Abstract
A glutamic acid residue in the first transmembrane domain of the human adenosine A2A receptor was mutated to glutamine. Radioligand binding studies on COS-7 cell membranes expressing either the wild-type or the mutant receptor revealed that the affinity of the prototypic agonist CGS21680 (2-[4-[(2-carboxyethyl)phenyl]ethylamino]-5′-N-ethylcarboxamidoadenosine) for the mutant receptor was 15-fold lower than for the wild-type receptor. This was confirmed in functional studies with intact cells. The EC50 values of CGS21680 for the stimulation of cAMP production differed in a similar way. Antagonists of various chemical structure were equally effective on both mutant and wild-type receptors, thus the mutation selectively diminishes agonist affinity. We propose an indirect perturbation of the binding site, perhaps through a proton transfer mechanism as suggested by molecular modelling.
Keywords: Adenosine A2A receptor, human, Mutation, Glutamic acid, CGS21680, XAC, Amiloride
1. Introduction
Adenosine receptors belong to the superfamily of G-protein-coupled receptors. Four subtypes, A1, A2A, A2B and A3, have been identified both on pharmacological and molecular biological grounds. Adenosine A2A receptors appear to be involved in important physiological processes, such as cardiovascular homeostasis, and brain function related to dopaminergic pathways (for a review see Jacobson et al., 1992).
On the basis of the primary sequence of the adenosine A2A receptor we constructed a three-dimensional model for this receptor, using the atomic coordinates of bacteriorhodopsin as a template (IJzerman et al., 1994). From this model it was suggested that a glutamic acid residue present in the first transmembrane domain and highly specific for adenosine receptors could be involved in ligand binding and activation, being more relevant for agonists than for antagonists.
In the present study we mutated this negatively charged glutamic residue in the human adenosine A2A receptor (Furlong et al., 1992) for an (uncharged) glutamine (E13Q), and analysed its effects on ligand binding and receptor activation.
2. Materials and methods
2.1. Materials
Human adenosine A2A receptor cDNA (expression vector pSVL-A2a) was kindly provided by Dr. Marlene Jacobson (Merck Research Labs, West Point, PA, USA). Taq polymerase for the polymerase chain reaction (PCR) was purchased from Perkin Elmer (Emeryville, CA, USA). All enzymes used in this study were obtained from New England Biolabs (Beverly, MA, USA). Oligonucleotides used were synthesized by Bioserve Biotechnologies (Laurel, MD, USA). The following compounds were gifts that are gratefully acknowledged: CGS21680 (2-[4-[(2-carboxyethyl)phenyl]ethylamino]-5′-N-ethylcarboxamido-adenosine) and CGS15943 (9-chloro-2-(furyl)[l,2,4]tri-azolo[l,5-c]quinazolin-5-amine) from Ciba Geigy (Summit, NJ, USA); amiloride from Merck, Sharp & Dohme (Haarlem, Netherlands). XAC (8-[4-[[[[(2-aminoethyl)amino]carbonyl]methyl]oxy]phenyl]-1,3-dipropylxanthine) was obtained from RBI (Natick, MA, USA). [3H]CGS21680 (41 Ci/mmol) and [3H]XAC (109 Ci/mmol; custom synthesis) were from DuPont NEN (Dordrecht, Netherlands). All other reagents which were obtained from standard commercial sources were of analytical grade.
2.2. Methods
2.2.1. Plasmid construction, site-directed mutagenesis and transient expression of wild-type and mutant receptors in COS-7 cells
This protocol has been described before (Kim et ah. 1995), except that the receptor proteins were untagged.
2.2.2. Membrane preparation and radioligand binding assay
Cells were scraped into ice-cold lysis buffer (4 ml of 50 mM Tris, pH 6.8 at room temperature, containing 10 mM MgCl2). Harvested cells were homogenized using a Polytron homogenizer and then spun at 27 000 × g for 15 min. The pellet (plasma membranes) was resuspended in the same buffer, incubated with 2 U/ml adenosine deaminase at 37°C for 30 min, quickly frozen and kept at −80°C until use.
Competition binding experiments were performed with both a tritiated agonist ([3H]CGS21680) and a tritiated antagonist ([3H]XAC) as radioligands. The [3H]CGS21680 binding assay was done as follows. Each test tube contained 50 μl of radioligand (final concentration approximately 5 nM), 50 μl of competitor/buffer (50 mM Tris, pH 6.8 at 25°C, 10 mM MgCl2) and 100 μl membrane suspension (20 μg of protein). The mixtures were incubated at 25°C for 90 min, filtered and washed three times with 3 ml of ice-cold buffer each using a Brandel cell harvester. The [3H]XAC binding assay was performed as follows. Each test tube contained 50 μl of radioligand (final concentration 1.5–2.0 nM), 50 μl of competitor or buffer (50 mM Tris, pH 7.4 at 25°C), 50 μl of GTP (1 mM) or buffer, 50 μl of NaCl (1 M) or buffer and 100 μl of membrane suspension (20 μg of protein). The mixtures were incubated at 25°C for 90 min and filtered over Whatman GF/B filters using a Brandel cell harvester. The filters were presoaked for 2 h in 0.3% polyethyleneimine, and washed three times with 3 ml of ice-cold buffer to which 0.01% polyethyleneimine was added. All filters were dried for 45 min at 70°C and subsequently counted in an LKB Rackbeta 1214 liquid scintillation spectrometer after the addition of 3 ml Emulsifier Safe counting liquid (Packard).
Saturation binding experiments were done with [3H]XAC only, according to a similar protocol as described above, with [3H]XAC concentrations ranging from 0.3 to 50 nM.
2.2.3. cAMP generation and assay
cAMP levels were determined by measuring the conversion of [3H]ATP to cyclic [3H]AMP. One day after transfection, cells were transferred from 100-mm dishes into 6-well dishes (about 3 × 105 cells/well) and incubated with culture media containing 2 μCi/ml [3H]adenine. After 24 h the cultures were washed and incubated with 1 ml/well Hank’s balanced salt solution containing 0.1 mM rolipram and 2 U/ml adenosine deaminase for 15 min at 37°C. The cells were incubated with different concentrations of the agonist CGS21680 (in culture media) for 30 min at 37°C. The reaction was terminated by aspiration of the media and addition of 1 ml of ice-cold 5% trichloroacetic acid containing 1 mM ATP and 1 mM cAMP. After 30 min incubation at 4°C, cell lysates were eluted through sequential chromatography on Dowex and alumina columns. Cyclic AMP formation was expressed as fold-stimulation of conversion of [3H]ATP into cyclic [3H]AMP (Weiss et al., 1985).
2.2.4. Data analysis
All saturation, displacement and dose-response curves were analysed with the PRISM software package (Graph-Pad Software, San Diego, CA, USA).
3. Results
3.1. Radioligand binding studies
In a first series of experiments membranes from both wild-type and mutant receptors were incubated with [3H]CGS21680. At 5 nM of the radioligand no specific binding was detected on cell membranes with the E13Q mutant. Ample receptor binding (approximately 50% of total binding at 5 nM of the radioligand) was present on the cell membranes with the wild-type receptor. Similar experiments with the antagonist [3H]XAC as the radioligand (2 nM) revealed almost identical receptor binding for both the wild-type and the mutant receptor (approximately 70% of total binding). These findings prompted us to perform full saturation binding experiments, yielding similar KD and Bmax values for [3H]XAC on both receptors (wild-type: KD = 25 ± 3 nM, Bmax = 6.4 ±1.1 pmol/mg protein; E13Q: KD = 30 ± 4 nM, Bmax = 8.4 ± 2.0 pmol/mg protein). These data led us to use the radiolabeled antagonist [3H]XAC for further experiments.
In Table 1 the affinities of adenosine receptor ligands for the wild-type and mutated adenosine A2A receptor are summarized. The agonist CGS21680 was significantly less (15-fold) potent on the mutant receptor both in the absence and presence of GTP. The addition of GTP as such shifted the affinity of CGS21680 to higher IC50 values (approximately 2-fold) both on the wild-type and the mutant receptor. A further inclusion of NaCl in the assay decreased affinity even more although to a different extent for the two receptors, resulting in a smaller shift (3.6) between the wild-type and the mutant receptor. Neither GTP nor NaCl influenced [3H]XAC binding significantly.
Table 1.
Approximated IC50 values (μM) of agonists and antagonists obtained in radioligand binding studies with [3H]XAC on the wild-type (WT) and E13Q mutant of the cloned human adenosine A2A receptor (n = 2, with individual IC50 values differing less than 30%: CGS21680 alone: n = 3, IC50 ± S.E.M.)
| Compounds | WT | E13Q | Shifta |
|---|---|---|---|
| Agonist | |||
| CGS21680 | 0.85 ± 0.22 | 13.0 ± 4.3 | 15 |
| CGS21680 +GTPb | 1.65 | 24.8 | 15 |
| CGS21680 +GTPb+NaClc | 10.5 | 37.9 | 3.6 |
| Antagonists | |||
| XAC | 0.018 | 0.042 | 2.3 |
| CGS 15943 | 0.0022 | 0.0027 | 1.2 |
| Amiloride | 52 | 46 | 0.9 |
IC50. E13Q/IC50. WT.
GTP concentration (final): 1 mM.
NaCl concentration (final): 1 M.
The affinities of the three antagonists studied, XAC, CGS15943 and amiloride, did not differ between the two receptors. CGS 15943 was most potent, in the low nanomolar range, whereas amiloride had micromolar affinity. The addition of both GTP and NaCl (only studied for XAC) did not influence the affinity of this antagonist (data not shown).
3.2. Cyclic AMP production
CGS21680 dose-dependently increased the cAMP production in the cells transfected with either the wild-type or the E13Q mutant cDNA. In both cases the maximal stimulation achieved was 4–5-fold over control values. However, the EC50 values of CGS21680 were significantly different, i.e. 2.1 ± 0.8 nM (n = 3) for the wild-type and 17 nM (n = 2.9–25 nM) for the mutant receptor, respectively.
4. Discussion
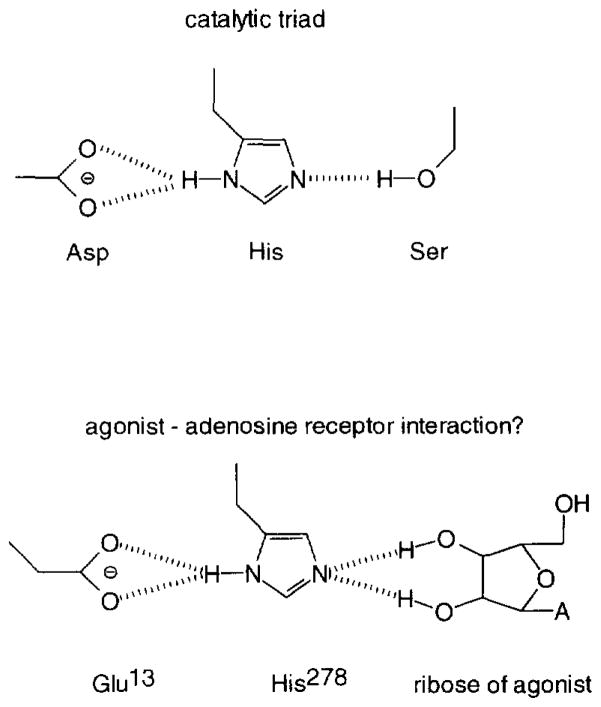
In the present study the interaction between the human adenosine A2A receptor and its ligands was studied by mutational analysis. In a previous molecular modelling study (IJzerman et al., 1994) it was suggested that a glutamic acid residue (E13) in transmembrane α-helix I was involved in the interaction of the receptor with agonists rather than antagonists. It was speculated that this glutamic acid residue could play a role in some form of proton transfer based on structural analogies with the catalytic triad in the enzyme family of serine proteases (Fig. 1).
Fig. 1.
Upper panel: Catalytic triad as found in serine proteases. Proton transfer is possible from a serine, via a histidine, to an aspartic acid residue along the dashed lines. Lower panel: Proposed mechanism of activation of the adenosine A2A receptor by a proton transfer from (the ribose moiety of) an agonist ligand (A, adenine or modified adenine base), via histidine 278 in helix VII, to glutamic acid 13 in helix I along the dashed lines.
The mutation of glutamic acid to glutamine (E13Q) eliminates the negative charge of the residue but it largely preserves other characteristics such as size and hydrogen bonding capacity. The results obtained with the wild-type and the mutant receptors in both the radioligand binding (Table 1) and the functional (cAMP) studies agree to a great extent. The prototypic agonist CGS21680 had diminished affinity for the mutant receptor, whereas the antagonists considered (XAC, CGS 15943 and amiloride) did not differentiate between wild-type and mutant receptors.
The 15-fold lower affinity of CGS21680 observed in the binding studies could explain the absence of specific binding when the tritiated form of CGS21680 was used as the radioligand on the mutant receptor. The receptor affinities of CGS21680 reported in Table 1 reflect the concomitant presence of high- and low-affinity states of the receptor due to the use of an antagonist radioligand. Due to the limited amounts of receptor protein available we decided not to perform detailed binding experiments to explore this well-known phenomenon further. The addition of GTP (1 mM) caused a small, only 2-fold, reduction in affinity due to a presumed shift in the ratio of high- and low-affinity states of the receptor. Such a small effect of GTP was anticipated, since a similar small shift had been observed previously with another antagonist radioligand, [3H]KF17837S (8-(3,4-dimethoxystyryl)-1,3-dipropyl-7-methylxanthine), binding to rat striatal membranes (Nonaka et al., 1994). The GTP sensitivity was similar for the wild-type and the mutant receptor. This finding suggests that both receptors recognize the G-protein equally well. A further addition of NaCl (1 M), known to remarkably reduce agonist affinity even further (Linden et al., 1988), was more influential on the wild-type than on the mutant receptor. It may therefore be that the negatively charged glutamic acid, as present in the wild-type receptor, is neutralized to some extent by the positive sodium ions, thereby regulating agonist affinity. The neutral glutamine would not favour a strong interaction with the sodium ion. It has been documented for various other G-protein-coupled receptors that another, highly conserved, acidic residue (mostly an aspartic acid) in the second transmembrane domain is also critical for allosteric sodium ion regulation (see overview in Ceresa and Limbird, 1994).
Antagonist binding was hardly influenced by the mutation. Both XAC (in saturation as well as displacement studies) and CGS15943 were and remained bound in the nanomolar range. The diuretic amiloride has been shown to be a modestly potent adenosine receptor antagonist (Garritsen et al., 1991). Its chemical structure bears a positively charged guanidino group, unlike most other adenosine receptor antagonists. It is not likely, however, that this positive charge is close to the glutamic acid residue, since amiloride’s affinity for the mutant was virtually unchanged.
Functional studies on the intact cells revealed that CGS21680 was capable of stimulating both the wild-type and the mutant receptor, yielding an almost equal extent of cAMP production. However, the agonist had again lower potency on the mutant receptor, in agreement with the radioligand binding studies. Apparently, E13 is involved in agonist recognition, but it does not seem to be essential for the coupling of receptor and G-protein. It remains to be seen whether this residue is an integral part of the ligand binding site or that its influence is of a more indirect nature. The ideas presented in the modelling study (IJzerman et al., 1994) were based on the experimentally determined structure of bacteriorhodopsin. It has to be kept in mind that there is virtually no sequence homology between this bacterial proton pump and G-protein-coupled receptors. Its mammalian homolog rhodopsin, however, is a G-protein-coupled receptor, and its overall structure has been elucidated too (Schertler et al., 1993). Its appearance is that of bacteriorhodopsin, although the helices might be more tilted relative to each other. The spatial characteristics of the ligand binding domain, being more or less in the middle of the receptor cavity, are probably not very different between rhodopsin and bacteriorhodopsin (Baldwin, 1993). Mutation studies based on the above model were subsequently performed (Kim et al., 1995) and proved largely confirmatory, suggesting that the binding site (defined as all amino acids within 4Å of the agonist) does not include the glutamic acid residue. A histidine residue in helix VII (H278 in the human adenosine A2A receptor) appears crucial for the binding of the agonist molecule, probably by virtue of hydrogen bonding to the 2′- and 3′-hydroxy groups present in the ribose moiety. Since helices I and VII are adjacent, residues E13 and H278 are in close proximity. In that case, a proton transfer could be a communication mechanism between the binding site histidine and E13. This hypothesis resembles the catalytic triad in serine proteases, where a proton transfer is possible between a serine (the ribose hydroxy groups here), a histidine and an aspartic (here glutamic) acid residue (Fig. 1).
In conclusion, the combination of molecular modelling and molecular biological techniques is a powerful means to study ligand-receptor interactions. It identified a glutamic acid residue in helix I of the adenosine receptor as critical in the recognition of agonists. It is our belief that this strategy, in which such various techniques are integrated, is pivotal for a better understanding of drug action.
Acknowledgments
The authors wish to thank Dr. Scott Rivkees (Indiana University, Indianapolis, IN, USA) for helpful discussions.
References
- Baldwin JM. The probable arrangement of the helices in G protein-coupled receptors. EMBO J. 1993;12:1693. doi: 10.1002/j.1460-2075.1993.tb05814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceresa BP, Limbird LE. Mutation of an aspartate residue highly conserved among G-protein-coupled receptors results in nonreciprocal disruption of α2-adrenergic receptor-G-protein interactions. J Biol Chem. 1994;269:29557. [PubMed] [Google Scholar]
- Furlong TJ, Pierce KD, Selbie LA, Shine J. Molecular characterization of a human brain adenosine A2 receptor. Mol Brain Res. 1992;15:62. doi: 10.1016/0169-328x(92)90152-2. [DOI] [PubMed] [Google Scholar]
- Garritsen A, IJzerman AP, Tulp MTM, Cragoe EJ, Jr, Soudijn W. Receptor binding profiles of amiloride analogues provide no evidence for a link between receptors and the Na+/H+ exchanger, but indicate a common structure on receptor proteins. J Recept, Res. 1991;11:891. doi: 10.3109/10799899109064686. [DOI] [PubMed] [Google Scholar]
- IJzerman AP, Van der Wenden EM, Van Galen PJM, Jacobson KA. Molecular modeling of adenosine receptors. II. The ligand binding site on the rat A2a receptor. Eur J Pharmacol Mol Pharmacol, Sect. 1994;268:95. doi: 10.1016/0922-4106(94)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson KA, Van Galen PJM, Williams M. Adenosine receptors: pharmacology, structure-activity relationships, and therapeutic potential. J Med Chem. 1992;35:407. doi: 10.1021/jm00081a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wess J, Van Rhee AM, Schöneberg T, Jacobson KA. Site-directed mutagenesis identifies residues involved in ligand recognition in the human A2a adenosine receptor. J Biol Chem. 1995;270:13987. doi: 10.1074/jbc.270.23.13987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden J, Patel A, Earl CQ, Craig RH, Daluge SM. 125I-labeled 8-phenylxanlhine derivatives: antagonist radioligands for adenosine A1 receptors. J Med Chem. 1988;31:745. doi: 10.1021/jm00399a010. [DOI] [PubMed] [Google Scholar]
- Nonaka H, Mori A, Ichimura M, Shindou K, Yanagawa T, Shimada J, Kase H. Binding of [3H]KFI7837S, a selective adenosine A2 receptor antagonist, to rat brain membranes. Mol Pharmacol. 1994;46:817. [PubMed] [Google Scholar]
- Schertler GFX, Villa C, Henderson R. Projection structure of rhodopsin. Nature (London) 1993;362:770. doi: 10.1038/362770a0. [DOI] [PubMed] [Google Scholar]
- Weiss S, Sebben M, Garcia-Sainz JA, Bockaert J. D2-dopamine receptor-mediated inhibition of cyclic AMP formation in striatal neurons in primary culture. Mol Pharmacol. 1985;27:595. [PubMed] [Google Scholar]