SUMMARY
The regulation of intracellular vesicular trafficking is mediated by specific families of proteins that are involved in vesicular budding, translocation, and fusion with target membranes. We purified a vesicle-associated protein from hepatic microsomes using sequential column chromatography and partially sequenced it. Oliogonucleotides based on these sequences were used to clone the protein from a rat liver cDNA library. The clone encoded a novel protein with a predicted mass of 137 kDa (p137). The protein had an N terminus WD repeat motif with significant homology to Sec31p, a member of the yeast COPII coat that complexes with Sec13p. We found that p137 interacted with mammalian Sec13p using several approaches: co-elution through sequential column chromatography, co-immunoprecipitation from intact cells, and yeast two-hybrid analysis. Morphologically, the p137 protein was localized to small punctate structures in the cytoplasm of multiple cultured cell lines. When Sec13p was transfected into these cells, it demonstrated considerable overlap with p137. This overlap was maintained through several pharmacological manipulations. The p137 compartment also demonstrated partial overlap with ts045-VSVG protein when infected cells were incubated at 15°C. These findings suggest that p137 is the mammalian orthologue of Sec31p.
Keywords: Sec31p, Sec13p, COP II, Vesicular traffic
INTRODUCTION
The secretory pathway in eukaryotic cells carries newly synthesized cargo to many cellular destinations. The discrete organelles composing this pathway are temporally and spatially connected by small vesicular carriers, whose number, extent and nature are areas of active investigation. Key components in this pathway include a class of proteins that form cytosolic coats which promote vesicle formation on donor membranes and also seem to be involved in selecting cargo and membrane machinery from these compartments.
Three classes of cytoplasmic coats have been identified. (1) Clathrin, interacting with different adaptor proteins, participates in endocytosis, lysosome biogenesis and as yet unidentified vesicular transport processes that arise in the trans-Golgi region of cells (reviewed by Marsh and McMahon, 1999; Lowe and Kreis, 1998). (2) The COPI coatomer is involved in retrograde traffic within the Golgi and from the cis-Golgi region to the endoplasmic reticulum (ER). It may also participate in anterograde transport from the ER (reviewed by Aridor and Balch, 1999). (3) COPII coats mediate anterograde transport of cargo out of the ER. The components of COPII in yeast are the small GTPase Sar1p and two protein complexes, Sec23/24p and Sec13/31p (reviewed by Barlowe, 1998). Mammalian homologs of several COPII proteins have been identified and characterized. These include homologs of Sar1p (Kuge et al., 1994), Sec23p (Paccaud et al., 1996), Sec24p (Aridor et al., 1998), and Sec13p (Swaroop et al., 1994; Shaywitz et al., 1995). Although a mammalian Sec13/31p complex has been partially purified by others, the mammalian orthologue of Sec31p has not been identified (Aridor et al., 1998). In this study, we report the cloning and sequencing of a novel membrane-associated mammalian protein, p137. Based on several independent lines of evidence, we propose that p137 is the putative orthologue of yeast Sec31p. p137 has sequence and structural homology to the N-terminal domain of yeast COPII protein Sec31p; it interacts with mammalian Sec13p, another COPII component; and its subcellular localization puts p137 at ER exit sites together with Sec13p.
MATERIALS AND METHODS
Materials
All chemical reagents were purchased from Sigma Chemical Co. (St Louis, MO) unless otherwise noted. Purified Sec13/31p complex (Barlowe et al., 1994) was provided by Randy Schekman (University of California-Berkeley, Berkeley, CA).
Antibodies
Affinity purified antibody 212 to synapsin I site 2 was provided by Paul Greengard (Matovcik et al., 1994). Rabbit antiserum to human Sec13p has been described (Pryer et al., 1993). Rabbit antibody to vesicular stomatitis virus glycoprotein (VSVG) was provided by Carolyn Machamer (Johns Hopkins Medical School, Baltimore, MD). Mouse monoclonal antibody to mannosidase II (ascites, clone 53FC3) was purchased from Berkeley Antibody Company (Richmond, CA). Mouse anti-T7 antibody was purchased from Novagen (Madison, WI). FITC-labeled goat anti-mouse and Cy3-labeled donkey anti-rabbit antibodies were purchased from Jackson Laboratories (West Grove, PA). Mouse anti-His antibody was purchased from Pharmacia (Piscataway, NJ).
Biochemical analysis
Samples for SDS-PAGE were solubilized in Laemmli’s buffer with 5% β-mercaptoethanol and separated using acrylamide concentrations from 8% to 12% as described (Laemmli, 1970).
Immunoblotting
Proteins separated by SDS-PAGE were transferred to nitrocellulose or Immobilon membrane. The membranes were processed for immunoblot analysis as described (Burnette, 1981). Mouse anti-p137 and rabbit anti-p137 were used at 1:1000 and 1:10,000, respectively. Antibodies 212 and anti-Sec13p were used at a 1:3000 dilution. The primary antibodies were then detected by enhanced chemiluminescence (ECL; Amersham, Arlington Heights, IL) or an alkaline phosphatase colorimetric reaction (Promega, Madison, WI) following the manufacturers’ protocols.
Purification of p137
Livers from male and female Sprague-Dawley rats were placed in ice-cold 0.3 M sucrose, 25 mM PIPES, pH 6.8, 1 mM dithiothreitol (DTT), and protease inhibitors (final concentrations: 10 μM antipain, 2 μg/ml aprotinin, 5 mM benzamidine, 50 μM leupeptin, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 μM E-64) at 1:10 mass to volume and homogenized with 10 strokes in a Brendler homogenizer. All subsequent steps were performed at 4°C or on ice unless otherwise noted. The homogenate was centrifuged at 1000 g for 10 minutes, the resulting supernatant was centrifuged at 10,000 g for 10 minutes, and the subsequent supernatant centrifuged at 100,000 g for 1 hour to yield a microsomal pellet. The pellet was resuspended at a ratio of 2–4 ml per gram liver in 0.3 M sucrose made 70 mM Na2HPO4, pH 7.3, 250 mM NaCl, 3 mM EDTA, incubated for one hour and centrifuged (100,000 g for 1 hour). Analysis of the supernatant and pellet for p137 protein content indicated that >90% of p137 was released. The supernatant fraction (typically 30–50 ml from 20 g of liver) was diluted 8-fold with water and applied to a 2.6 × 10 cm Bio-Gel HTP hydroxyapatite column (Bio-Rad, Hercules, CA) equilibrated in HTP buffer (10 mM Na2HPO4, pH 7.3, 1 mM DTT, 5 mM benzamidine) and eluted with a 500 ml linear gradient (HTP buffer to HTP with 250 mM Na2HPO4 and 1.2 M NaCl). Fractions containing p137 were identified by immunoblot, pooled, concentrated (Centricon10, Amicon, Beverly, MA) and applied to a 2.6 × 100 cm Sephacryl S-300 HR (Pharmacia, Piscataway, NJ) column in 25 mM Tris-HCl, pH 7.8, 100 mM NaCl, 1 mM DTT, 5 mM benzamidine. Fractions from the gel filtration column containing p137 were pooled, diluted 4-fold with 10 mM Tris-HCl, pH 7.8, 1 mM DTT, 5 mM benzamidine, and separated on a Mono Q HR 5/5 FPLC column (Pharmacia, Piscataway, NJ) using a 20 ml linear gradient (0.0 to 1.0 M NaCl with 10 mM Tris-HCl, pH 7.8).
Peak fractions eluted from the S300 column were separated by SDS-PAGE, transferred to Immobilon, stained with Ponceau S and the 170 kDa band was excised and subjected to amino acid sequencing. Peptide 1 was the N terminus of the protein and other peptides were generated by tryptic digestion of the 170 kDa band followed by purification on HPLC. All analyses were performed at the Protein Sequencing Facility at Rockefeller University.
Cloning and sequencing of p137
Based on a partial amino acid sequence, a PCR product of 275 nucleotides was obtained following amplification from a human brain cDNA library (Gibco BRL, Grand Island, NY) using Amplitaq (Perkin Elmer, Norwalk, CT) and degenerate primers, 5′-(C/I)(C/T)-(C/I)TCACCAATCATGAACCC-3′ and 5′-AIGGTITT(T/G)CC(A/G)-AT(A/G)GG(A/G)CG-3′. Random labeled (Boehringer Mannheim, Arlington Heights, IL) [32P]dCTP PCR product (Fig. 1B, probe 1) was used to screen a rat liver cDNA library (λgt10; Clontech, Palo Alto, CA). The first screen produced several partial length clones, two of which included poly(A) tails (Fig. 1B, clones 1A and 9A). EcoRI fragments of purified phage DNA were subcloned into pBluescript (Stratagene, La Jolla). Rescreening the same library with an [32P]dCTP ApaI fragment of library clone 1A (Fig. 1B, probe 2) resulted in the identification of a partially overlapping clone (Fig. 1B, clone 18B). A 5′ RACE kit (Gibco BRL) was used to identify the remainder of the clone. Using the adaptor oligo AP1 and the sequence-specific oligonucleotide, 939- (5′-AGGACACCACTGG-ATATCAAAGC-3′), based on clone 18B, amplification was performed with the Marathon-ready rat liver cDNA (Gibco BRL) as template. Products of the RACE reactions were subcloned into pCRII (Invitrogen, Carlsbad, CA), a TA cloning vector, after treating the samples with 1 unit of Amplitaq polymerase (Perkin-Elmer).
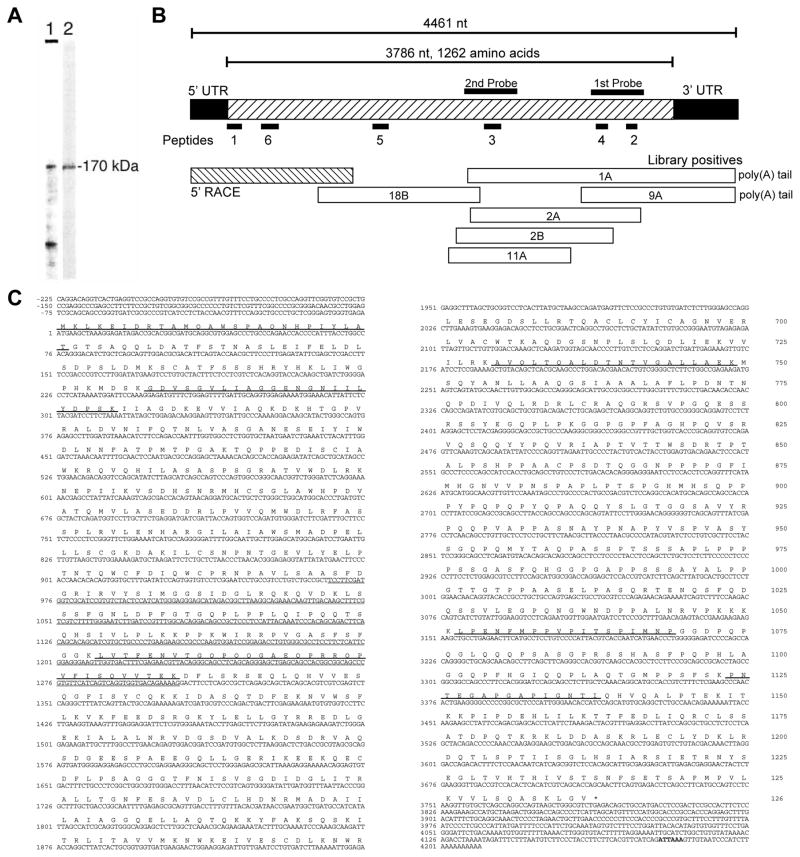
Fig. 1.
Purification and cloning of p137. (A) SDS-PAGE (8% acrylamide) of the peak fraction from the S300 gel chromatography step: (1) Coomassie Blue; (2) immunoblot with antibody 212. The protein band corresponding to 170 kDa was subjected to partial amino acid sequencing. (B) Cloning strategy used to obtain the complete p137 cDNA. The coding region spans 3786 nucleotides, not including the stop codon (hatched box). The 5′ and 3′ untranslated regions include an additional 672 nucleotides (solid boxes). The locations of the six peptides obtained from the purified protein are indicated by the solid bars numbered 1–6. The two probes used to screen the rat liver cDNA library are indicated by solid bars labeled ‘1st probe’ and ‘2nd probe’. Clones 1A, 2A, and 9A were obtained from the initial screen of a rat liver cDNA library and clones 2B, 11A, and 18B from the second screen of the same library. The small hatched box indicates the 5′ RACE product. (C) cDNA and amino acid sequence of p137. Shown is the nucleotide sequence of the combination of overlapping clones that encompass the coding region and the adjacent 5′- and 3′-noncoding regions, with 1 representing the first nucleotide of the coding region. The deduced amino acid sequence of p137 is shown above the nucleotide sequence. Nucleotide sequences encoding the six peptides obtained from the purified protein are underlined. The sequence of the fusion protein used for antibody production is indicated with a dash underline. A possible polyadenylation signal sequence (ATTAAA) is shown in bold. These sequence data are available from GenBank/EMBL/DDBJ under accession number AFO34582.
Plasmids containing cDNA inserts were isolated by alkaline lysis of minipreps and sequenced by the Sanger dideoxy termination method using Sequenase 2.0 (Amersham) and [35S]dATP. Complete bi-directional sequence was obtained by sequencing overlapping restriction endonuclease fragments as well as using internal sequence-specific primers.
The full-length p137 cDNA has a coding region of 3786 nucleotides, flanked by 270 bp of 5′ untranslated sequence and 411 bp of 3′ untranslated sequence (Fig. 1C). A probable polyadenylation signal sequence, ATTAAA, was found preceding a poly(A) tail. Translation of the open reading frame predicts a 1262 amino acid protein with a molecular mass of 137 kDa.
Northern analysis
Human organ northern blots (MTN I and II) (Clontech) were hybridized with a [32P]CTP labeled probe (Fig. 1B, probe 1).
Production of antibodies
Polyclonal antibodies specific for p137 were made in rabbits and mice. A pET fusion protein containing an N-terminal 6×His tag and a T7 tag was made using amino acids (aa) 323-436 of p137 (Fig. 1C). The region was amplified from cloned template using oligonucleotides 967+ (5′-TCCTTCGATGGTCGCATCCG-3′) and 1308- (5-TCAGCAGTCCTTTTCTGTCACCACCTG-3′), subcloned into pBluescript and sequenced. A stop codon and a cysteine residue were added to the antisense oligonucleotide, which was ligated into SalI-EagI pET-28b (Novagen, Madison, WI). After transformation into BL21 (DE3) competent cells and induction, the expressed protein was isolated using Ni-NTA according to the manufacturer’s instructions (Novagen). The purified protein was conjugated to maleimide-activated keyhole limpet hemocyanin (KLH: Pierce, Rockford, IL).
Rabbit antibodies were produced by Covance (Denver, PA) and antiserum from rabbit 2549 was used (bleeds 2 to 5). For production of mouse anti-p137 antibodies, the protein-KLH conjugate was emulsified in Freund’s complete adjuvant (primary injection, 100 μg) or incomplete adjuvant (3 boosts, 50 μg each) and injected intraperitoneally into female Balb/C mice. Mouse ascites containing anti-p137 antibodies was prepared by intraperitoneal injection of immunized mice with SP2/0 myeloma cells (Luo and Lin, 1997).
Yeast two-hybrid
Two-hybrid screens were performed with positive interactions determined by testing for β-galactosidase expression as described (Fields and Ok-kyu, 1989). The strains used were CKY556 (MATa ura3-52, leu2-3, 112, his3, trp1, GAL+ with pSH 18–34 [URA3]) or L40. Gal4 fusion constructs were made in pGADGH and represent aa 1-490 of yeast Sec31p (pDS131) (Shaywitz et al., 1997), aa 1-470 of p137 (WD), and aa 702-1262 of p137 (PRD). LexA fusion constructs were made in pBTM116 and represent the entire sequence of yeast Sec13p (pDS138) and human Sec13p (pDS168) (Shaywitz et al., 1997). The entire sequence of yeast Sec23p (pDS72) and murine Sec23 (pDS73) were made in pGILDA, a LexA fusion vector with an inducible GAL1 promoter instead of the ADH1 promoter, as described (Shaywitz et al., 1997).
The two p137 GAL4 fusion constructs containing the WD domain and the PRD region were amplified from cloned template using oligonucleotides, –2+ (5′-CAATGAAGCTAAAGGAGATAGACCG-3′) and 1410– (5′-CTTCTCGAAGTCAGTCTGGGACG-3′) and 2104+ (5′-GGACCAAAGCTCAAGATGGTAGC-3′) and 3789– (5′-TGAGACGCCCAGCTTACTGGC-3′), respectively. The amplified products were subcloned into pCRII (Invitrogen), sequenced to confirm identity with the original clone, and ligated into EcoRI digested dephosphorylated plasmid pGADGH.
Transfection constructs
To make the Sec13-GFP and T7-Sec13p constructs, the entire sequence of human Sec13p was amplified from cloned template using oligonucleotides, –2+ (5′-TGATGGTGTCAGTAATTAACACTGTG-3′) and 979– (5′-CTGCTCGTTCTGCTGGCCCTC-3′). The amplified product was subcloned into pCRII, sequenced to confirm identity with the original clone, and ligated into EcoRI digested dephosphorylated plasmid, pEGFP-N3 (Clontech) for Sec13p-GFP (pCS172). For the T7-Sec13p construct, the T7 tag was added by first ligating into EcoRI digested dephosphorylated pET-28b, then digesting with XbaI and NotI, and ligated into dephosphorylated-pEGFP-N3 digested with NheI and NotI.
Cell culture and transfection
WIF-B cells (a hepatic cell line; Shanks et al., 1994; Ihrke et al., 1993) were grown on glass coverslips in modified Ham’s F12 medium (Gibco BRL) supplemented with HAT (0.01 mM hypoxanthine, 40 nM aminopterin, 0.002 mM thymidine) and 5% fetal bovine serum (FBS). Normal rat kidney (NRK) cells were grown on glass coverslips in DMEM (Gibco BRL) supplemented with 2 mM L-glutamine and 10% FBS. Clone 9 cells were grown in Ham’s F12 medium supplemented with 2 mM glutamine and 10% FBS.
Clone 9 cells were transfected with Sec13p-GFP using the calcium phosphate method. After overnight incubation, they were washed and examined. NRK cells were transfected with a plasmid encoding the tsO45 mutant of VSVG as described (Weisz and Machamer, 1994) and the T7-Sec13p plasmid using Lipofectin (Gibco BRL) according to the manufacturer’s instructions.
Immunoprecipitation
A rat liver S100 fraction was solubilized in 1% Triton X-100 with protease inhibitors, and the extract incubated first with rabbit polyclonal antibody 2549, then immunoprecipitated with Protein A-Sepharose.
Immunofluorescence microscopy
NRK, WIF-B, and Clone 9 cells were cells were fixed/permeabilized in methanol or 0.5% paraformaldehyde/0.05% saponin and processed as described (Matovcik et al., 1994). Paraformaldehyde was used in all experiments with a GFP-tagged probe. Primary antibodies were used at the following dilutions: mouse anti-p137, 1:100; rabbit anti-p137, 1:1000; rabbit anti-VSVG, 1:300; mouse anti-T7, 1:1000. Primary antibodies were detected with FITC-labeled (1:500), Texas Red-labeled secondary antibody (1:500) or Cy3-labeled (1:3000) secondary antibodies. The labeled cells were examined by epifluorescence using an Axioplan universal microscope (Carl Zeiss Inc., Thornwood, NY). Digital images were collected using the Microcomputer Imaging Device image analysis system (Imaging Research Inc., St Catharines, Ontario, Canada). For confocal visualization (anti-p137/anti-VSVG double-labeled cells only), samples were analyzed using a model MRC600 laser scanning confocal microscope (Biorad, Hercules, CA) and images were collected from 0.8 μm optical sections. Digitized images were assembled and labeled in Adobe Photoshop (Adobe Systems Inc., San Jose, CA). For experiments with brefeldin A (10 μg/ml), cells were placed in serum-free Ham’s F12 medium (Gibco BRL) in the presence or absence of brefeldin A for 1 hour at 37°C. For experiments with nocodazole (33 μM), cells were transfected with T7-Sec13p plasmid and 24 hours later incubated in the presence or absence of the drug for 1 hour in serum-free medium. VSV-G ts045-transfected cells were either kept at the nonpermissive temperature (40°C) or shifted to 32°C or 15°C for 1 hour. After methanol fixation, cells were doubled labeled with primary antibodies followed by FITC-labeled anti-rabbit and Cy3-labeled anti-mouse secondary antibodies.
RESULTS
Sequencing of p170 reveals a novel 137 kDa protein with homology to yeast Sec31p
Previous work from our laboratory had identified two vesicle-associated proteins of ~85 kDa and ~170 kDa that were present in epithelial tissues and were recognized by a poly-specific antibody called ab 212 (Matovcik et al., 1994). We partially purified the major 170 kDa immunoreactive protein band in rat liver. Protein staining and immunoblot analysis of the fractions from the final chromatographic step (see Materials and Methods) demonstrated that the 170 kDa band was ~25% of the total protein (Fig. 1A). A second major protein staining band of ~35 kDa was also present. We obtained partial sequence of five tryptic fragments and of the amino terminus of the 170 kDa protein, including the initiating methionine (Fig. 1B). A search of GenBank’s databases with the first four of the peptide sequences revealed that two (peptides 2 and 4) were found within a single entry, N28605, in the expressed sequence tag database (dbest). We subsequently obtained the full-length sequence of the 170 kDa protein (Fig. 1C). The clone encoded a novel protein with a deduced mass of 137 kDa that was named p137.
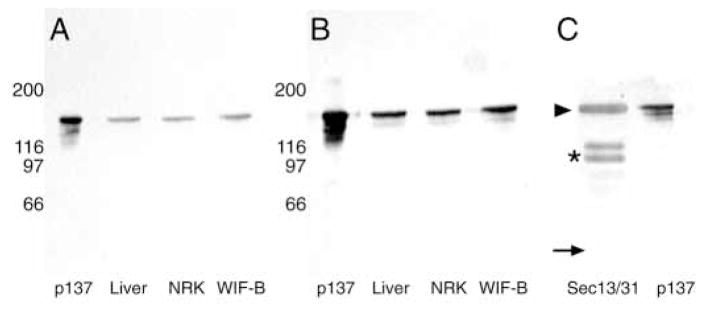
The discrepancy between the predicted sizes based on sequence and mobility in SDS-PAGE was investigated. Rabbit and mouse antibodies to p137 were generated using a recombinant peptide of aa 323-436. Both antibodies recognized a single band in total homogenates of rat liver, WIF-B, and NRK cells; the partially purified protein used to clone p137 co-migrated with the approximately 170 kDa bands detected in other tissues (Fig. 2A and B). The antibodies were generated against a region in p137 that shares homology with yeast Sec31p. As shown in Fig. 2C, anti-p137 antibodies also recognized yeast Sec31p. The fact that Sec31p and p137 have the same predicted molecular masses and comigrated in SDS-PAGE supports the idea that they behave anomalously under the conditions of SDS-PAGE employed here. Furthermore, this similar behavior in SDS-PAGE suggested that they were related.
Fig. 2.
Anti-p137 antibodies detect a 170 kDa protein in rat liver, WIF-B cells, NRK cells and isolated yeast Sec13/13p complexes that comigrates with purified p137. Purified p137 (lane 1) and total homogenates of rat liver (lane 2, 20 μg), WIF-B cells (lane 3, 20 μg), and NRK cells (lane 4, 20 μg) were separated by SDS-PAGE (8–12% acrylamide) and subjected to immunoblot analysis using mouse anti-p137 (A) or rabbit anti-p137 (B) antibodies. Both antibodies detect a 170 kDa band that comigrates with purified p137. Purified yeast Sec13/31p complex and p137 were subjected to immunoblot analysis using rabbit anti-p137 (C). The antibody recognizes both p137 and yeast Sec31p (arrowhead), but not yeast Sec13p (arrow); the two additional bands detected are Sec31p degradation products (asterisk).
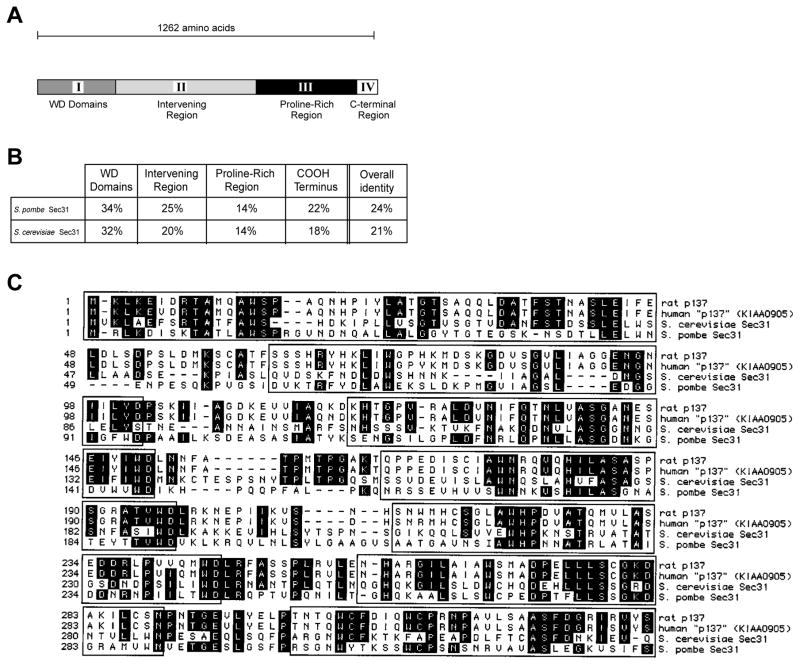
The p137 protein can be divided into four domains (Fig. 3A). The N terminus consists of repeating WD domains that extend to aa 332. The second region stretches from aa 333-807. The third domain is proline-rich (21.6%), extends from aa 808-1145 and contains 10.8% serine, 9.1% glutamine and 5.7% threonine. The fourth domain extends from aa 1146 to the end of the protein and begins with a cluster of potential phosphorylation sites. Throughout the molecule, there are additional potential phosphorylation sites for protein kinase C, calcium/calmodulin-dependent protein kinase II, and tyrosine kinase.
Fig. 3.
Domain structure of p137 protein and homology to Sec31p. (A) The deduced protein sequence of p137 can be divided into four domains: a WD-repeat region (I), an intervening region (II), a proline-rich region (III) and a C-terminal region (IV). (B) Percentage identity of p137 with Sec31p from S. pombe and S. cerevisiae. (C) Comparison of the WD-repeat region of rat liver p137 to KIAA0905 (human), S. pombe and S. cerevisiae Sec31p. Conserved amino acid sequences are shaded. WD domains are boxed.
Although our initial search of the GenBank databases revealed that p137 was a novel protein, more recent entries include the nucleotide sequence for the same protein cloned from a human brain library (KIAA0905). The protein of known function with the greatest degree of homology to p137 is Sec31p, a member of the ER-Golgi COPII coat (reviewed by Barlowe et al., 1994). Overall, p137 is 24% identical to Sec31p from Schizosaccharomyces pombe and 21% identical to Sec31p from Saccharomyces cerevisiae (Fig. 3B). The WD region of p137 shows the greatest degree of homology with yeast Sec31p; it is 34% identical to the S. pombe protein and 32% identical to the S. cerevisiae protein (Fig. 3C). To assess the statistical significance of alignments, 50 comparisons between p137 and Sec31p (S. cerevisiae) were made in a randomized order using the GAP program of GCG software (Madison, WI). The mean (+ standard deviation) of the comparisons (quality score Q = 8.9 + 5.3) was significantly less that the quality score of p137 directly compared to Sec31p (S. cerevisiae) (Q = 221, Z score = 4). Similarly, p137 compared to randomized Sec31p (S. pombe) (n = 50, Q = 8.6 + 4.8) was significantly less than p137 directly compared to Sec31p (S. pombe) (Q = 539, Z = 111). This analysis supports the idea that p137 is significantly related to Sec31p from S. cerevisiae and S. pombe.
Northern blots were generated by probing poly(A) selected mRNA from human tissues with a cDNA probe generated from the C-terminal region of the p137 clone. As shown in Fig. 4, the probe labeled a major 4.3 kb band in all tissues. Northern blot analysis of rat tissues (brain, kidney, small intestine, heart) also demonstrated a major 4.3 kb band as well as less prominent larger and smaller bands (not shown).
Fig. 4.
p137 mRNA expression. p137 mRNA expression was analyzed in a human organ northern blot. Sizes of the RNA markers (kb) are indicated on the left. Message encoding p137 is expressed as a major 4.3 kb band.
p137 associates with Sec13p and Sec23p
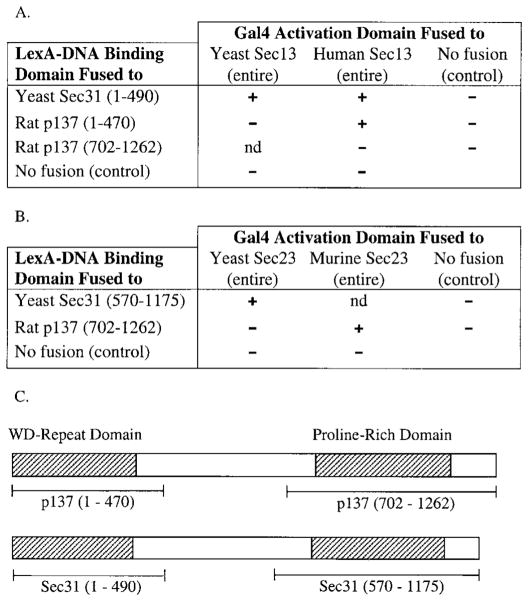
We next examined the association of p137 with several components of COPII. First, we compared the elution patterns of the mammalian Sec13p with p137 through the sequential steps of p137 purification and found that p137 consistently eluted with Sec13p (Fig. 5). We then determined whether p137 was associated with Sec13p in intact cells using non-denaturing immunoprecipitation. Using the rabbit anti-p137 antibody, we were able to specifically co-immunoprecipitate endogenous Sec13p (Fig. 6). T7-human Sec13p expressed in transiently transfected Clone 9 cells was also found in a complex that could be immunoprecipitated by anti-p137 (not shown). We then turned to the yeast two-hybrid system to further explore the interaction between p137 and Sec13p. Based on β-galactosidase expression, we confirmed that the WD domain of yeast Sec31 interacted with both yeast and human Sec13p (Fig. 7A and Shaywitz et al., 1997). Importantly, the WD region of p137 interacted with human Sec13p (Fig. 7A) but not with yeast Sec13p. We also found that the proline-rich region of p137 (aa 702-1262) interacted with murine Sec23p but not with yeast Sec23p (Fig. 7B). As a control, we determined that p137 by itself did not activate β-galactosidase expression.
Fig. 5.
Co-elution of p137 and Sec13p by column chromatography. The supernatant of the microsomal fraction was applied sequentially to hydroxyapatite, Sephacryl S300, and MonoQ HR 5/5 columns. The eluted fractions were solubilized and separated on SDS-PAGE (8% acrylamide), then subjected to immunoblot analysis using antibody 212 (to detect p137) and anti-human Sec13 antibody. The elution profiles for both protein bands coincide at each chromatography step.
Fig. 6.
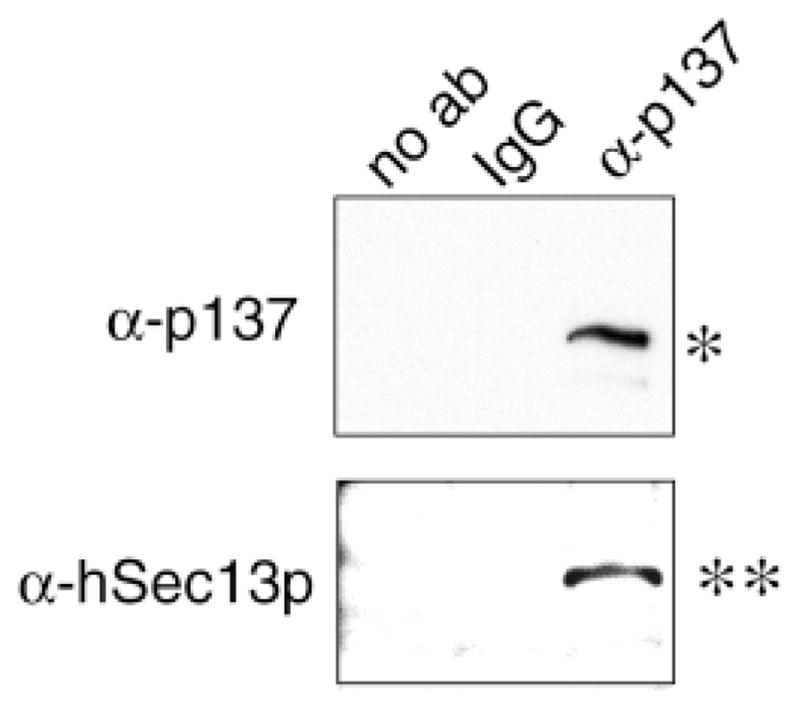
Anti-p137 co-immunoprecipitates Sec13p. Non-ionic detergent extracts prepared from a rat liver S100 fraction were immunoprecipitated with the rabbit anti-p137 (third lane), non-immune IgG (second lane) or no antibody (first lane), separated on SDS-PAGE (8 to 12% acrylamide) and transferred to Immobilon. The blot was probed with anti-p137 (top panel) and anti-human Sec13p (bottom panel). Single bands at the appropriate mobilities were detected only when specific antibody to p137 was used in the immunoprecipitation step.
Fig. 7.
p137 interacts with mammalian Sec13p and Sec23p but not yeast Sec13p and Sec23p in a yeast two-hybrid assay. (A–B) Two hybrid interactions among Sec31p, p137, Sec13p and Sec23p were detected by β-galactosidase assay. (C) Gene fragments of Sec31 and p137 are indicated in the diagrams and by the amino acid numbers included in parentheses. nd, not done.
p137 and human Sec13p co-localize in intracellular structures
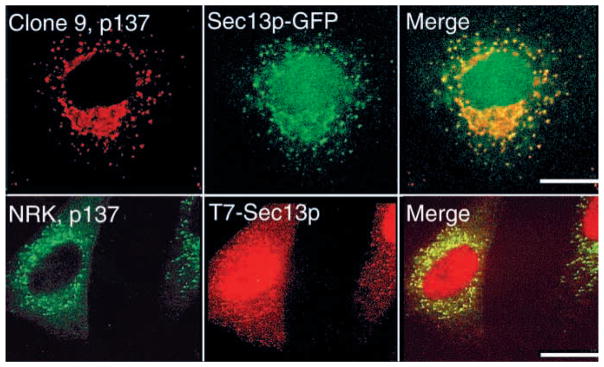
Having established a biochemical association between p137 and Sec13p, we were interested in comparing their subcellular localizations using double-label immunofluorescence. In both Clone 9 and NRK cells, antibodies to p137 labeled similar small punctate structures that were distributed throughout the cytoplasm but most concentrated in a juxtanuclear region (Fig. 8). Sec13p was localized to small cytoplasmic vesicles that exhibited considerable overlap with p137 immunoreactivity. Some exogenously expressed Sec13p was also found over the nucleus.
Fig. 8.
The vesicular compartment containing p137 colocalizes with transfected Sec13p. Clone 9 cells (top row) or NRK cells (bottom row) were transfected with Sec13p-GFP or Sec13p with a T7 tag, respectively, fixed, then incubated with anti-p137 alone (Clone 9) or together with anti-T7 antibody (NRK). Sec13p was localized to cytoplasmic vesicles in both cell lines; it was also found over the nucleus in some cells. p137 was distributed to small cytoplasmic vesicles, but not over the nucleus. Most of the vesicular labeling for p137 and Sec13p overlapped (merged images). Bars, 10 μm.
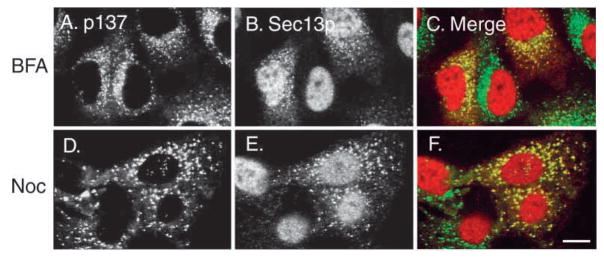
We next used a pharmacological approach to explore the intracellular distributions of p137 and Sec13p. The fungal metabolite brefeldin A (BFA) causes changes in the morphology of selected organelles in the secretory pathway as a consequence of its inhibition of COPI coatomer binding to membranes (Lippincott-Schwartz et al., 1991). When cells were treated with BFA, the Golgi membrane protein mannosidase II, whose normal localization is juxtanuclear, became diffusely distributed, consistent with its redistribution into the ER (data not shown). However, under these conditions, both the p137 protein and Sec13p remained together and associated with small punctate structures (Fig. 9A–C). These results are consistent with experiments showing that the COPII coat in yeast (Barlowe, 1994) and mammalian cells (Shaywitz et al., 1995) is insensitive to BFA and remains membrane-associated while the COPI coat redistributes to the cytosol (Orci et al., 1991). Fig. 9D–F also shows that the two proteins remained together after nocodazole treatment; however, the structures they were found in appeared larger than those in untreated cells (compare Fig. 9D–F with Fig. 8, NRK cell row) and more scattered throughout the cell. It is well-known that microtubule depolymerization leads to fragmentation and dispersal of the Golgi and ER-to-Golgi intermediate compartments, although the mechanism is still being debated (Lippincott-Schwartz et al., 1998; Shima et al., 1998). Nonetheless, the parallel responses of the two proteins under different experimental conditions suggested to us that p137 and Sec13p were co-localized in the same structure.
Fig. 9.
p137 and Sec13p remain associated during different experimental treatments. (A–C) Cells transfected with T7-Sec13p plasmid were treated with 10 μg/mL brefeldin A (BFA) for 1 hour at 37°C, then fixed and labeled with mouse anti-p137 antibody (A, p137) and anti-T7 antibody (B, Sec13p). This treatment redistributed the Golgi protein mannosidase II into the ER (not shown). In the presence of brefeldin A, p137 and Sec13p remain co-localized and associated with vesicles (C, merge). (D–F) Cell transfected with T7-Sec13p plasmid were treated with 33 μM nocodazole for 1 h, then fixed and labeled with the same antibodies as described in A. This treatment depolymerized microtubules, as demonstrated by staining with anti-tubulin antibody (not shown); however, p137 (D) and Sec13p (E) remained co-localized (F) but became more widely distributed in the cytoplasm than in control cells.
To provide more direct evidence that p137 localized to punctate structures derived from the ER, we used a temperature-sensitive viral protein, tsO45 VSVG (Bergmann, 1989). In cells expressing tsO45 VSVG at 40°C, newly synthesized protein is trapped in the ER. At 32°C, VSVG rapidly exits the ER and moves to the Golgi and then to the plasma membrane. When cells are incubated at 15°C, the protein preferentially accumulates in pre-Golgi intermediates that consist of vesicular elements localized at or near ER exit sites (Bannykh et al., 1996; Scales et al., 1997). We reproduced these findings. In cells expressing VSVG, the protein remained in the ER at 40°C (Fig. 10B). Incubation at 32°C led to a redistribution to the Golgi and plasma membrane (Fig. 10E). Importantly, p137 remained associated with punctate structures under both conditions (Fig. 10A and D). However, at 15°C, the distribution of p137 partially overlapped with that of VSVG (Fig. 10G–I). Notably, the structures containing both p137 and VSVG at 15°C appeared to be larger than those observed to contain p137 under physiologic conditions. This observation is consistent with the finding that cargo may accumulate in an enlarged intermediate compartment containing aggregates of COPII coated vesicles (Aridor et al., 1998). Nonetheless, our results suggest that p137 is associated with ER-derived vesicles mediating anterograde transport between the ER and Golgi.
Fig. 10.
p137 colocalizes with a marker of ER-derived transport vesicles. NRK cells were transfected at the restrictive temperature (40°C) with a plasmid encoding tsO45 VSVG. The cells were kept at 40°C (top panel) or shifted to either 32°C (middle panel) or 15°C (bottom panel) for 1 hour. The cells were then fixed and labeled with mouse anti-p137 (A,D,G) and rabbit anti-VSVG (B,E,H). As shown in the merged images (C,F,I) at 15°C but not at 40°C or 32°C, p137 overlaps with VSVG in pre-Golgi structures. In the merged image: green, VSVG; red, p137; yellow, areas of colocalization. Arrows in G and H indicate an area of colocalization that is also shown in the magnified inset (I).
DISCUSSION
In this study, we have identified a novel membrane-associated mammalian protein, p137. Based on three independent lines of evidence, we propose that the ubiquitously expressed p137 is a component of the mammalian COPII coat and is the putative orthologue (i.e. functional homologue) of Sec31p. p137 has sequence and structural homology to the N-terminal domain of yeast COPII protein Sec31p; it interacts biochemically with mammalian Sec13p, the 33 kDa COPII protein that forms a functional complex with Sec31p in yeast; and it localizes morphologically with human Sec13p at ER exit sites in mammalian cells.
Similarities between p137 and yeast Sec31ps
The p137 and yeast Sec31p sequences share common features in their domain organization. Their N-terminal regions are most homologous, both containing seven WD domains. This protein motif consists of tryptophan and aspartic acid residues separated by intervals of approximately 40 amino acid residues (Komachi and Johnson, 1997). Variable numbers of repeats form a compact β-propeller structure on which other proteins bind to form macromolecular complexes in cells (Smith et al., 1999). In yeast and mammalian cells, Sec13p, the cytosolic binding partner of Sec31p, consists entirely of WD domains (Garcia-Higuera et al., 1999) and interacts with the WD region of Sec31p (Shaywitz et al., 1997). However, compared to p137, both yeast Sec31p’s contain additional amino acids between the third and fourth WD domain and at the beginning of the fifth WD domain. These additional residues may account for the lack of interaction observed between the N-terminal regions of mammalian p137 and yeast Sec13p in the yeast two-hybrid assay.
Both yeast Sec31p and mammalian p137 also contain large proline-rich regions at approximately the same locations in their sequences. Yeast two-hybrid analysis and in vitro binding assays have shown that yeast Sec23p and Sec24p interact with Sec31p in this region, but the interacting sites have not been defined (Shaywitz et al., 1997). Other proteins not yet identified may also modulate COPII vesicle formation or fate through binding to motifs in this region. A protein with 98% identity to p137 has recently been cloned from a human brain library (KIAA0905 protein: accession #BAA74928). Interestingly, the clone lacks two amino acid sequences in the proline-rich region (aa 834-865 and aa 1018-1031). Unpublished work from our laboratory indicates that these differences represent p137 splice variants. We speculate that the different isoforms may play interesting roles in the function of p137, perhaps by modulating binding proteins.
Both yeast Sec31p and mammalian p137 also contain multiple potential phosphorylation sites in their C-terminal regions. Furthermore, both proteins are phosphorylated in vivo (Buccione et al., 1996; Fabbri et al., 1994). In yeast, this modification is on serine residues and seems to play a role in COPII coat formation, since phosphatase-treated complexes are biochemically stable but less efficient than their phosphorylated counterparts in in vitro assembly assays (Salama et al., 1997). In mammalian cells, two pharmacological modulators of protein kinase C, calphostin C and phorbol myristyl acetate, have been reported to reduce or increase, respectively, the number of ER exit sites (Fabbri et al., 1994). The findings support the possibility that phosphorylation also plays a regulatory role in mammalian COPII assembly. Further work is needed to determine if mammalian p137 is the target of these agents.
The p137/Sec13p complex
Similar to the interaction of Sec31p and Sec13p in yeast, p137 forms a complex with mammalian Sec13p. In our study, partial purification of rat liver p137 through multiple chromatographic steps yielded a macromolecular complex that also contained mammalian Sec13p. The estimated mass of the complex, 500 to 1000 kDa, is similar to that of yeast Sec13/31p, which has been reported as ~700 kDa by size exclusion chromatography (Salama et al., 1997). In vitro assembly assays have clearly shown that this complex is a functional unit of yeast COPII coat assembly (Salama et al., 1993, 1997), but its stoichiometry is presently unclear. Using glycerol gradient centrifugation, Salama et al. (1993) reported a size for Sec13/31p of only 166 kDa, which is consistent with that of a globular dimer but not with the much larger size estimated by gel filtration. As noted by Salama et al. (1993), these differences in the apparent molecular mass of the p13/31p complex suggest that it is elongated. Previous studies have reported that Sec31p interacts with Sec13p through not only its WD domain (Shaywitz et al., 1997) but also a C-terminal region surrounding residue aa 1239, whose mutation in the sec31-1 strain yields a protein with lowered affinity for Sec13p (Salama et al., 1997). To reconcile these findings, it will important to visualize the purified complex using high-resolution microscopy. Such an approach should yield information on the stoichiometry and shape of the p13/31p complex and the mode by which the two β-propeller proteins interact.
In this study, we also found endogenous p137 associated with human Sec13p in intact cells. Biochemically they could be co-immunoprecipitated and morphologically they exhibited extensive overlap in the cytoplasm of several different cell types. Their intracellular distribution is consistent with that of the ER exit sites or export complexes described recently by others (Bednarek et al., 1995; Bannykh et al., 1996). These structures have been shown to contain other COPII components (Barlowe et al., 1994), and membrane cargo (Aridor et al., 1998), but not resident ER molecules. They are postulated to be a dynamic intermediate that mediates anterograde transport between the ER and early Golgi (Allan and Balch, 1999; Lippincott-Schwartz et al., 1998).
What is the function of Sec31p?
Matsuoka et al. (1998) have recently reported that synthetic liposomes containing acidic phospholipids support the (albeit, inefficient) formation of recognizable coated vesicles using only three yeast COPII components. In order-of-addition assays, Sec13/31p acts last and is required for formation of a coated vesicle. However, Sec13/31p also seems to stabilize the membrane association of Sec23/24p in vitro, even though the latter complex is able to associate with putative cargo receptors and targeting machinery in the absence of Sec13/31p (but in the presence of GTP-Sar1p; Kuehn et al., 1998; Aridor et al., 1998; Springer and Schekman, 1998). Furthermore, Sec31p has been shown to interact genetically and biochemically with Sec16p, a peripheral protein that is tightly associated with the ER membrane and serves as a platform for efficient coat formation (Shaywitz et al., 1997). Thus, Sec31p has many binding partners within the COPII vesicle itself and could serve as a regulator of budding.
Could Sec31p have additional roles? Although this issue has not yet been explored in yeast or mammalian cells, studies indicate that several homologues of Sec13p seem to regulate the post-Golgi transport of amino acid permeases in yeast independently of Sec31p (Roberg et al., 1998). Thus, there must be cytoplasmic pools of such homologues that are not complexed to Sec31p. Is this also true for Sec31p? The existence of p137 isoforms hints at additional roles; however, further work will be needed to identify other binding partners.
Acknowledgments
The authors thank Jakub Svoboda for performing the two hybrid analysis between p137 and Sec23p, Jonathan Pevsner for the sequence comparisons, and Lelita Braiterman for critical reading of the manuscript. A.H. was supported by NIGMS R0129185, E.R.K. by NIGMS MSTP Award T 32 07309 and F.G. by a Yale Liver Center Pilot Project Award, NIDDK RO154021 and a VA Merit Award.
References
- Allan BB, Balch WE. Protein sorting by directed maturation of Golgi compartments. Science. 1999;285:63–66. doi: 10.1126/science.285.5424.63. [DOI] [PubMed] [Google Scholar]
- Aridor M, Weissman J, Sergei B, Nuoffer C, Balch W. Cargo selection by the COPII budding machinery during export from the ER. J Cell Biol. 1998;141:875–893. doi: 10.1083/jcb.141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aridor M, Balch WE. Integration of endoplasmic reticulum signaling in health and disease. Nature Med. 1999;5:745–751. doi: 10.1038/10466. [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Rowe T, Balch WE. The organization of endoplasmic reticulum export complexes. J Cell Biol. 1996;135:19–35. doi: 10.1083/jcb.135.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazazola M, Amherdt M, Schekman R. COPII: a membrane coat formed by sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Barlowe C. COPII and selective export from the endoplasmic reticulum. Biochim Biophys Acta. 1998;1404:67–76. doi: 10.1016/s0167-4889(98)00047-0. [DOI] [PubMed] [Google Scholar]
- Bednarek SY, Ravazzola M, Hosobuchi M, Amherdt M, Perrelet A, Schekman R, Orci L. COPI- and COPII-coated vesicles bud directly from the endoplasmic reticulum in yeast. Cell. 1995;83:1183–1196. doi: 10.1016/0092-8674(95)90144-2. [DOI] [PubMed] [Google Scholar]
- Bergmann JE. Using temperature-sensitive mutants of VSV to study membrane protein biogenesis. Meth Cell Biol. 1989;35:85–110. doi: 10.1016/s0091-679x(08)61168-1. [DOI] [PubMed] [Google Scholar]
- Buccione R, Bannykh S, Santone I, Baldassarre M, Facchiam F, Bozzi Y, Di Tullio G, Mironov G, Luini A, De Matteis MA. Regulation of constitutive exocytic transport bymembrane receptors: a biochemical and morphological study. J Biol Chem. 1996;271:3523–3533. doi: 10.1074/jbc.271.7.3523. [DOI] [PubMed] [Google Scholar]
- Burnette WN. ’Western Blotting’: Electrophoretic transfer of protein from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981;112:195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Fabbri M, Bannykh S, Balch WE. Export of protein from the endoplasmic reticulum is regulated by a diacylglycerol/phorbol ester binding protein. J Biol Chem. 1994;269:26848–26857. [PubMed] [Google Scholar]
- Fields S, Ok-kyu S. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Garcia-Higuera I, Fenoglio J, Li Y, Lewis C, Panchenko MP, Reiner O, Smith TF, Neer EJ. Folding of proteins with WD-repeats: comparison of six members of the WD-repeat superfamily to the G Protein β subunit. Biochemistry. 1999;35:13985–13994. doi: 10.1021/bi9612879. [DOI] [PubMed] [Google Scholar]
- Ihrke G, Neufeld EB, Meads T, Shanks MR, Cassio D, Laurent M, Schroer TA, Pagano RE, Hubbard AL. WIF-B cells: an in vitro model of studies of hepatocyte polarity. J Cell Biol. 1993;123:1761–1775. doi: 10.1083/jcb.123.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komachi K, Johnson AD. Residues in the WD repeats of Tup1 required for interaction with alpha2. Mol Cell Biol. 1997;17:6023–6028. doi: 10.1128/mcb.17.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn MJ, Herrmann JM, Schekman R. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 1998;391:187–190. doi: 10.1038/34438. [DOI] [PubMed] [Google Scholar]
- Kuge O, Dascher C, Orci L, Rowe T, Amherdt M, Plutner H, Ravazzola M, Tanigawa G, Rothman JE, Balch WE. Sar1 promotes vesicle budding from the endoplasmic reticulum but not Golgi compartments. J Cell Biol. 1994;125:51–65. doi: 10.1083/jcb.125.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Yuan L, Tipper C, Amherdt M, Orci L, Klausner RD. Brefeldin A’s effects on endosomes, lysosomes, and TGn suggest a general mechanism for regulating organelle structure and membrane traffick. Cell. 1991;67:601–616. doi: 10.1016/0092-8674(91)90534-6. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Cole NB, Donaldson JG. Building a secretory apparatus: role of ARF1/COPI in Golgi biogenesis and maintenance. Histochem Cell Biol. 1998;109:449–462. doi: 10.1007/s004180050247. [DOI] [PubMed] [Google Scholar]
- Lowe M, Kreis TE. Regulation of membrane traffic in animal cells by COPI. Biochim Biophys Acta. 1998;1404:53–66. doi: 10.1016/s0167-4889(98)00046-9. [DOI] [PubMed] [Google Scholar]
- Luo W, Lin SH. Generation of moderate amounts of polyclonal antibodies in mice. Biotechniques. 1997;23:630–632. doi: 10.2144/97234bm16. [DOI] [PubMed] [Google Scholar]
- Marsh M, McMahon HT. The structural era of endocytosis. Science. 1999;285:215–220. doi: 10.1126/science.285.5425.215. [DOI] [PubMed] [Google Scholar]
- Matovcik LM, Karapetian O, Czernik A, Marino C, Kinder B, Gorelick F. Antibodies to an epitope on synapsin I recognize an endosome-associated protein in non-neural tissues. Eur J Cell Biol. 1994;65:327–340. [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Orci L, Tagaya M, Amherdt M, Perreiet A, Donaldson JG, Lippincott-Schwartz J, Klausner RD, Rothman JE. Brefeldin A, a drug that blocks secretion, prevents the assembly of non-clathrin-coated buds on Golgi cisternae. Cell. 1991;64:1183–1195. doi: 10.1016/0092-8674(91)90273-2. [DOI] [PubMed] [Google Scholar]
- Paccaud JP, Reith W, Carpentier JL, Ravazzola M, Amherdt M, Schekman R, Orci L. Cloning and functional characterization of mammalian homologues of the COPII component Sec23. Mol Biol Cell. 1996;7:1535–1546. doi: 10.1091/mbc.7.10.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Salama NR, Schekman R, Kaiser CA. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum in vitro. J Cell Biol. 1993;120:865–875. doi: 10.1083/jcb.120.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg KJ, Bickel S, Rowley N, Kaiser CA. Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics. 1998;147:1569–1584. doi: 10.1093/genetics/147.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama N, Yeung T, Schekman R. The Sec13p complex and reconstitution of vesicle budding from the ER with purified cytosolic proteins. EMBO J. 1993;2:4073–4082. doi: 10.1002/j.1460-2075.1993.tb06091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salama NR, Chuang JS, Schekman RW. Sec31 encodes an essential component of the COPII coat required for transport vesicle budding from the endoplasmic reticulum. Mol Biol Cell. 1997;8:205–217. doi: 10.1091/mbc.8.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scales SJ, Pepperkok R, Kreis TE. Visualization of ER-to-golgi transport in living cells reveals sequential mode of action for COPII and COPI. Cell. 1997;90:1137–1148. doi: 10.1016/s0092-8674(00)80379-7. [DOI] [PubMed] [Google Scholar]
- Shanks MR, Cassio D, Lecoq L, Hubbard A. An improved polarized rat hepatoma hybrid cell line: Generation and comparison with its hepatoma relatives and hepatocytes in vivo. J Cell Sci. 1994;107:813–825. doi: 10.1242/jcs.107.4.813. [DOI] [PubMed] [Google Scholar]
- Shaywitz DA, Orci L, Ravazzola M, Swaroop A, Kaiser CA. Human SEC13Rp functions in yeast and is located on transport vesicles budding from the endoplasmic reticulum. J Cell Biol. 1995;128:769–777. doi: 10.1083/jcb.128.5.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–25416. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- Shima D, Cabrera-Poch N, Pepperkok R, Warren G. An ordered inheritance strategy for the Golgi apparatus: visualization of mitotic disassembly reveals a role for the mitotic spindle. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat, a common architeture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- Springer S, Schekman R. Nucleation of COPII vesicular coat complex by endoplasmic reticulum to Golgi vesicle SNAREs. Science. 1998;281:698–700. doi: 10.1126/science.281.5377.698. [DOI] [PubMed] [Google Scholar]
- Swaroop A, Yang-Feng TL, Liu W, Gieser L, Barrow LL, Chen KC, Agarwal N, Meisler MH, Smith DI. Molecular characterization of a novel humangene, sec13R, related to the yeast secretory pathway gene, sec13, and mapping to a conservative linkage group on human chromosome 3p24-p25 and mouse chromosome 6. Hum Mol Genet. 1994;3:1281–1286. doi: 10.1093/hmg/3.8.1281. [DOI] [PubMed] [Google Scholar]
- Weisz OA, Machamer CE. Meth Cell Biol. A. Vol. 43. Baltimore: Academic Press, Inc; 1994. Use of recombinant vaccinia virus vectors for cell biology; pp. 137–159. [DOI] [PMC free article] [PubMed] [Google Scholar]