Abstract
Improving seed related traits remains key objective in lentil breeding. In recent years, genomic resources have shown great promise to accelerate crop improvement. However, limited genomic resources in lentil greatly restrict the use of genomics assisted breeding. The present investigation aims to build an intraspecific genetic linkage map and identify the QTL associated with important seed relevant traits using 94 recombinant inbreds (WA 8649090 × Precoz). A total of 288 polymorphic DNA markers including simple sequence repeat (SSR), inter simple sequence repeat (ISSR) and random amplified polymorphic DNA (RAPD) were assayed on mapping population. The resultant genetic linkage map comprised 220 loci spanning 604.2 cM of the lentil genome, with average inter-marker distance of 2.74 cM. QTL mapping in this RIL population uncovered a total of 18 QTL encompassing nine major and nine minor QTL. All major QTL were detected for seed related traits viz., seed diameter (SD), seed thickness (ST), seed weight (SW) and seed plumpness (SP) across two locations. A considerable proportion of the phenotypic variation (PV) was accounted to these QTL. For instance, one major QTL on LG5 controlling SW (QTL 15) explained 50% PV in one location, while the same QTL accounted for 34.18% PV in other location. Importantly, the genomic region containing multiple QTL for different seed traits was mapped to a 17-cM region on LG5. The genomic region harbouring QTL for multiple traits opens up exciting opportunities for genomics assisted improvement of lentil.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0438-7) contains supplementary material, which is available to authorized users.
Keywords: Lentil, Molecular marker, Genome, Linkage map, QTL, Trait
Introduction
Lentil (Lens culinaris ssp. culinaris) is a self-pollinating grain legume grown across the Indian subcontinent, northern Africa, western Asia, southern Europe, North and South America and Australia (Akibode and Maredia 2011). It is an ancient crop that is believed to have originated in the Near East and later spread all through the Mediterranean Basin and central Asia (Cubero et al. 2009). Globally, 4.88 million tons of lentil is harvested annually, and the leading lentil-producing countries are Canada, India, Australia, Turkey and Nepal (FAOSTAT 2014). It is a diploid crop (2n = 2x = 14) with a large genome size of 4063 Mbp (Arumuganathan and Earle 1991). Lentil remains an excellent source of plant-based protein (up to 26%) to vegetarian people worldwide, especially in the developing world (https://www.pinterest.com/pin/556053885216362537/).
In recent years, crop genomics has witnessed remarkable developments and plenty of genomic resources have been generated in grain legume crops (Bohra et al. 2014). However, lentil still suffers from a dearth of genomic resources compared to the other well-researched legume crops such as soybean and common bean. Molecular markers, genetic linkage maps and QTL are the important genomic tools that remain central to any molecular breeding programme. Among the various marker systems, simple sequence repeat (SSR) marker still remains a preferred category of DNA markers given their greater abundance, co-dominant inheritance and multilocus nature (Gupta and Varshney 2000), though the trend is increasingly shifting towards high-density marker assays including single nucleotide polymorphism (SNP) (Fedoruk et al. 2013).
To understand the genetic architecture of important traits, QTL mapping provides a well-established means to allow the identification of molecular markers associated with the desirable traits. The marker trait associations (MTAs) can subsequently be harnessed in marker-assisted selection (MAS) schemes (Bohra 2013). In the context, linkage maps are useful for predicting the position of QTL/gene within the genome. Several genetic linkage maps have been constructed in lentil (see Bohra et al. 2014).
Seed traits viz. seed size and seed weight represent prime target traits in lentil breeding and manifest quantitative inheritance (Verma et al. 2015). Among legumes, researchers have performed mapping of QTL for seed weight/size in different crops including soybean (Sun et al. 2012), mungbean (Isemura et al. 2012), chickpea (Hossain et al. 2010; Upadhyaya et al. 2006) etc. In lentil, several agronomic traits such as plant height, days to flowering, winter hardiness, pod dehiscence, growth habit and yield have been genetically dissected using both inter-and intra-specific populations (Tar’an et al. 2003; Kahraman et al. 2004; Fratini et al. 2007; Tullu et al. 2008). Similarly, QTL for resistance to diseases like ascochyta blight, anthracnose and stemphylium blight have also been mapped (Ford et al. 1999; Rubeena and Taylor 2003; Tullu et al. 2006). Mapping of seed weight was also reported (Abbo et al. 1991; Verma et al. 2015).
Notwithstanding the plenty of QTL in lentil, the deployment of MTAs in lentil breeding has been limited. Seed diameter (SD), seed thickness (ST) and seed weight (SW) hold immense relevance to market class and consumers acceptance. Similarly, seed plumpness (SP) is also an important trait that influences the dehulling efficiency of the seed during milling. Precise and rapid improvement of these traits using MAS could be cost-effective and time-saving. In view of this, the present research aims to develop a genetic linkage map for cultivated lentil, and identify the DNA markers linked to the genes/QTL controlling seed-related traits using an F8 recombinant inbred line (RIL) population.
Materials and methods
Plant materials
A RIL population comprising 94 individuals derived from an intraspecific cross of Lens culinaris ssp. culinaris (WA 8649090 × Precoz) was used for the construction of genetic linkage map. The parents differed from each other significantly with respect to various agro-morphological and seed related traits. The RILs were grown in greenhouse during 2014–2015 for collecting leaf samples for DNA extraction.
This mapping population was phenotyped for nine important quantitatively inherited traits in which four were seed related characters viz. SD (mm), ST (mm), SP and SW (g). Besides seed traits, phenotypic observations were also recorded on days to flowering (DF), number of primary branches (PB), days to maturity (DM), plant height (PHT, cm) and seed per plant (SPP). The material was raised with two replications at two different locations i.e. Palampur (L1) during 2014–2015 and Akrot (L2) during 2011–2012 in Himachal Pradesh, India. The data on PHT, DM and PB were recorded only in one location (L1). The recommended cultivation practices were followed. The location-wise data of all these traits are provided in Supplementary Table 1.
Traits SD (mm) and ST (mm) were measured by averaging ten seeds selected randomly using scale and vernier calliper, respectively. SP was calculated as the ratio of ST and SD. Phenotypic data were also recorded for DF as number of days from planting to appearance of first flower. Similarly, DM were calculated as days from sowing to 75% maturity of the plants, while PHT was measured in cm by centimeter scale. For SW (g), 100 seeds were weighed with the help of electronic balance.
SSR analysis
SSRs were assayed on mapping parents to search for DNA polymorphism. The polymorphic SSRs were then used for genotyping the mapping population. For amplification of genomic DNA, a reaction mixture of 12.5 µl volume was prepared using 7.15 µl of sterilized distilled water, 1.0 µl template DNA (25 ng/µl), 0.5 µl of forward and 0.5 µl of reverse primer (5 µM), 1.0 µl MgCl2 (25 mM), 1.25 µl 10 × PCR buffer (10 mM Tris–HCl, 50 mM KCl, pH 8.3), 1.0 µl dNTP mix (0.2 mM each of dATP, dGTP, dCTP and dTTP) and 0.1 µl Taq polymerase (5U/µl). The amplifications were carried out in Gene Amp PCR System 9700® (Applied Biosystems, CA, USA) and 2720 Thermal Cycler (Applied Biosystems, CA, USA).
The amplified products were electrophoresed in 3% agarose gel (HiMedia, Mumbai, India) and stained with ethidium bromide (0.5 µg/ml). The gels were visualized and photographed using the Gel-Documentation Unit (Bio-Rad, Hercules, CA, USA).
RAPD and ISSR analyses
Due to the lack of adequate SSR polymorphism, RAPD and ISSR primers were also screened to gain the sufficient number of polymorphic DNA markers. A total of 250 RAPD decamer primers (Operon Technologies, Alameda, CA, USA) and 30 ISSR primers (15–23 nucleotides in length) were screened on mapping parents. The primers that produced easy-to-score and polymorphic fragments were then used to genotype the mapping population. Information on polymorphic markers generated from additional 42 RAPD primers (kindly supplied by Fred Muehlbauer, USDA, ARS, USA) was also used in the study. For amplification of genomic DNA, a reaction mixture of 12.5 µl volume was prepared using 7.15 µl of sterilized distilled water, 1.0 µl template DNA (25 ng/µl), 1.0 µl primer (RAPD/ISSR), 1.0 µl MgCl2 (25 mM), 1.25 µl 10X PCR buffer (10 mM Tris–HCl, 50 mM KCl, pH 8.3), 1.0 µl dNTP mix (0.2 mM each of dATP, dGTP, dCTP and dTTP) and 0.1 µl Taq polymerase (5U/µl). The amplification was carried out in a 2720 Thermal Cycler (Applied Biosystems, CA, USA).
PCR products were mixed with 2 µl of gel loading dye (0.25% bromophenol blue and 40% sucrose), resolved on 1.5% agarose gel for RAPDs and 1.8% gel for ISSRs and electrophoresed at 100 V for 90 min in 1× Tris acetate-EDTA (TAE) buffer (40 mM Tris, 40 mM Acetic acid Glacial, 1 mM EDTA, pH 8.0). The gels were stained with ethidium bromide (0.5 µg/ml) and the PCR products were visualized and photographed using the Gel-Documentation Unit (Bio- Rad, Hercules, CA, USA).
Linkage analysis
Genotyping data were used to perform linkage analysis using JoinMap v. 4.0 (Van Ooijen 2006). Marker order was determined using the regression mapping algorithm with maximum recombination frequency of 0.4 at minimum logarithm of odd (LOD) score of 3 and jump threshold of 5. The Kosambi mapping function was used to calculate the map distance (Kosambi 1944). To check segregation distortion, Chi square (χ2) values were calculated using “Locus genotype frequency” function of JoinMap v. 4.0. Final linkage maps were drawn using MapChart v. 2.5 (Voorrips 2002).
QTL analysis
Genotypic and phenotypic data scored on the RIL population were subjected to QTL analysis by using QTL Cartographer v. 2.5 (Wang et al. 2012) (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). Composite interval mapping (CIM) was performed by selecting Model 6 with the default parameters such as 10 cM window size, control marker number 5, and backward regression method.
Results
Parental polymorphism and population genotyping
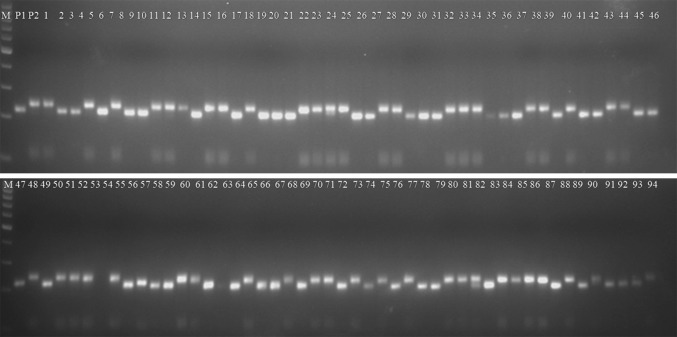
Three different types of marker systems viz. SSR, RAPD and ISSR were employed in the present investigation. The SSR markers were Lens specific and also from Trifolium pratense. Of the total 725 SSR screened on the parental combination, 47 polymorphic SSRs yielded scorable amplicons of expected size. The amplicons generated using SSR primer in parents and RIL are shown in Fig. 1. In parallel, ISSR markers were also screened on parental genotypes to detect DNA polymorphism. Three of the polymorphic ISSRs yielded 5 clear, scorable and reproducible fragments (1.67 markers/primer). The size of the RAPD markers varied from approximately 300–2400 bp.
Fig. 1.
Genotyping profile of SSR marker (SSR-113). The gel image illustrates the segregation pattern of SSR 113 in mapping parents and recombinant inbred individuals. Lane M, 100 bp standard DNA ladder; lane P1, WA 8649090; lane P2, Precoz; lanes 1–94, mapping individuals
Construction of an intraspecific genetic linkage map
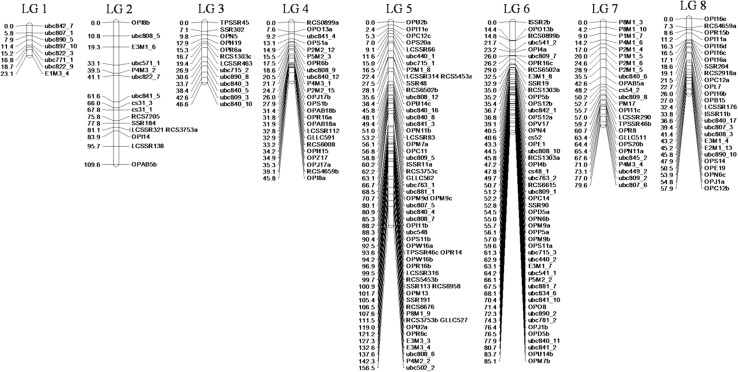
Of the 288 markers (223 RAPD, 5 ISSR, 60 SSR) assayed on parents and RIL, 220 were placed on to the linkage map (Fig. 2), whereas 68 (51 RAPD, 2 ISSR and 15 SSR) remained unlinked (Table 1).
Fig. 2.
Intraspecific genetic linkage map of lentil. A total of 220 loci were placed onto eight LGs. The map was developed from a RIL population (WA 8649090 × Precoz)
Table 1.
Distribution of mapped loci on the linkage map
| Linkage groups(LGs) | Marker | |||
|---|---|---|---|---|
| RAPD | ISSR | SSR | Total | |
| LG1 | 8 | – | – | 8 (3.63%) |
| LG2 | 11 | – | 5 | 16 (7.27%) |
| LG3 | 9 | – | 4 | 13 (5.90%) |
| LG4 | 19 | – | 5 | 24 (10.90%) |
| LG5 | 39 | 1 | 17 | 57 (25.90%) |
| LG6 | 43 | 1 | 7 | 51 (23.18%) |
| LG7 | 22 | – | 3 | 25 (11.36%) |
| LG8 | 21 | 1 | 4 | 26 (11.81%) |
| Total | 172 (78.18%) | 03 (1.36%) | 45 (20.45%) | 220 |
The present genetic linkage map was constructed at LOD score of 2.5 with a maximum recombination value of 0.40. The map comprised a total of 220 markers including 172 RAPDs, 3 ISSRs and 45 SSRs. Of the total 172 mapped RAPDs, 90 markers were mapped previously in the same population by Kahraman et al. (2004). The linkage analysis established eight linkage groups (LG1-8) spanning a total map length of 604.2 cM with individual LGs varying between 23.1 cM and 156.5 cM. The number of mapped loci per LG ranged from 8 to 57. The average inter marker distance was 2.74 cM (Table 2).
Table 2.
Salient features of the intraspecific genetic linkage map of lentil
| Linkage groups (LGs) | Length (cM) | Markers mapped | Average marker spacing (cM) | Largest inter-marker distance (cM) | Smallest inter-marker distance (cM) |
|---|---|---|---|---|---|
| LG1 | 23.1 | 8 | 2.89 | 4.4 | 1.6 |
| LG2 | 109.6 | 16 | 6.85 | 13.9 | 1.6 |
| LG3 | 46.6 | 13 | 3.58 | 4 | 1.4 |
| LG4 | 45.8 | 24 | 1.91 | 6.7 | 0.1 |
| LG5 | 156.5 | 57 | 2.75 | 14.2 | 0.2 |
| LG6 | 85.1 | 51 | 1.67 | 2.8 | 0.1 |
| LG7 | 79.6 | 25 | 3.18 | 2.6 | 1 |
| LG8 | 57.9 | 26 | 2.23 | 3.1 | 0.2 |
| Total | 604.2 | 220 | 2.74 |
RAPD markers were distributed across all LGs in the genome. Similarly, SSRs were mapped on all LGs except LG1. ISSR markers were mapped on three different LGs (LG8, LG5 and LG6). The distribution of DNA markers across different LGs was unequal and the size of the LG did not necessarily reflect the number of mapped loci. For instance, LG6 with 51 markers covered 85.10 cM with an average marker spacing of 1.67 cM, whereas LG4 spanning a distance of 45.8 cM harboured 24 markers with average spacing of 1.91 cM.
Concerning the marker segregation pattern, of the 68 unlinked markers, nine RAPDs (OPA 10, OPAB6, ubc841_9, E2M1_9, cs54_2, ubc204_1, E2M1_14, ubc502_1, P8M1_8), seven SSRs (SSR 207, LcSSR70, SSR183, SSR309, TPSSR15, RCS5704, RCS7182) and one ISSR (ISSR2a) deviated significantly from the Mendelian segregation of 1:1 (P < 0.01). Similarly, one SSR (RCS6021a) and three RAPDs (P4M3_4, OPU14a, E3M1_3) deviated significantly at (P < 0.05). The remaining 47 unlinked markers (39 RAPDs, one ISSR and 7 SSRs) segregated in Mendelian fashion. On the other hand, 42 (20 RAPD, 21 SSR and 1 ISSR) distorted markers could be successfully placed onto the linkage map.
QTL mapping
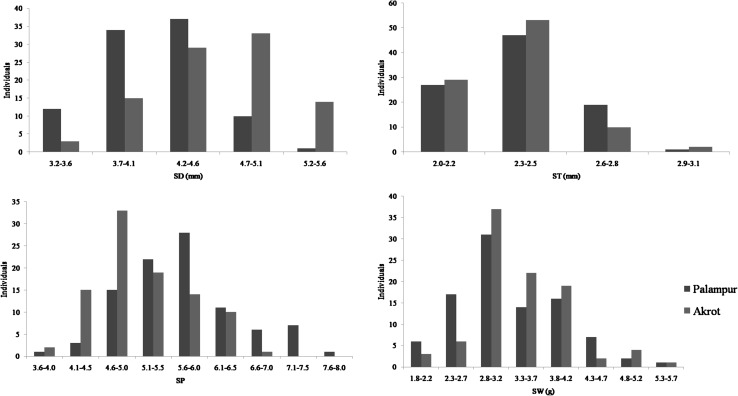
The two parents (WA 8649090 and Precoz) differed significantly with regards to the quantitative traits measured here. Precoz showed higher SD of 5.2 mm compared to 3.7 mm for WA 8649090 (Fig. 3). The RIL had a minimum SD of 3.2 mm and a maximum of 5.6 mm with an average value of 4.40 mm (Table 3). The parents were highly contrasting for DF with Precoz showing average DF value of 88 days in two locations, whereas WA 8649090 was late maturing with average DF value of 172. DF in RIL ranged from 83 to 145.25 with average of 114.12 in two locations. Meager difference was observed for ST (mm) with WA 8649090 and Precoz showing the ST values of 2.3 and 2.2 mm, respectively. In RIL, ST ranged from 2.0 to 3.0 mm with an average of 2.50 mm. Due to variations in both SD and ST, difference in SP score was observed. The genotype WA 8649090 exhibited SP value of 6.2, whereas Precoz with more seed diameter had a plumpness score of 4.2. SP values in RIL varied from 3.6 to 7.8 with an average of 5.7. A significant difference was recorded for 100-SW (g) between the two parents (WA 8649090: 2.6 g and Precoz: 4.63 g), while 100-SW in RIL ranged from 1.82 to 5.50 g with an average of 3.66 g.
Fig. 3.
Frequency distribution of different seed related traits in parents and RIL in two locations
Table 3.
Trait mean and range of parents and RI individuals
| S. no | Traits | P1 | P2 | RILs | ||
|---|---|---|---|---|---|---|
| (WA 8649090) | (Precoz) | Minimum | Maximum | Average | ||
| 1 | DF | 172 | 88 | 83 | 145 | 114.12 |
| 2 | SD (mm) | 3.7 | 5.2 | 3.2 | 5.6 | 4.40 |
| 3 | ST (mm) | 2.3 | 2.2 | 2 | 3.0 | 2.50 |
| 4 | SP | 6.2 | 4.2 | 3.6 | 7.8 | 5.7 |
| 5 | SW (g) | 2.6 | 4.63 | 1.82 | 5.5 | 3.66 |
Analysis of different genetic parameters also showed a significant difference in the RIL for these traits (Table 4). For all traits, phenotypic values greater than that of the higher parent and lower than that of the lower parent were observed. We observed a higher level of heritability for traits viz. DF, SD, ST, SW and SP with percent heritability of 97.56, 98.23, 99.28, 70.27 and 88.31, respectively. Concerning correlations among different traits, significant positive correlations were recorded of ST with DF and SP with that of SW. By contrast, significant but negative correlations were observed for SD with DF, and of SW with ST.
Table 4.
Genetic parameters in RIL for different traits
| Traits | Heritability (%) | GA | % GA | PCV | GCV | GM | CV |
|---|---|---|---|---|---|---|---|
| DF | 97.56 | 42.42 | 33.3 | 16.57 | 16.36 | 127.99 | 2.59 |
| SD (mm) | 98.23 | 113.52 | 120.18 | 59.39 | 58.86 | 94.46 | 7.91 |
| ST (mm) | 99.28 | 337.11 | 149.25 | 72.98 | 72.71 | 225.87 | 6.2 |
| SW (g) | 70.27 | 0.08 | 19.26 | 13.3 | 11.15 | 0.42 | 7.25 |
| SP | 88.31 | 0.09 | 19.92 | 10.95 | 10.29 | 0.46 | 3.74 |
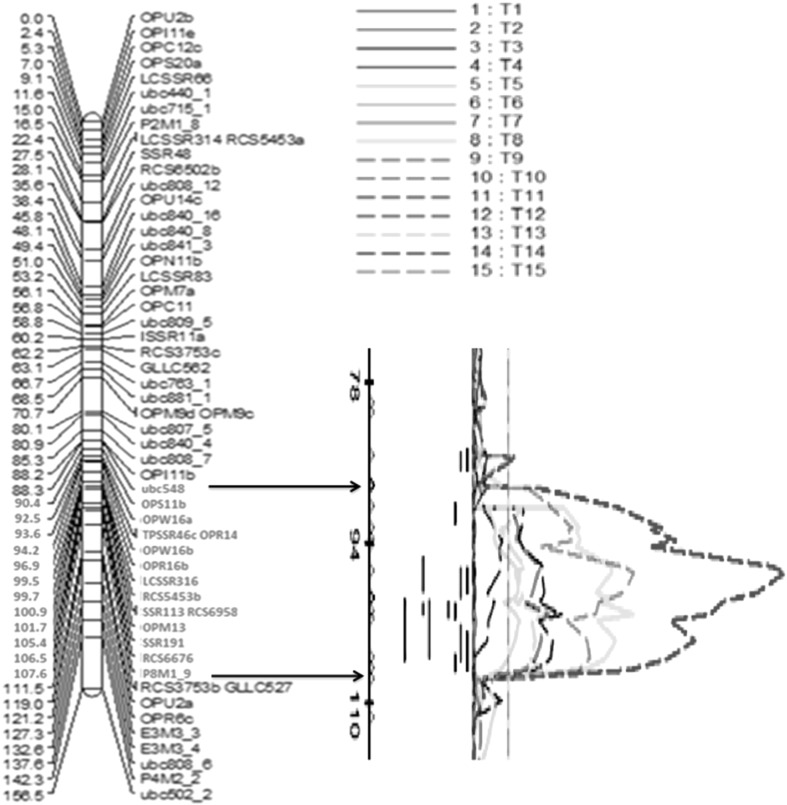
QTL analysis uncovered a total of nine QTL (QTL 2, 3, 4, 5, 6, 8, 9, 11 and 15) for seed size related traits that explained more than 10% of the phenotypic variation (PV) (Fig. 4). Remaining nine QTL with PVs ranging between 2.5 and 9.8% were obtained for traits SD, SW, SP, ST, PHT, and SPP. No significant QTL could be detected for DF, DM and PB. Concerning major QTL, two QTL for SD i.e. QTL 2 in L1 and L2 and QTL 3 (in L2) accounted for 35.3, 14.6 and 12.6% PV, respectively. Similarly, three major QTL for ST i.e. QTL 4 (in L2 with 11.7% PV), QTL 5 (in L1 and L2 with 24.0 and 20.3%, PV respectively) and QTL 6 (in L1 and L2 with 18.6 and 17.3% PV, respectively) were mapped on three different LGs (Table 5). Three major QTL were detected for SP on two LGs viz. on LG5, QTL 8 with 18.3 and 17.0% PV in L1 and L2, respectively and QTL 9 (in L2) with 15% PV, and QTL 11 (L2) explaining 10.2% PV on LG7. Similarly, one major QTL for SW on LG5 (QTL 15) explained 50.0% and with 34.2% PV in L1 and L2, respectively.
Fig. 4.
Genomic region on LG5 harbouring multiple QTL. Five major effect QTL controlling seed related traits were discovered on LG5, of which three QTL were mapped within a genomic region of 90-107 cM. Codes of the traits used for QTL analysis from T1-T15 are as follows: T1 (DF, L1), T2 (DF, L2), T3 (PB, L1), T4 (DM, L1), T5 (PHT, L1), T6 (SPP, L1), T7 (SPP, L2), T8 (SD, L1), T9 (SD, L2), T10 (ST, L1), T11 (ST, L2), T12 (SP, L1), T13 (SP, L2), T14 (SW, L1), T15 (SW, L2)
Table 5.
QTL for seed-related traits in RIL (WA8649090 × Precoz) across two locations viz. Palampur (L1) and Akrot (L2), Himachal Pradesh, India
| Traits | LG | aQTL | Position (cM) | Marker interval | Maximum LOD | Additive effect | %PV |
|---|---|---|---|---|---|---|---|
| SD(mm)(L2) | LG5 | QTL 1 | 9.16 | OPS20a–ubc440_1 | 2.6 | −0.0138 | 7.2 |
| SD(mm)(L1) | LG5 | QTL 2 | 96.2 | OPW16b–LcSSR316 | 12 | −0.031 | 35.3 |
| SD(mm)(L2) | LG5 | QTL 2 | 96.2 | OPW16b–LcSSR316 | 6.5 | −0.019 | 14.6 |
| SD(mm)(L2) | LG5 | QTL 3 | 148 | ubc808_6–ubc502_2 | 2.6 | 0.018 | 12.6 |
| ST(mm)(L2) | LG2 | QTL 4 | 97.71 | LcSSR138–OPAB5b | 3.8 | −0.009 | 11.75 |
| ST(mm)(L1) | LG6 | QTL 5 | 54 | SSR90–OPN6b | 7 | 0.0107 | 24 |
| ST(mm)(L2) | LG6 | QTL 5 | 54 | SSR90–OPN6b | 6 | 0.0094 | 20.3 |
| ST(mm)(L1) | LG8 | QTL 6 | 33.84 | LcSSR176–ubc840_17 | 6.5 | 0.0093 | 18.62 |
| ST(mm)(L2) | LG8 | QTL 6 | 33.84 | LcSSR176–ubc840_17 | 5.6 | 0.0098 | 17.36 |
| ST(mm)(L1) | LG8 | QTL 7 | 41.38 | ubc807_3–E3M1_4 | 2.6 | 0.0053 | 6.2 |
| SP(L1) | LG5 | QTL 8 | 100.9 | RCS5453b–OPM13 | 7 | 0.361 | 18.3 |
| SP(L2) | LG5 | QTL 8 | 100.9 | RCS5453b–OPM13 | 6.5 | 0.0314 | 17 |
| SP(L2) | LG5 | QTL 9 | 150 | ubc808_6–ubc502_2 | 3.5 | −0.029 | 15 |
| SP(L2) | LG5 | QTL 10 | 156.33 | P4M2_2–ubc502_2 | 2.6 | −0.0236 | 9.8 |
| SP(L2) | LG7 | QTL 11 | 39.53 | ubc840_6–OPAB5a | 2.6 | 0.0249 | 10.22 |
| SP(L1) | LG8 | QTL 12 | 33.84 | LcSSR176–ubc840_17 | 3 | 2.38 | 8 |
| SW(g)(L1) | LG4 | QTL 13 | 23.71 | P4M3_1–OPJ17b | 5.5 | −0.1964 | 6.4 |
| SW(g)(L1) | LG5 | QTL 14 | 58.8 | OPC11–ISSR11a | 2.6 | −0.1333 | 2.5 |
| SW(g)(L1) | LG5 | QTL 15 | 98.8 | OPR16b–RCS5453b | 22 | −0.55 | 50 |
| SW(g)(L2) | LG5 | QTL 15 | 98.8 | OPR16b–RCS5453b | 12 | −0.4 | 34.18 |
| SPP(L1) | LG5 | QTL 16 | 63.12 | ISSR11a–GLLC562 | 2.6 | −47.28 | 7.7 |
| SPP(L1) | LG8 | QTL 17 | 26.03 | OPL7–OPB15 | 3.2 | −64.13 | 9.35 |
| PHT(cm)(L1) | LG3 | QTL 18 | 14.87 | OPN5–OPR6a | 2.8 | −2.94 | 7.48 |
aIn each location, QTL with the highest PV is shown in cases where more than one QTL is detected within the same marker interval
The QTL associated with seed related traits were flanked by different markers, viz. QTL 2 (in L1 and L2) for SD was flanked by the DNA markers OPW16b and LcSSR316, while QTL 3 for the same trait (L2) contained within the marker interval ubc808_6-ubc502_2 on LG5. Three QTL for ST, QTL 4 (in L2) flanked by the markers LcSSR138 and OPAB5b, QTL 5 (in L1 and L2) flanked by the markers SSR90 and OPN6b, and QTL 6 (in L1 and L2) flanked by the markers LcSSR176 and ubc840_17 were detected on LG2, LG6 and LG8, respectively. Three QTL for the trait SP were detected on two different LGs viz. LG5 and LG7, of which QTL 8 (in L1 and L2) and QTL 9 (in L2) were located within the marker intervals RCS5453b-OPM13 and ubc808_6-ubc502_2, respectively, whereas QTL 11 (in L2) on LG7 was flanked by the markers ubc840_6 and OPAB5a. For the trait SW, QTL 15 was detected on LG5 in both locations (L1 and L2) within the marker interval OPR16b-RCS5453b.
Of the total five major-effect QTL detected on LG5, three were located with a 17-cM region, which included QTL 2 for SD, QTL 8 for SP and QTL 15 for SW. Importantly all these QTL were detected in both locations L1 and L2 (Fig. 4). On the other hand, QTL 3 for SD and QTL 9 for SP located at 148 cM and 150 cM, respectively were obtained only in one location (L2) (Table 5). In total, nine QTL were mapped on LG5, with the QTL for SW (L1) showing the highest PV of 50%.
Discussion
Biparental QTL mapping is a standard method to discover the genomic regions that are tightly associated with economically significant traits. DNA markers and linkage maps are essential prerequisite for the identification of these candidate genomic segments that harbour the genes/QTL influencing the traits of interest. To date, various linkage maps have been constructed in lentil using inter-specific as well as intra-specific experimental populations (see Bohra et al. 2014). Different kinds of mapping populations like F2 as well as RIL have been employed for the development of genetic linkage maps by exploiting DNA marker systems such as RAPD, AFLP, ISSR (Eujayl et al. 1998; Rubeena and Taylor 2003; Duran et al. 2004; Kahraman et al. 2004, 2010; Tullu et al. 2008), SSR (Hamwieh et al. 2005;Verma et al. 2015) and SNP (Kaur et al. 2013). In the present investigation, we could map 220 loci (45 SSRs, 3 ISSRs and 172 RAPDs) onto eight LGs, varying in lengths from 23.1 cM to 156.5 cM. The map spanned a total of 604.2 cM of the lentil genome with an average marker density of 2.74 cM. The genetic map allowed assignment of 23 new cross-genera SSR markers from T. pratense (reported by Sato et al.2005) out of total 45 SSR markers placed onto the linkage map. We could not compare mapping positions/order of the loci in the current linkage map with that of earlier published maps (Hamwieh et al. 2005; Phan et al. 2007;Saha et al. 2010; Gupta et al.2012). The positive and negative additive effects of a QTL controlling a particular trait imply towards an increase in the phenotypic value of the trait by the alleles contributed respectively by Precoz and WA 8649090.
With regard to marker segregation pattern, several DNA markers used in the current analysis showed deviation from Mendelian inheritance. Sixty-six markers showed segregation distortion of the total 288 DNA markers assayed in the population. Though more pronounced in case of inter-specific crosses (Eujayl et al. 1998), instances of segregation distortion were also observed in cultivated crosses in lentil. For instance, Rubeena and Taylor (2003) observed a segregation distortion of about 14% (17 out of 118 markers) in an intraspecific F2 population of lentil. Similarly, segregation distortions up to varying degrees were reported in lentil mapping populations (Duran et al. 2004; Hamwieh et al. 2005; Phan et al. 2007; Tullu et al. 2008; Saha et al. 2010; Gupta et al. 2012; Verma et al. 2015).
Detection of genetic determinants underlying seed related traits via QTL mapping is a promising way to facilitate targeted trait improvement in lentil. Given the context, we analyzed the phenotypic and genotypic data recorded on a RIL population in order to map important traits such as seed related characters (SD, ST, SW and SP). It becomes imperative to mention here that a set of five major-effect QTL was mapped on LG5 across two locations. Most importantly, the genomic region on LG5 contained multiple QTL associated with different traits. The QTL contained within this genomic region control important traits like SW (QTL 15 in L1 and L2), SD (QTL 2 in L1 and L2) and SP (QTL 8 in L1 and L2). For SW, QTL 15 at L1 explained the highest PV of 50%. Also, the QTL located within this 17 cM region explained considerable PV ranging between 14.60 and 50%. More recently, Verma et al. (2015) reported QTL for SW on LG4. Occurrence of such genomic regions that contain a range of QTL controlling multiple traits has been reported in case of other grain legume crops. For example, Varshney et al. (2014) reported a hot spot QTL in chickpea through analyzing data on two RIL populations, and notably, the QTL detected within this region explained PV up to 58%.
Likewise, Tahir and Muehlbauer (1995) also obtained three QTL for seed weight on LG1, 4 and 5. As reported in chickpea by Varshney et al. (2013), 100-seed weight QTL was found to be consistent across environments out of the QTL for 12 traits contained within the QTL hotspot region. Knowledge of genomic segments that harbour QTL for multiple traits offers exciting opportunities for genomics assisted crop improvement. The recent example includes the introgression of QTL hotspot into an elite yet drought susceptible chickpea cultivar (Varshney et al. 2013), thereby demonstrating the relevance of such genomic regions in crop breeding.
In summary, the current study offers valuable supplements to genomic toolkit of lentil breeder. A 220-loci genetic linkage map with 604.2 cM length was built. Most importantly, we could identify a set of large-effect QTL within a 17-cM genomic segment on LG5. This genomic region, after confirmation in independent experiments, could serve as a promising candidate for future lentil genomics and breeding.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
RJ and TRS acknowledge financial support from the department of biotechnology (DBT), India. Special thanks are due to Dr. Fred Muehlbauer (Grain Legume Genetics & Physiology Research Unit, USDA-ARS, Washington State University, Pullman, USA) for providing experimental population.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Rintu Jha and Abhishek Bohra have contributed equally to this work.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-017-0438-7) contains supplementary material, which is available to authorized users.
References
- Abbo S, Ladizinsky G, Weeden NF. Genetic-analysis and linkage study of seed weight in lentil. Euphytica. 1991;58:259–266. doi: 10.1007/BF00025258. [DOI] [Google Scholar]
- Akibode CS, Maredia M (2011) Global and regional trends in production, trade and consumption of food legume crops. Report submitted to the Standing Panel on Impact Assessment (SPIA) of the CGIAR Science Council, FAO, Rome
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–218. doi: 10.1007/BF02672069. [DOI] [Google Scholar]
- Bohra A. Emerging paradigms in genomics-based crop improvement. Sci World J. 2013;585467:17. doi: 10.1155/2013/585467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohra A, Pandey MK, Jha UC, Singh B, Singh IP, Datta D, Chaturvedi SK, Nadarajan N, Varshney RK. Genomics assisted breeding in four major pulse crops of developing countries: present status and prospects. Theor Appl Genet. 2014;127:1263–1291. doi: 10.1007/s00122-014-2301-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero JI, Pérez de la Vega M, Fratini R. Origin, phylogeny, domestication and spread. In: Erskine W, Muehlbauer FJ, Sarker A, Sharma B, editors. The lentil: botany, production and use. Wallingford: CABI press; 2009. pp. 13–33. [Google Scholar]
- Duran Y, Fratini R, Garcia P, Perez de la Vega M. An inter subspecific genetic map of Lens. Theor Appl Genet. 2004;108:1265–1273. doi: 10.1007/s00122-003-1542-3. [DOI] [PubMed] [Google Scholar]
- Eujayl I, Baum M, Powell W, Erskine W, Pehu E. A genetic linkage map of Lentil (Lens sp.) based on RAPD and AFLP markers using recombinant inbred lines. Theor Appl Genet. 1998;97:83–89. doi: 10.1007/s001220050869. [DOI] [Google Scholar]
- FAOSTAT (2014) FAOSTAT database. http://faostat.fao.org/
- Fedoruk MJ, Vandenberg A, Bett KE. Quantitative trait loci analysis of seed quality characteristics in lentil using single nucleotide polymorphism markers. Plant Genome. 2013;6:37–39. doi: 10.3835/plantgenome2013.05.0012. [DOI] [PubMed] [Google Scholar]
- Ford R, Pang ECK, Taylor PWJ. Genetics of resistance to ascochyta blight (A. lentis) of lentil and the identification of closely linked RAPD markers. Theor Appl Genet. 1999;98:93–98. doi: 10.1007/s001220051044. [DOI] [Google Scholar]
- Fratini R, Duran Y, Garcia P, Perez de la Vega M. Identification of quantitative trait loci (QTL) for plant structure, growth habit and yield in lentil. Span J Agric Res. 2007;5:348–356. doi: 10.5424/sjar/2007053-255. [DOI] [Google Scholar]
- Gupta PK, Varshney RK. The development and use of microsatellite markers for genetics and plant breeding with emphasis on bread wheat. Euphytica. 2000;113:163–185. doi: 10.1023/A:1003910819967. [DOI] [Google Scholar]
- Gupta M, Verma B, Kumar N, Chahota RK, Rathour R, Sharma SK, Bhatia S, Sharma TR. Construction of intersubspecific molecular genetic map of lentil based on ISSR, RAPD and SSR markers. J Genet. 2012;91:279–287. doi: 10.1007/s12041-012-0180-4. [DOI] [PubMed] [Google Scholar]
- Hamwieh A, Udupa SM, Choumane W, Sarker A, Dreyer F, Jung C, Baum M. A genetic linkage map of Lens sp. based on microsatellite and AFLP markers and the localization of Fusarium vascular wilt resistance. Theor Appl Genet. 2005;110:669–677. doi: 10.1007/s00122-004-1892-5. [DOI] [PubMed] [Google Scholar]
- Hossain S, Ford R, McNeil D, Pittock C, Panozzo JF. Inheritance of seed size in chickpea (Cicer arietinum L.) and identification of QTL based on 100-seed weight and seed size index. Aust J Crop Sci. 2010;4:125–134. [Google Scholar]
- Isemura T, Kaga A, Tabata S, Somta P, Srinives P, Himizu S, Jo P, Vaughan DA, Tomooka N. Construction of a genetic linkage map and genetic analysis of domestication related traits in mung bean (Vigna radiata) PLoS ONE. 2012;7:e41304. doi: 10.1371/journal.pone.0041304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahraman A, Kusmenoglub I, Aydinc N, Aydogan A, Erskine W, Muehlbauer FJ. QTL mapping of winter hardiness genes in lentil. Crop Sci. 2004;44:13–22. doi: 10.2135/cropsci2004.1300. [DOI] [Google Scholar]
- Kaur S, Cogan NO, Stephens A, Noy D, Butsch M, Forster JW, Materne M. EST-SNP discovery and dense genetic mapping in lentil (Lens culinaris Medik.) enable candidate gene selection for boron tolerance. Theor Appl Genet. 2013;127:703–713. doi: 10.1007/s00122-013-2252-0. [DOI] [PubMed] [Google Scholar]
- Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. doi: 10.1111/j.1469-1809.1943.tb02321.x. [DOI] [Google Scholar]
- Phan HT, Ellwood SR, Hane JK, Ford R, Materne M, Oliver RP. Extensive macrosynteny between Medicago truncatula and Lens culinari ssp. culinaris. Theor Appl Genet. 2007;114:549–558. doi: 10.1007/s00122-006-0455-3. [DOI] [PubMed] [Google Scholar]
- Rubeena Ford R, Taylor PWJ. Construction of an intraspecific linkage map of lentil, Lens culinaris ssp. culinaris. Theor Appl Genet. 2003;107:910–916. doi: 10.1007/s00122-003-1326-9. [DOI] [PubMed] [Google Scholar]
- Saha GC, Sarker A, Chen W, Vandemark GJ, Muehlbauer FJ. Identification of markers associated with genes for rust resistance in Lens culinaris Medik. Euphytica. 2010;175:261–265. doi: 10.1007/s10681-010-0187-y. [DOI] [Google Scholar]
- Sato S, Isobe S, Asamizu E, Ohmido N, Kataoka R, Nakamura Y, Kaneko T, Sakurai N, Okumura K, Klimenko I, Sasamoto S, Wada T, Watanabe A, Kohara M, Fujishiro T, Tabata S. Comprehensive structural analysis of the genome of red clover (Trifolium pratense L.) DNA Res. 2005;12:301–364. doi: 10.1093/dnares/dsi018. [DOI] [PubMed] [Google Scholar]
- Sun YN, Pan JB, Shi XL, Du XY, Wu Q, Qi ZM, Jiang HW, Xin DW, Liu CY, Hu GH, Chen QS. Multi-environment mapping and meta-analysis of 100-seed weight in soybean. Mol Biol Rep. 2012;39:9435–9443. doi: 10.1007/s11033-012-1808-4. [DOI] [PubMed] [Google Scholar]
- Tahir M, Muehlbauer FJ. Association of quantitative trait loci with isozyme markers in lentil (Lens culinaris L.) J Genet Breed. 1995;49:145–150. [Google Scholar]
- Tar’an B, Buchwaldt L, Tullu A, Banniza S, Warkentin TD, Vandenberg A. Using molecular markers to pyramid genes for resistance to ascochyta blight and anthracnose in lentil (Lens culinaris Medik) Euphytica. 2003;134:223–230. doi: 10.1023/B:EUPH.0000003913.39616.fd. [DOI] [Google Scholar]
- Tullu A, Tar’an B, Breitkreutz C, Banniza S, Warkentin TD, Vandenberg A, Buchwaldt L. A quantitative-trait locus for resistance to ascochyta blight (Ascochyta lentis) maps close to a gene for resistance to anthracnose (Colletotrichum truncatum) in lentil. Can J Plant Pathol. 2006;28:588–595. doi: 10.1080/07060660609507337. [DOI] [Google Scholar]
- Tullu A, Tar’an B, Warkentin TD, Vandenburg A. Construction of an intraspecific linkage map and QTL analysis for earliness and plant height in lentil. Crop Sci. 2008;48:2254–2264. doi: 10.2135/cropsci2007.11.0628. [DOI] [Google Scholar]
- Upadhyaya HD, Kumar S, Gowda CLL, Singh S. Two major genes for seed size in chickpea (Cicer arietinum L.) Euphytica. 2006;147:311–315. doi: 10.1007/s10681-005-9013-3. [DOI] [Google Scholar]
- Van Ooijen J (2006) JoinMap 4. Software for the calculation of genetic linkage maps in experimental populations, Kyazma BV, Wageningen
- Varshney RK, Gaur PM, Chamarthi SK, Krishnamurthy L, Tripathi S, Kashiwagi J, Singh VK, Thudi M, Jaganathan D. Fast-track introgression of “QTL-hotspot” for root traits and other drought tolerance trait in JG 11, an elite and leading variety of chickpea (Cicer arietinum L.). Plant. Genome. 2013;6:3. [Google Scholar]
- Varshney RK, Thudi M, Nayak SN, Gaur PM, Kashiwagi J, Krishnamurthy L, Jaganathan D, Koppolu J, Bohra A, Tripathi S, Rathore A, Jukanti AK, Jayalakshmi V, Vemula A, Singh SJ, Yasin M, Sheshshaye MS, Viswanatha KP. Genetic dissection of drought tolerance in chickpea (Cicer arietinum L.) Theor Appl Genet. 2014;127:445–462. doi: 10.1007/s00122-013-2230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma P, Goyal R, Chahota RK, Sharma TR, Abdin MZ, Bhatia S. Construction of a genetic linkage map and identification of QTLs for seed weight and seed size traits in lentil, Lens culinaris Medik. PLoS ONE. 2015;10:e0139666. doi: 10.1371/journal.pone.0139666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wang S, Basten CJ, Zeng ZB (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.