Abstract
Tepsin is currently the only accessory trafficking protein identified in adaptor-related protein 4 (AP4) coated vesicles originating at the trans-Golgi network (TGN). The molecular basis for interactions between AP4 subunits and motifs in the tepsin C-terminus have been characterized, but the biological role of tepsin remains unknown. We determined X-ray crystal structures of the tepsin ENTH and VHS/ENTH-like domains. Our data reveal unexpected structural features that suggest key functional differences between these and similar domains in other trafficking proteins. The tepsin ENTH domain lacks helix0, helix8, and a lipid binding pocket found in epsin1/2/3. These results explain why tepsin requires AP4 for its membrane recruitment and further suggest ENTH domains cannot be defined solely as lipid binding modules. The VHS domain lacks helix8 and thus contains fewer helices than other VHS domains. Structural data explain biochemical and biophysical evidence that tepsin VHS does not mediate known VHS functions, including recognition of dileucine-based cargo motifs or ubiquitin. Structural comparisons indicate the domains are very similar to each other, and phylogenetic analysis reveals their evolutionary pattern within the domain superfamily. Phylogenetics and comparative genomics further show tepsin within a monophyletic clade that diverged away from epsins early in evolutionary history (~1,500 million years ago). Together, these data provide the first detailed molecular view of tepsin and suggest tepsin structure and function diverged away from other epsins. More broadly, these data highlight the challenges inherent in classifying and understanding protein function based only on sequence and structure.
Keywords: membrane trafficking, vesicle coats, protein structure, evolution
Graphical Abstract
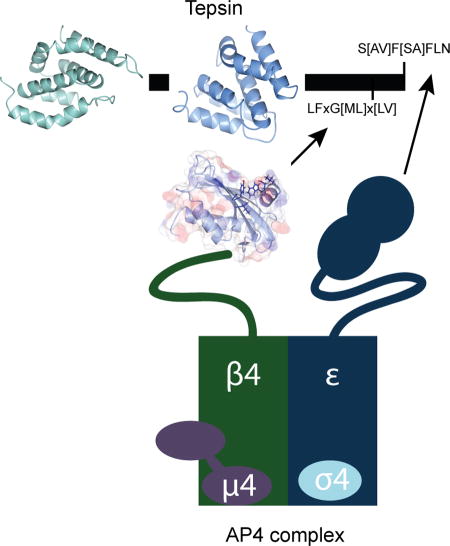
Introduction
Large multi-subunit coat protein complexes drive vesicle or tubule formation at specific organelle membranes. The mammalian AP (Assembly Polypeptide) adaptor protein complex family (APs 1–5, COPI F-subcomplex) is a family of heterotetrameric complexes implicated in Golgi-ER and post-Golgi trafficking pathways. Coat components are recruited by lipids1 and membrane-associated small G-proteins2,3; they recognize and incorporate transmembrane protein cargo4–8 into forming vesicles or tubules; and they recruit accessory proteins and additional machinery9 required for assembly and maturation10. The molecular structures, assembly mechanisms, and cellular functions of non-clathrin associated AP complexes remain poorly understood. The AP4 complex (ε/β4/µ4/σ4 subunits) is recruited to the trans-Golgi network (TGN) by the small GTPase, Arf1, in its GTP-bound state11. AP4 has been implicated in polarized cargo sorting in epithelial cells12 and neurons13. While AP4 is ubiquitously expressed, the most striking phenotypes are associated with the brain and central nervous system. A β4 knockout mouse exhibits mis-sorting of glutamate receptors from the somatodendritic region to axons 13 but shows no significant anatomical abnormalities. In contrast, human patients with mutations in any of the four AP4 subunits14–17 suffer from the hereditary spastic paraplegias (14, reviewed in18). AP4 thus plays a key role in neurological development and function, but the underlying mechanism for this role remains unclear. Identifying and understanding the protein components and molecular mechanisms required to form AP4 coats remain paramount to uncover the function of AP4 both in the cell and in human brain disease.
Tepsin (Figure 1A) was the first accessory protein identified in AP4 coated vesicles19; it is a member of the epsin family of post-Golgi trafficking proteins. In mammals, epsins serve as key accessory proteins and cargo adaptors in clathrin coated vesicle (CCV) formation at the plasma membrane and TGN. Members of the epsin family are defined by the presence of an Epsin N-Terminal Homology (ENTH) domain followed by a mostly unstructured C-terminus. The ENTH domain has been identified and characterized in both mammals20 and yeast21. Structural, biochemical, and cell biological data indicate that ENTH domains interact directly with phosphoinositide head groups22 and with SNARE proteins23–25. The unstructured C-termini of many epsins contain ubiquitin interacting motifs (UIMs)26 and short linear 27,28 or secondary structural motifs29 for binding AP1/AP2 or clathrin. Epsins thus interact directly with membranes in multiple ways: they directly bind phosphoinositides and cargo, and indirectly bind other trafficking machinery. In contrast, tepsin lacks motifs for binding ubiquitin, clathrin, AP1, or AP2. Tepsin instead contains two short linear motifs in its C-terminus that directly and specifically bind the AP4 ε and β4 appendage domains30,31. Tepsin is the only epsin known to depend upon its AP complex for membrane recruitment19 and is unique among family members in possessing a second internal folded module, the VHS/ENTH-like domain19.
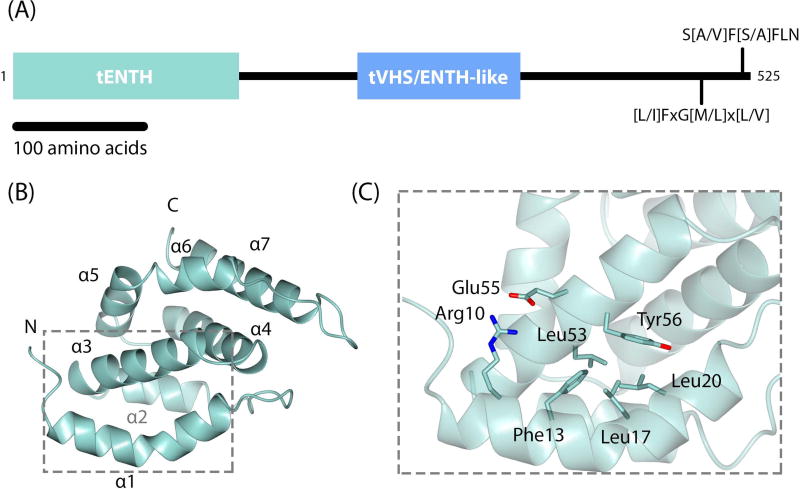
Figure 1. Tepsin architecture and crystal structure of tepsin ENTH domain.
(A) Overall domain architecture of human tepsin, which contains an N-terminal ENTH (tENTH) and internal VHS/ENTH-like (tVHS) domains. The unstructured C-terminus contains two motifs for binding C-terminal appendage domains of AP4 ε and β4 subunits. (B) Crystal structure of human tENTH residues 1–136 at 1.38 Å resolution. tENTH contains only seven α-helices; it lacks both helix0 and helix8 found in other ENTH domains. (C) View of key interactions that facilitate packing between helices α1 and α3 in tENTH, including an ion pair (Arg10 and Glu55) and multiple hydrophobic interactions.
We determined high resolution X-ray crystal structures of the human tepsin ENTH (tENTH) and the horse tepsin VHS/ENTH-like (tVHS) domains. For clarity, we have shortened the name of the VHS/ENTH-like domain to tVHS. Both domain structures reveal important differences from other members of the ENTH/ANTH/VHS superfamily, because they lack key helices at both the N- and C-termini. tENTH lacks both helix0 and helix8. Loss of helix0 precludes formation of a lipid binding pocket and explains why tENTH cannot interact directly with phosphoinositide head groups, which we confirm biochemically. Our structural data suggest the tENTH domain could exist as a dimer or tetramer, but we have not found evidence of oligomer formation in solution. The tVHS structure lacks the C-terminal helix8 that differentiates VHS from ENTH domains within the ENTH/ANTH/VHS superfamily. Important functions of other VHS domains include cargo motif or ubiquitin recognition and binding, but the structure explains our experimental observations that tVHS cannot perform any of these known functions. One striking observation from our data is that the tepsin ENTH and VHS domains are structurally similar. Our phylogenetic analyses indicate that tepsin forms a monophyletic clade within the epsin family but do not support the idea that tepsin ENTH and VHS domains arose from a gene duplication event. Our structural data provide the first detailed molecular view of tepsin, and evolutionary data suggest that ENTH and VHS domains, as well as tepsin itself, have evolved to serve different functions over the course of evolution.
Results
Structure of the tepsin ENTH domain
Other ENTH domains, including those in epsin1 and epsinR, can be expressed and purified in the absence of the first 10–12 residues that constitute helix022,25. Initial attempts to purify the tepsin ENTH domain with and without its putative helix0 failed. However, a construct containing the full predicted ENTH domain (residues 1–136) was successfully purified, indicating the N-terminus was absolutely required to obtain soluble protein (data not shown). We determined a high resolution structure (beyond 1.4 Å) of human tENTH residues 1–136 using molecular replacement methods by placing individual α-helices into the density (further details in Methods). The crystal form contained two molecules in the asymmetric unit; both copies show clear and well-ordered density from residues 3–133 in chain A and residues 4–133 in chain B. There is no significant difference between the two copies; they overlay with an r.m.s.d. of 0.524 Å. Following several rounds of refinement, we obtained a model with excellent geometry (0% Ramachandran and rotamer outliers, 98% Ramachandran favored; Table S1).
The tENTH domain is all α-helical and contains seven helices connected by loops of varying length (Figure 1B; Table S1). The first α-helix, which we have named helix α1 (discussed further below), extends from residues 7–24 and contains a ‘kink’ in the middle resulting from the presence of a proline at residue 18. From the structure, we can explain our biochemical result that deleting the first helix results in completely insoluble protein. Helix α1 packs against and makes important contacts with specific residues in helix α3; residues Phe13, Leu17, and Leu20 in helix α1 make hydrophobic interactions with Leu53 and Tyr56 in helix α3, while Arg10 in helix1 forms a salt bridge with Glu55 in helix3 (distance=2.9 Å) (Figure 1C).
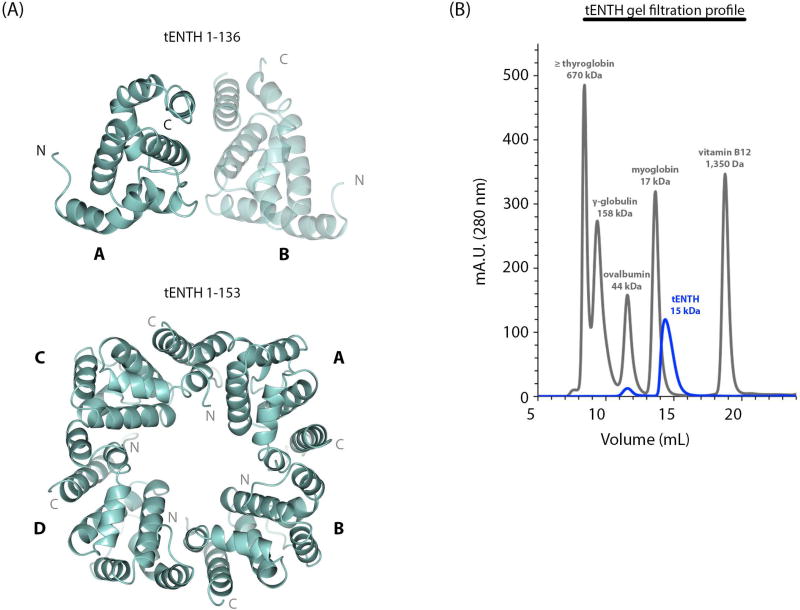
ENTH domains normally contain nine α-helices (discussed further below), but both secondary structural prediction servers (Network Protein Sequence Analysis32) and our crystal structure indicate tENTH contains only seven helices. To test this experimentally, we also crystallized and determined the structure of a second construct containing tepsin residues 1–153 (Figure S2). The electron density indicated no additional α-helical or other secondary structure located beyond helix α7 (Figure S2). Together these data suggest that additional amino acids beyond residue 136 in the longer construct are disordered and the tepsin ENTH domain is indeed smaller than other published ENTH structures by two α-helices.
The tENTH 1–136 construct crystallized as a dimer with two molecules in the asymmetric unit (Figure 2A). In this crystallographic dimer, residues in the loop between helices α1 and α2, together with helices α4 and α7, interact to form the interface between chains A and B; this dimer buries 725 Å2 of solvent accessible surface area as calculated by PISA33. PISA did not identify this dimer as a potential biological interface but instead identified a tetramer with four-fold circular symmetry (4,637 Å2 buried surface area). The longer tENTH construct (residues 1–153) contains one copy in the asymmetric unit, and four monomers pack to form a tetramer with four-fold circular symmetry in the crystal lattice (Figure 2B; 4,765 Å2 buried solvent accessible surface area). The tetramer observed in the tENTH 1–153 lattice is the same tetramer predicted by PISA based on the tENTH 1–136 structure.
Figure 2. tENTH is a multimer in crystals but a monomer in solution.
(A) Top panel: The tENTH 1–136 construct crystallized as a dimer (labeled A and B) in the asymmetric unit. Bottom panel: The tetramer observed in the crystal lattice of the tENTH 1–153 structure. (B) Gel filtration profile of the tENTH domain (blue trace) with standards (grey trace), which are consistent with tENTH existing as a monomer in solution.
Based on these data, we tested whether tENTH can form a dimer and/or tetramer in solution. We turned to gel filtration using a Superdex75 analytical column (10/300 GL, GE Healthcare) using protein standards for comparison. Multiple runs indicated the tENTH domain ran as a monomer around its predicted molecular weight of 15 kDa (Figure 2B), close to the myoglobin standard peak (17 kDa). We therefore find no evidence that tENTH exists as an oligomer in solution but cannot rule out the possibility that multimers may form when tepsin is concentrated on membranes in the cell.
Structural and functional comparison of ENTH domains
ENTH domains have been characterized as lipid binding modules34. Biochemical and structural data from multiple ENTH domains indicate helix α0 is an amphipathic helix that is conformationally dynamic and becomes ordered only in the presence of its binding partner. The epsin1 ENTH binds the phosphoinositide Ptd(Ins)4,5P2 22 at the plasma membrane, where the first eighteen residues of the ENTH domain form helix α0 only in the presence of the PIP head group. Helix α0 folds and contributes basic residues to a binding pocket that specifically accommodates the Ptd(Ins)4,5P2 head group. Helix α0 has thus been hypothesized to facilitate or drive membrane curvature by inserting partially into the membrane22. Several ENTH structures have been determined in the absence and presence of phosphoinositide and protein binding partners. Examples include both epsin1/2 and epsinR ENTHs: yeast Ent1 (PDB: 5LOZ), zebrafish epsin1 (PDB: 5LP0), and mammalian epsin1 (1EDU, 1H0A, 1EYH); yeast Ent3 (PDB: 3ONK, 3ONL) and mammalian epsinR (PDB: 2QY7). These ENTHs adopt compact globular structures containing nine α-helices, and helix0 is often removed to improve domain solubility when producing proteins in vitro.
The first notable difference between tENTH and other ENTH domains is its size. Our data indicate tENTH contains only seven helices, which we have numbered helix α1-α7. In contrast to other ENTHs, first helix in tENTH is required for protein folding and solubility. The extensive contacts between tENTH helix α1 and helix α3 preclude conformational flexibility of the first helix in our structure; we predict this helix can never be disordered and thus cannot be defined as an amphipathic helix0. Overlaying tENTH with epsin1 ENTH (Figure 3A) indicates that helices α1–7 overlay nicely (r.m.s.d. 1.56 Å), except that tENTH helix α1 is longer by seven residues. The tENTH domain overlays with a variety of ENTH domains and is structurally most similar to yeast Ent1 (Figure S3A), as measured by r.m.s.d. scoring in CCP4 Superpose35.
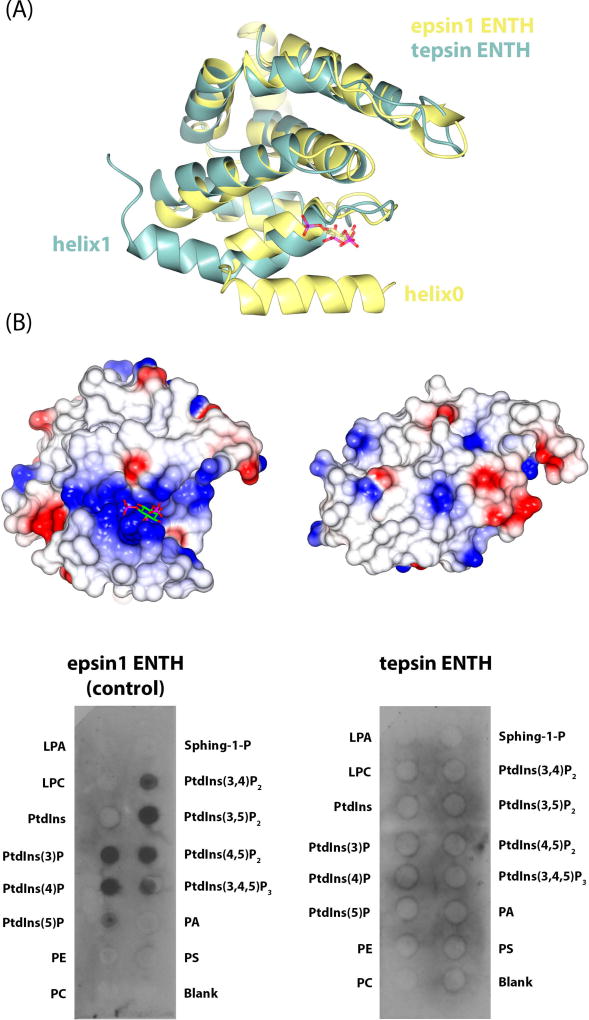
Figure 3. Structural and functional comparison of epsin1 and tepsin ENTH domains.
(A) Overlay of epsin1 ENTH (yellow; PDB: 1H0A) and tENTH (green) domains. Tepsin lacks the amphipathic helix0 found in other ENTHs and instead contains an elongated helix1. (B) Equivalent electrostatic surface views of epsin1 ENTH (left) and tENTH (right). Epsin1 contains a basic binding pocket to accommodate a phosphoinositide head group, while tepsin lacks this binding pocket because of the absence of helix0. Biochemical data using recombinant ENTH domains on PIP strips confirms the structural predictions that epsin1 ENTH recognizes phosphoinositides, while tepsin ENTH does not.
A second consequence of the missing helix α0 is that tENTH lacks a basic binding pocket for binding a phosphoinositide (Figure 3B). The tENTH surface is mostly hydrophobic in character with no major basic (or acidic) pockets or regions, while in contrast, the epsin1 ENTH possesses a highly basic binding pocket. We also considered the possibility that our domain might be similar to the N-terminal portion of an ANTH domain. A sequence alignment using ClustalW36 (data not shown) between the tENTH and CALM ANTH (residues 1–260) revealed that none of the Ptd(Ins)4,5P2 binding residues in CALM (K38, K40, H41) are conserved between the two domains. We also tested experimentally whether the tENTH domain could bind a panel of phosphoinositide head groups (Figure 3B). We incubated recombinant purified tENTH-H6 protein with PIP strips™, using epsin1 ENTH-H6 as a positive control, and blotted against the 6xHis tag. As predicted by our structure, the tepsin ENTH does not bind any phosphoinositide head groups (Figure 3B). Finally, the CALM ANTH domain contains a short helix α0 that does not bind a phospholipid head group directly but has been shown to insert into membranes 9. Structural overlays (data not shown) again indicate tENTH is different: tENTH contains an elongated helix α1, as opposed to the short helix α0 plus α1 in CALM. Although one face of tENTH helix α1 is hydrophobic, nearly all these residues participate in packing interactions with helix α3 (cf. Figure 1) to stabilize the structure of the domain.
Recently, ENTH domains in yeast Ent1 and zebrafish epsin1 have been reported to bind very weakly to ubiquitin (~2mM KD)37. We thus tested whether recombinant tENTH could interact with 15N-labelled mono-ubiquitin in an NMR chemical shift perturbation experiment, because NMR is the most sensitive method for detecting weak interactions. We detect no interaction between tENTH and ubiquitin (Figure S3B).
Structure of the tepsin VHS-like domain
Multiple attempts to crystallize human tepsin VHS-like (tVHS) domain failed. We instead used sequence alignments (Figure S4A) to identify and undertake trials with five other species that contained 75–90% sequence identity to the human domain: cow (Bos taurus), mouse (Mus musculus), horse (Equus caballus), rat (Rattus norvegicus), and marmoset (Callithrix jacchus). We synthesized five constructs (GenScript), then expressed and purified these proteins in E. coli. We obtained crystals from the horse domain (residues 306–437), which diffracted beyond 1.85 Å. However, we could not obtain a solution to the phase problem using molecular replacement in Phaser38 with a variety of VHS structures as input models. Instead, we expressed and purified a horse VHS-like derivative that contained two selenomethione residues at positions 319 and 379; these crystals diffracted beyond 2 Å. We used AutoSol in PHENIX39 for automated experimental SAD phasing and automated model building (full details in Methods). We obtained excellent initial maps and a preliminary model containing 100 residues. We then used this model to obtain a molecular replacement solution for our native crystals, which diffracted to slightly higher resolution. The horse domain is 80% identical to its human counterpart and thus provides an excellent mammalian model.
The tVHS domain (Figure 4A; Figure S4) is an all α-helical protein containing six helices, which we have numbered α1-α5/6 and α7 (discussed in next section). The most notable feature in both the native and SeMet structures is that we do not observe density for the first twelve residues. Secondary structure prediction programs indicated tVHS contains seven α-helices, and all our constructs were designed to include this predicted helix but it is completely disordered in our structures. Instead, bioinformatics analysis using PSIPRED 40 suggests the first helix may constitute a ‘disordered helix’. VHS domains normally contain eight α-helices, so like its ENTH counterpart, the tepsin VHS domain contains fewer α-helices than expected. However, the first α-helix observed in the density clearly corresponds to helix α1 found in other VHS domains (discussed below) based on structural comparisons. We used circular dichroism spectroscopy thermal denaturation experiments (data not shown) to ascertain whether or not the α-helix is folded in solution. We conducted experiments using our crystallization construct (tVHS residues 306–437) and a shorter version (residues 321–437) that lacked the predicted helix. We could find no evidence to support an additional folded helix; if the helix ever folds, it may do so when the domain encounters a binding partner in the cell.
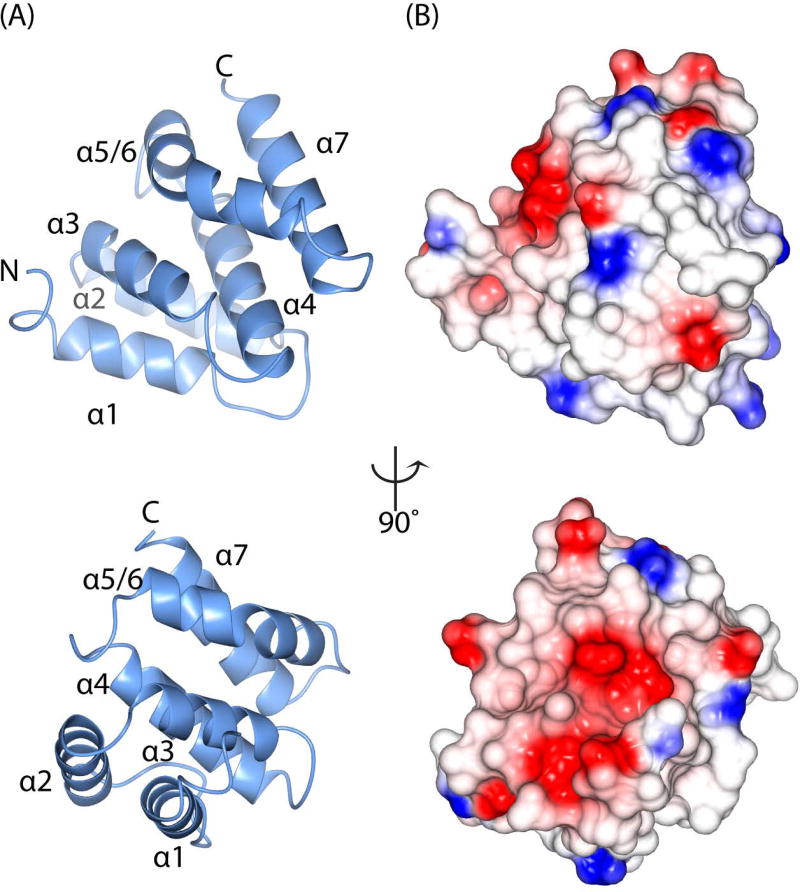
Figure 4. Crystal structure of the tepsin VHS/ENTH-like domain.
(A) Ribbon diagrams of tVHS domain structure determined at 1.8 Å resolution; the views are rotated by 90 degrees. Unlike other VHS domains, tVHS contains only six α-helices. For consistency with other VHS domains, the helices are labelled α1–4, α5/6 (see Results), and α7. (B) Electrostatic surface representations of tVHS, shown in the same orientations as (A). tVHS contains a deep acidic pocket or groove on one surface.
Structural and functional comparison of VHS domains
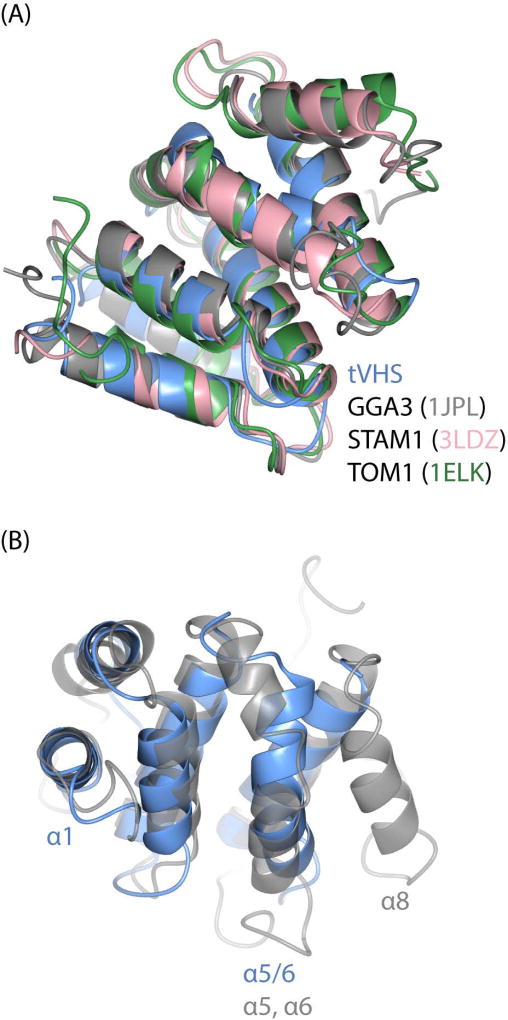
VHS domains are found in other trafficking proteins, including the Golgi-localized, γ adaptin ear homology domain, Arf-binding protein (GGA) adaptor protein; Hrs/STAM subunits of the ESCRT-0 complex; and Tom1. Here we compare published VHS domains to tVHS and discuss functional implications. Overall, the first notable difference in tepsin is the position of the VHS-like domain in the middle of the protein. All other VHS domains are found at the N-terminus of proteins, and published VHS structures contain a right-handed superhelix of eight helices numbered α1-α8. However, our tVHS domain contains only six helices. In general, the tVHS structure overlays well with other VHS domains at helices α1–4 (Figure 5A, Figure S5); note that superpositions indicate the tVHS contains helix α1, so a possible disordered helix would be a new feature in VHS domains. Other VHS domains then have a short helix α5 followed by a flexible loop and helix α6. tVHS instead has one long, ‘kinked’ helix that we have named α5/6, because the first half overlays with helix α5 and the second half with helix α6 in other VHS domains (Figure 5B). tVHS helix α5/6 contains a proline at position 406, which causes the kinked structure. tVHS then overlays well once again at helix α7, and it lacks helix α8 altogether, based on both secondary structural predictions (data not shown) and our structural data.
Figure 5. Structural comparison of VHS domains.
(A) Overlay of VHS domain structures from tepsin, GGA3 (PDB: 1JPL), STAM1 (PDB: 3LDZ), and TOM1 (PDB: 1ELK). (B) Overlay of tepsin and GG3 VHS domains to highlight key structural differences; view is rotated 90 degrees relative to (A). tVHS aligns well with other VHS domains at helices 1–4. tVHS contains a bent α-helix (α5/6), while other VHS domains contain two separate helices (α5, α6). Helix8 is absent in tVHS.
GGA adaptor proteins
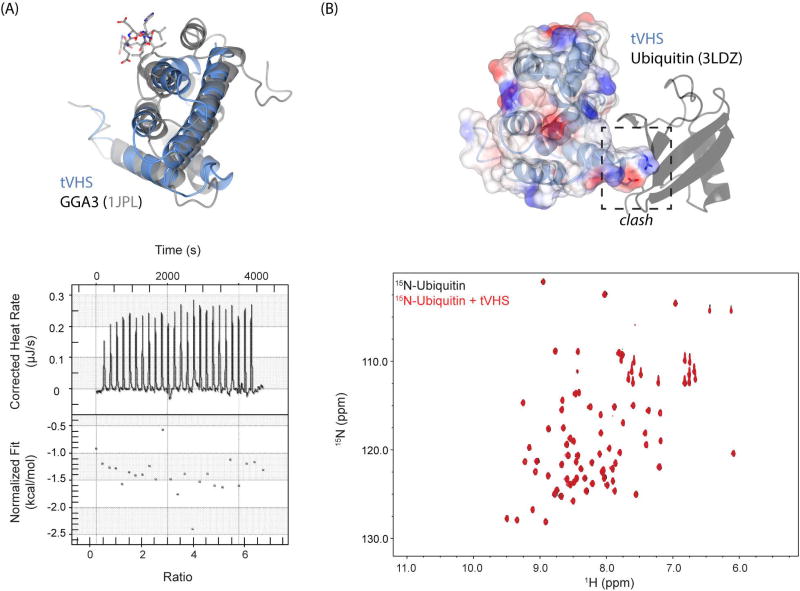
GGAs are monomeric adaptor proteins that localize to the TGN. The VHS domain of GGAs binds short acidic dileucine motifs (DxxL[L/I])41 found in the cytosolic portion of transmembrane protein cargo; one example is recognition of the cation-independent mannose-6-phosphate receptor (CI-MPR) by GGA1 and GGA3. One might predict that cargo recognition and binding is a possible function of the tepsin VHS domain within the AP4 coat. However, our structure explains why tVHS cannot recognize dileucine-based cargoes. In the GGA3 VHS (PDB ID: 1JPL), helices α6 and α8 are required for motif binding: both helices provide side chains that engage the dileucine motif (buried surface area of 542 Å2), especially the conserved Leu residues, in shallow hydrophobic pockets. Because tVHS lacks helix α8 (Figure 5B; 6A), it cannot provide enough surface area to engage a dileucine motif. Indeed, we confirmed this experimentally using a model dileucine cargo peptide in isothermal titration calorimetry experiments with recombinant purified tVHS domain (Figure 6A).
Figure 6. tVHS structure explains functional divergence from other VHS domains.
(A) Top panel: overlay of tepsin (blue) and GGA3 (grey) VHS domains. The acidic dileucine motif recognized by GGA3 is shown in grey stick figures. GGA3 VHS helix8 contributes key surface area to accommodate dileucine motif binding; loss of this helix prevents binding by tVHS. Bottom panel: ITC run with recombinant tVHS protein and 10x molar excess of a model acidic dileucine motif (DSVIL), demonstrating no binding. (B) tVHS ribbon and transparent electrostatic surface modeled by overlaying the STAM1-Ub structure (PDB: 3LDZ). tVHS lacks key residues that form a Ub binding surface, and an arginine present in tVHS would clash with ubiquitin. Bottom panel: HSQC experiment of 15N-labeled ubiquitin (black) spectrum overlaid with a spectrum collected in the presence of a ten times molar excess of tVHS domain. We observe no chemical shifts, suggesting that tVHS cannot bind ubiquitin.
Hrs/STAM
Hrs and STAM constitute two subunits of the ESCRT-0 complex, which targets ubiquitinated proteins to the proteasome for degradation. Hrs and STAM VHS domains bind mono-ubiquitin (Ub) in vitro42, and published surface plasmon resonance experiments42 have suggested all VHS domains may bind mono-ubiquitin with a range of affinities (from low millimolar to high micromolar). We thus considered the possibility that tepsin interacts with Ub. We tested experimentally whether recombinant tVHS could interact with 15N-labelled Ub in an NMR chemical shift perturbation experiment (Figure 6B), because this technique can detect weak (high millimolar) interactions. We observed no binding between tVHS and Ub; in contrast, STAM1 VHS binds Ub with a KD ~220 µM42. Our structure again explains why tVHS cannot engage Ub: an arginine at tVHS residue 344 would directly clash with Ub (Figure 6B).
Structural comparison of tepsin ENTH & VHS-like domains
Once we determined both structures, a surprising feature emerged. Based on r.m.s.d. values, the tepsin ENTH and VHS-like domains appear more similar to each other than to most other domains in either family (Figure S5). In other words, at the structural level, tVHS “looks” more like certain ENTH domains than VHS domains. That tVHS appears similar to ENTHs might be explained by the lack of helix0 in many published ENTH structures. For example, deposited epsinR structures either lack helix0 altogether (PDB: 2QY7) or fail to resolve helix0 in the absence of its phosphoinositide (PDB: 1XGW). We also fail to resolve the first predicted helix in our tVHS construct, and tepsin is unusual in having an internal folded domain at all. Together, these data raise the question of whether the tVHS domain is actually an ENTH domain that arose from a gene duplication event. To address this question, we also tested and verified biochemically that recombinant tVHS does not bind a panel of phosphoinositides (data not shown). Furthermore, our tVHS structure reveals a very acidic patch (Figure 4B) that would strongly repel most anionic phospholipid head groups found in membranes.
In yeast, ENTH and ANTH proteins have been shown to interact with each other in the presence of a phosphoinositide ligand43. We considered the possibility that tENTH and tVHS domains could function using a similar principle, although unlike the yeast proteins, we find neither tENTH nor tVHS domains bind phosphoinositides on their own. We tested for direct binding using pulldown experiments (GST-tENTH with tVHS-H6 and GST-VHS with tENTH-H6). We observed no interactions between these domains at either the Coomassie or Western levels (data not shown).
Phylogenetics and comparative genomics
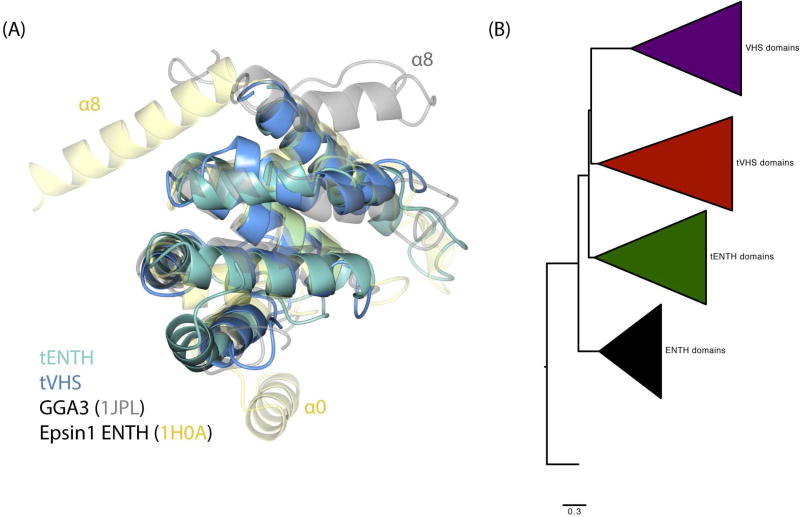
Our structural, biochemical, and biophysical data raised several questions about tepsin evolution. We thus decided to use phylogenetics and comparative genomics first to analyze how tepsin ENTH and VHS domains evolved, and second, to understand the evolution of tepsin within the epsin family. Our structural data strengthened the possibility that the tENTH and tVHS domains arose from a gene duplication event. In addition, the smaller size of both domains might imply they constitute a ‘minimal core’ structure of the ENTH/ANTH/VHS superfamily (Figure 7A), in which helix0 and/or helix8 are absent. We conducted phylogenetic analyses on ENTH and VHS domains from a variety of species representing five eukaryotic supergroups (Figure 7B). We used ANTH domains as an outgroup44: ANTH domains are about twice the size of ENTH or VHS domains but they contain the same core of 6–8 α-helices. Our analysis reveals that ENTH, tENTH, VHS, and tVHS domains each form monophyletic clades as expected, with the VHS and tVHS domains more closely related to one another than to other domains. The tVHS domains in two species (Aplysia californica and Crassostrea gigas) appear to be particularly divergent and do not occur in a consistent location on the phylogeny (Figure S7). The phylogenetic analysis further reveals that VHS domains are generally acquiring mutations at a higher rate than ENTH domains, with non-tepsin VHS domains showing the highest within-group genetic distance (Table S9).
Figure 7. Evolution of ENTH and VHS domains.
(A) Structural overlay of tENTH, tVHS, epsin1 ENTH, and GGA3 VHS domains. All four domains demonstrate a conserved structural core containing helices α1–7. (B) Phylogenetic tree of ENTH, tepsin ENTH (tENTH), VHS, and tepsin VHS-like (tVHS) domains, with ANTH domains as an outgroup.
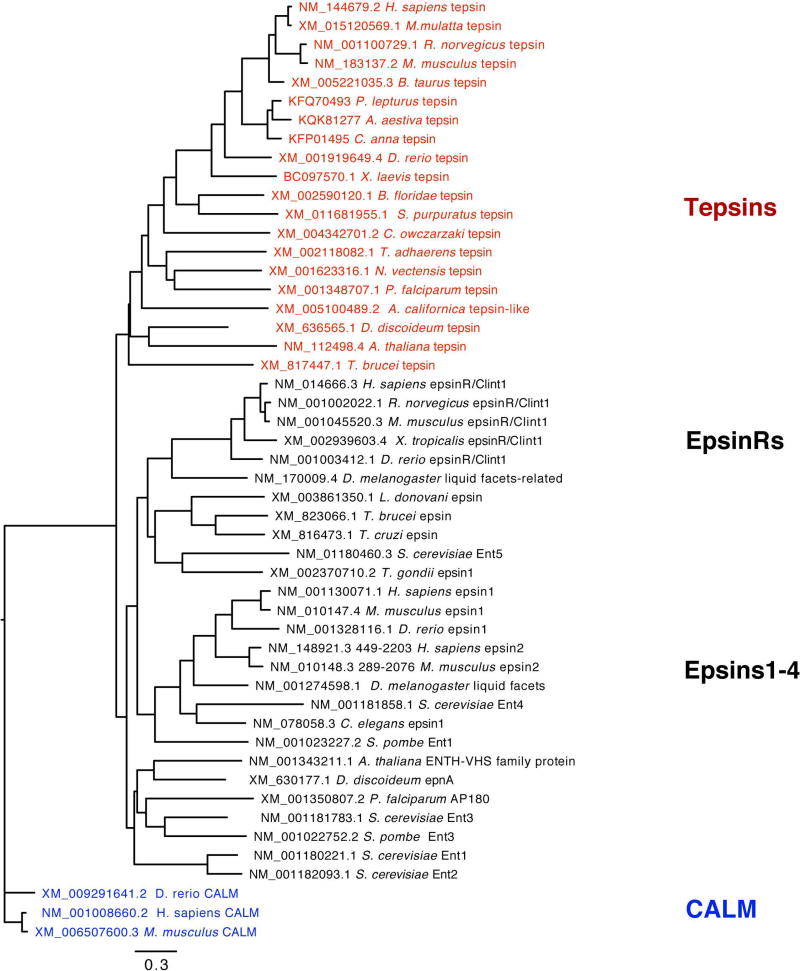
We also conducted a phylogenetic analysis across the epsin family to learn how tepsin fits into the tree. Previously published trees 44,45 did not include tepsin, likely because it had not yet been discovered or annotated in most genomes. We obtained sequences representing species across the five eukaryotic supergroups, again using CALM/PI-CALM (an ANTH-containing protein) as an outgroup44. Our tree (Figure 8, Figure S8) reveals tepsin forms its own monophyletic clade within the family.
Figure 8. Epsin family phylogenetic tree.
Tepsin forms a monophyletic clade that likely diverged early in the evolutionary history of eukaryotes. (CALM sequences, which contain ANTH domains, were used as an outgroup.)
Discussion
Our phylogenetic data do not support the idea of a straightforward gene duplication event giving rise to the unique second folded domain in tepsin; they instead suggest a more complicated duplication event. It is not possible to interpret whether our domain tree supports the idea of tepsin domains constituting a ‘minimal core’ structure. One might argue that a common ancestor contained only seven helices: tepsin domains maintained this structure, while other ENTHs or VHS domains picked up additional helices (helix0 and/or helix8) as they evolved to undertake different functions. Alternatively, the ancestral domain may have contained nine helices in which a single loss of helix0 occurred in tENTHs and a loss of helix8 in both tepsin domains. Both scenarios are equally parsimonious on the current tree. But many other permutations are possible, and it is also possible we have not fully sampled the landscape.
In contrast, our phylogenetic data reveal a much clearer picture of tepsin in the context of the epsin family. This tree indicates tepsin diverged away from other epsins early in the evolutionary history of eukaryotes (~1,500 million years ago), and we predict that tepsin diverged away to support different biological functions. The monophyletic tepsin clade includes sequences from plants, animals, and algae; it is thus parsimonius to suggest a secondary loss of tepsin in yeast and insects as opposed to multiple evolutionary origins of tepsin-like proteins. Our phylogenetic analysis is consistent with our biochemical and biophysical data demonstrating how both tepsin domains fail to undertake known functions of other ENTH or VHS domains. Other ENTH and VHS domains are implicated in phosphoinositide recognition22,46; ubiquitin binding37,42 binding; or dileucine-based cargo motif binding47. We predict both tepsin domains will likely engage a protein partner. One possible binding partner for the ENTH domain is a SNARE protein; epsinR specifically recognizes the SNARE Vti1b23,24. There is currently no evidence that tepsin ENTH engages a SNARE protein19,48. However, we note that extension of tENTH helix α1 compared to other ENTH domains could provide additional surface area for engaging a SNARE or other protein binding partner.
The VHS domain has been proposed to interact directly with AP4, based on co-immunoprecipitation (IP) experiments using a variety of deletion constructs in HeLa cells31. We could find no evidence for an interaction in HeLa cell lysates using a C-terminally GFP-tagged construct containing the ENTH and VHS domains (residues 1–356; Figure S6); a construct containing only the VHS domain (residues 225–356) repeatedly failed to express upon transfection, while our positive control efficiently immunoprecipitated AP4. The micromolar interactions between specific tepsin motifs and the AP4 appendage domains 30,31 are likely most important. If there is a tertiary interaction between tVHS and AP4, it must be very weak (beyond high millimolar) and likely only relevant in an assembled coat when avidities are high.
Our structures raise broader questions about protein classification, specifically regarding the differences between ENTH and VHS domains. ENTH and VHS domains show clear differences when classified at the sequence level, and one anticipates that structural evidence would explain these differences. Previously published structures suggest two concrete structural differences: ENTHs contain helix0, while VHS domains do not; and the position of helix8 differs between ENTH and VHS domains. However, tepsin domains lack all features that normally differentiate ENTHs from VHS domains. Surprisingly, phylogenetic data indicate tepsin VHS sequences cluster with VHS domains, while tENTH and ENTH domains are ancestral to this VHS/tVHS clade. It is difficult for us to rationalize or explain precisely what features generally differentiate ENTH from VHS domains in light of new structural data from tepsin. Others have noted the importance of sampling broadly across evolutionary space to understand gene and protein function. One relevant trafficking example is the role of ENTH/ANTH proteins in trypanosomes (T. brucei). Here, TbCALM and TbEpsinR function with clathrin but independently of AP249. Our data further support the idea that we miss important features and functions by sampling narrowly across eukaryotic lineages. Perhaps we cannot fully classify tepsin domains, or any protein domain, in the absence of functional data.
Materials and methods
Reagents
Unless otherwise noted, all chemicals were purchased from Sigma (St. Louis, MO, USA). The following antibodies were used in this study: rabbit anti-AP4 β, rabbit anti-AP4 ε (for western blotting and immunoprecipitation; both in-house; Hirst et al., 1999), mouse anti-AP4 ε (for immunofluorescence; 612019; BD Transduction Labs), rabbit anti-clathrin (in-house; Simpson et al., 1996), rabbit anti-GFP (gift from Matthew Seaman, Cambridge Institute for Medical Research, UK), and rabbit anti-tepsin (in-house; Borner et al., 2012). Horseradish peroxidase (HRP)–conjugated secondary antibodies were purchased from Sigma-Aldrich, and fluorescently labelled secondary antibodies were from Invitrogen. For Western blotting of immunoprecipitates where the protein band of interest was close to an IgG band, protein-A-HRP (BD Biosciences) was used in the place of HRP-conjugated secondary antibody.
Molecular biology and cloning
All GST-fusion proteins of the tepsin ENTH domain were sub-cloned from full-length tepsin19 into the BamHI/SalI sites of pGEX-6P-1. Because there are no tryptophan residues in the ENTH domain, a single tryptophan was added at the C-terminus to facilitate light absorbance measurements and estimates of protein concentration. The following constructs were made in this way: GST-tENTH (residues 1–136W); GST-tENTH-H6 (residues 1–136W with a C-terminal 6xHis tag; and GST-tENTHlong (residues 1–153W). Horse tepsin VHS-like domain (NCBI reference sequence: XM_001489994.4) was synthesized by Genscript and sub-cloned into BamHI/SalI sites of pGEX-6P-1.
Protein expression and purification
Human tepsin ENTH and horse VHS-like domain constructs were expressed in BL21(DE3)pLysS cells (Invitrogen) for 16–20 hr at 22°C after induction with 0.4 mM IPTG. Native horse tVHS-like domain was expressed in BL21(DE3)pLysS cells (Invitrogen) for 16–20 h at 22°C after induction with 0.4 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG) at OD600=1.0. Selenomethionine labeled (SeMet) tVHS-like domain was expressed in BL21(DE3)pLysS cells (Invitrogen) with incorporation by metabolic inhibition. Briefly, cells were grown in minimal media to OD600=0.3, when amino acid supplements (Lys, Phe, Thr, Leu, Ile, Val, and SeMet) were added to induce metabolic inhibition of methionine synthesis and supply selenomethionine. Cells were induced with 0.4 mM IPTG at OD600=0.8, and expression occurred for 16 h at 22°C.
Tepsin ENTH was purified in 20 mM Tris (pH 8.5), 250 mM NaCl, 2 mM βME. Human tepsin VHS-like was purified in 20 mM Tris (pH 8.7), 200 mM NaCl, 2 mM βME. Native horse tVHS was purified in 20 mM HEPES, pH 7.5, 200 mM NaCl, 2 mM DTT buffer. SeMet horse tVHS was purified in 20 mM HEPES, pH 7.5, 200 mM NaCl, 10 mM DTT buffer. AEBSF protease inhibitor (Calbiochem) was used at all early stages of purification. Cells were lysed by a disruptor (Constant Systems Limited, Daventry, UK), and proteins were affinity purified using glutathione sepharose (GE Healthcare) in relevant purification buffers. GST-fusion proteins were cleaved overnight at 4°C using in-house recombinant GST-3C protease and eluted in batch. All proteins were further purified by gel filtration on a Superdex S200 preparative or analytical column (GE Healthcare).
Crystallization, structure determination, and structural comparisons
tENTH domains
Purified tENTH (residues 1–136W) was concentrated to 3–5 mg/ml and crystallized in 12% (w/v) PEG3350, 4% (v/v) tacsimate pH 5.0. Crystallization trays were set up using 200nL drops on a Mosquito robot (TTP LabTech). Crystals were harvested directly from 96-well plates into 500nL drops in well buffer plus 25% glycerol for cryo-protection. Data were collected at Diamond Light Source, beamline I04-1, on a Pilatus 2M detector and integrated using Xia2 and Mosflm. Crystals diffracted beyond 1.4 Å resolution and were of space group C2 2 21 a=80.8 Å, b=84.8 Å, c= 80.9 Å, α= 90°, β=90°, γ=90°, in Pointless (50). There were two molecules in the asymmetric unit, and no translational NCS was detected in either Scala or Phaser. Because of the high resolution, the structure was determined by molecular replacement in Phaser by individually placing α-helices in the density to identify the seven α-helices in the ENTH domain. Initial building was done in ARP/wARP50. Rounds of refinement and rebuilding were undertaken in phenix.refine51 and Coot52, respectively. The longer tENTH construct comprising residues 1–153W was concentrated to 3 mg/ml and crystallized in 150 mM MES monohydrate pH 6.0, 15% (w/v) PEG6000. Crystallization trays were set up and harvested as described for the tENTH 1–136W construct. Data were collected at Advanced Photon Source (APS) beamline 21-ID-F (0.979 Å) using a Rayonix MX300 CCD detector. Crystals diffracted to 1.8 Å resolution and were of spacegroup P4 21 2 with cell dimensions a= 90.2 Å, b=90.2 Å, c= 42.8 Å, α=90°, β=90°, 90°. The structure was determined using molecular replacement in Phaser with the tENTH 1–136 structure as a model. Rounds of refinement and rebuilding were undertaken in phenix.refine and Coot, respectively. All structure figures were generated using CCP4MG53.
tVHS-like domain
Native tVHS-like domain was concentrated to 5–7 mg/ml and spin-filtered (Millipore). SeMet tVHS-like domain was concentrated to 8.75 mg/ml and spin-filtered (Millipore). A Mosquito robot (TTP Labtech, Cambridge, MA) was used to set the protein in sitting drops. Multiple hits were obtained from the PEG/Ion HT screen (Hampton Research, Aliso Viejo, CA). The plates were stored and imaged at room temperature by Rock Imager (Formulatrix, Bedford, MA). Crystals were cryoprotected using 25% glycerol or perfluoropolyether cryo oil (Hampton Research) by flash freezing in liquid nitrogen. The best native data were collected from crystals grown in 0.2M sodium tartrate dibasic dehydrate, 20% w/v polyethylene glycol 3,350, pH 7.2, and cryoprotected with perfluoropolyether cryo oil. The best SeMet data were collected from crystals grown in the same condition, using 25% glycerol as a cryo-protectant.
Native data were collected at the Advanced Photon Source (APS) beamline 21-ID-G (0.97857 Å) using a MAR300 CCD detector. SeMet data were collected on beamline 21-ID-D (0.97910 Å) using a Dectris Eiger 9M detector. Native crystals diffracted to 1.85 Å and were of space group P61. The unit cell dimensions were a = 58.828 Å, b = 59.929 Å, c = 69.078 Å and α = 90°, β = 90°, γ = 120°. SeMet crystals diffracted to 1.95 Å and were of space group P61. The unit cell dimensions were a = 58.775 Å, b = 58.775 Å, c = 69.522 Å, α = 90°, β = 90°, γ = 120°.
Both native and SeMet data were integrated using HKL2000, then processed further using either the CCP4 or Phenix suites. We used Autosol in Phenix for automated SAD phasing and model building; the initial model contained 109 of 133 residues. Additional rounds of model building was undertaken in Coot with iterative rounds of refinement in phenix.refine. The native structure was determined by molecular replacement using the final SeMet model. Final refinement and validation runs were performed using phenix.refine and MolProbity.
Despite extensive manual inspection of the density maps, twelve N-terminal residues (306–317 in the horse sequence) could not be placed in the electron density. Although secondary structural prediction programs suggested these residues formed a helix, we observe no visible density that would allow us to confidently place these N-terminal residues. We suspect this disordered region contributes to our R-factors being higher than expected at this resolution.
Structural comparisons
Superpose (CCP4) was used to compare structures of ENTH and VHS domains deposited in the PDB. The SSM algorithm was used to align the structures; and to determine the RMSD, Q-score, and number of residues aligned between structures.
Immunoprecipitations and Western blotting
PIP strips™ (Life Technologies) were used to probe ENTH and VHS domain binding to phosphoinositides. PIP strips were blocked for 1 hour at 23°C and then incubated with 2 µg/mL of Epsin1-H6, tENTH-H6 or tVHS-H6 recombinant purified proteins for 1 hour at 23°C. PIP strips were subsequently washed, and incubated with a 1:3000 dilution of anti-6X His tag® [GT359] HRP antibody (abcam®) at 4°C overnight. PIP strips were visualized using Amersham ECL Western blotting detection reagents (GE Healthcare).
HeLa cells (ATCC®) were grown on T-75 cm2 flasks to 80–90% cell density. Cells were then transfected with EGFP-N1, tepsin FL (1–525)-GFP, tENTH (1–136)-GFP, or tENTH+tVHS (1–356)-GFP using LipfectamineTM 3000 Reagent (Invitrogen). Cells were imaged and harvested 24hrs post transfection. Cells were resuspended and lysed in 1 mL 10 mM HEPES pH 7.5, 150 mM NaCl, 0.5% NP-40, 0.02 mg/mL AEBSF-HCl (EMD Millipore), and one cOmplete™ Mini EDTA-free Protease Inhibitor Cocktail (Roche). Cell slurry was incubated on ice for 30 min and mixed every ten minutes. Cell slurry was centrifuged at 20,000 × RCF for 15 min, remove and save soluble fraction. GFP-Trap_A resin (ChromoTek, gta-20) batch equilibrated with 10 mM HEPES pH 7.5, 100 mM NaCl, 0.5 mM EDTA, 0.5% NP-40. Resin slurry (60 uL) was added to each soluble cell fraction. Samples were then incubated and rotated for 1 hour at 4° C. Samples were centrifuged at 2500 × RCF for 5 min. Supernatant was removed, and resin was washed with 1 mL of wash buffer for a total of three washes. After final removal of supernatant fraction, 80uL 2XSDS loading buffer was added to each sample and boiled at 95°C for 10 min. Boiled samples were centrifuged at 5000 × RCF for 5 min, then supernatant was remove and transferred to a new tube. The samples were used for western blotting and probed with anti-GFP-HRP at 1:5000 (abcam) and anti-AP4ε at 1:1000 (gift from Margaret Robinson lab).
Isothermal titration calorimetry
ITC experiments were conducted on a NanoITC instrument (TA Instruments) at 20°C. A model dileucine peptide (DSVIL) was dissolved in 20 mM sodium phosphate (pH 6.5), 200 mM NaCl, 0.5 mM TCEP. A PD-10 desalting column (GE Healthcare) was used to buffer exchange human tVHS (residues 220-360) into the same buffer as the peptide. Incremental titration was performed with an initial baseline of 180 s and injection intervals of 180 s. Peptide was titrated in to a ratio of 6x molar excess. Data were analyzed in NanoAnalyze (TA Instruments).
NMR spectroscopy
Uniformly enriched 15N-labeled mono-ubiquitin (gift from Natalja Pashkova) was diluted in 50 mM sodium phosphate pH 7.0, 1 mM DTT to 30 µM, with 10% v/v D2O. Samples containing a ten times molar excess of either human tVHS (residues 220-360) or human tENTH (1-136W) were prepared by diluting in 50 mM sodium phosphate pH 7.0, 1 mM DTT to 30 µM, with 10% v/v D2O. Standard two-dimensional 15N-1H HSQC NMR spectra were collected at 25°C on a 800-MHz Bruker Avance III spectrometer with a CPTCI triple resonance cryoprobe (Bruker BioSpin). Data were processed in Topspin 3.2 (Bruker BioSpin), with zero filling in the indirect dimension and squared sine bell apodization in both dimensions.
Phylogenetic analyses
To construct phylogenetic trees, we first assembled nucleotide sequences that encode tepsin and epsin proteins. Sequences were identified as tepsin or epsin based on (1) references in the published literature 19,44,45; (2) annotations and sequence names in UNIPROT or NCBI (tepsin annotations also include ENTHD2, and epsin annotations also include clint1, epn, and ent); and (3) sequence homology to known proteins, assessed via tBlastn). We also included CALM/PI-CALM (an ANTH-containing protein) sequences from three species as an outgroup clade 44. In total, we compiled 50 sequences for this analysis. Sequences were aligned using the E-INS-i algorithm in MAFFT 55, which is optimized for sequences with sections of homology that are potentially separated by stretches of non-homology. We made 100 bootstrap replicates of the aligned sequences using seqboot in the Phylip package 56, built a maximum-likelihood tree for each replicate using dnaml, and obtained a consensus sequence using consense. To further refine this tree and to estimate branch lengths, we used the consensus phylogeny as a starting tree for 100 iterations of the PASTA (Practical Alignments using SATé and TrAnsitivity) algorithm57. On each iteration, this algorithm splits the tree into subsets, aligns the subsets with the MAFFT algorithm, merges the subsets with the MUSCLE algorithm58, and re-analyzes the tree with the RaxML algorithm (GTR-GAMMA model) 59, ultimately returning the tree that optimizes the maximum-likelihood score. In addition, we compiled nucleotide sequences from specific domains: ENTH and VHS domains from epsin sequences, tENTH and tVHS domains from tepsin sequences, and ANTH domains from CALM/PI-CALM sequences. We constructed a phylogeny for 121 domain-specific sequences using the same methods described above.
Supplementary Material
Table S1. Tepsin ENTH domain structure determination. Data collection and refinement statistics for tENTH crystal structures. Values in parentheses represent the highest resolution shell.
Figure S2. The tepsin ENTH domain lacks helix8. (A) 2mFobs-mFcalc density for tENTH construct 1–136 showing residues Leu111-Ser133 from the final model. (B) Equivalent view for tENTH construct 1–153. No additional density was observed after residue Asp134 in the longer construct, supporting the idea that helix α7 is the final helix in the tepsin ENTH domain.
Figure S3. Structural and functional comparison of ENTH domains. (A) The tepsin ENTH structure was compared to other ENTH domains found in a variety of species and deposited in the PDB. Analysis was carried out using Superpose (CCP4). (B) HSQC spectrum of 15N-labelled ubiquitin (black peaks) and 15N-labelled ubiquitin with a ten-fold molar excess of tENTH (red peaks). No chemical shifts are observed, implying that tENTH does not bind ubiquitin in vitro.
Figure S4. Tepsin VHS-like domain structure determination. (A) Sequence alignments of tVHS across five species that demonstrate 70–90% sequence identity with human tVHS domain. We cloned, expressed, and purified these constructs to determine the most suitable protein for crystallization. (B) Data collection and refinement statistics for native and selenomethione-labeled horse tVHS domain.
Figure S5. Structural comparison of tepsin VHS/ENTH-like domain. The tepsin VHS-like domain was compared to both ENTH and VHS domains from a variety of species and depositing in the PDB. Analysis was carried out using Superpose (CCP4). Based on RMSD values and Q-scores, these data suggest tVHS may be more similar to ENTH than to VHS domains.
Figure S6. The tepsin N-termius is insufficient for AP4 immunoprecipitation. HeLa cells were transfected with empty vector (EGFP-N1) or tepsin constructs corresponding to the ENTH domain (tepsin-1-136-GFP); both N-terminal domains (tepsin 1-356-GFP); or full length tepsin (tepsinFL-GFP). Only full-length tepsin efficiently immunopreciptated AP4 from lysates, as judged by immunoblotting against the ε subunit. We were unable to transfect a construct containing on the tepsin VHS domain.
Figure S7. Phylogenetic analysis of tepsin ENTH and VHS-like domains. Maximum-likelihood phylogenetic tree of ANTH (outgroup), ENTH, tENTH, tVHS, and VHS domains. Note that the tVHS domains for Aplysia californica and Crassostrea gigas appear to have diverged significantly from other tVHS domains and do not reliably form a clade with the other tVHS sequences. The tree with the best maximum likelihood score, pictured here, places these sequences outside of the VHS and tVHS clade. NCBI/GenBank accession numbers are included for reference.
Figure S8. Phylogenetic analysis of the epsin family. Maximum-likelihood phylogenetic tree of tespsin, epsin, and PI-CALM (outgroup) sequences. Sequences annotated as tepsin, epsin, and PI-CALM each form respective monophyletic clades. NCBI/GenBank accession numbers are included for reference.
Table S9. Genetic distance between members of different ENTH/ANTH/VHS domains. Genetic distance was calculated for each pair of sequences as the proportion of differences (number of nucleotide differences divided by the number of sites compared). Gaps were not considered in the distance calculation (pairwise deletion of gaps).
Acknowledgments
The authors thank Linton Traub (Pittsburgh) for the rat epsin1 GST-ENTH-H6 construct; Natalja Pashkova and Rob Piper for 15N-labeled ubiquitin; Jenny Hirst, Alexandra Davies, and Margaret Robinson for AP4 antibodies; and LS-CAT beamline scientists Elena Kondrashkina and Zdzislaw Wawrzak for assistance with remote data collection. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). The crystallization trials described here used the Vanderbilt robotic crystallization facility supported by NIH S10 RR026915. TLA, MNF, AM, AKK, and LPJ are supported by NIH 1R35GM119525, and LPJ is a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. MNF was supported in part by NIH Molecular Biophysics 2T32GM008320-26.
Footnotes
The authors declare they have no conflicts of interest.
References
- 1.Höning S, Ricotta D, Krauss M, et al. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol Cell. 2005;18(5):519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 2.Yu X, Breitman M, Goldberg J. A structure-based mechanism for Arf1-dependent recruitment of coatomer to membranes. Cell. 2012;148(3):530–542. doi: 10.1016/j.cell.2012.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren X, Farías GG, Canagarajah BJ, Bonifacino JS, Hurley JH. Structural basis for recruitment and activation of the AP-1 clathrin adaptor complex by Arf1. Cell. 2013;152(4):755–767. doi: 10.1016/j.cell.2012.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owen DJ, Evans PR. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science (80-) 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. http://www.ncbi.nlm.nih.gov/pubmed/9812899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelly BT, McCoy AJ, Späte K, et al. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature. 2008;456(7224):976–979. doi: 10.1038/nature07422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson LP, Kelly BT, McCoy AJ, et al. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell. 2010;141(7):1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly BT, Owen DJ. Endocytic sorting of transmembrane protein cargo. Curr Opin Cell Biol. 2011;23:1–9. doi: 10.1016/j.ceb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Jackson LP, Lewis M, Kent H, et al. Molecular Basis for Recognition of Dilysine Trafficking Motifs by COPI. Dev Cell. 2012;23(6):1255–1262. doi: 10.1016/j.devcel.2012.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller SE, Mathiasen S, Bright NA, et al. CALM Regulates Clathrin-Coated Vesicle Size and Maturation by Directly Sensing and Driving Membrane Curvature. Dev Cell. 2015;33(2):163–175. doi: 10.1016/j.devcel.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson MS. Forty Years of Clathrin-coated Vesicles. Traffic. 2015 doi: 10.1111/tra.12335. [DOI] [PubMed] [Google Scholar]
- 11.Boehm M, Aguilar RC, Bonifacino JS. Functional and physical interactions of the adaptor protein complex AP-4 with ADP-ribosylation factors (ARFs) EMBO J. 2001;20(22):6265–6276. doi: 10.1093/emboj/20.22.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simmen T, Höning S, Icking A, Tikkanen R, Hunziker W. AP-4 binds basolateral signals and participates in basolateral sorting in epithelial MDCK cells. Nat Cell Biol. 2002;4(2):154–159. doi: 10.1038/ncb745. [DOI] [PubMed] [Google Scholar]
- 13.Matsuda S, Miura E, Matsuda K, et al. Accumulation of AMPA receptors in autophagosomes in neuronal axons lacking adaptor protein AP-4. Neuron. 2008;57(5):730–745. doi: 10.1016/j.neuron.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 14.Abou Jamra R, Philippe O, Raas-Rothschild A, et al. Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am J Hum Genet. 2011;88(6):788–795. doi: 10.1016/j.ajhg.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdollahpour H, Alawi M, Kortüm F, et al. An AP4B1 frameshift mutation in siblings with intellectual disability and spastic tetraplegia further delineates the AP-4 deficiency syndrome. Eur J Hum Genet. 2015;23(2):256–259. doi: 10.1038/ejhg.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardies K, May P, Djemie T, et al. Recessive loss-of-function mutations in AP4S1 cause mild fever-sensitive seizures, developmental delay and spastic paraplegia through loss of AP-4 complex assembly. Hum Mol Genet. 2015:1–10. doi: 10.1093/hmg/ddu740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tüysüz B, Bilguvar K, Koçer N, et al. Autosomal recessive spastic tetraplegia caused by AP4M1 and AP4B1 gene mutation: expansion of the facial and neuroimaging features. Am J Med Genet A. 2014;164A(7):11677–1685. doi: 10.1002/ajmg.a.36514. [DOI] [PubMed] [Google Scholar]
- 18.Hirst J, Irving C, Borner GHH. Adaptor protein complexes AP-4 and AP-5: new players in endosomal trafficking and progressive spastic paraplegia. Traffic. 2013;14(2):153–164. doi: 10.1111/tra.12028. [DOI] [PubMed] [Google Scholar]
- 19.Borner GHH, Antrobus R, Hirst J, et al. Multivariate proteomic profiling identifies novel accessory proteins of coated vesicles. J Cell Biol. 2012;197(1):141–160. doi: 10.1083/jcb.201111049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenthal JA, Chen H, Slepnev VI, et al. The epsins define a family of proteins that interact with components of the clathrin coat and contain a new protein module. J Biol Chem. 1999;274(48):33959–33965. doi: 10.1074/jbc.274.48.33959. http://www.ncbi.nlm.nih.gov/pubmed/10567358. [DOI] [PubMed] [Google Scholar]
- 21.Kay BK, Yamabhai M, Wendland B, Emr SD. Identification of a novel domain shared by putative components of the endocytic and cytoskeletal machinery. Protein Sci. 1999;8(2):435–438. doi: 10.1110/ps.8.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ford MGJ, Mills IG, Peter BJ, et al. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419(6905):361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 23.Hirst J, Miller SE, Taylor MJ, von Mollard GF, Robinson MS. EpsinR is an adaptor for the SNARE protein Vti1b. Mol Biol Cell. 2004;15(12):5593–5602. doi: 10.1091/mbc.E04-06-0468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chidambaram S, Müllers N, Wiederhold K, Haucke V, von Mollard GF. Specific interaction between SNAREs and epsin N-terminal homology (ENTH) domains of epsin-related proteins in trans-Golgi network to endosome transport. J Biol Chem. 2004;279(6):4175–4179. doi: 10.1074/jbc.M308667200. [DOI] [PubMed] [Google Scholar]
- 25.Miller SE, Collins BM, McCoy AJ, Robinson MS, Owen DJ. A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature. 2007;450(7169):570–574. doi: 10.1038/nature06353. [DOI] [PubMed] [Google Scholar]
- 26.Aguilar RC, Watson HA, Wendland B. The yeast Epsin Ent1 is recruited to membranes through multiple independent interactions. J Biol Chem. 2003;278(12):10737–10743. doi: 10.1074/jbc.M211622200. [DOI] [PubMed] [Google Scholar]
- 27.Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the β2-adaptin appendage domain. EMBO J. 2000;19(16):4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brett TJ, Traub LM, Fremont DH. Accessory protein recruitment motifs in clathrin-mediated endocytosis. Structure. 2002;10(6):797–809. doi: 10.1016/s0969-2126(02)00784-0. http://www.ncbi.nlm.nih.gov/pubmed/12057195. [DOI] [PubMed] [Google Scholar]
- 29.Edeling MA, Mishra SK, Keyel PA, et al. Molecular switches involving the AP-2 beta2 appendage regulate endocytic cargo selection and clathrin coat assembly. Dev Cell. 2006;10(3):329–342. doi: 10.1016/j.devcel.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 30.Frazier MN, Davies AK, Voehler M, et al. Molecular Basis for the Interaction Between AP4 β4 and its Accessory Protein, Tepsin. Traffic. 2016;17(4):400–415. doi: 10.1111/tra.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mattera R, Guardia CM, Sidhu SS, Bonifacino JS. Bivalent Motif-Ear Interactions Mediate the Association of the Accessory Protein Tepsin with the AP-4 Adaptor Complex. J Biol Chem. 2015;290(52):30736–30749. doi: 10.1074/jbc.M115.683409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Combet C, Blanchet C, Geourjon C, Deléage G. NPS@: Network Protein Sequence Analysis. TIBS. 2000;25(3):147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Itoh T, De Camilli P. BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane-cytosol interfaces and membrane curvature. Biochim Biophys Acta. 2006;1761(8):897–912. doi: 10.1016/j.bbalip.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst D. 2004;60(12):2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 36.Sievers F, Wilm A, Dineen D, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levin-Kravets O, Tanner N, Shohat N, et al. A bacterial genetic selection system for ubiquitylation cascade discovery. Nat Meth. 2016;13(11):945–952. doi: 10.1038/nmeth.4003. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser Crystallographic Software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, Afonine PV, Bunkóczi G, et al. PHENIX: a comprehensive, Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buchan DWA, Minneci F, Nugent TCO, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res. 2013;41:W349–57. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misra S, Puertollano R, Kato Y, Bonifacino JS, Hurley JH. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature. 2002;415(6874):933–937. doi: 10.1038/415933a. [DOI] [PubMed] [Google Scholar]
- 42.Ren X, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010;29(6):1045–1054. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skruzny M, Brach T, Ciuffa R, Rybina S, Wachsmuth M, Kaksonen M. Molecular basis for coupling the plasma membrane to the actin cytoskeleton during clathrin-mediated endocytosis. Proc Natl Acad Sci U S A. 2012;109(38):E2533–42. doi: 10.1073/pnas.1207011109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Craene J-O, Ripp R, Lecompte O, Thompson JD, Poch O, Friant S. Evolutionary analysis of the ENTH/ANTH/VHS protein superfamily reveals a coevolution between membrane trafficking and metabolism. BMC Genomics. 2012;13(1):297. doi: 10.1186/1471-2164-13-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabernet-Castello C, Dacks JB, Field MC. The single ENTH-domain protein of trypanosomes; endocytic functions and evolutionary relationship with epsin. Traffic. 2009;10(7):894–911. doi: 10.1111/j.1600-0854.2009.00910.x. [DOI] [PubMed] [Google Scholar]
- 46.Boal F, Mansour R, Gayral M, et al. TOM1 is a PI5P effector involved in the regulation of endosomal maturation. J Cell Sci. 2015;128(4):815–827. doi: 10.1242/jcs.166314. [DOI] [PubMed] [Google Scholar]
- 47.Misra S, Puertollano R, Kato Y, Bonifacino JS, Hurley JH. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature. 2002;415(6874):933–937. doi: 10.1038/415933a. [DOI] [PubMed] [Google Scholar]
- 48.Hein MY, Hubner NC, Poser I, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163(3):712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 49.Manna PT, Gadelha C, Puttick AE, Field MC. ENTH and ANTH domain proteins participate in AP2-independent clathrin-mediated endocytosis. J Cell Sci. 2015;128(11):2130–2142. doi: 10.1242/jcs.167726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Cryst D Crystallogr. 2001;57(10):1445–1450. doi: 10.1107/s0907444901014007. http://www.ncbi.nlm.nih.gov/pubmed/11567158. [DOI] [PubMed] [Google Scholar]
- 51.Afonine P, Grosse-Kunstleve RW, Adams PD. phenix.refine. CCP4 Newsl. 2005;42 Contribution 8. [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: the CCP4mg molecular-graphics software. Acta Cryst D. 2011;67(4):386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.6. Distrib by author. 2005 Department. [Google Scholar]
- 57.Mirarab S, Nguyen N, Warnow T. PASTA: ultra-large multiple sequence alignment. Proc RECOMB. 2014 doi: 10.1089/cmb.2014.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Tepsin ENTH domain structure determination. Data collection and refinement statistics for tENTH crystal structures. Values in parentheses represent the highest resolution shell.
Figure S2. The tepsin ENTH domain lacks helix8. (A) 2mFobs-mFcalc density for tENTH construct 1–136 showing residues Leu111-Ser133 from the final model. (B) Equivalent view for tENTH construct 1–153. No additional density was observed after residue Asp134 in the longer construct, supporting the idea that helix α7 is the final helix in the tepsin ENTH domain.
Figure S3. Structural and functional comparison of ENTH domains. (A) The tepsin ENTH structure was compared to other ENTH domains found in a variety of species and deposited in the PDB. Analysis was carried out using Superpose (CCP4). (B) HSQC spectrum of 15N-labelled ubiquitin (black peaks) and 15N-labelled ubiquitin with a ten-fold molar excess of tENTH (red peaks). No chemical shifts are observed, implying that tENTH does not bind ubiquitin in vitro.
Figure S4. Tepsin VHS-like domain structure determination. (A) Sequence alignments of tVHS across five species that demonstrate 70–90% sequence identity with human tVHS domain. We cloned, expressed, and purified these constructs to determine the most suitable protein for crystallization. (B) Data collection and refinement statistics for native and selenomethione-labeled horse tVHS domain.
Figure S5. Structural comparison of tepsin VHS/ENTH-like domain. The tepsin VHS-like domain was compared to both ENTH and VHS domains from a variety of species and depositing in the PDB. Analysis was carried out using Superpose (CCP4). Based on RMSD values and Q-scores, these data suggest tVHS may be more similar to ENTH than to VHS domains.
Figure S6. The tepsin N-termius is insufficient for AP4 immunoprecipitation. HeLa cells were transfected with empty vector (EGFP-N1) or tepsin constructs corresponding to the ENTH domain (tepsin-1-136-GFP); both N-terminal domains (tepsin 1-356-GFP); or full length tepsin (tepsinFL-GFP). Only full-length tepsin efficiently immunopreciptated AP4 from lysates, as judged by immunoblotting against the ε subunit. We were unable to transfect a construct containing on the tepsin VHS domain.
Figure S7. Phylogenetic analysis of tepsin ENTH and VHS-like domains. Maximum-likelihood phylogenetic tree of ANTH (outgroup), ENTH, tENTH, tVHS, and VHS domains. Note that the tVHS domains for Aplysia californica and Crassostrea gigas appear to have diverged significantly from other tVHS domains and do not reliably form a clade with the other tVHS sequences. The tree with the best maximum likelihood score, pictured here, places these sequences outside of the VHS and tVHS clade. NCBI/GenBank accession numbers are included for reference.
Figure S8. Phylogenetic analysis of the epsin family. Maximum-likelihood phylogenetic tree of tespsin, epsin, and PI-CALM (outgroup) sequences. Sequences annotated as tepsin, epsin, and PI-CALM each form respective monophyletic clades. NCBI/GenBank accession numbers are included for reference.
Table S9. Genetic distance between members of different ENTH/ANTH/VHS domains. Genetic distance was calculated for each pair of sequences as the proportion of differences (number of nucleotide differences divided by the number of sites compared). Gaps were not considered in the distance calculation (pairwise deletion of gaps).