformed into E. coli DH5α. The recombinant clones were screened and identified using PCR, restriction endonuclease digestion analysis, and sequencing. The different repeat promoter expression vectors were constructed from DNA fragments that were in accordance with the known promoter fragments (Fig. 1).
Fig. 1.
The recombined PBI121vector containing tandem repeat sequences of the Ccs gene promoter. PL0, PBI121vector carrying full-length Ccs promoter fragments with a W box and AACA motif; PL1, PBI121 vector carrying full-length Ccs promoter fragments without a W box and AACA motif; PL2, PBI121 vector carrying two repeat units of the promoter fragment; PL2-1, PBI121 vector carrying one repeat unit of the promoter fragment; PL3, PBI121 vector carrying zero repeat units of the promoter fragment
Construction of cloning vectors from Ccs gene promoters with various numbers of repeats
A combination of the PBI121 plasmid, the expression vector (PBI121), and the cloning vector carrying the target sequence were digested with double-strand exonucleases and then incubated for 10 h at 37 °C in a water bath. The enzyme product was recovered using a DNA extraction kit [TaKaRa Biotechnology (Dalian) Co., Ltd]. The target gene fragments were ligated to the PBI121 expression vector with T4 DNA ligase (Supplementary Fig. 1–5) while incubated at 16 °C overnight and then transformed into E. coli DH5α. The target gene fragments were screened and identified by colony PCR.
Construction of the plant expression vector
The enzymatic reaction system is presented in Supplementary Table 2. The plant expression vectors (i.e., PL0, PL1, PL2, PL2-1, and PL3) were constructed by cloning the fusion gene into the PBI121 plasmid. The enzymatic reaction system is presented in Supplementary Table 3. The expression vectors were constructed and confirmed by restriction enzymes digests followed by fragments sequencing. The correct target fragments were extracted and the plasmids were named PL3, PL2, PL1, PL2-1, and PL0. The recombinant promoter expression vectors were introduced into Agrobacterium tumefaciens GV3101 using the freeze–thaw method (Wang et al. 2013). The promoter regions from the target repetitive units were detected by PCR, and the bacterial cultures were preserved at −80 °C for further use.
The preparation of Capsicum and tomato materials
Capsicum and tomato seeds were germinated as previously described (Tian et al. 2014). The germinated seeds were sown into wells on a plate. When three to four true leaves were present, the seedlings were transplanted into plastic pots and maintained under field conditions until green fruits developed. These potted Capsicum and tomato plants were then moved into controlled climate conditions.
Transient expression of the GUS reporter gene in Capsicum and tomato fruits with various numbers of promoter repeats
A solution of Agrobacterium (20 μL) containing the Ccs gene promoter fragment was transferred into 20 mL of LB liquid medium (50 mg/L Kan, 50 mg/L Gen, 10 mg/L Rif) for overnight incubation at 28 °C while being centrifuged at 35×g (Kapila et al. 1997). After overnight incubation, the Agrobacterium culture was centrifuged at 2504×g at room temperature conditions for 10 min. The concentration of the bacterial suspension was diluted with 10 mL of penetration buffer solution (0.5% d-glucose, 50 mM MES, 2 mM Na3PO4·12H2O, 0.1 mM AS) (Sparkes et al. 2006) and the resulting supernatant was discarded. These steps were repeated in order to remove as much excess LB liquid medium as possible. Then, the absorbency at 600 nm of 2.5 mL of the resuspended Agrobacterium solution containing the Ccs gene promoter fragments was measured. The solution was then diluted with penetration buffer to OD600 = 0.5 for subsequent genetic transformation.
Capsicum and tomato plants were placed in controlled climate conditions with high intensity irradiation for 1 h prior to injection in order to allow the plants to acclimatize to the new environment. Green fruits of same age were selected and tagged. Fruit stalks were selected as the injection location and the Agrobacterium solution (0.8 mL) was injected into Capsicum and tomato fruits with a 1 mL syringe. The bacterial culture was introduced slowly into Capsicum and tomato fruits. Five fruits were selected for each bacterial culture carrying a different Ccs gene promoter fragment, and each treatment was repeated five times. After inoculation with Agrobacterium, Capsicum and tomato plants were transferred into a controlled environment at 28 °C with a 14 h day/10 h night photoperiod for two days. These processed materials were divided into two parts, one for GUS histochemical staining and the other for the determination of GUS activities.
GUS histochemical staining
Capsicum and tomato pericarp tissue was smooth and very dense. To facilitate GUS histochemical staining, the experimental tomato materials were divided into thin layers and placed into a sterilized Petri dish containing GUS-staining liquid (Supplementary Table 4). The Petri dishes were sealed with plastic film and promptly placed in a homoeothermic incubator at 37 °C for 24 h. The GUS-staining solution was removed and replaced with 70% ethyl alcohol to remove excess pigments and was then incubated at 37 °C for 6–8 h. After the initial incubation, the 70% ethyl alcohol was replaced with 90% ethyl alcohol and incubated at 37 °C for 10 h. The 90% ethyl alcohol was removed, and changes in chlorophyll were observed. Then, the plates were flushed with 90% ethyl alcohol continuously for an additional 10–12 h until no chlorophyll remained. The material was then placed on a light box and photographs were taken (Subramanyam et al. 2013).
The determination of GUS activities
To prepare a GUS enzyme extract, 0.1 g of tomato fruit that had been previously treated with GUS histochemical stain was ground in liquid nitrogen using a mortar and pestle. GUS extraction buffer was added to the mortar, and grinding was continued until the material became a uniform powder. The powder was then poured into a microcentrifuge tube and shaken for 5 min. Then the powder was centrifuged at 12,000 rpm for 10 min, and the resulting supernatant was removed and stored at 4 °C.
To conduct the enzymatic reaction, 4-methylumbelliferone (4-MU) was used for fabrication of the fluorescence standard curve. Then 1 mM 4-methylumbelliferyl glucuronide (MUG) was added into the extraction buffer to form the reaction buffer. The reaction buffer was placed in a water bath at 37 °C for 10 min, and 20 µL of GUS extraction buffer was added into the 200 µL reaction buffer volume and mixed thoroughly. Then, 100 µL of mixture buffer was removed immediately and added to 900 µL of 0.2 M Na2CO3 to terminate GUS activity. The remaining reaction mixture volume was reacted at 37 °C for 30 min, and then 900 µL of 0.2 M Na2CO3 was added to terminate the reaction. The fluorescence intensity, 4-MU content, and GUS activity were then measured (Yang et al. 2000).
Statistical analysis
SAS 6.12 software (SAS Institute, Gary, NC, USA) was used for data analysis. Duncan’s multiple-range test was used, and Least significant ranges (LSR) analysis was conducted at P < 0.05.
Results and discussion
DNA extraction of pepper fruits
The extracted DNA was not contaminated with RNA, protein, or phenol and the target bands were clearly visible (Supplementary Fig. 6).
PCR amplification of various promoter fragments
The PL0 bands (full-length promoter fragments) and PL3 bands (proximal promoter fragments) were clearly visible on 1% agarose gel electrophoresis. The PL0 fragment was about 1000 bp, which was consistent with the target fragment length (Fig. 2). Sequence analysis of different repeat units of the Ccs promoter had revealed that there were no frameshifts and/or base mutations. This verified that the promoters of different repeat unit lengths (i.e., PL0 and PL3) were obtained successfully (Fig. 2); however, the PL1 and PL2 bands were faint (Fig. 2), and PCR amplification was therefore repeated for PL1 and PL2 (Fig. 3). PL0 was used as a template to re-amplify PL1 and PL2, and the target bands for PL1 and PL2 were thus obtained. Sequence analysis of PL1 and PL2 revealed that there were no frameshift and/or base mutations. This showed that promoters of intermediate repeat units lengths (i.e., PL1 and PL2) were successfully obtained (Fig. 3).
Fig. 2.
Different specific tandem repeat regions for PCR amplification
Fig. 3.
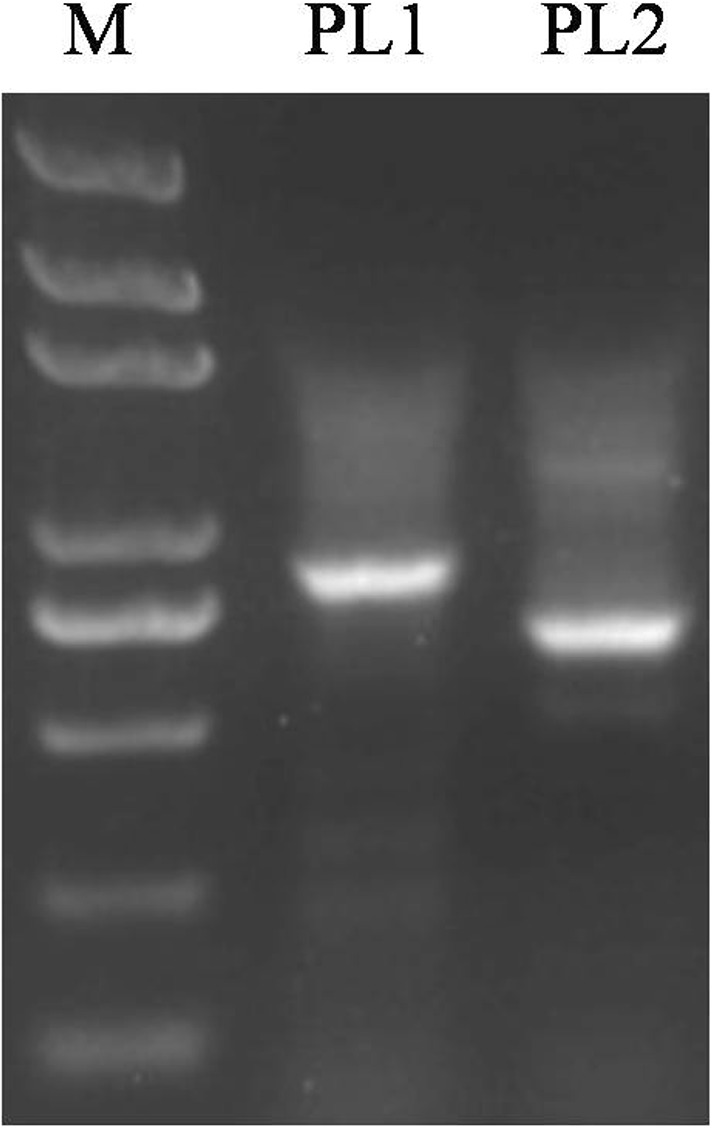
PCR amplification of the PL1 and PL2 tandem repeat regions
The same method was followed, and PL2 PCR amplicons were used as the template to amplify PL2-1, thus obtaining the PL2-1 target band. PL2-1was digested ligated to the pMD19-T vector, and transformed into E. coli DH5α. Sequence analysis of PL2-1 revealed that there were no frameshift and/or nucleotide base mutations (Fig. 4).
Fig. 4.
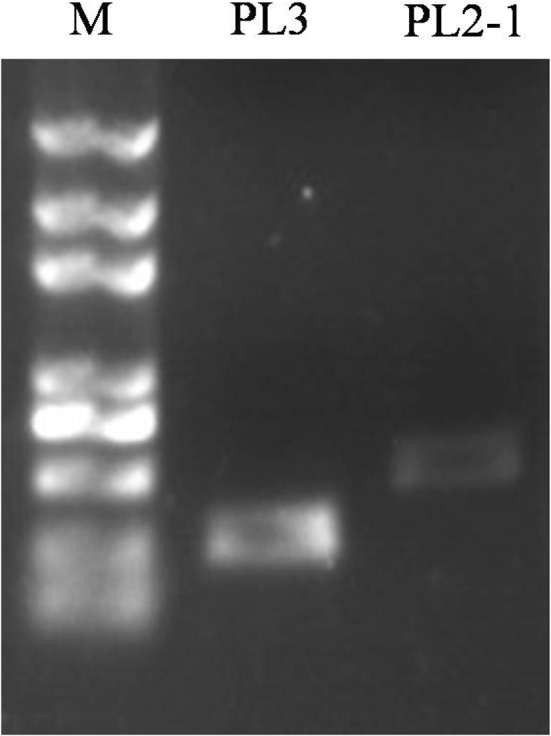
PCR amplification of the PL2-1 and PL3 tandem repeat regions
Detection of expression vectors from promoter repeats
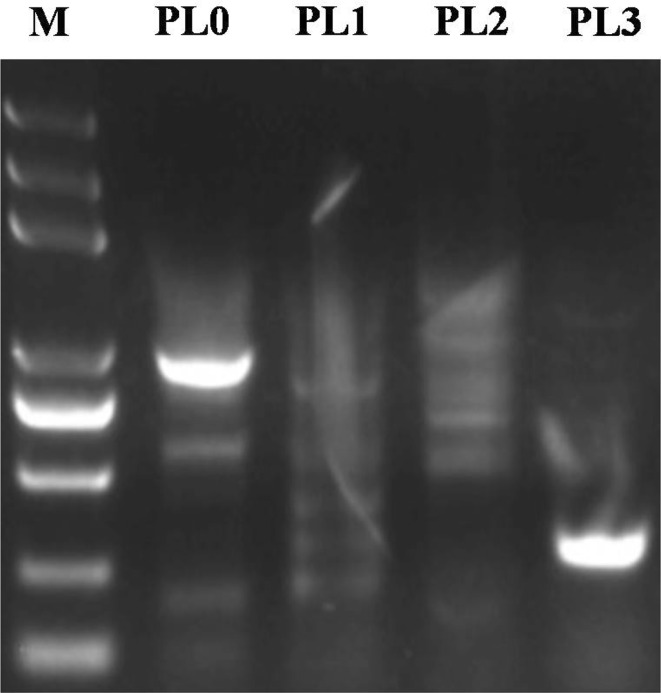
Five expression vectors carrying different repeat units were digested with BamHI and HindIII, respectively, and their banding patterns were analyzed after 1% agarose gel electrophoresis. Their lengths were approximately 1000, 900, 700, 500, and 380 bp, respectively (Fig. 5). Sequencing results of the DNA bands were in accordance with the known different repeat units of the Ccs promoter. Sequence analysis of expression vectors carrying different repeat units revealed that there were no frameshift and/or nucleotide base mutations. This confirmed that the expression vectors were successfully constructed (Fig. 5).
Fig. 5.
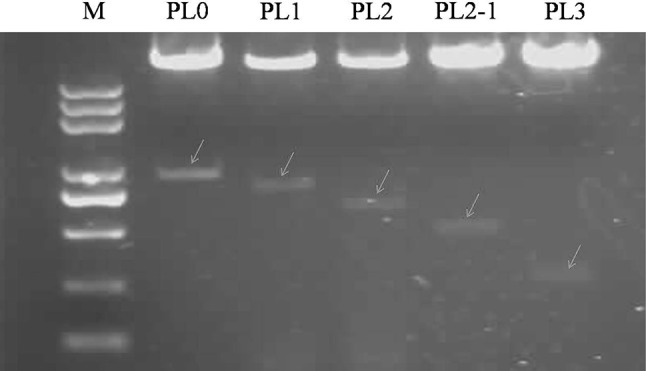
Expression vectors digested with BamHI and HindIII. Expression vectors carrying different repeat units (i.e., PL0, PL1, PL2, PL2-1, and PL3) were digested with BamHI and HindIII, respectively, and were analyzed using 1% agarose gel electrophoresis. The lengths of these bands were approximately 1000, 900, 700, 500, and 380 bp, respectively
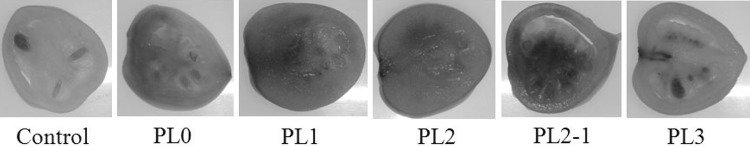
Transient expression in mature green Capsicum fruits
Each partial and full-length Ccs promoter was expressed normally in mature green Capsicum fruit (Fig. 6).
Fig. 6.
Expression analysis of tandem repeat promoter regions in pepper fruits at the green ripening stage. Control, the control group (PBI121 vector without any Ccs promoter repeat units); PL0, PBI121 vector carrying full-length Ccs promoter fragments with a W box and AACA motif; PL1, PBI121 vector carrying full-length Ccs promoter fragments without a W box and AACA motif; PL2, PBI121 vector carrying two repeat units of the promoter fragment; PL2-1, PBI121 vector carrying one repeat unit of the promoter fragment; and PL3, PBI121 vector carrying zero repeat units of the promoter fragment. The GUS reporter gene in mature, green pepper fruits was expressed by all containing Ccs promoter fragments containing repeat units (PL0, PL1, PL2, and PL2-1)
Transient expression various promoters mediated by Agrobacterium in mature green tomatoes
Each variable repeat Ccs promoter (i.e., PL0, PL1, PL2, and PL2-1) was expressed normally in mature green tomato fruits except for PL3; the fruit carrying PL3 did not properly stain blue because there was no repetition of the Ccs gene promoter in PL3. These highly similar repetitive sequences (i.e., PL1, PL2, PL2-1, and PL0) were able to promote Ccs expression in the fruits. No clear differences were observed among the various levels of blue staining in PL0, PL1, PL2, and PL2-1 (Fig. 7).
Fig. 7.
Expression analysis of different specific tandem repeats in the promoter region in ‘Micro-Tom’ fruits. Control, the control group (PBI121 vector without any Ccs promoter repeat units); PL0, PBI121 vector carrying full-length Ccs promoter fragments with a W box and AACA motif; PL1, PBI121 vector carrying full-length Ccs promoter fragments without a W box and AACA motif; PL2, PBI121 vector carrying two repeat units of the promoter fragment; PL2-1, PBI121 vector carrying one repeat unit of the promoter fragment; PL3: PBI121 vector carrying zero repeat units of the promoter fragment. The GUS reporter gene in mature, green tomato fruits was expressed by all plants containing Ccs repeat units (PL0, PL1, PL2, and PL2-1)
A comparison of Figs. 6 and 7, reveals that pepper and tomato yielded similar expression results. The ‘Micro-Tom’ tomato cultivar has a short life cycle as it is a dwarf plant, thus, the fruiting period is early and uniform, ‘Micro-Tom’ fruits are therefore an ideal material for studying transient expression.
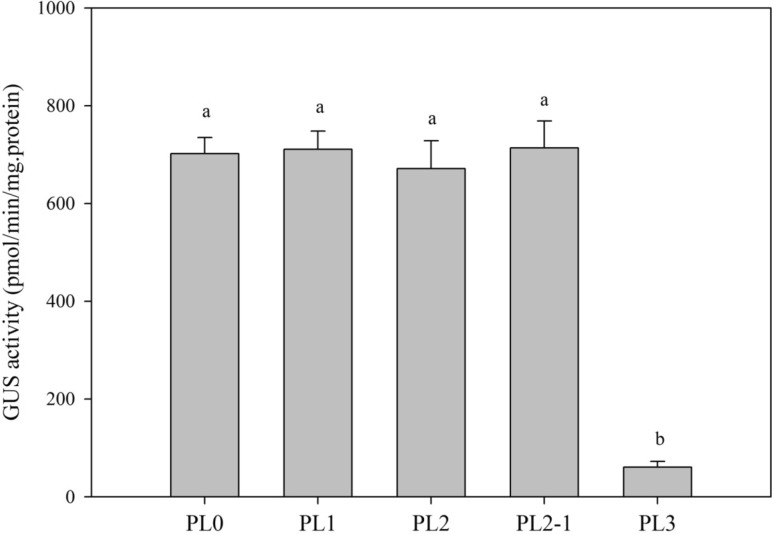
Activity analysis of different repetitive units
To further confirm the differences among PL0, PL1, PL2, and PL2-1 constructs, an activity analysis of GUS expression for constructed with different numbers of promoter repeats was conducted. Promoters consisting of each of the various repetitive units were active, and there were no obvious differences among PL0, PL1, PL2, and PL2-1. However, the activity of PL3 was extremely low (Fig. 8).
Fig. 8.
Analysis of GUS activity in ‘Micro-Tom’ fruits for different specific repeat sequences of the Ccs gene promoter. PL0, PBI121 vector carrying full-length Ccs promoter fragments with a W box and AACA motif; PL1, PBI121 vector carrying full-length Ccs promoter fragments without a W box and AACA motif; PL2, PBI121 vector carrying two repeat units of the promoter fragment; PL2-1, PBI121 vector carrying one repeat unit of the promoter fragment; and PL3, PBI121 vector carrying zero repeat units of the promoter fragment. Activity of the GUS reporter gene was observed in plants with all numbers of Ccs promoter repeat units; however, there were no obvious differences among PL0, PL1, PL2, and PL2-1 plants in expression level, while GUS expression in PL3 plants was extremely low
Promoter function studies have primarily focused on the construction of deletions, mutations, and function verification of promoter regions. Usually, the expression level of the GUS reporter gene decreases as promoters are shortened (Li et al. 2012; Liang et al. 2012). Han et al. (2007) constructed a series of deletion fragments of the LRP16 promoter and conducted a promoter activity analysis that showed that the LRP16 promoter was a typical type II promoter with a maximally active region 200–600 bp upstream (Han et al. 2007). Liang et al. (2012) found that all deletion fragments of a promoter could be detected from the activity of the GUS reporter gene, but the activity of deletion fragments differed with construct lengths. Li et al. (2012) found that GUS expression activity increased as the 5′ terminal deletions typically increased GUS activity. Similarly, Li et al. (2013) revealed that the expression of a gene became weaker with the decreases in promoter length. Our results indicated that the GUS activity associated with the Ccs promoter repeats were not related to promoter length. No apparent differences were observed in the activity of GUS between plants with the deletion of the full-length promoter, PL0 (which included three repeat units), and the promoter with the fewest repeats PL2-1 (with one repeat unit; Fig. 8).
In our experiment, the expression level of the GUS reporter gene did not decrease with the deletion promoter deletion size. The different repeat units of the Ccs gene promoter were not obviously associated with differences in GUS activity. Previous studies focusing on the Ccs gene promoter did not report differences in effects associated with the number of repeat units in the promoter region. Any sequence deletion in a promoter changes the structure and function of the promoter (Han et al. 2007; Liang et al. 2012; Li et al. 2013); however, every repeat unit of the Ccs gene promoter in Capsicum was a relatively complete repetitive sequence, although the partial sequences did not compose the full-length promoter. This complicated our results as there were no obvious differences in GUS activity among the repetitive units of the Ccs gene promoter. Specific repetitions of the Ccs gene promoter appear to be redundant. Each deletion in the series of partial Ccs gene promoters contained at least one complete repeat unit. The expression level of the GUS reporter gene showed that a single repeat was sufficient to ensure Ccs gene expression, in other words, each repeat was an adequate substitute that ensured normal Ccs gene expression for color formation in pepper fruits. However, for other gene promoters, totally lacking repeated units, the partial deletions of promoter sequences rendered their structure incomplete in comparison with a single-repeat Ccs gene promoter, which thus impaired promoter function (Liang et al. 2012; Li et al. 2013). Our study provides the basis for further research on cis-regulatory elements of the Ccs promoter and the effect of Ccs expression on pepper fruit color.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Schematic representation of recombinant PBI121 vectors carrying Ccs gene promoter fragments (JPEG 590 kb)
DNA extraction of pepper fruits (JPEG 17 kb)
Primers were used in plasmid construction (DOCX 13 kb)
The reaction system (DOCX 13 kb)
The ligase reaction system (DOCX 13 kb)
Histochemical staining reactants for assaying GUS activity (DOCX 13 kb)
Acknowledgements
The authors acknowledge the following organizations for their financial support: National Key Research and Development Program of China (No. 2016YFD0101900), National Natural Science Foundation of China (No. 31272163); Innovation Scientists and Technicians Troop Construction Projects of Henan Province (C20150054); Science and Technology Development Project of Henan Province (162102110084); Key Scientific Research Projects in Universities of Henan Province (16A210010).
Abbreviations
- Ccs
Capsanthin/capsorubin synthase
- GUS
β-Glucuronidase
- PL0
Vector carrying full-length Ccs promoter fragments with W box and AACA motif
- PL1
Vector carrying full-length Ccs promoter fragments without W box or AACA motif
- PL2
Vector carrying two repeat units of the promoter fragment
- PL2-1
Vector carrying one repeat unit of the promoter fragment
- PL3
Vector carrying no repeat units of the promoter fragment
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0437-8) contains supplementary material, which is available to authorized users.
Shi-Lin Tian and Zheng Li authors have contributed equally to this work.
References
- Han WD, Zhao YL, Wu ZQ, Meng YG, Zang L, Mu YM. Estrogen receptor α (ERα) target gene LRP16 interacts with ERα and enhances receptor’s transcriptional activity. Chin J Cancer Res. 2007;19:233–237. doi: 10.1007/s11670-007-0233-z. [DOI] [PubMed] [Google Scholar]
- Kapila J, Rycke RD, Van Montagu M, Angenon G. An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 1997;122(1):101–108. doi: 10.1016/S0168-9452(96)04541-4. [DOI] [Google Scholar]
- Li Y, Sun Y, Yang QC, Kang JM, Zhang TJ, Gruber MY, Fang F. Cloning and function analysis of an alfalfa (Medicago sativa L.) zinc protein promoter MsZPP. Mol Biol Rep. 2012;39:8559–8569. doi: 10.1007/s11033-012-1712-y. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang S, Gui XL, Chang XB, Gong ZH. A further analysis of the relationship between yellow ripe-fruit color and the capsanthin-capsorubin synthase gene in pepper (Capsicum spp.) indicated a new mutant variant in C. annuum and a tandem repeat structure in promoter region. PLoS ONE. 2013;8(4):e61996. doi: 10.1371/journal.pone.0061996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Xia H, Wu S, Ma FW. Genome-wide identification and expression profiling of dehydrin gene family in Malusdomestica. Mol Biol Rep. 2012;39:10759–10768. doi: 10.1007/s11033-012-1968-2. [DOI] [PubMed] [Google Scholar]
- Segal E, Barash Y, Simon I, Friendman N, Koller D. From promoter sequence to expression: Aprobabilistic framework. RECOMB. 2002;5:263–272. doi: 10.1145/565196.565231. [DOI] [Google Scholar]
- Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu Rev Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1(4):2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- Subramanyam K, Rajesh M, Jaganath B, Vasuki A, Theboral J, Elayaraja D, Karthik S, Manickavasagam Ganapathi MA. Assessment of factors influencing the Agrobacterium mediated in plantaseed transformation of brinjal (Solanumm elongena L.) Appl Biochem Biotech. 2013;17:450–468. doi: 10.1007/s12010-013-0359-z. [DOI] [PubMed] [Google Scholar]
- Tian SL, Li L, Chai WG, Shah SNM, Gong ZH. Effects of silencing key genes in the capsanthin biosynthetic pathway on fruit color of detached pepper fruits. BMC Plant Biol. 2014;14:314. doi: 10.1186/s12870-014-0314-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian SL, Li L, Shah SNM, Gong ZH. The relationship between red fruit colour formation and key genes of capsanthin biosynthesis pathway in Capsicum annuum. Biol Plant. 2015;59(3):507–513. doi: 10.1007/s10535-015-0529-7. [DOI] [Google Scholar]
- Wang JE, Liu KK, Li DW, Zhang YL, Zhao Q, He YM, Gong ZH. A novel peroxidase CaPOD gene of pepper is involved in defense responses to phytophtoracapsici Infection as well as abiotic stress tolerance. Int J Mol Sci. 2013;14:3158–3177. doi: 10.3390/ijms14023158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M. In vivo analysis of plant promoters and transcription factors by Agroinfiltration of tobacco leaves. Plant J. 2000;22:543–551. doi: 10.1046/j.1365-313x.2000.00760.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of recombinant PBI121 vectors carrying Ccs gene promoter fragments (JPEG 590 kb)
DNA extraction of pepper fruits (JPEG 17 kb)
Primers were used in plasmid construction (DOCX 13 kb)
The reaction system (DOCX 13 kb)
The ligase reaction system (DOCX 13 kb)
Histochemical staining reactants for assaying GUS activity (DOCX 13 kb)