Abstract
The Polycomb group proteins are transcriptional repressors that are critically important for the control of stem cell activity and the maintenance of cellular identity during stem cell differentiation into specialized cell types. Polycomb proteins interact with each other to form chromatin-associated repressive complexes of two general types (Polycomb Repressive Complexes 1 and 2, PRC1/2) leading to chromatin compaction and gene silencing. However, the roles of the distinct components of the PRC2 complex in the control of skin development and keratinocyte differentiation remain obscure. In this issue, Dauber et al. demonstrate that conditional ablations of three essential subunits of the Polycomb Repressive Complex 2 (EED, Suz12, or Ezh1/2) in the epidermal progenitor result in very similar skin phenotypes including the premature acquisition of a functional epidermal barrier, formation of ectopic Merkel cells, and defective postnatal hair follicle development. These data demonstrate that in the skin epithelium, EED, Suz12, and Ezh1/2 function largely as subunits of the PRC2 complex, which is important in the context of the data demonstrating the possibility for their independent activities in other cell types or in cancer cells. The data of Dauber et al. provide an important platform for further analyses to fully understand the complexity of repressive action of the Polycomb genes in the control of epidermal development and regeneration, as well as open new avenue for analyses of the role of distinct Polycomb components in the control of alterations of the gene expression programs in keratinocytes in the disorders of epidermal differentiation, such as psoriasis, or in epidermal cancers.
Epigenetic mechanisms play an important role in the control of cellular functions in living organisms and are considered as a driving force of the phenotypic plasticity and evolutionary adaptation (Feinberg, 2013). Epigenetic regulatory machinery operates at several levels including modulation of covalent DNA/histone modifications, as well as through higher-order chromatin remodeling to establish the long-range topological interactions between the genes and their enhancer elements in three-dimensional (3D) nuclear space (Bickmore and van Steensel, 2013).
Epigenetic regulators exhibit both activating and repressive effects on chromatin in keratinocytes: histone demethylase Jmjd3, ATP-dependent chromatin remodeler Brg1 and genome organizer Satb1 promote terminal keratinocyte differentiation, while DNA methyltransferase DNMT1, histone deacetylases HDAC1/2, Polycomp components Bmi1, Ezh1/2 and Cbx4 stimulate proliferation of progenitor cells via repression of the genes encoding cell-cycle inhibitors, as well as inhibit premature activation of terminal differentiation-associated genes (reviewed in Botchkarev et al. 2012, Eckert, 2011; Frye and Benitah, 2012; Perdigoto et al., 2014; Adam and Fuchs, 2016).
The Polycomb group proteins are transcriptional repressors that play a critically important role in the control of stem cell activity and the maintenance of cellular identity during stem cell differentiation into specialized cell types (Schwartz and Pirrotta, 2014; Simon and Kingston, 2009; Zhang et al., 2015). By repressing their target genes, Polycomb proteins are involved in the regulation of multiple cellular processes in development, pluripotency, cancer and senescence. Polycomb proteins interact with each other to form chromatin-associated repressive complexes of two general types (Polycomb Repressive Complex 1 and 2, PRC1 and PRC2) leading to chromatin compaction and gene silencing. The canonical PRC1 complex contains Ring1A or Ring1B, Pcgf2 or Pcgf4, and one of the Cbx and Phc proteins, while PRC2 consist of H3K27 methyltrasferases Ezh1 or Ezh2, Suz12, one of the Eed variant, and might also contain additional subunits (Bantignies and Cavalli, 2011; Simon and Kingston, 2009; Zhang et al., 2015) (Fig. 1). The existence of multiple variants of core PRC subunits and incorporation of various additional proteins into the complexes provides numerous variants, which activity remains to be, as yet, poorly understood.
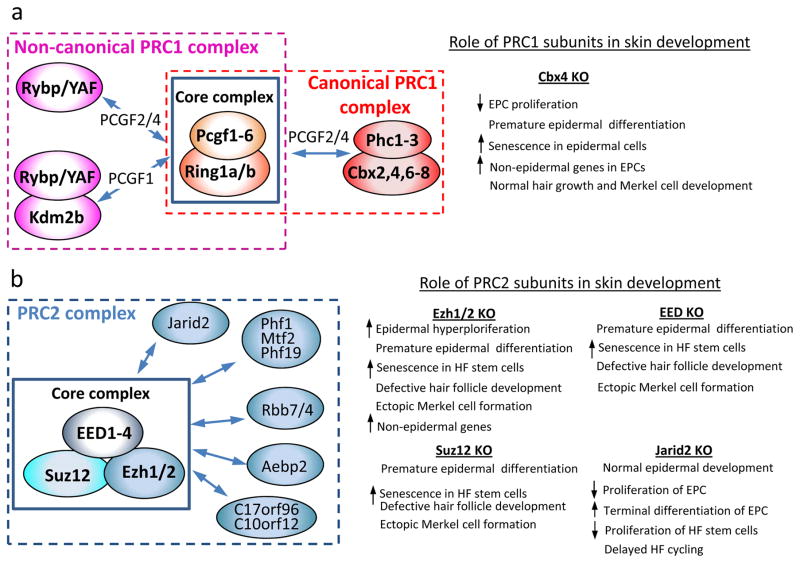
Figure 1. Polycomb group complexes in mammalian skin.
PcG proteins form two major complexes, PRC1 (a) and PRC2 (b). Both complexes contain core subunits that interact with other accessory proteins and define the complete composition of each subcomplex. The accessory proteins regulate recruitment to specific chromatin domains and/or modulate the catalytic activity of the core complex. (a) Association of PRC1 core subunits Ring1a/b and Pcgf2/4 with one of Phc and Cbx proteins defines the canonical PRC1 complex, while a complex with Rybp/YAF and/or Kdm2b forms the non-canonical PRC1. (b) Three core subunits of PRC2 can associate with different proteins at the same time.
Recent data reveal that binding of the non-canonical PRC1 complex containing histone demethylase KDM2B, PCGF1 and RING/YY1 binding protein (RYBP) promotes basal ubiquitylation of the H2A at lysine 119 (H2AK119) at unmethylated CpG-rich DNA regions, which is sufficient to recruit the PRC2 complex (reviewed in Schwartz and Pirotta, 2014; Zhang et al., 2015).. In turn, the PRC2 component Ezh1/Ezh2 histone methyltrasferases promotes tri-methylation of the Lysine 27 at histone H3 (H3K27) followed by targeting the Cbx proteins as a part of the canonical PRC1 complex to H3K27me3, which result in further increase of the H2AK119 ubiquitylation catalyzed by the PRC1 component Ring1B (reviewed in Schwartz and Pirotta, 2014; Zhang et al., 2015). Polycomb-dependent repressive mechanisms play critical roles in the control of skin development and keratinocyte differentiation. In the epidermis, the Polycomp components Bmi1, Ezh1/2 and Jarid2 stimulate proliferation of the progenitor cells via repression of the genes encoding cell-cycle inhibitors including the INK4A-INK4B locus, as well as inhibit premature activation of terminal differentiation-associated genes (Ezhkova et al., 2009; Luis et al., 2011). Also, Ezh1/2 restricts differentiation of the epidermal progenitor cells (EPCs) by repressing the Sox2 gene, which, in turn, promotes Merkel cell-specific differentiation (Bardot et al. 2013). In normal human skin, the PRC1 component Cbx4 protein protects epithelial stem cells from senescence through PRC-dependent repression of the Ink4a locus, as well as controlling their differentiation through PRC-independent mechanisms (Luis et al., 2011). In addition, Cbx4 plays a crucial role in maintaining the epithelial identity and proliferative activity in keratinocytes via repression of the selected non-epidermal lineage and cell cycle-inhibitor genes (Mardaryev et al., 2016).
Despite the fact that the roles of the histone methyltransferases Ezh1/2 in the control of skin and hair follicle development have been identified previously (Ezhkova et al., 2009, 2011; Bardot et al., 2013), the functions of two other key PRC2 subunits (EED and Suz12) remain obscured. In this issue, Dauber et al. described skin phenotypes of three conditional knockout mouse lines, in which the essential PRC2 subunits EED, Suz12, or Ezh1/2 are conditionally ablated in the embryonic epidermal progenitors that give rise to the epidermis, hair follicles (HFs), and Merkel cells. Dauber et al. showed that all three mouse strains with loss-of-function of EED, Suz12, or Ezh1/2 show very similar skin phenotypes including premature acquisition of a functional epidermal barrier, formation of ectopic Merkel cells, and defective postnatal hair follicle development. These data demonstrate that in the skin epithelium, EED, Suz12, and Ezh1/2 function largely as subunits of the PRC2 complex, which is important in the context of the data demonstrating the possibility for their independent activities in other cell types or in cancer cells (reviewed in Conway et al., 2015).
One of the intriguing findings uncovered by Dauber at el. is a differential response of epidermal and hair follicle progenitor cells to PRC2 loss in postnatal skin. Similar to Ezh1/2 deletion (Ezhkova et al., 2011), loss of neither EED nor Suz12 affects proliferation of epidermal progenitor cells, while hair follicle progenitor cells show a decline in cell proliferation and an increased apoptosis linked to up-regulation of senescence-associated INK4a/b locus genes. Such striking contrast in cell behaviour of these two epithelial progenitor cell populations suggests that there is a mechanism that minimizes or compensates the loss of PRC2 function in the epidermal progenitor cells. The ability of epidermal progenitor cells to withstand the PRC2 loss is likely rooted to unique features of their microenvironment, rather than to their intrinsic properties, as the Ezh1/2-depleted epidermal progenitor cells behave similar to hair follicle progenitor cells if taken out of their niche and cultured in vitro (Ezhkova et al., 2011). Dissecting the role of niche in the resistance to PRC2 loss in epidermal progenitor cells will undoubtedly bring new insights into functional differences in the behaviour between these two stem/progenitor cells compartments in skin homeostatic and injury-induced regeneration, as well as pathological conditions with uncontrolled epithelial cell proliferation and differentiation.
Thus, the data of Dauber et al. provide an important platform for further analyses to fully understand the complexity of repressive action of the Polycomb genes in the control of epidermal development, regeneration and stem cell activity. In particular, these data open new avenue for analyses of the role of distinct Polycomb components in the control of alterations of the gene expression programs in keratinocytes in the disorders of epidermal differentiation, such as psoriasis, or in epidermal cancers. These studies are important in the context of the recently developed approaches for pharmacological modulation of the Ezh2 activity: indeed, several small molecules inhibiting Ezh2 activities have been developed, and some of them are under clinical trials for distinct neoplastic conditions (Simo-Riudalbas and Esteller, 2015). Thus, understanding of Polycomb-dependent mechanisms controlling epidermal differentiation and skin stem cell activity and their alterations in different pathological skin conditions will help to progress towards the development of novel approaches for the treatment of skin disorders by targeting distinct Polycomb proteins.
References
- Adam RC, Fuchs E. The Yin and Yang of chromatin dynamics in stem cell fate selection. Trends Genet. 2016;32:89e100. doi: 10.1016/j.tig.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantignies F, Cavalli G. Polycomb group proteins: repression in3D. Trends Genet. 2011;27:454e64. doi: 10.1016/j.tig.2011.06.008. [DOI] [PubMed] [Google Scholar]
- Bardot ES, Valdes VJ, Zhang J, Perdigoto CN, Nicolis S, Hearn SA, et al. Polycomb subunits Ezh1 and Ezh2 regulate the Merkel cell differentiation program in skin stem cells. EMBO J. 2013;32:1990e2000. doi: 10.1038/emboj.2013.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickmore WA, van Steensel B. Genome architecture: domain organization of interphase chromosomes. Cell. 2013;152:1270e84. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- Botchkarev VA, Gdula MR, Mardaryev AN, Sharov AA, Fessing MY. Epigenetic regulation of gene expression in keratinocytes. J Invest Dermatol. 2012;132:2505e21. doi: 10.1038/jid.2012.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol. 2015;37:42e8. doi: 10.1016/j.ceb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Dauber KL, Perdigoto CN, Valdes VJ, Santoriello FJ, Cohen I, Ezhkova E. Dissecting the roles of polycomb repressive complex 2 subunits in the control of skin development. J Invest Dermatol. 2016;136:1647e55. doi: 10.1016/j.jid.2016.02.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert RL, Adhikary G, Rorke EA, Chew YC, Balasubramanian S. Polycomb group proteins are key regulators of keratinocyte function. J Invest Dermatol. 2011;131:295e301. doi: 10.1038/jid.2010.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Lien WH, Stokes N, Pasolli HA, Silva JM, Fuchs E. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485e98. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, et al. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122e35. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP. The epigenetic basis of common human disease. Trans Am Clin Climatol Assoc. 2013;124:84e93. [PMC free article] [PubMed] [Google Scholar]
- Frye M, Benitah SA. Chromatin regulators in mammalian epidermis. Semin Cell Dev Biol. 2012;23:897e905. doi: 10.1016/j.semcdb.2012.08.009. [DOI] [PubMed] [Google Scholar]
- Luis NM, Morey L, Mejetta S, Pascual G, Janich P, Kuebler B, et al. Regulation of human epidermal stem cell proliferation and senescence requires polycomb-dependent and -independent functions of Cbx4. Cell Stem Cell. 2011;9:233e46. doi: 10.1016/j.stem.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Mardaryev AN, Liu B, Rapisarda V, Poterlowicz K, Malashchuk I, Rudolf J, et al. Cbx4 maintains the epithelial lineage identity and cell proliferation in the developing stratified epithelium. J Cell Biol. 2016;212:77e89. doi: 10.1083/jcb.201506065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdigoto CN, Valdes VJ, Bardot ES, Ezhkova E. Epigenetic regulation of epidermal differentiation. Cold Spring Harb Perspect Med. 2014;4:281e300. doi: 10.1101/cshperspect.a015263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Ruled by ubiquitylation: a new order for polycomb recruitment. Cell Rep. 2014;8:321e5. doi: 10.1016/j.celrep.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Simo-Riudalbas L, Esteller M. Targeting the histone orthography of cancer: drugs for writers, erasers and readers. Br J Pharmacol. 2015;172:2716e32. doi: 10.1111/bph.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697e708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Zhang T, Cooper S, Brockdorff N. The interplay of histone modifications—writers that read. EMBO Rep. 2015;16:1467e81. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]