Abstract
Rationale
It is now recognized that macrophages residing within developing and adult tissues are derived from diverse progenitors including those of embryonic origin. Although the functions of macrophages in adult organisms are well studied, the functions of macrophages during organ development remain largely undefined. Moreover, it is unclear whether distinct macrophage lineages have differing functions.
Objective
To address these issues, we investigated the functions of macrophage subsets resident within the developing heart, an organ replete with embryonic-derived macrophages.
Methods and Results
Using a combination of flow cytometry, immunostaining, and genetic lineage tracing, we demonstrate that the developing heart contains a complex array of embryonic macrophage subsets that can be divided into chemokine (C-C motif) receptor 2− and chemokine (C-C motif) receptor 2+ macrophages derived from primitive yolk sac, recombination activating gene 1+ lymphomyeloid, and Fms-like tyrosine kinase 3+ fetal monocyte lineages. Functionally, yolk sac–derived chemokine (C-C motif) receptor 2− macrophages are instrumental in coronary development where they are required for remodeling of the primitive coronary plexus. Mechanistically, chemokine (C-C motif) receptor 2− macrophages are recruited to coronary blood vessels at the onset of coronary perfusion where they mediate coronary plexus remodeling through selective expansion of perfused vasculature. We further demonstrate that insulin like growth factor signaling may mediate the proangiogenic properties of embryonic-derived macrophages.
Conclusions
Together, these findings demonstrate that the embryonic heart contains distinct lineages of embryonic macrophages with unique functions and reveal a novel mechanism that governs coronary development.
Keywords: flow cytometry, fms-like tyrosine kinase 3, macrophages, monocytes, yolk sac
It is now recognized that tissue macrophages comprised a heterogeneous group of cells derived from distinct developmental lineages including monocyte-derived and embryonic-derived subsets.1,2 Monocyte-derived macrophages originate from definitive hematopoietic progenitors and are recruited to tissues through established chemokine signaling pathways.3,4 In contrast, embryonic-derived macrophages reside within tissues independent of monocyte input and originate from several potential lineages including primitive yolk sac, recombination activating gene 1 (Rag1)+ lymphomyeloid, and fetal liver monocyte progenitors.5–10
Previous studies have established that the earliest macrophages are derived from a bipotent erythromyeloid progenitor located within the early yolk sac.9 This lineage is distinct from other embryonic lineages in that it does not require the transcription factor MYB.9 Yolk sac–derived macrophages are thought to migrate outside of the yolk sac during early embryonic periods and reside within a subset of developing organs including the brain, dermis, and heart. Within the brain, yolk sac–derived macrophages differentiate into microglia where they persist throughout life independent of blood monocyte input. Intriguingly, although yolk sac–derived macrophages occupy the early embryonic dermis, they seem to represent a transient lineage as they are largely replaced by other embryonic macrophage lineages.7 Interestingly, yolk sac–derived macrophages exist within the adult heart in concert with other macrophage lineages including adult monocyte-derived subsets.1,11,12
More recent studies have focused attention away from early yolk sac lineages and suggested that the majority of developing organs contain macrophages derived from later progenitors including fetal monocytes.13 These studies have demonstrated that fetal monocytes arise from a distinct yolk sac–derived erythromyeloid progenitor that migrates from the yolk sac to the liver during midgestation.13 For example, although the lung contains a small yolk sac–derived macrophage population, the majority of alveolar macrophages are derived from fetal monocyte-derived macrophages that enter the tissue during neonatal periods.14 In addition, Langerhans cells residing within the skin are also derived from fetal monocytes emigrating from the fetal liver.7 Despite intensive investigation, the exact macrophage composition of many organs remains to be fully elucidated. Moreover, it is not yet clear whether macrophages derived from Rag1+ lymphomyeloid progenitors and those derived from hemogenic endothelium or aorto-gonad-mesonephros progenitors significantly contribute to tissue resident macrophages.15
Although the roles of monocyte-derived macrophages are well described, the endogenous functions of embryonic-derived macrophages remain largely undefined. We have recently demonstrated that macrophages derived from early embryonic progenitors are essential for tissue repair and functional restoration of the injured neonatal heart.11 However, it remains unclear whether embryonic macrophages play important roles during normal tissue development. Furthermore, no studies to date have addressed whether distinct embryonic macrophage subsets might have differing functions.
To define physiological functions of embryonic-derived macrophages, we focused on the developing heart, an organ that harbors significant numbers of embryonic-derived macrophages.11,12,16 We demonstrate that the developing heart contains a complex array of embryonic macrophage subsets that can be divided into chemokine (C-C motif) receptor 2 (CCR2)− and CCR2+ macrophage subsets derived from distinct lineages. We further show that yolk sac–derived CCR2− macrophages are proangiogenic and are required for coronary development where they mediate remodeling of the primitive coronary plexus by promoting the selective expansion of perfused vasculature. Together, our findings indicate that the developing heart contains distinct lineages of embryonic macrophages with differing functions that are critical for proper organ development.
Methods
Mouse Strains
Mice were bred and maintained at the Washington University School of Medicine and the University of Oxford, and all experimental procedures were done in accordance with the animal use oversight committee. Mouse strains utilized included reverse orientation splice acceptor (Rosa) 26-td, Fms-like tyrosine kinase (Flt) 3-Cre, colony-stimulating factor 1 receptor (CSF1R)-murine estrogen receptor Cre (MerCre), LysM-Cre, CX3C chemokine receptor 1 (CX3CR1)GFP, Rag1-Cre, Rosa26-yellow florescent protein, Csf1op, Ccr2GFP, and Ccr2−/−.4,5,12,17–22 All mice were genotyped according to established protocols, and littermates were used as controls. Macrophage lineage tracing was performed by mating Rosa26-td and Rosa26-yellow florescent protein mice with Flt3-Cre, CSF1R-MerCre mice, or Rag1-Cre. Embryonic macrophages were labeled by treating pregnant CSF1R-MerCre/Rosa26-td mice with 0.1-mg tamoxifen/g body weight (Sigma) by oral gavage at E7.5.
Immunohistochemistry and Vascular Imaging
Embryonic and adult hearts were harvested and washed in PBS. Tissues were then fixed in 2% paraformaldehyde overnight at 4°C, embedded in OCT, infiltrated with sucrose, and frozen. Twelve-micrometer cryosections were then obtained and subjected to immunostaining. Primary antibodies used included platelet endothelial cell adhesion molecule 1:200 (BD), smooth muscle actin 1:400 (Sigma), bromodeoxyuridine 1:100 (Abcam), CD68 1:400 (Serotec), platelet-derived growth factor receptor-β 1:200 (R&D), and lymphatic vessel endothelial hyaluronan receptor 1 1:500 (Abcam). Immunofluorescence was visualized on a Zeiss confocal microscopy system. Bromodeoxyuridine-positive cells, blood vessel density, and macrophages were quantified by examining at least 4 similarly oriented sections from 4 independent samples in blinded fashion.
Whole-mount platelet endothelial cell adhesion molecule staining was performed as previously described.23 After staining, E13.5 and E17.5 hearts were frozen in OCT and 12-μm cryosections obtained. Quantitative imaging was performed using Zeiss Axiovision software. At least 5 sections from 4 independent biological specimens were examined in blinded fashion per experimental group.
For postnatal coronary imaging, anesthetized mice were injected IV with 5 mg/kg of fluorescein isothiocyanate (FITC)-dextran (Sigma 150 000 MW) 5 minutes before tissue harvest. The hearts were then fixed and imaged on a Zeiss Discovery V.12 stereomicroscope. For embryonic coronary imaging, E14.5 embryos were dissected in warmed PBS with the placenta and umbilical cord left intact. The umbilical vein was then injected with 50 μL of FITC-lectin (Vector Laboratories) using a 33-gauge needle. Embryonic hearts were then harvested, fixed, cryosections costained, and imaged on a Zeiss confocal microscope. Endocardial staining was utilized to confirm successful FITC-lectin injection.
Flow Cytometry, Microarray, and Reverse Transcription Polymerase Chain Reaction Assays
For flow cytometry experiments, mice were euthanized by cervical dislocation. Hearts were then perfused with cold PBS, finely minced, and digested with Collagenase D (Roche) and DNAse I (CalBiochem or Sigma) for 15 to 45 minutes at 37°C. The digested material was filtered through 40- to 50-μmol/L filters and pelleted by centrifugation (400g for 5 minutes at 4°C) in HBSS supplemented with 2% FCS+0.2% BSA or DMEM supplemented with 0.5% FCS. Red blood cells were lysed in ACK lysis buffer (Invitrogen) or ammonium chloride (Stemcell Technologies) and then resuspended in FACS buffer (PBS containing 2% to 5% FCS and 2 mmol/L EDTA). Samples were incubated with Fc block (BD), labeled with fluorescently conjugated antibodies (Online Table 1) for 25 to 30 minutes at 4°C, and washed twice in FACS buffer. Flow cytometry analysis was done on a BD LSRII analyzer and data analyzed using FlowJo software. After gating on CD45+ cells, doublets were excluded, live cells were analyzed using DAPI (4′,6-diamidino-2-phenylindole) dead/live exclusion, and macrophages identified based on CD64 and F4/80 expression as previously described.11
Cell sorting was performed using a MoFlo flow cytometry instrument (Beckman Coulter). Cytospin preparations and flow cytometry analysis were utilized to measure sample purity. For RNA isolation, macrophages were directly sorted into the QLT buffer and RNA isolated using the RNeay micro kit (Qiagen) per manufacturer’s instructions. Gene expression profiling was performed using microarray analysis. RNA was amplified using the WTA (Sigma) system and hybridized to Agilent 8X60 gene chips. Data analysis was performed using Partek genome suite software. For reverse transcription polymerase chain reaction experiments, cDNA was synthesized using the high-capacity cDNA reverse transcription kit (Ambion). Quantitative reverse transcription polymerase chain reaction was performed on an ABI 7200 machine using SYBR green-based assays. SYBR green primers were obtained from IDT. All samples were normalized to Gapdh and then scaled relative to controls, where control samples were set at a value of 1. Thus, results for all experimental samples were graphed as relative expression compared with control.
For macrophage culture experiments, samples were sorted directly into DMEM 10% FBS. Macrophages were plated at a density of 20 000 cells per well of a 96-well plate and allowed to adhere for 24 hours. To obtain macrophage conditioned media, cell were incubated in DMEM 1% FBS for 48 hours.
Matrigel Angiogenesis and Scratch Assays
To determine whether embryonic CCR2− macrophage conditioned media was sufficient to stimulate angiogenesis in vitro, mouse coronary endothelial cells24 were stimulated with control media or macrophage conditioned media for 24 hours and plated on growth factor reduced Matrigel (BD) at a density of 2×104 cells/mL in a 24-well plate format. Photographs were acquired 8 hours after plating and the number of tube segments/10× field quantified. For endothelial scratch assays, mouse coronary endothelial cells were seeded on 12-well plates and grown to 90% confluency. A pipette tip was used for wounding. Twenty four hours after wounding, the cells were fixed, stained with 1% crystal violet, and photographs obtained. The wounded area was examined for the presence of crystal violet staining. Four independent samples were included in each experimental group, and assays were repeated at least 2× to ensure technical accuracy. Insulin like growth factor (IGF) 1, IGF2, pleiotrophin, and platelet factor 4 protein was obtained from R&D and utilized at concentrations recommended by the manufacturer. IGF receptor 1 inhibitor was obtained from Tocris.
Statistical Analysis
Data are expressed as mean±SD. Student t test or ANOVA was used for comparisons between groups. P values of <0.05 were considered significant. Bonferroni correction was performed when multiple hypotheses were tested. In these cases, the type I error (α=0.05) was split equally among each test.
Antibodies Used for Flow Cytometry
Antibodies used for flow cytometry is shown in the Table.
Table.
Antibodies Used for Flow Cytometry
| Antibody | Clone | Supplier |
|---|---|---|
| CD45 | 30-F11 | Biolegend |
| CD45.2 | 104 | eBioscience |
| CD64 | X54-5/7.1 | Biolegend |
| F4/80 | C1:A3-1 and BM8 | eBioscience |
| MHC-II (I-Ab) | AF6-120.1 | Biolegend |
| CCR2 | 475301 | R&D |
| MerTK | 108928 | R&D |
| Ly6G | 1A8 | Biolegend |
| Ly6C | HK1.4 | Biolegend |
| CD11c | N418 | Biolegend |
| Siglec F | E50-2440 | BD Biosciences |
| CD11b | M1/70 | eBiosceince |
| CD16/CD32 | 2.4G2 | BD Biosciences |
| Tie2 | Tek4 | Biolegend |
Results
To establish the timing and precise composition of macrophage subsets that reside within the developing heart, we utilized established macrophage reporter mice in combination with flow cytometry. Analysis of CX3CR1-green florescent protein (GFP) mice18 demonstrated that macrophages were first detected in the heart on embryonic day 12.5 (E12.5) and persist within the ventricular myocardium throughout development (Figure 1A). Flow cytometry revealed that the vast majority of CD45+ immune cells within the developing heart coexpressed CX3CR1-GFP+ and the macrophage antigen F4/80 and increased in abundance between E12.5 and E14.5 (Figure 1B and 1C).
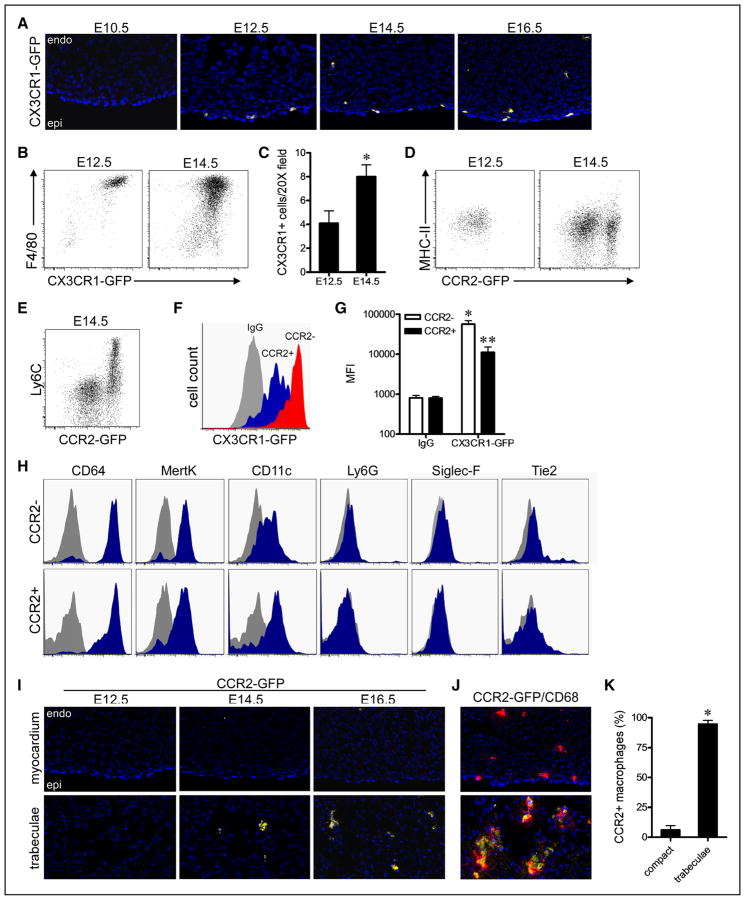
Figure 1. Distinct chemokine (C-C motif) receptor 2 (CCR2)− and CCR2+ macrophages populate the embryonic heart.
A, CX3C chemokine receptor 1 (CX3CR1)-green florescent protein (GFP) embryos demonstrating that macrophages are present in the embryonic heart beginning after E10.5. DAPI (4′,6-diamidino-2-phenylindole), blue; CX3CR1-GFP, yellow; B, Flow cytometry analysis of gated CD45+ cells from the embryonic heart showing that all CD45+ cells express CX3CR1-GFP and F4/80. C, Quantification of the number of CX3CR1-GFP+ macrophages per 20× field at E12.5 and E14.5. *P<0.05. D, Flow cytometry demonstrating that at E12.5 the embryonic heart contains a single CCR2− MHC-IIlow macrophage subset, whereas at E14.5, the embryonic heart contains CCR2− MHC-IIlow and CCR2+ major histocompatibility complex (MHC)-IIlow macrophage subsets. E, CCR2+ macrophage coexpress Ly6C. F and G, Quantification of mean florescent intensity (MFI) demonstrating that CCR2− macrophages express CX3CR1-GFP to a greater extent to that of CCR2+ macrophages. *P<0.05 compared with control, **P<0.05 compared with all other groups. H, CCR2− and CCR2+ macrophages express CD64, MertK, and CD11c. Gray: isotype control, blue: designated antibody. I, Analysis of CCR2-GFP embryos revealing that CCR2+ macrophages are specifically located adjacent to the endocardial trabeculae at E14.5 and E16.5. J and K, Immunostaining for CCR2-GFP (yellow) and CD68 (red) demonstrating that CCR2+ macrophages exclusively reside within the endocardial trabeculae and CCR2− macrophages in the compact myocardium. *P<0.05. A, I, and J: ×20 magnification. Each experiment included at least 4 independent biological replicates. Endo indicates endocardium; and Epi, epicardium.
To decipher whether the developing heart contains a single macrophage population or distinct macrophage subsets, we explored several markers previously reported to distinguish between macrophage populations. Among the tested markers, CCR2, which we previously found to identify distinct macrophage subsets in the adult heart,11,12,16 readily identified 2 distinct macrophage subsets in the embryonic heart. Flow cytometric analysis of CCR2-GFP mice12 revealed that CCR2− macrophages exclusively populated the developing heart at E12.5. In contrast, at E14.5, the heart contained distinct CCR2− and CCR2+ macrophage subsets, the latter of which expressed the monocyte marker Ly6C (Figure 1D and 1E). CD68 immunostaining confirmed that CX3CR1-GFP and CCR2-GFP specifically labeled macrophages in the developing heart (Online Figure IA and IB). Quantification of mean florescent intensity revealed that CCR2− macrophages expressed higher levels of CX3CR1-GFP than CCR2+ macrophages (Figure 1F and 1G). CCR2− and CCR2+ macrophage subsets expressed similar levels of the macrophage markers CD64, MertK, and CD11c, but did not express markers of neutrophils (Ly6G) or eosinophils (Siglec F). Of note, neither CCR2− nor CCR2+ macrophages expressed Tie2, a marker recently reported to be expressed by primitive erythromyeloid macrophage progenitors25 (Figure 1H; Online Figure IC). Immunostaining analysis demonstrated that CCR2− and CCR2+ macrophages were localized within different regions of the embryonic heart. CCR2− macrophages were found almost exclusively within the myocardial wall, whereas CCR2+ macrophages were located almost entirely within the trabecular projections of the endocardium (Figure 1I–1K).
To determine whether CCR2− and CCR2+ embryonic cardiac macrophages represent functionally distinct populations, we purified each macrophage subset using flow cytometry (Figure 2A) and performed microarray gene expression profiling. Principal component and hierarchical clustering analyses revealed that CCR2− and CCR2+ macrophage subsets represented distinct populations with significantly different gene expression profiles (Figure 2B and 2C). Using a 1.5-fold change cutoff, 674 genes were differentially expressed in CCR2− and CCR2+ macrophages, respectively (Figure 2D). Pathway analysis demonstrated that many of the differentially expressed genes belonged to pathways implicated in wound healing, inflammation, cell death, and metabolic processes (Online Figure IIA). Further delineation of genes associated with monocyte and macrophage function demonstrated that CCR2− macrophages differentially expressed transcripts previously associated with yolk sac–derived and resident macrophage subsets including Cx3cr1, Lyve1, Emr1, Cd207, and Ccl12.9,26 In contrast, CCR2+ macrophages differentially expressed transcripts associated with monocytes and monocyte-derived macrophages including Ly6c, Cxcr2, Sell, Irf5, and Nr4a1 (Figure 2E; Online Figure IIB).9,11,12,27,28
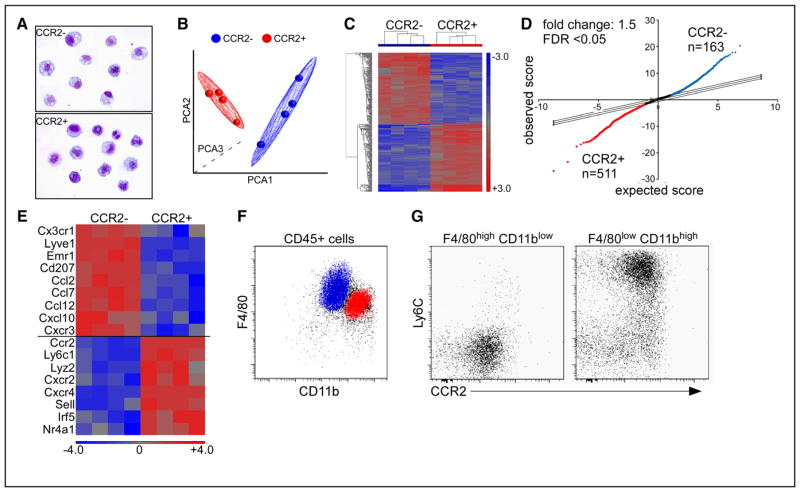
Figure 2. Gene expression profiling of embryonic cardiac macrophages.
A, Cytospin specimens of chemokine (C-C motif) receptor 2 (CCR2)− and CCR2+ macrophage subsets sorted by flow cytometry demonstrating robust purification of cell populations. B and C, Principal components analysis (PCA) (B) and hierarchical clustering (C) analyses showing that CCR2− and CCR2+ macrophages have distinct gene expression profiles. D, Significance analysis of microarrays plot highlighting the number of genes upregulated in CCR2− (blue, n=163) and CCR2+ (red, n=511) macrophages. E, Heat map depicting monocyte and macrophage-associated genes that distinguish embryonic CCR2− and CCR2+ macrophages. F and G, Flow cytometry at E14.5 revealing the surface phenotype of CCR2− macrophages (blue): F4/80highCD11blowLy6Cneg and CCR2+ macrophages (red): F4/80lowCD11bhighLy6Cpos. Microarray experiments included 4 biologically independent samples each of which containing 4 to 6 E14.5 hearts. FDR indicates false discovery rate.
Based on our gene expression profiling data, we hypothesized that CCR2− macrophages are derived from yolk sac progenitors, whereas CCR2+ macrophages are derived from fetal monocyte progenitors. To test this hypothesis, we performed detailed flow cytometry and genetic lineage tracing. Previous studies have revealed that within the embryo F4/80highCD11blow macrophages are derived from yolk sac progenitors, whereas F4/80lowCD11bhigh macrophages are of fetal monocyte origin.9 Consistent with this notion, detailed flow cytometric analysis showed that embryonic cardiac macrophages can be divided into F4/80highCD11blow and F4/80lowCD11bhigh subsets. The F4/80highCD11blow subset contains exclusively CCR2−Ly6C− macrophages suggesting that these cells are of yolk sac origin, whereas the F4/80lowCD11bhigh subset predominately consists of CCR2+Ly6C+ macrophages consistent with fetal monocyte origin (Figure 2F and 2G).
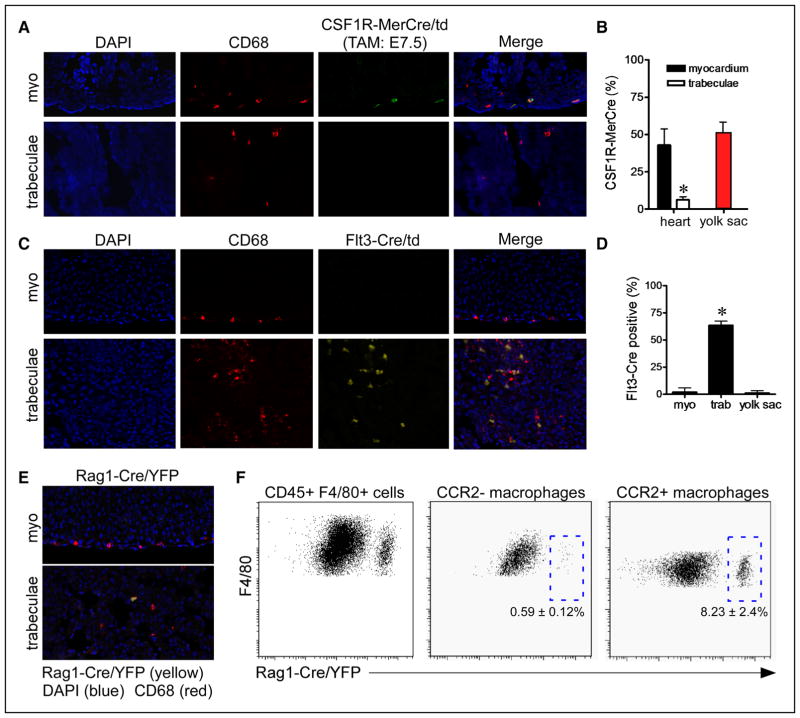
To more directly define the developmental origin of embryonic cardiac macrophages, we performed genetic lineage tracing. We took advantage of the exclusive localization of CCR2− (compact myocardium) and CCR2+ (trabeculae) macrophages and utilized immunostaining techniques to decipher-specific contributions to each of these macrophage subsets. Administration of tamoxifen to pregnant Rosa26-tdCsf1R-MerCre mice19 at E7.5 (approximate timing of yolk sac hematopoiesis) demonstrated labeling of macrophages located within the compact myocardium (which are CCR2−; Figure 1J and 1K), but not macrophages located within the trabeculae (which are CCR2+; Figure 1J and 1K) at E14.5. Importantly, the percentage of macrophages located within the compact myocardium that were labeled by Rosa26-tdCsf1R-MerCre mice was similar to that of macrophages located within the yolk sac (Figure 3A and 3B; Online Figure IIIA). Together, these data support the conclusion that CCR2− macrophages are derived from yolk sac progenitors. To define the origins of trabecular (CCR2+) macrophages, we performed genetic lineage tracing with Flt3-Cre4 and Rag1-Cr2e5 mice to examine the contribution of definitive hematopoietic and the recently described lymphomyeloid lineages, respectively. Analysis of Rosa26-tdFlt3-Cre mice at E14.5 revealed significant contribution to the CCR2+ macrophage population suggesting that they are largely derived from fetal monocyte progenitors (Figure 3C and 3D; Online Figure IIIB). Immunostaining analysis of Rosa26-yellow florescent proteinRag1-Cre mice at E14.5 demonstrated infrequent labeling of macrophages located within the trabeculae, suggesting that only a minority of CCR2+ macrophages are derived from Rag1+ lymphomyeloid progenitors (Figure 3E). Consistent with this conclusion, flow cytometric analysis confirmed that <10% of CCR2+ macrophages express yellow florescent protein (Figure 3F).
Figure 3. Developmental origins of embryonic cardiac macrophages.
A, Genetic lineage tracing using colony-stimulating factor 1 receptor (Csf1R)-murine estrogen receptor Cre (MerCre); reverse orientation splice acceptor (Rosa) 26-td (green) demonstrating that tamoxifen administration at E7.5 exclusively labels a subset of CD68+ macrophages (red) located within the compact myocardium and does not label CD68+ macrophages located in the trabeculae. Immunostaining was performed at E14.5. B, Quantification of the percent of total macrophages labeled by Csf1R-MerCre; Rosa26-td reveals that cardiac macrophages located within the myocardium are labeled at a frequency similar to yolk sac macrophages. Five independent hearts were included in the quantitative analysis. C, Fms-like tyrosine kinase (Flt) 3-Cre lineage tracing at E14.5 (yellow) demonstrating exclusive labeling of CD68+ macrophages (red) located in the trabeculae. D, Quantification of CD68 and Flt3-Cre/Rosa26td staining showing that >60% of macrophages located within the trabeculae are Flt3-Cre/Rosa26td positive. In contrast, only rare cells were labeled in the compact myocardium and yolk sac. Five independent hearts were included in the quantitative analysis. E, Recombination activating gene 1 (Rag1)-Cre lineage tracing at E14.5 (yellow) revealing infrequent labeling of CD68+ macrophages (red) located within the trabeculae. F, Flow cytometry analysis of CD45+, CD11b+, F4/80+ cells demonstrating that Rag1-Cre labels <10% of chemokine (C-C motif) receptor 2 (CCR2)+ macrophages. Quantitation is based on 3 independent pools of Rag1-Cre; Rosa26-yellow florescent protein (YFP) embryonic hearts. A, C, and E, ×20 magnification. *P<0.05 compared with all other groups. DAPI indicates 4′,6-diamidino-2-phenylindole.
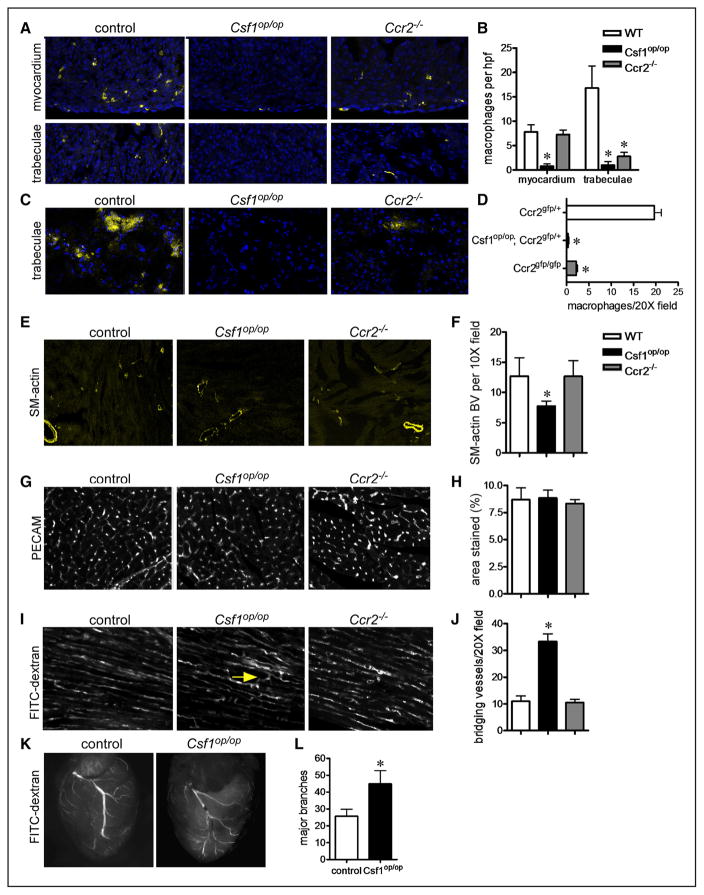
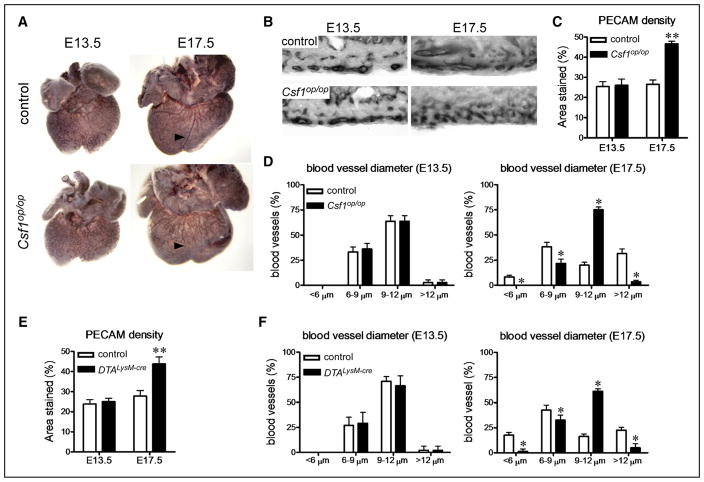
To decipher whether embryonic macrophages are functionally required for heart development, we examined control, Csf1op/op,20 and Ccr2−/−29 embryos. Compared with controls, Csf1op/op embryos lacked all macrophages, whereas Ccr2−/− embryos had a specific reduction in CCR2+ macrophages located within the trabecular myocardium (Figure 4A–4D). Based on recent data implicating embryonic-derived macrophages in coronary angiogenesis,11,30 we focused on coronary development. Smooth muscle actin immunostaining at postnatal day 21 (P21) demonstrated that compared with controls and Ccr2−/− hearts, Csf1op/op hearts contained fewer smooth muscle–coated coronary arterioles (Figure 4E and 4F). Detailed examination of the microvasculature revealed that although Csf1op/op hearts contained similar capillary density compared with control and Ccr2−/− hearts (Figure 4G and 4H), the microvasculature of Csf1op/op hearts displayed exaggerated branching and connectivity (Figure 4I and 4J). Consistent with aberrant microvascular patterning, imaging of P21 hearts perfused with FITC-dextran revealed that Csf1op/op hearts displayed excessive coronary artery branching compared to control and Ccr2−/− hearts (Figure 4K–4L). Together, these data indicate that macrophages are essential for coronary patterning at the microvascular and macrovascular level and suggest that yolk sac–derived CCR2− macrophages mediate this developmental process.
Figure 4. Embryonic cardiac macrophages are essential for coronary patterning.
A and B, CD68 immunostaining (A) and quantification (B) at E16.5 demonstrating that Csf1op/op embryos lack all macrophage subsets, whereas Ccr2−/− embryos specifically have decreased numbers of macrophages within the trabeculae. C and D, chemokine (C-C motif) receptor 2 (CCR2)-green florescent protein (GFP) reporter imaging at E16.5 demonstrating marked reductions in the number of CCR2+ macrophages (yellow) in Csf1op/op and Ccr2−/− hearts. E and F, Smooth muscle (SM) actin immunostaining at P21 showing reduced number of smooth muscle+ arterioles in Csf1op/op hearts. G and H, Platelet endothelial cell adhesion molecule (PECAM) immunostaining at P21 showing unaffected capillary density between genotypes. I, Compressed z-stack images of hearts perfused with fluorescein isothiocyanate (FITC)-dextran demonstrating increased connections (arrow: bridging blood vessels) within the microvasculature. Photographs were obtained from areas of the heart where cardiomyocytes were oriented in parallel. J, Quantification of bridging blood vessels. K and L, FITC dextran perfusion at P21 revealing excessive coronary branching in Csf1op/op mice. Major branches were defined as primary and secondary branches arising from the left anterior descending artery. E, ×10 magnification. A, C, G, and I: ×20 magnification. *P<0.05 compared with controls. Each experiment included at least 5 hearts per genotype. WT indicates wild-type.
To define the mechanistic basis by which macrophages contribute to coronary development, we performed whole-mount platelet endothelial cell adhesion molecule/CD31 immunostaining on control and Csf1op/op embryonic hearts. Compared with controls, Csf1op/op hearts displayed normal coronary plexus development at E13.5, but aberrant coronary patterning at E17.5, suggesting a defect in coronary remodeling (Figure 5A). Quantitative analysis of coronary plexus density and patterning revealed that at E13.5 control and Csf1op/op hearts had similar coronary density and distribution of blood vessel sizes. However, at E17.5, Csf1op/op hearts displayed increased coronary vessel density compared with controls and failure of the vascular plexus to remodel into blood vessels of varying diameter. Notably, Csf1op/op hearts displayed blood vessel diameters that were indistinguishable from those of the E13.5 primitive vascular plexus with reduced numbers of small and large diameter blood vessels and increased numbers of medium diameter blood vessels (Figure 5B–5D). Examination of an independent model of macrophage deficiency (Rosa26-DTALysM-Cre)17,31 revealed a phenotype identical to that of Csf1op/op mice (Figure 5E and 5F). Consistent with a specific role for CCR2− macrophages, platelet endothelial cell adhesion molecule immunostaining of Ccr2−/− hearts revealed normal coronary plexus development and remodeling (Online Figure IV).
Figure 5. Embryonic macrophages function to remodel the developing coronary vascular plexus.
A, Platelet endothelial cell adhesion molecule (PECAM) whole-mount immunostaining demonstrating normal coronary plexus development at E13.5 and aberrant coronary patterning at E17.5 in Csf1op/op hearts. Arrowhead denotes expected location of a mature coronary vessel. B, Cryosections of PECAM-stained embryonic hearts demonstrating increased capillary density and failure of the coronary plexus to remodel into blood vessels of varying diameters at E17.5 in Csf1op/op hearts. C and D, Quantification of coronary density (C) and blood vessel diameter (D) in control and Csf1op/op hearts. Csf1op/op hearts have increased coronary density at E17.5 and display failure to remodel the vascular plexus into larger and smaller blood vessels. E and F, PECAM immunostaining demonstrating increased coronary vascular plexus density (E) and defective coronary plexus remodeling (F) in reverse orientation splice acceptor (Rosa) 26-DTALysM-Cre hearts at E17.5. Each experiment included at least 5 hearts per genotype. B, ×20 magnification. *P<0.05 compared with controls.
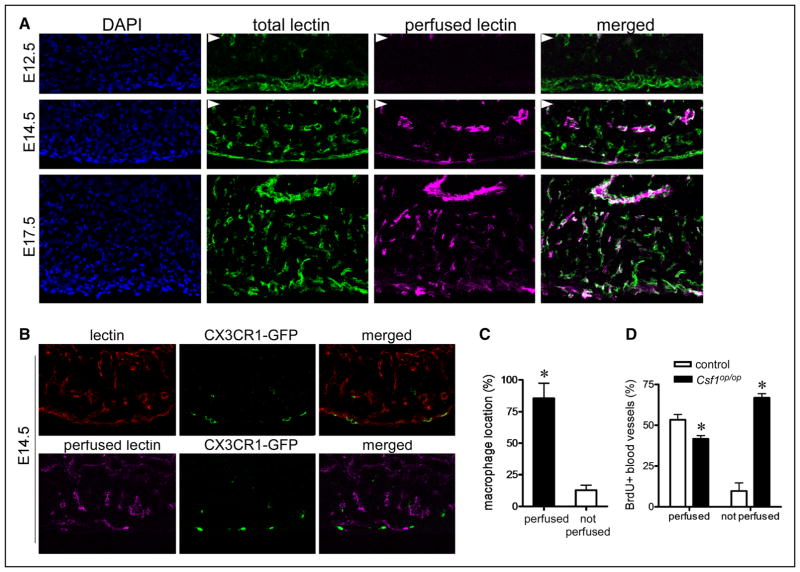
To understand how embryonic macrophages might govern coronary remodeling, we examined the precise localization of macrophage with respect to the coronary vasculature. Previous work has suggested that the coronary plexus develops in the absence of blood flow and that the onset of coronary perfusion occurs coincident with coronary plexus remodeling.17,31 Indeed, analysis of E14.5 hearts obtained from embryos perfused with rhodamine conjugated lectin and subsequently stained after sectioning with FITC-conjugated lectin demonstrated that coronary perfusion begins in a subset of coronary vasculature at ≈E14.5. By E17.5, the entire coronary system is perfused (Figure 6A). Intriguingly, examination of CX3CR1-GFP embryos revealed that although macrophages are located adjacent to coronary endothelial cells, they are selectively associated with perfused coronary vasculature (Figure 6B and 6C). Based on these data, we hypothesized that macrophages may govern coronary remodeling by selectively interacting with perfused coronary vasculature. Consistent with this notion, analysis of coronary endothelial proliferation in control and Csf1op/op hearts revealed that although control hearts displayed selective proliferation of perfused vasculature, hearts that lacked macrophages demonstrated reduced proliferation of perfused vasculature and marked increased proliferation of nonperfused vasculature (Figure 6D). Other important contributors to coronary remodeling including pericyte and smooth muscle cell differentiation32–34 were grossly unaffected by macrophage deficiency (Online Figure V).
Figure 6. Chemokine (C-C motif) receptor 2 (CCR2)− embryonic macrophages are selectively associated with perfused coronary plexus vasculature.
A, Simultaneous imaging of total coronary vasculature (green, fluorescein isothiocyanate-lectin staining) and perfused coronary vasculature (magenta, perfused rhodamine-lectin) demonstrating the onset of coronary perfusion at approximately E14.5. Arrowheads denote the endocardium. B, Perfused (magenta) and total (red) lectin imagining at E14.5 revealing that CX3C chemokine receptor 1 (CX3CR1)-green florescent protein (GFP)+ macrophages are selectively associated with perfused coronary vasculature. C, Quantification of the percent of macrophages localized adjacent to perfused and nonperfused coronary plexus vasculature. D, Bromodeoxyuridine (BrdU) immunostaining showing reduced proliferation of perfused coronary vasculature and markedly increased proliferation of nonperfused vasculature in Csf1op/op hearts. Each experiment included at least 5 hearts. A and B, ×20 magnification. *P<0.05 compared with controls.
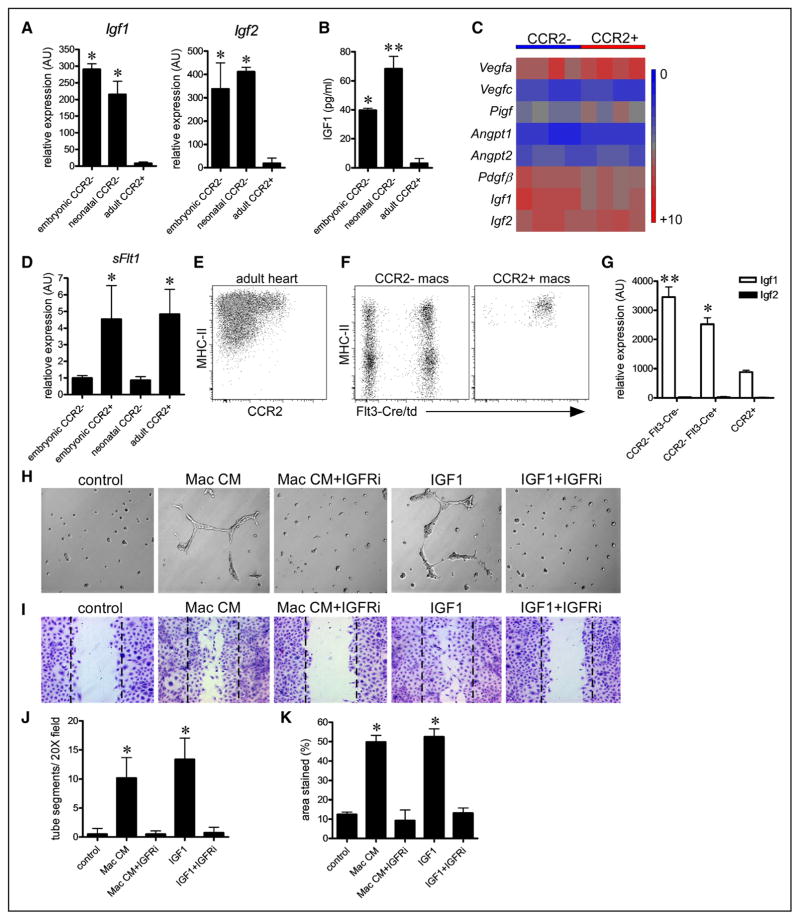
To identify potential macrophage-derived effector molecules that mediate coronary remodeling, we utilized our microarray expression data to identify secreted growth factors that are expressed by embryonic macrophages and have previously been implicated in angiogenesis. Among the identified genes, quantitative reverse transcription polymerase chain reaction assays revealed that insulin-like growth factors 1 and 2 (Igf1, Igf2), Ptn, and Pf4 were strongly expressed in embryonic CCR2− cardiac macrophages. Importantly, these factors were also expressed in neonatal CCR2− cardiac macrophages, a population that we have previously shown to be proangiogenic. In contrast, minimal expression of Igf1, Igf2, Ptn, and Pf4 was found in adult CCR2+ cardiac macrophages, a subset that lacks proangiogenic activity11 (Figure 7A; Online Figure VIA). To test whether these growth factors possess proangiogenic activity toward coronary endothelial cells, we performed a set of in vitro assays. Matrigel tube and endothelial cell migration assays demonstrated that only IGF1 and IGF2 possessed proangiogenic activity toward cultured coronary endothelial cells (Online Figure VIB–VIE), suggesting that IGF ligands may be one of the relevant macrophage effector molecules. Consistent with these findings, IGF1 ELISA revealed that conditioned media obtained from embryonic and neonatal CCR2− macrophage populations contained higher concentrations of IGF1 protein than conditioned media obtained from adult CCR2+ macrophages (Figure 7B).
Figure 7. Insulin like growth factor (IGF) ligands constitute a proangiogenic cardiac macrophage-derived signal.
A, Quantitative reverse transcription polymerase chain reaction (RT-PCR) revealing selective expression of Igf1 and Igf2 mRNA in embryonic chemokine (C-C motif) receptor 2 (CCR2)− and neonatal CCR2− macrophages. B, IGF1 ELISA demonstrating that conditioned media obtained from embryonic CCR2− and neonatal CCR2− macrophages contains higher concentrations of IGF1 protein compared with conditioned media obtained from adult CCR2+ macrophages. n=4 for each macrophage population. *P<0.05 compared with adult CCR2+ macrophages. **P<0.05 compared with all other groups. C, Heat map depicting proangiogenic growth factor expression in embryonic CCR2− and CCR2+ macrophages. D, Quantitative RT-PCR demonstrating that embryonic and adult CCR2+ macrophages express higher levels of the angiogenesis inhibitor, soluble Flt1. E and F, Flow cytometry of adult cardiac macrophages revealing the presence of CCR2− and CCR2+ macrophage populations. Lineage tracing shows that CCR2− macrophages can be divided into embryonic-derived (Flt3-Cre negative) and monocyte-derived (Flt3-Cre positive) subsets, whereas CCR2+ macrophages are exclusively monocyte derived. Macrophages were identified as CD45+ CD64+ Ly6G− cells. G, Quantitative RT-PCR demonstrating increased expression of Igf1 in CCR2− macrophages compared with CCR2+ macrophages. CCR2− embryonic-derived macrophages express slightly higher levels of Igf1 mRNA compared with CCR2− monocyte-derived macrophages. Igf2 expression could not be detected in adult cardiac macrophages. *P<0.05 to adult CCR2+ macrophages. **P<0.05 compared with all other groups. G, Matrigel angiogenesis assays demonstrating that inhibition of IGF signaling suppresses the ability of CCR2− embryonic macrophage conditioned media to stimulate endothelial cell tube formation in vitro. H, Endothelial scratch assays revealing that inhibition of IGF signaling suppresses the ability of CCR2− embryonic macrophage conditioned media to stimulate endothelial cell migration in vitro. I and J, Quantitative analysis of Matrigel angiogenesis (I) and scratch assays (J). Each experiment included at least 5 biological replicates. H and I, ×20 magnification. *P<0.05 compared with vehicle control.
Examination of classic proangiogenic growth factor expression revealed that both CCR2− and CCR2+ embryonic macrophages expressed significant and comparable levels of vascular endothelial growth factor-a and platelet derived growth factor-β mRNA. CCR2− and CCR2+ macrophages expressed minimal levels of vascular endothelial growth factor-c, placenta growth factor, angiopoietin 1, and angiopoietin 2 (Figure 7C). Intriguingly, embryonic and adult CCR2+ macrophages expressed significantly higher levels of the angiogenesis inhibitor, soluble Flt1 than CCR2− macrophage subsets (Figure 7D).
To determine whether IGF1 and IGF2 expression was conserved in adult cardiac macrophage subsets that possess proangiogenic activity, we purified CCR2− and CCR2+ macrophage populations using flow cytometry. CCR2− macrophages were further divided into embryonic-derived (Flt-Cre negative) and monocyte-derived (Flt3-Cre positive) macrophage subsets (Figure 7E and 7F). Consistent with a previous study11 demonstrating that CCR2− macrophages resident within the adult heart are proangiogenic, CCR2− macrophage subsets expressed higher levels of Igf1 mRNA than CCR2+ macrophages. Of note, embryonic-derived CCR2− macrophages expressed slightly higher levels of Igf1 mRNA than monocyte-derived CCR2− macrophages. Igf2 mRNA expression could not be detected in CCR2− or CCR2+ adult cardiac macrophages (Figure 7G).
Together, the above data suggested that cardiac macrophage-derived IGF signals may mediate the proangiogenic activities of embryonic cardiac macrophages. To decipher whether IGF signaling is necessary for the proangiogenic activity of embryonic macrophages within the heart, we focused on CCR2− macrophages as they are located in close proximity to the coronary vasculature and are required for coronary development. We examined whether inhibition of IGF1 receptor signaling abrogated the proangiogenic effects of CCR2− embryonic macrophages in vitro. Consistent with the proangiogenic potential of CCR2− embryonic macrophages, conditioned media obtained from this macrophage subset was sufficient to stimulate coronary endothelial cell tube formation and migration in vitro. However, addition of an IGF1 receptor–specific inhibitor abolished the ability of CCR2+ embryonic macrophage conditioned media to stimulate coronary endothelial cell tube formation and migration (Figure 7H–7K) implicating IGF ligands as a potentially relevant proangiogenic signal by which CCR2− embryonic macrophages might mediate coronary development.
Discussion
Together, our results reveal that functionally distinct lineages of embryonic macrophages populate the developing heart and are required for proper cardiac development. Specifically, we demonstrate that the developing heart contains distinct lineages of embryonic macrophages with contributions from primitive yolk sac, fetal monocyte, and lymphomyeloid progenitors. Functionally, yolk sac–derived CCR2− macrophages are essential for coronary maturation where they promote remodeling of the primitive coronary plexus through the selective expansion of perfused coronary vasculature. In contrast, CCR2+ macrophages are derived from fetal monocytes including from Rag1+ lymphomyeloid progenitors and seem to be dispensable for heart development. Collectively, these data define the composition of cardiac embryonic-derived macrophages, determine when each lineage takes residence within the myocardium, and identify a novel role for embryonic-derived macrophages in heart development. In addition, we provide evidence supporting a new mechanism that mediates coronary plexus remodeling and functional maturation of the coronary vasculature. Finally, our results implicate IGF signaling as a potential macrophage-derived mediator of coronary angiogenesis.
In contrast to previous studies reporting that developing tissues primarily contain a single embryonic macrophage lineage (ie, yolk sac or fetal monocyte derived),6,7,13,25 our findings indicate that within the heart multiple and distinct embryonic macrophage lineages coexist. Interestingly, CCR2− and CCR2+ macrophages entered the heart at different developmental time points. CCR2− macrophages were first apparent within the heart at E12.5 and were localized within the subepicardial space, whereas CCR2+ macrophages were observed within the endocardium beginning at E14.5. After entering the developing heart, CCR2− and CCR2+ macrophages seemed to occupy separate niches. Namely, CCR2− yolk sac–derived macrophages were closely associated with the coronary vasculature, whereas CCR2+ macrophages derived from fetal monocytes and Rag1+ progenitors were located in close proximity to the endocardial trabeculae. The exact mechanisms that govern recruitment of CCR2− and CCR2+ macrophages to the developing heart and signals that control their localization within the myocardium are not yet clear, but undoubtedly will be the topic of future investigation.
Transcriptional profiling of CCR2− and CCR2+ embryonic-derived cardiac macrophages supported the concept that these macrophage subsets may have distinct functions. Consistent with previous literature, our findings demonstrate a role for CCR2− embryonic-derived macrophages in vascular growth and maturation. Previous studies focused on mouse hindbrain and retina development have revealed an essential role for microglia in CNS vascular branching.35–37 Mice deficient in microglia (Csf1op/op and Pu.1−/−) had reduced vascular branch points in the ventricular and retina vascular plexus. Within these systems, vascular endothelial growth factor signaling did not seem to influence microglia numbers, location, or function. Instead, zebrafish studies implicated that macrophages may assist in bridging anastomotic connections between endothelial tip cells.35 Intriguingly, within the postnatal deep retinal vascular plexus microglia suppress angiogenic sprouting through a WNT-FLT1–dependent pathway,38 indicating that the effects of primitive macrophages on the developing vasculature may be context dependent.
In contrast to the roles of microglia in CNS vascular development, within the developing heart, CCR2− macrophages did not stimulate branching of the primitive capillary plexus. Instead, they were essential for remodeling of the vascular plexus into a mature vascular tree containing vessels of differing diameters. In contrast to coronary plexus development, less is known about cellular molecular mechanisms that govern remodeling of the primitive coronary plexus. Several studies have indicated that signaling pathways that regulate the differentiation of pericytes and smooth muscle cells (transforming growth factor-β, platelet-derived growth factor receptor-α, NOTCH1, NOTCH3, WNT, myocardin-related transcription factor) are essential for coronary remodeling.32–34,39,40 Our findings identify a novel role for embryonic macrophages in this important process.
Mechanistically, CCR2− macrophages regulated coronary remodeling in a potentially flow-dependent manner. Specifically, macrophages preferentially promoted the expansion of perfused blood vessels at the expense of blood vessels receiving no or minimal perfusion. In the absence of macrophages, perfused coronary plexus vasculature proliferated at slower rates. Importantly, vasculature not receiving perfusion continued to proliferate at high rates accounting for the expanded vascular network in Csf1op/op mice. These data suggest that macrophages directly or indirectly regulate coronary endothelial cell proliferation. Although we did not observe defects in pericyte or smooth muscle cell differentiation, it remains possible that macrophages may interact with vascular supporting cells to promote branching and growth of perfused vasculature.
Using a gene expression-based approach, we identified IGF1 and IGF2 as potential mediators by which CCR2− cardiac macrophages mediate coronary remodeling. Consistent with this notion, IGF1 and IGF2 were sufficient to stimulate coronary endothelial tube formation in vitro. Moreover, IGF signaling was necessary for the proangiogenic effects of CCR2− macrophages in vitro. Consistent with previous literature, IGF signaling was a potent stimulus for coronary endothelial cell migration.41 Based on these data, we hypothesize that macrophage-derived IGF signals may selectively mediate the selective expansion of perfused coronary vasculature by stimulating endothelial cell migration to sites of vascular perfusion and subsequent incorporation of endothelial cells into areas of active remodeling. In addition, we demonstrated that CCR2− macrophages resident within the adult heart also express significant levels of IGF1 and have previously been shown to possess proangiogenic activity.11 Intriguingly, both embryonic-derived and monocyte-derived macrophages expressed IGF1, suggesting that monocytes may be able to take on an embryonic macrophage-like phenotype. These data support but do not confirm the possibility that IGF ligands constitute the relevant proangiogenic signals secreted by CCR2− cardiac macrophages. Future studies will be required to determine whether macrophage-derived IGF signals are necessary for coronary remodeling in vivo using conditional gene targeting.
Although we demonstrated that CCR2− macrophages are critical regulators of coronary vessel maturation, the exact function of CCR2+ macrophages was not readily apparent. Specifically, we were unable to identify defects in heart development using CCR2-deficient mice. Because Ccr2−/− mice displayed reduced (but not absent) numbers of CCR2+ macrophages, our conclusions may be limited as it is possible that a small number of CCR2+ macrophages is sufficient to perform their endogenous functions. Few tools exist to address the endogenous functions of CCR2+ macrophages derived from fetal monocyte and Rag1+ progenitors. Myb−/− mice contain only yolk sac–derived macrophages, but display mortality during midgestation (E15.5–E16.5),9,42 precluding their use to address the functions of CCR2+ macrophages during later stages of heart development. It is also plausible that CCR2+ macrophages may play a specific role in the context of embryonic cardiac injury, a possibility that was not examined in this study.
Several important questions remain, including how are CCR2− macrophages selectively recruited to perfused vasculature? Recent studies have revealed that perfused coronary endothelium expresses the Notch ligand Jagged-1, which is required for pericyte differentiation into smooth muscle cells at the onset of coronary perfusion.40 Intriguingly, within the retina, microglia express significant levels of Notch1, which is required for recruitment of microglia to sites of vascular branching.43 These data indicate that Notch signaling is essential for macrophage trafficking. Future studies will be required to determine whether Notch signals are responsible for migration of CCR2− macrophages to perfused coronary vasculature.
In conclusion, our findings indicate that multiple lineages of embryonic-derived macrophages exist within the developing heart, and that CCR2− yolk sac–derived macrophages are essential regulators of coronary development. In conjunction with previous work identifying an essential proangiogenic role for yolk sac–derived macrophages in the context of neonatal cardiac injury,11 our study suggests that regulation of coronary growth may represent a conserved function of primitive yolk sac–derived cardiac macrophages present in the developing and adult heart. Future studies will be required to determine whether embryonic-derived macrophages resident in the adult heart similarly mediate coronary angiogenesis and arteriogenesis under physiological and pathological conditions.
Supplementary Material
Novelty and Significance.
What Is Known?
Distinct subsets of macrophages derived from embryonic and definitive hematopoietic progenitors reside within various tissues of the body including the heart.
The exact origins of cardiac embryonic-derived macrophages, timing by which these cells enter the heart, and their normal functions remain poorly defined.
What New Information Does This Article Contribute?
Identified the relevant subsets and respective developmental origins of macrophages that reside within the fetal heart.
Deciphered the function of embryonic macrophages during heart development.
Demonstrated that yolk sac–derived macrophages have a conserved role in regulating coronary angiogenesis possibly through regulation of insulin like growth factor 1 signaling
Recently, an important paradigm has emerged in the macrophage field: organs are replete with resident macrophages of embryonic origin, distinct from monocyte-derived macrophages. The literature is now filled with publications and reviews outlining various embryonic macrophage lineages. However, the functional relevance of each of these cell types remains to be elucidated. We have taken this important next step and defined the exact origins of embryonic-derived macrophages within the heart and delineated their respective functions. We demonstrate that the developing heart contains a complex array of embryonic macrophage subsets derived from early yolk sac and fetal liver progenitors. Within the developing heart, yolk sac–derived macrophages are instrumental in coronary development where they are required for remodeling and patterning of the primitive coronary plexus. In contrast, macrophages derived from lymphomyeloid and fetal monocyte progenitors and seem to be dispensable for heart development. Mechanistically, yolk sac–derived macrophages are recruited to the developing coronary vasculature at the onset of blood vessel perfusion where they mediate vascular remodeling and maturation through selective expansion of perfused vasculature. Together, these findings demonstrate that the embryonic heart contains distinct lineages of embryonic macrophages with unique functions and reveals a conserved role for yolk sac–derived macrophages in coronary angiogenesis.
Acknowledgments
K.J. Lavine wrote the article, designed, and performed the experiments. J. Leid performed the Matrigel angiogenesis assays, endothelial cell migration assays, and assisted in the remainder of the experiments. J. Carrelha and H. Boukarabila performed the Rag1-Cre lineage tracing experiments. S.E. W. Jacobsen assisted in experimental design, data interpretation and article writing. S. Epelman assisted in experimental design and data interpretation.
Sources of Funding
This project was made possible by funding provided from the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (CHII2015-462), Foundation of Barnes-Jewish Hospital (8038–88), Burroughs Foundation Welcome Fund, and a programm grant (H4RPLK0) from the Medical Research Council UK to SEJ. K.J. Lavine was supported by National Institutes of Health (NIH) T32 HL007081, K08 HL123519, and the Oliver Langenberg Physician-Scientist Training Program. J. Carrelha was partially supported by a Marie Curie Early Stage Researcher Fellowship.
Nonstandard Abbreviations and Acronyms
- CCR2
chemokine (C-C motif) receptor 2
- CSF1R
colony-stimulating factor 1 receptor
- CX3CR1
CX3C chemokine receptor 1
- FITC
fluorescein isothiocyanate
- GFP
green florescent protein
- IGF
insulin like growth factor
- Rag1
recombination activating gene 1
- ROSA
reverse orientation splice acceptor
Footnotes
The online-only Data Supplement is available with this article at http://circres.ahajournals.org/lookup/suppl/doi:10.1161/CIRCRESAHA.115.308270/-/DC1.
Disclosures
None.
References
- 1.Epelman S, Lavine KJ, Randolph GJ. Origin and functions of tissue macrophages. Immunity. 2014;41:21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol. 2013;14:986–995. doi: 10.1038/ni.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ajami B, Bennett JL, Krieger C, McNagny KM, Rossi FM. Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci. 2011;14:1142–1149. doi: 10.1038/nn.2887. [DOI] [PubMed] [Google Scholar]
- 4.Boyer SW, Schroeder AV, Smith-Berdan S, Forsberg EC. All hematopoietic cells develop from hematopoietic stem cells through Flk2/Flt3-positive progenitor cells. Cell Stem Cell. 2011;9:64–73. doi: 10.1016/j.stem.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Böiers C, Carrelha J, Lutteropp M, et al. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell. 2013;13:535–548. doi: 10.1016/j.stem.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330:841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoeffel G, Wang Y, Greter M, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med. 2012;209:1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samokhvalov IM, Samokhvalova NI, Nishikawa S. Cell tracing shows the contribution of the yolk sac to adult haematopoiesis. Nature. 2007;446:1056–1061. doi: 10.1038/nature05725. [DOI] [PubMed] [Google Scholar]
- 9.Schulz C, Gomez Perdiguero E, Chorro L, Szabo-Rogers H, Cagnard N, Kierdorf K, Prinz M, Wu B, Jacobsen SE, Pollard JW, Frampton J, Liu KJ, Geissmann F. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 10.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, Hume DA, Perlman H, Malissen B, Zelzer E, Jung S. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lavine KJ, Epelman S, Uchida K, Weber KJ, Nichols CG, Schilling JD, Ornitz DM, Randolph GJ, Mann DL. Distinct macrophage lineages contribute to disparate patterns of cardiac recovery and remodeling in the neonatal and adult heart. Proc Natl Acad Sci U S A. 2014;111:16029–16034. doi: 10.1073/pnas.1406508111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epelman S, Lavine KJ, Beaudin AE, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40:91–104. doi: 10.1016/j.immuni.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoeffel G, Chen J, Lavin Y, et al. C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity. 2015;42:665–678. doi: 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hashimoto D, Chow A, Noizat C, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Azzoni E, Conti V, Campana L, Dellavalle A, Adams RH, Cossu G, Brunelli S. Hemogenic endothelium generates mesoangioblasts that contribute to several mesodermal lineages in vivo. Development. 2014;141:1821–1834. doi: 10.1242/dev.103242. [DOI] [PubMed] [Google Scholar]
- 16.Heidt T, Courties G, Dutta P, Sager HB, Sebas M, Iwamoto Y, Sun Y, Da Silva N, Panizzi P, van der Laan AM, van der Lahn AM, Swirski FK, Weissleder R, Nahrendorf M. Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ Res. 2014;115:284–295. doi: 10.1161/CIRCRESAHA.115.303567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8:265–277. doi: 10.1023/a:1008942828960. [DOI] [PubMed] [Google Scholar]
- 18.Jung S, Aliberti J, Graemmel P, Sunshine MJ, Kreutzberg GW, Sher A, Littman DR. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol. 2000;20:4106–4114. doi: 10.1128/mcb.20.11.4106-4114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, Kaiser EA, Snyder LA, Pollard JW. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature. 2011;475:222–225. doi: 10.1038/nature10138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz LD, Nishikawa S. The murine mutation osteopetrosis is in the coding region of the macrophage colony stimulating factor gene. Nature. 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 21.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavine KJ, White AC, Park C, Smith CS, Choi K, Long F, Hui CC, Ornitz DM. Fibroblast growth factor signals regulate a wave of Hedgehog activation that is essential for coronary vascular development. Genes Dev. 2006;20:1651–1666. doi: 10.1101/gad.1411406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbieri SS, Weksler BB. Tobacco smoke cooperates with interleukin-1beta to alter beta-catenin trafficking in vascular endothelium resulting in increased permeability and induction of cyclooxygenase-2 expression in vitro and in vivo. FASEB J. 2007;21:1831–1843. doi: 10.1096/fj.06-7557com. [DOI] [PubMed] [Google Scholar]
- 25.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, Rodewald HR. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gautier EL, Shay T, Miller J, et al. Immunological Genome Consortium. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Courties G, Heidt T, Sebas M, et al. In vivo silencing of the transcription factor IRF5 reprograms the macrophage phenotype and improves infarct healing. J Am Coll Cardiol. 2014;63:1556–1566. doi: 10.1016/j.jacc.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hilgendorf I, Gerhardt LM, Tan TC, Winter C, Holderried TA, Chousterman BG, Iwamoto Y, Liao R, Zirlik A, Scherer-Crosbie M, Hedrick CC, Libby P, Nahrendorf M, Weissleder R, Swirski FK. Ly-6Chigh monocytes depend on Nr4a1 to balance both inflammatory and reparative phases in the infarcted myocardium. Circ Res. 2014;114:1611–1622. doi: 10.1161/CIRCRESAHA.114.303204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boring L, Gosling J, Chensue SW, Kunkel SL, Farese RV, Jr, Broxmeyer HE, Charo IF. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J Clin Invest. 1997;100:2552–2561. doi: 10.1172/JCI119798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aurora AB, Porrello ER, Tan W, Mahmoud AI, Hill JA, Bassel-Duby R, Sadek HA, Olson EN. Macrophages are required for neonatal heart regeneration. J Clin Invest. 2014;124:1382–1392. doi: 10.1172/JCI72181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S, Wu Y, Capecchi MR. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- 32.Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circ Res. 2010;106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riley PR, Smart N. Vascularizing the heart. Cardiovasc Res. 2011;91:260–268. doi: 10.1093/cvr/cvr035. [DOI] [PubMed] [Google Scholar]
- 34.Pérez-Pomares JM, de la Pompa JL. Signaling during epicardium and coronary vessel development. Circ Res. 2011;109:1429–1442. doi: 10.1161/CIRCRESAHA.111.245589. [DOI] [PubMed] [Google Scholar]
- 35.Fantin A, Vieira JM, Gestri G, Denti L, Schwarz Q, Prykhozhij S, Peri F, Wilson SW, Ruhrberg C. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 2010;116:829–840. doi: 10.1182/blood-2009-12-257832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arnold T, Betsholtz C. The importance of microglia in the development of the vasculature in the central nervous system. Vasc Cell. 2013;5:4. doi: 10.1186/2045-824X-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rymo SF, Gerhardt H, Wolfhagen Sand F, Lang R, Uv A, Betsholtz C. A two-way communication between microglial cells and angiogenic sprouts regulates angiogenesis in aortic ring cultures. PLoS One. 2011;6:e15846. doi: 10.1371/journal.pone.0015846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stefater JA, III, Lewkowich I, Rao S, et al. Regulation of angiogenesis by a non-canonical Wnt-Flt1 pathway in myeloid cells. Nature. 2011;474:511–515. doi: 10.1038/nature10085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trembley MA, Velasquez LS, de Mesy Bentley KL, Small EM. Myocardin-related transcription factors control the motility of epicardium-derived cells and the maturation of coronary vessels. Development. 2015;142:21–30. doi: 10.1242/dev.116418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volz KS, Jacobs AH, Chen HI, Poduri A, McKay AS, Riordan DP, Kofler N, Kitajewski J, Weissman I, Red-Horse K. Pericytes are progenitors for coronary artery smooth muscle. Elife. 2015;4 doi: 10.7554/eLife.10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bach LA. Endothelial cells and the IGF system. J Mol Endocrinol. 2015;54:R1–13. doi: 10.1530/JME-14-0215. [DOI] [PubMed] [Google Scholar]
- 42.Mucenski ML, McLain K, Kier AB, Swerdlow SH, Schreiner CM, Miller TA, Pietryga DW, Scott WJ, Jr, Potter SS. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 1991;65:677–689. doi: 10.1016/0092-8674(91)90099-k. [DOI] [PubMed] [Google Scholar]
- 43.Outtz HH, Tattersall IW, Kofler NM, Steinbach N, Kitajewski J. Notch1 controls macrophage recruitment and Notch signaling is activated at sites of endothelial cell anastomosis during retinal angiogenesis in mice. Blood. 2011;118:3436–3439. doi: 10.1182/blood-2010-12-327015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.