Abstract
Rab GTPases are central elements of the vesicular transport machinery. An emerging view is that downstream effectors of these GTPases are multiprotein complexes that include nucleotide exchange factors to ensure coupling between GTPase activation and effector function. We have previously shown that Rab5, which regulates various steps of transport along the early endocytic pathway, is activated by a complex consisting of Rabex-5, a Rab5 nucleotide exchange factor, and the effector Rabaptin-5. We postulated that the physical association of these two proteins is necessary for their activity in Rab5-dependent endocytic membrane transport. To evaluate the functional implications of such complex formation, we have reconstituted it with the use of recombinant proteins and characterized its properties. First, we show that Rabaptin-5 increases the exchange activity of Rabex-5 on Rab5. Second, Rab5-dependent recruitment of Rabaptin-5 to early endosomes is completely dependent on its physical association with Rabex-5. Third, complex formation between Rabaptin-5 and Rabex-5 is essential for early endosome homotypic fusion. These results reveal a functional synergy between Rabaptin-5 and Rabex-5 in the complex and have implications for the function of analogous complexes for Rab and Rho GTPases.
INTRODUCTION
A well-established first step in endocytosis is the delivery of molecules internalized from the plasma membrane via clathrin-coated vesicles (CCV) into the early endosome. Initial generation of transport vesicles at the plasma membrane is mediated by the recruitment of soluble adaptor AP2 complexes (Kirchhausen, 1999) and subsequent clathrin assembly, resulting in the formation of clathrin-coated pits (Schmid, 1997). Coat propagation and fission of CCV from the plasma membrane are regulated by a series of protein factors and lipids (Kirchhausen, 2000), including the small GTPase Rab5 that plays a role in ligand sequestration (McLauchlan et al., 1998). Rab5 also mediates the tethering and fusion of CCV with early endosomes as well as homotypic fusion between early endosomes (Gorvel et al., 1991; Bucci et al., 1992; Li et al., 1994) in concert with the soluble N-ethylmaleimide–sensitive factor attachment receptor machinery (McBride et al., 1999). Finally, Rab5 modulates the motility of early endosomes along microtubules (Nielsen et al., 1999). A number of Rab5 effectors have been characterized, including early endosome-associated protein-1 (EEA1) (Mills et al., 1998; Simonsen et al., 1998; Christoforidis et al., 1999a), the phosphatidylinositol 3-kinases hVPS34-p150 and p85α-p110β (Christoforidis et al., 1999b), the Rabenosyn-5/VPS45 complex (Nielsen et al., 2000), Rabaptin-5 (Stenmark et al., 1995; Horiuchi et al., 1997), and the related Rabaptin-5β (Gournier et al., 1998), but several others remain to be identified and functionally characterized (Christoforidis and Zerial, 1999). This high complexity supports the view that different Rab effectors are probably required to regulate distinct downstream events.
Rabaptin-5 was initially identified as a Rab5 effector in a two-hybrid screen (Stenmark et al., 1995). It binds Rab5 in a GTP-dependent manner and is an essential and rate-limiting component for both homotypic early endosome and heterotypic CCV-early endosome fusion (Stenmark et al., 1995; Horiuchi et al., 1997; Vitale et al., 1998). The regulation of endocytosis during apoptosis by caspase-3 cleavage of Rabaptin-5 (Cosulich et al., 1997; Swanton et al., 1999) underlines the critical importance of this protein in the Rab5 pathway. Rabaptin-5 specifically interacts not only with Rab5 through its carboxyl terminus, but also with Rab4 through a distinct binding domain located at the amino terminal end of the protein (Vitale et al., 1998). Recruitment of endogenous cytosolic Rabaptin-5 to early endosomes occurs in a Rab5-dependent manner (Stenmark et al., 1995). However, Rabaptin-5 is not functional by itself, because it inhibits early endosome fusion in vitro (Horiuchi et al., 1997). An explanation for this effect was later provided by the finding that in cytosol Rabaptin-5 is stably bound to Rabex-5 (Horiuchi et al., 1997), a guanine nucleotide exchange factor (GEF) for Rab5. By forming a complex with Rabaptin-5, Rabex-5 was therefore proposed to physically couple activation of Rab5 to the recruitment and activity of the effector. Recently, it was shown that this property is shared by HOPS, a protein complex that contains an exchange factor and acts as an effector for Ypt7p in homotypic vacuole fusion (Seals et al., 2000; Wurmser et al., 2000). This suggests that the necessity to activate Rab proteins concomitantly with Rab effector function may be common to different trafficking steps. However, neither the functional requirement for the existence of Rab effectors in complex with exchange factors nor their mechanism of action has been demonstrated. It becomes therefore important to understand the significance and the functional properties of the Rabaptin-5/Rabex-5 complex. Here, to pursue this molecular dissection, we have expressed recombinant Rabaptin-5 and Rabex-5 and compared their roles with respect to nucleotide exchange for Rab5, membrane recruitment, and activity in early endosome fusion with individually expressed proteins or as a complex.
MATERIALS AND METHODS
Plasmids
The plasmid pGEM UEP (Vitale et al., 1998) was digested with NcoI and XbaI to release the Rabaptin-5 cDNA and ligate it into the his6-tagged pFAST BAC HTa expression vector (Life Technologies, Karlsruhe, Germany) digested with the same enzymes. To generate recombinant Rabex-5 baculoviruses, Rabex-5 was amplified by polymerase chain reaction (PCR) from the pGEM T-Rabex-5 (Horiuchi et al., 1997) with the use of the 5′ BamHI oligo GAC GGATCC CAT ATG AGC CTT AAG TCT GAA CGC and the 3′ SP6 polycloning site oligo. This PCR product was then digested with BamHI and SalI and ligated into pMAL-C2 (New England Biolabs, Frankfurt, Germany) treated with the same enzymes. The construct was confirmed by sequencing to avoid PCR-induced mutations. The pMAL-C2 Rabex-5 plasmid was subsequently digested with BamHI and HindIII to subclone Rabex-5 into the BamHI/HindIII digested pFAST BAC1 or pFAST BAC HTb (Life Technologies) to yield both the untagged (pFAST BAC1 Rabex-5) or his6-tagged (pFAST BAC HTb Rabex-5) constructs, respectively.
As templates for in vitro transcription/translation (see below), pMAL-C2 Rabex-5 was digested with BamHI/HindIII and Rabex-5 subcloned into pGEM-1 (Promega, Mannheim, Germany) containing a myc epitope and digested with the same enzymes to generate pGEM myc3-Rabex-5. The vector pGEM myc-UEP (i.e., pGEM myc-Rabaptin-5) vector has been previously described (Vitale et al., 1998).
Expression and Purification of Recombinant and Native Rabaptin-5/Rabex-5 Complex
Expression of the recombinant proteins in insect cells with the use of baculoviruses was achieved as described in detail elsewhere (Lippe et al., 2001). The individual proteins were expressed as his6-tagged proteins in High Five insect cells according to the manufacturer instructions with the use of pFAST BAC HTa-Rabaptin-5 or pFAST BAC1-Rabex-5. To produce recombinant Rabaptin-5/Rabex-5 complex, High Five cells were coinfected with his6-tagged Rabaptin-5 (pFAST BAC HTa-Rabaptin-5) and untagged Rabex-5 (pFAST BAC1-Rabex-5) baculoviruses (Lippe et al., 2001). In all cases, a single-step purification on a Ni-NTA agarose (Qiagen, Hilden, Germany) column was achieved as outlined in Lippe et al. (2001). The purification of the native cytosolic Rabaptin-5/Rabex-5 was achieved as previously reported in detail (Horiuchi et al., 1997).
Nucleotide Exchange Assay
Incubation mixtures were subjected to a filter binding assay, as described in Ridley et al. (1993). Briefly, 1.5 μM recombinant Rab5 produced in Escherichia coli or High Five insect cells (Alexandrov et al., 1994; Cremers et al., 1994) was incubated at 30°C in exchange buffer (20 mM HEPES, pH 7.4, 2 mM β-mercaptoethanol, 150 mM NaCl, 10% glycerol, 2.5 mM MgCl2) alone, with 100 nM Rabex-5 or with 100 nM Rabaptin-5/Rabex-5 complex in the presence of 5 μM [32P]GTPγ for the indicated times. The exchange reaction was stopped by adding 2 ml of ice-cold exchange buffer and filtered through nitrocellulose filters (2-cm diameter BA85; Schleicher & Schuell, Dassel, Germany). The filters were washed twice with 4 ml of the above-mentioned ice-cold buffer and then dried. The filter-bound radioactivity was counted with a beta scintillation counter.
Recruitment of 35S-Labeled Rabaptin-5/Rabex-5 onto Early Endosomes
When indicated, Rabaptin-5 and Rabex-5 under the control of the T7 promoter (see “Plasmids”) were transcribed and translated in vitro in presence of [35S]methionine with the use of a T7 polymerase TNT-coupled reticulocyte lysate kit (Promega, Madison, WI) exactly as per manufacturer's instructions. The quality of the preparations was analyzed by SDS-PAGE and autoradiography. The samples were quantified on a Fujifilm FLA2000 Phosphor Fluorescent Imager and the counts corrected for methionine content.
Aliquots of early endosomes were incubated in the presence of normalized (i.e., equimolar) 35S-labeled molecules (Rabaptin-5, Rabex-5, Rabaptin-5 plus Rabex-5, or preformed Rabaptin-5/Rabex-5 complex), ATP-regenerating system, 1 mM GTP, or 1 μM GDI and 1 mM GDP in the in vitro fusion assay buffer for 30 min at 37°C. Subsequently, the reaction was adjusted to 42% (wt/vol) sucrose and overlayed with 35 and 10% sucrose (wt/vol) in 3 mM imidazole pH 7.4 containing 1 mM GTP or GDP. After centrifugation at 215 000 × g for 3 h at 4°C (SW60 rotor), floated membranes were collected from the 35–10% interphase, diluted with the SIM buffer (250 mM sucrose, 3 mM imidazole, 1 mM MgCl2 pH 7.4), and pelleted by centrifugation at 350,000 × g for 30 min at 4°C in a TLA 100.4 rotor. The pellets were resuspended in loading buffer and analyzed by SDS-PAGE and fluorography or immunoblotting.
Immunoprecipitations
For coimmunoprecipitation studies of baculovirus-expressed recombinant proteins, 2 μl of crude rabbit polyclonal preimmune serum, α Rabaptin (L1–46) or α Rabex (3399) antibody was added to 5–10 μl of recombinant protein diluted to 150 μl in lysis buffer (20 mM Tris-Cl pH 7.6, 120 mM NaCl, 4 mM MgCl2, 1% NP-40, and protease inhibitors). It should be noted that the antibodies are specific for their respective antigens and that they do no cross react (Horiuchi et al., 1997; Lippe et al., 2001). After a 15-min incubation on a wheel at 4°C, 20 μl of protein A agarose (Roche, Mannheim, Germany) pre-equilibrated in lysis buffer was added and the samples incubated for a further hour. The unbound material was kept and the beads successively washed three times in buffer B (10 mM Tris-Cl pH 7.5, 150 mM NaCl, 2 mM EDTA, 0.2% NP-40), twice in buffer C (10 mM Tris-Cl pH 7.5, 500 mM NaCl, 2 mM EDTA, 0.2% NP-40) and once in buffer D (10 mM Tris-Cl pH 7.5). Unbound and bead bound material was analyzed by SDS-PAGE and Western blot as described above.
An alternative protocol was used for the coimmunoprecipitation of in vitro transcribed/translated recombinant proteins to obtain a complete immunoprecipitation and mimic the conditions used for recruitment. The modifications included the use of 5 μl of crude antiserum, equimolar amounts of 35S-labeled proteins incubated for 0–30 min under the conditions used for recruitment, 25 μl of protein A agarose beads, and a 1 h 30 min incubation of beads with samples.
In Vitro Fusion Assay
The fusion assay was described previously (Horiuchi et al., 1997). Briefly, enriched early endosomes fractions containing either biotinylated transferrin or sheep α human transferrin antibodies were isolated (Gorvel et al., 1991). They were mixed in the presence of fusion buffer (12.5 mM HEPES pH 7.4, 1.5 mM MgOAc, 3 mM imidazole, 1 mM dithiothreitol, 75 mM KOAc), ATP-regenerating mix (17.3 mM creatine phosphate, 87 μg/ml creatine kinase, and 2.2 mM ATP), unlabeled holo-transferrin as quencher, and 3 mg/ml complete HeLa cytosol or immunodepleted of its Rabaptin-5/Rabex-5 complex content. To deplete cytosol, protein A agarose beads were first saturated with 10 mg/ml bovine serum albumin for 10 min at 4°C, quickly washed twice in KEHM buffer (50 mM KCl, 10 mM EGTA, 50 mM HEPES pH 7.4, 2 mM MgCl2) and incubated for 1 h with 20 μl of crude preimmune or the L1–46 α Rabaptin-5-specific antiserum (Stenmark et al., 1995) in a total of 750 μl in KEHM. The beads were then washed twice for 10 min at 4°C to remove unbound antibodies and resuspended in 100 μl of cytosol. After a 30-min incubation, the beads were spun down and the depleted cytosol recovered. The depletion was analyzed by SDS-PAGE and Western blot with the use of L1–46 to detect Rabaptin-5 (Stenmark et al., 1995) or the affinity-purified rabbit polyclonal antibody 2263 to detect Rabex-5 (Horiuchi et al., 1997).
After a 25-min incubation at 37°C, the fusion reaction was stopped on ice with wash buffer (50 mM Tris-Cl pH 8.5, 100 mM NaCl, 1 g/l bovine serum albumin, 2% wt/vol Triton X-100). The immunocomplexes were retrieved with streptavidin-coated magnetic beads (Dynal, Hamburg, Germany) and labeled with a secondary α sheep antibody coupled to ruthenium trisbipyridine chelate (IGEN, Oxford, England). The fusion quantified with an Origen analyzer (IGEN).
Stoichiometry
To determine the stoichiometry of the Rabaptin-5/Rabex-5 complex, 4 × 10-cm dishes of subconfluent HeLa cells were incubated overnight in labeling medium (met−/cys− DMEM, 2% serum, 500 μCi of 35S translabel (ICN, Irvine, CA). After two washes with cold phosphate-buffered saline (PBS), the cells were scraped in PBS (1 ml/dish), pooled and resuspended in 1–1.5 ml. They were then broken with a syringe and a 27-gauge needle in presence of protease inhibitors. The nuclei and unbroken cells were removed by centrifugation and the cytosol was obtained by centrifugation at 100,000 × g for 30 min. The supernatant was split into four equal samples and precleared with preimmune serum or an antiserum specific for Rabaptin-5β to allow quantification of the Rabaptin-5α/Rabex-5 complex (Gournier et al., 1998). The samples were then immunoprecipitated as described above with affinity-purified anti-Rabaptin-5α or Rabex-5 antibodies. The samples were analyzed by SDS-PAGE, the gels treated with Entensify (DuPont, Bad Homburg, Germany), dried, and put into a phosphorimager cassette. They were finally quantified on a Fujifilm FLA2000 Phosphor Fluorescent Imager and the counts corrected for methionine and cysteine content.
RESULTS
Reconstitution of Recombinant Rabaptin-5/Rabex-5 Complex from Insect Cells
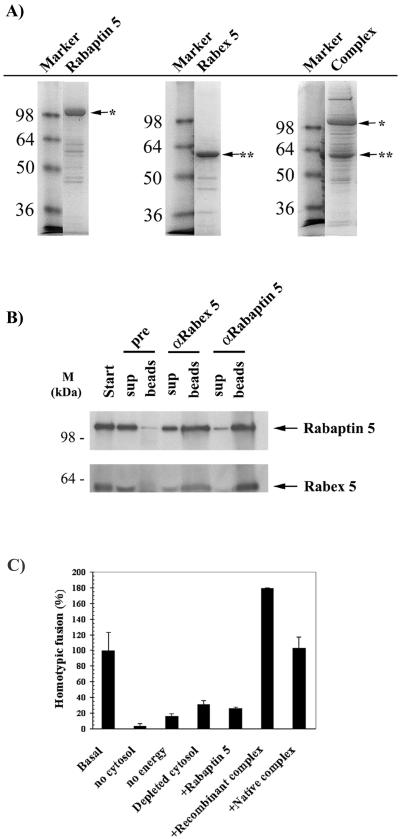
To analyze the functional properties of the Rabaptin-5/Rabex-5 complex in vitro, we reconstituted the complex from recombinant proteins expressed in insect cells with the use of the Bac-to-Bac baculovirus expression system. Histidine-tagged Rabaptin-5 was coexpressed in insect cells with untagged Rabex-5. The complex was then purified on a nickel agarose affinity column and analyzed by SDS-PAGE. Under these conditions, untagged Rabex-5 copurified with tagged Rabaptin-5, suggesting that the two proteins formed a complex in vivo (Figure 1A). As controls, histidine-tagged Rabaptin-5 or Rabex-5 was independently expressed, purified in parallel, and analyzed as for the complex (Figure 1A).
Figure 1.
Purification of functional recombinant Rabaptin-5/Rabex-5 complex. (A) Recombinant protein expression. Histidine-tagged Rabaptin-5 or Rabex-5 was expressed in High Five insect cells with the use of recombinant baculoviruses as described in MATERIALS AND METHODS. In addition, histidine-tagged Rabaptin-5 and untagged Rabex-5 baculoviruses were coexpressed to produce a complex in vivo. All recombinant proteins were purified in a single step via a nickel agarose affinity column. The purified proteins were then loaded onto a 12% SDS-PAGE gel and stained with Coomassie. The molecular mass markers are indicated to the left of the gels, whereas the position of the recombinant proteins (∗, Rabaptin-5; ∗∗, Rabex-5) is shown by arrows. (B) Coexpression of the two proteins results in the formation of a complex in vivo. The recombinant complex purified from insect cells coexpressing the two proteins was immunoprecipitated with preimmune, α Rabex-5 or α Rabaptin-5 serum as detailed in MATERIALS AND METHODS. Both the washed beads and the supernatants were loaded onto a 12% SDS-PAGE gel and Coomassie stained. A minimal immunoprecipitation was observed with the preimmune anti body. (C) Homotypic early endosome fusion assay. Endosomes were purified and incubated in the presence of cytosol and energy, unless otherwise indicated. In lanes 4–7, the endogenous native Rabaptin-5/Rabex-5 complex was immunodepleted from the cytosol. The rescue of the depleted cytosol by 100 nM recombinant Rabaptin-5 (lane 5) or Rabaptin-5/Rabex-5 complex purified from insect cells coexpressing the two proteins (lane 6) was compared with the rescue by 100 nM purified native Rabaptin-5/Rabex-5 complex (lane 7).
To confirm the interaction between Rabaptin-5 and Rabex-5 and rule out a nonspecific copurification of Rabex-5 on the nickel agarose column, antibodies specific for either Rabaptin-5 or Rabex-5 (Stenmark et al., 1995; Horiuchi et al., 1997) were used to coimmunoprecipitate the recombinant complex (as described in MATERIALS AND METHODS). As shown in Figure 1B, anti-Rabaptin-5 antibodies coprecipitated Rabex-5 and vice-versa, confirming their mutual binding. This interaction was further corroborated by the comigration of the two proteins by gel filtration chromatography on a Superose 6 column (our unpublished results).
Recombinant Rabaptin-5/Rabex-5 Complex Is Functional
The native Rabaptin-5/Rabex-5 complex plays a crucial role in the homotypic fusion of early endosomes (Horiuchi et al., 1997). Depletion of this complex from cytosol severely reduces the ability of the endosomes to fuse in vitro. Addition of purified native Rabaptin-5/Rabex-5 complex rescues this inhibition. To ascertain the activity of the recombinant Rabaptin-5/Rabex-5 complex, we tested its ability to rescue the inhibition of early endosome fusion after the depletion of the endogenous Rabaptin-5/Rabex-5 complex from cytosol (Figure 1C). As previously reported (Horiuchi et al., 1997), endosome fusion was dependent on endogenous cytosolic Rabaptin-5/Rabex-5 complex and addition of recombinant Rabaptin-5 alone to the depleted cytosol did not rescue fusion. In contrast, addition of recombinant complex purified from insect cells coexpressing the two proteins restored fusion to basal levels or beyond. The recombinant complex was in fact at least as active as the native complex obtained from bovine cytosol, indicating that it was fully functional.
Rabaptin-5 Enhances Rabex-5 Nucleotide Exchange Activity
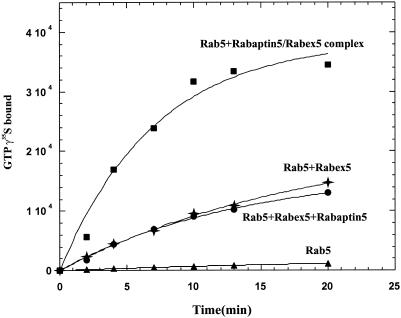
To further explore the functionality of the recombinant Rabaptin-5/Rabex-5 complex, we investigated its ability to catalyze the exchange of GDP for GTP on the Rab5 GTPase (Horiuchi et al., 1997). In particular, given the nucleotide exchange activity of Rabex-5 in the absence of Rabaptin-5 (Horiuchi et al., 1997), we wondered whether Rabaptin-5 might directly modulate this activity. To examine this possibility, we compared the nucleotide exchange activity of free and complexed Rabex-5 by incubation of Rab5 in the presence or absence of either Rabex-5 or equimolar amounts of Rabex-5 complexed to Rabaptin-5 or Rabaptin-5 and Rabex-5 added as separate molecules. The results shown in Figure 2 indicate that Rabaptin-5/Rabex-5 complex was threefold more active than Rabex-5 alone or in the presence of uncomplexed Rabaptin-5. This increase was not due to additive nucleotide exchange activities of Rabaptin-5 and Rabex-5 (Figure 2) consistent with our previous report that Rabaptin-5 has no detectable activity on its own (Horiuchi et al., 1997). These results indicate that the association of Rabaptin-5 to Rabex-5 stimulated the GEF activity of Rabex-5. It is possible that the stimulation of the enzymatic activity may be due to an increased stability of the GEF or to a proper folding of the protein when it is complexed to the Rab5 effector.
Figure 2.
Rabaptin-5 enhances Rabex-5 GEF activity. GDP to GTP nucleotide exchange on Rab5 was measured by filter binding assay in the presence of 5 μM [32P]GTPγs for the indicated times. This was achieved with 1.5 μM Rab5 alone (▴) or with 100 nM Rabex-5 (✦), 100 nM Rabex-5 and 100 nM Rabaptin-5 (●), or 100 nM Rabaptin-5/Rabex-5 complex (▪).
Recruitment of Rabaptin-5 Is Dependent on Its Association with Rabex-5
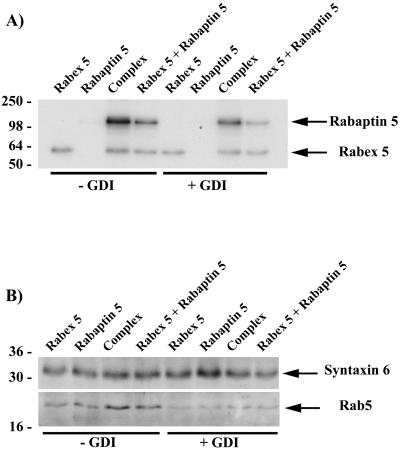
We previously proposed that the physical association of Rabaptin-5 to Rabex-5 might couple Rab5 activation and effector recruitment (Horiuchi et al., 1997). For instance, the Rabaptin-5/Rabex-5 association may be important for the clustering of active Rab5 proteins on the membrane, as previously proposed (Horiuchi et al., 1997). A central prediction of this hypothesis is therefore a dependence of Rab5 function on the recruitment of Rabaptin-5 and Rabex-5 as a complex rather than individual proteins. To test this, we examined whether recruitment of Rabaptin-5 onto early endosomes depends on its association to Rabex-5. To distinguish between proteins recruited from cytosol and endogenous Rabaptin-5 and Rabex-5 already present on the endosomes, we used equimolar amounts of [35S]methionine-labeled in vitro translated proteins. We examined the membrane recruitment of 1) Rabex-5 alone, 2) Rabaptin-5 alone, 3) Rabex-5 and Rabaptin-5 translated separately, and 4) cotranslated Rabaptin-5/Rabex-5 to facilitate complex formation. To confirm that the recruitment is dependent on Rab5, endosomes were incubated with either GTP or GDP and 1 μM RabGDI. The latter condition allows an efficient extraction of Rab5 from the endosomal membrane (Ullrich et al., 1994). After the incubation with the labeled proteins, membranes were floated and examined for the presence of the recruited proteins by SDS-PAGE and autoradiography. The results show a differential recruitment depending on the proteins and their association (Figure 3A). Surprisingly, free Rabex-5 was recruited onto endosomes and this recruitment was largely GDI-insensitive and, therefore, Rab5-independent. In contrast, incubation of early endosomes with Rabaptin-5 alone led to very poor Rabaptin-5 recruitment. A weak band was detectable only upon prolonged exposure. Early endosomes recruited both Rabaptin-5 and Rabex-5 when added as a complex. Importantly, addition of RabGDI significantly reduced this recruitment, concomitant with a decrease of Rab5 on the membranes (Figure 3B). The presence of Syntaxin 6, a transmembrane marker of early endosomes (Bock et al., 1997; Simonsen et al., 1999), indicates that similar amounts of endosomal membranes were loaded in the various lanes. Together, these results show that efficient Rabaptin-5 recruitment requires Rabex-5 and is dependent on the presence of Rab5 on the early endosomes.
Figure 3.
Recruitment of Rabaptin-5 and Rabex-5 on early endosomes. (A) Recruitment of the Rabaptin-5/Rabex-5 complex. The recruitment of Rabaptin-5, Rabex-5, and Rabaptin-5/Rabex-5 complex was evaluated by incubation of 35S-labeled in vitro transcribed/translated proteins with purified early endosomes for 30 min at 37°C. This was performed in the presence of 1 mM GTP or 1 μM GDI plus 1 mM GDP to determine the Rab5 dependence of the recruitment. After the recovery of the endosomes by flotation, the endosomes were pelleted and loaded onto a 12% SDS-PAGE gel and analyzed by autoradiography. (B) Western. Essentially, the above-described gels were cut below the 50-kDa molecular mass marker before drying and the lower part of the gels transferred to nitrocellulose. Removal of Rab5 by GDI was confirmed with the use of an α Rab5 antibody. As loading control, the amount of an integral protein present on endosomes was determined by Western with a Syntaxin 6-specific antibody.
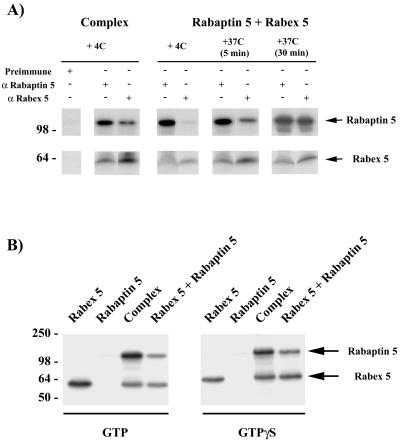
To determine whether the recruitment of Rabaptin-5 required direct association with Rabex-5, we compared the binding to endosomes of Rabaptin-5 and Rabex-5 either added together as individual proteins (equal molar amounts) or as preformed complex. Figure 3A indicates that incubation of the endosomes with Rabaptin-5 and Rabex-5 resulted in the recruitment of both proteins. However, the recruitment of Rabaptin-5 increased two- to threefold (256 ± 41%) when complexed to Rabex-5. The lower but significant recruitment of free Rabaptin-5 in presence of Rabex-5 translated in a separate reaction increased the possibility that a fraction of free Rabaptin-5 interacted with Rabex-5 and formed a complex during the course of the incubation. To verify this, we performed coimmunoprecipitation experiments under identical conditions (Figure 4A). Whereas the preformed complex could be coimmunoprecipitated, little or no complex was formed between the individually added Rabaptin-5 and Rabex-5 when incubated at 4°C. However, an increasing complex formation occurred when proteins were incubated for 5–30 min at 37°C. Consequently, Rabaptin-5 was most likely recruited on endosomes because it bound Rabex-5 beforehand. Because Rabaptin-5 directly binds to Rab5-GTP in biochemical and two-hybrid assays (Stenmark et al., 1995; Horiuchi et al., 1997), the possibility exists that Rab5 could first be activated by Rabex-5 and subsequently bind free Rabaptin-5. We therefore repeated the experiments in the presence of a large excess of guanosine-5′-O-(3-thio)triphosphate (GTPγS) (1 mM) to lock Rab5 into its active form. Under these conditions, rapid and efficient nucleotide loading on Rab5 takes place (Rybin et al., 1996), presumably due to the presence of sufficient amounts of Rabaptin-5 and Rabex-5 on the early endosome membrane. Rabaptin-5 alone was once again very poorly recruited onto the endosomes (Figure 4B) and a weak band was only detectable upon long exposure, indicating that it must be bound to Rabex-5 to be efficiently recruited. Thus, Rabaptin-5 could be stably recruited onto early endosomes solely as a complex and in a Rab5-dependent manner. In contrast, free Rabex-5 was recruited in a largely Rab5-independent manner.
Figure 4.
Rabaptin-5 recruitment is dependent on its association to Rabex-5. (A) In vitro formation of Rabaptin-5/Rabex-5 complex. To test whether Rabaptin-5 and Rabex-5 can form a complex in the test tube during the recruitment incubations, we pooled Rabaptin-5 and Rabex-5 for 5 or 30 min at 37°C in the conditions used for the recruitment (ATP-regenerating system, 1 mM GTP in the in vitro fusion assay buffer). The samples were then immunoprecipitated with preimmune, α Rabaptin-5 or α Rabex-5 serum (as described in MATERIALS AND METHODS). The controls included the incubation of a sample left at 4°C for 30 min (i.e., 0 min at 37°C) as well as the immunoprecipitation of Rabaptin-5/Rabex-5 complex preformed in vivo. (B) GTPγS-loaded Rab5 fails to recruit uncomplexed Rabaptin-5. To evaluate whether the recruitment of Rabaptin-5 is solely dependent on the activation of Rab5 by Rabex-5, we performed the recruitment experiments as in Figure 3 in the presence of 1 mM GTP or GTPγS.
Preformed Rabaptin-5/Rabex-5 Complex Is Required for Homotypic Fusion of Early Endosomes
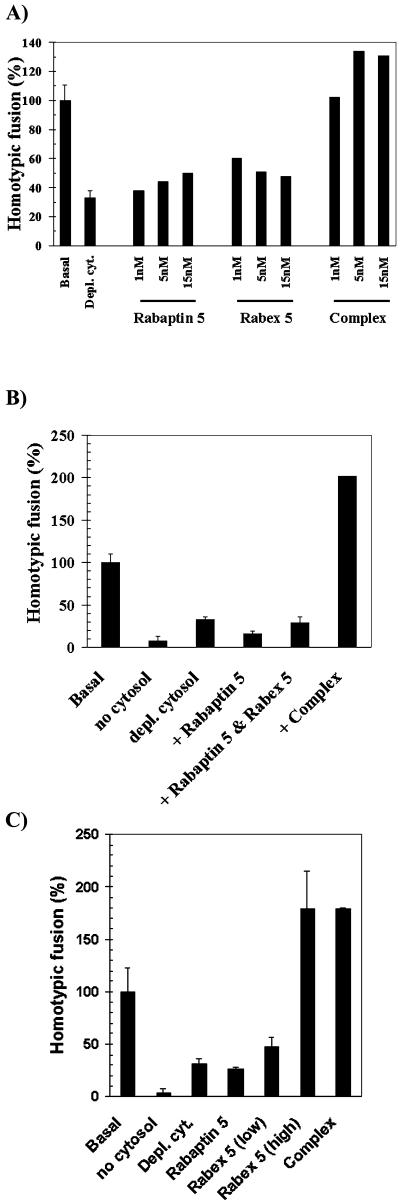
We demonstrated above that Rabaptin-5 enhances the nucleotide exchange activity of Rabex-5. We further showed that the association of Rabaptin-5 to Rabex-5 was necessary for Rabaptin-5 recruitment onto early endosomes in a Rab5-dependent manner. The next step was to evaluate the functional relevance of this requirement. We therefore determined whether the association of Rabaptin-5 with Rabex-5 is essential for the homotypic fusion of early endosomes. More specifically, we evaluated the ability of recombinant Rabaptin-5, Rabex-5, or Rabaptin-5/Rabex-5 complex to rescue the depletion of the endogenous native Rabaptin-5/Rabex-5 complex from the cytosol (Figure 5A). Normally, 95 nM Rabaptin-5 and 15 nM Rabex-5 are present in HeLa cytosol in a typical early endosome fusion assay (containing 3 mg/ml cytosol in a final 20-μl reaction). Depletion of the Rabaptin-5/Rabex-5 complex limited the fusion activity to 30% fusion compared with control cytosol. Addition of either Rabaptin-5 or Rabex-5 alone improved the fusion activity only to a limited extent. In contrast, addition of the complex completely restored and even stimulated the fusion activity in a concentration-dependent manner. To demonstrate that the rescue was dependent on the complex rather than on the presence of the two individual proteins, Rabaptin-5 and Rabex-5 were added individually or as preformed complex purified from insect cells coexpressing the two proteins. Interestingly, Figure 5B shows that although Rabaptin-5 did not support fusion in the presence of uncomplexed Rabex-5, an equivalent amount of Rabaptin-5/Rabex-5 complex fully restored fusion activity. Thus, even when added together, Rabaptin-5 and Rabex-5 could not promote early endosome fusion and only the preformed complex was functional.
Figure 5.
Rabaptin-5/Rabex-5 complex is essential for homotypic early endosome fusion. Rescue of cytosol depleted of endogenous Rabaptin-5/Rabex-5 complex was investigated in the homotypic early endosome fusion assay. (A) Rabaptin-5/Rabex-5 complex is needed for fusion. Low physiological concentrations of recombinant Rabaptin-5 (lanes 3–5), Rabex-5 (lanes 6–8), or complex (lanes 9–11) were used for the rescue. (B) Rabaptin-5/Rabex-5 complex must be preformed to be functional. Rabaptin-5 (150 nM) (lane 4), 150 nM Rabaptin-5, and 30 nM Rabex-5 (lane 5) or 150 nM Rabaptin-5/30 nM Rabex-5 complex (lane 6) was added to the test tubes. (C) Excess of Rabex-5 overrides the need for the Rabaptin-5/Rabex-5 complex. Rabaptin-5 (100 nM) (lane 4), 50 or 300 nM Rabex-5 (lanes 5–6), and 100 nM recombinant Rabaptin-5/Rabex-5 complex (lane 7) was, respectively, tested.
Excess Rabex-5 Can Override Need for Rabaptin-5/Rabex-5 Complex
Quantification of Rabaptin-5, Rabex-5, and Rab5 on endosomes by Western blot analyses with the use of recombinant proteins as standards revealed that 115 ± 61 pmol of Rab5 is present per milligram of suspension HeLa (sHeLa) protein in an early endosome-enriched fraction. In comparison, only 9 ± 2 pmol of Rabaptin-5 or 6 ± 2 of Rabex-5 is detectable per milligram of endosomes (Table 1). Thus, Rab5 is ∼15- to 20-fold more abundant on early endosomes than Rabaptin-5 or Rabex-5. Presumably, a local amplification of active Rab5 is necessary to spatially and temporally control Rab5 activity. We hypothesize this is achieved by the targeted recruitment of the Rabaptin-5/Rabex-5 complex. If true, the overall activation of Rab5 on the entire surface of endosomes, for instance, by an excess of Rabex-5, should override the necessity for a local recruitment of complex. We tested this possibility in our endosome fusion assay with the use of Rabaptin-5/Rabex-5–depleted cytosol. Figure 5C indicates that a threefold excess of Rabex-5 over the endogenous Rabex-5 levels (normally ∼15 nM in the assay) did not rescue the depletion of the complex. However, a 20-fold excess of Rabex-5 completely overrode the need for the complex. Moreover, it was equally efficient in restoring fusion as endogenous levels of Rabaptin-5/Rabex-5 complex. In contrast, free Rabaptin-5 did not rescue fusion. In fact, consistently with our previous results (Horiuchi et al., 1997), as little as a threefold excess completely abolished endosome fusion (our unpublished results). To evaluate whether Rabex-5 could as a free cytosolic molecule stimulate endosome fusion under physiological conditions, we metabolically labeled cells with [35S]met/cys and immunoprecipitated Rabaptin-5 or Rabex-5 with affinity-purified antibodies (in MATERIALS AND METHODS). After SDS-PAGE and quantification with a phosphorimager, the results indicated that free uncomplexed Rabex-5 is undetectable in cytosol (our unpublished results). Furthermore, the results showed that each mole of Rabex-5 was in fact complexed to 2 mol of Rabaptin-5 in suspension HeLa cytosol (2.1 ± 0.1; Table 1). This argues that under physiological conditions no excess Rabex-5 could override the need for Rabaptin-5 to support early endosome fusion.
Table 1.
Quantification of native Rabaptin 5/Rabex 5 complex
| A. Total amounts in cells | |||
|---|---|---|---|
| Rabaptin 5 | Rabex 5 | Rab5 | |
| Cytosol | 32 ± 13 pmol/mg | 5 ± 3 pmol/mg | 6 ± 3 pmol/mg |
| Early endosomes | 9 ± 2 pmol/mg | 6 ± 2 pmol/mg | 115 ± 61 pmol/mg |
| B. Stoichiometry of complex in cytosol | Rabaptin 5∶Rabex 5
|
|||
|---|---|---|---|---|
| Immunoprecipitation | Rabaptin 5 | Rabex 5 | Ratio | Average |
| α Rabaptin 5 | 1549 U | 258 U | 6.0∶1 | 5.9 ± 0.2∶1 |
| α Rabex 5 | 439 U | 212 U | 2.1∶1 | 2.1 ± 0.1∶1 |
A. Total Rabaptin-5, Rabex-5, and Rab5 in cells. Different quantities of sHeLa cytosol (10–700 μg) and enriched early endosomes (10 and 20 μg) were loaded on a 12% SDS-PAGE gel. In addition, 5–100 ng of recombinant Rabaptin-5, Rabex-5, or Rab5 isolated from insect cells were loaded as standards. The proteins were then transferred to nitrocellulose and probed by Western with Rabaptin-5, Rabex-5, or Rab5 specific antibodies. Several exposures on x-ray films were obtained and scanned, and the native proteins were quantified using a titration curve obtained with the recombinant standards. Only the exposures falling into the linear part of the titration curves were considered. The data are expressed per milligram of total sHeLa protein as measured with a Biorad protein assay kit. B. Stoichiometry of the native Rabaptin-5/Rabex-5 complex in cytosol. Subconfluent HeLa cells were metabolically labeled with 35S met/cys. Cytosol was prepared from the cells and split in four equal samples. Following a preclearing step, the complexed and free Rabaptin-5 or Rabex-5 molecules were immunoprecipitated with affinity-purified anti-Rabaptin-5α or Rabex-5 antibodies, respectively. The samples were analyzed by SDS-PAGE and were quantified on a Fujifilm FLA2000 Phosphor Fluorescent Imager. The arbitrary counts (units) were corrected for the methionine and cysteine contents of the proteins. “Ratio” refers to the signal obtained for Rabaptin-5 divided by the one obtained for Rabex-5 for the experiment shown in the table. To the right, “average” indicates the results from two independent experiments (± standard error).
DISCUSSION
The finding that the Rab5 effector Rabaptin-5 is bound to the exchange factor Rabex-5 in cytosol led us to suggest that complex formation between the two factors was a prerequisite for coupling nucleotide exchange to Rab5-dependent early endocytic membrane recruitment and fusion (Horiuchi et al., 1997). In this study, we have tested this hypothesis by investigating the functional properties of recombinant Rabaptin-5/Rabex-5 complex purified from insect cells. The complex was demonstrated to be functional by two different criteria, namely, activation of Rab5 by nucleotide exchange and homotypic fusion of early endosomes. This implies that the native Rabaptin-5 complex found in bovine cytosol has been fully reconstituted, consists of its two critical components, Rabaptin-5 and Rabex-5, and is active.
Since the identification of the Rabaptin-5/Rabex-5 complex (Stenmark et al., 1995; Horiuchi et al., 1997), various multiprotein complexes have been implicated in the regulation or as effectors of Rab GTPases in distinct transport reactions. For example, in yeast, the HOPS complex functions as an effector for Ytp7p in homotypic vacuole fusion and acts as a GEF for this small GTPase (Seals et al., 2000; Wurmser et al., 2000). In addition, the report that Sec2p (TerBush et al., 1996), the Sec4p exchange factor (Walch-Solimena et al., 1997), is a member of a large multiprotein complex (Nair et al., 1990) that is distinct from the Sec4p effector complex Exocyst (Guo et al., 1999) underlines the importance of the regulation of Rab proteins by complexes rather than individual proteins. In yeast Saccharomyces cerevisiae, transport from the Golgi apparatus to prevacuolar endosomes is thought to be mediated by the Rab5 homologue Ypt51p/Vps21p and a class D vps complex containing the effector Vac1p (Burd et al., 1997). Despite the lack of a Rabaptin-5 homologue in yeast, it would be interesting to see whether Vps9p, the homologue of Rabex-5 (Burd et al., 1996; Horiuchi et al., 1997), is complexed to other class D vps factors (Burd et al., 1997). Similarly, it would be interesting to see whether the transport protein particle (TRAPP) and Ric1/Rgp1 complexes that exchange nucleotides on Ypt1p/Ypt31p/32p and Ypt6p, respectively (Jones et al., 2000; Siniossoglou et al., 2000; Wang et al., 2000), also include effector functions.
The incorporation of GEFs and effectors within stable protein complexes has the advantage to couple Rab activation to downstream effector function. We have here demonstrated that Rabaptin-5 and Rabex-5 functionally cooperate but this synergy is conditional upon complex formation. First, Rabaptin-5 stimulated the basal GEF activity of Rabex-5 by threefold in the complex. This may result from an actual increase of Rabex-5 nucleotide exchange activity upon binding to Rabaptin-5. Alternatively, it may be that Rabaptin-5 stabilizes Rabex-5 in its active folded state. This latter possibility is interesting because it suggests that Rabaptin-5 may be a chaperone for the exchange factor. To examine these two possibilities, in-depth kinetic and folding studies will be necessary. Unfortunately, this is at present difficult given the limited yields of the Rabaptin-5/Rabex-5 complex (Lippe et al., 2001). Irrespective of the mechanism of this synergy, it is clear that Rabaptin-5 alone is not functional and that Rabaptin-5 and Rabex-5 mutually benefit from their association. Second, although Rabaptin-5 directly interacts with Rab5-GTP in two-hybrid or biochemical assays (Stenmark et al., 1995), it can only be efficiently recruited onto endosomes when associated to Rabex-5. In retrospect, this explains the previously reported 10-fold weaker recruitment of recombinant Rabaptin-5 on early endosomes compared with cytosolic Rabaptin-5 complexed to Rabex-5 (Stenmark et al., 1995; Horiuchi et al., 1997). A constant nucleotide exchange activity of Rabex-5 on Rab5 may be required for a stable membrane recruitment of Rabaptin-5 or the affinity of Rabaptin-5 for Rab5 may be increased upon Rabex-5 binding. Third, addition of free Rabaptin-5 together with uncomplexed Rabex-5 failed to support fusion under physiological conditions, i.e., when present at the same concentration as the endogenous proteins in cytosol, indicating that preformed complex was also essential for the Rab5-mediated fusion of early endosomes in vitro. Therefore, although Rabex-5 alone retained nucleotide exchange activity on Rab5, Rabex-5, and Rabaptin-5 must be bound to one another to be fully functional. The association of Rabaptin-5 with Rabex-5 thus has an important impact on Rab5 activation, effector recruitment, and function.
Given the Rab5-dependent recruitment of the complex, the function of Rabaptin-5 may be to position Rabex-5 on early endosomes to create a cluster of active Rab5 on the membrane (Horiuchi et al., 1997). The 2:1 molar ratio of Rabaptin-5 and Rabex-5 in the complex may serve to further amplify this clustering effect. In addition, oligomerization of the Rabaptin-5/Rabex-5 complex with EEA1 and other components (McBride et al., 1999) would further promote the formation of a patch of active Rab5 and of Rab5 effectors on the endosomal membrane (McBride et al., 1999; Roberts et al., 1999; Sonnichsen et al., 2000). An excess of Rabex-5 may artificially overcome the need for local recruitment of the Rabaptin-5/Rabex-5 complex by activating Rab5 in a spatially unrestricted manner (Figure 5B). However, the data indicate that such excess is not physiological. The use of such amplification systems involving GEF/effector complexes is not limited to Rab proteins but extends to other GTPases. Like Rabaptin-5, the Rac1 and Cdc42 effector PAK stimulates the endogenous nucleotide exchange activity of its associated GEF (PIX) and binding of PAK to PIX is essential for the membrane recruitment (and activation) of the effector on the membrane (Manser et al., 1998). However, the function of Rabaptin-5 goes beyond that of a membrane adaptor for Rabex-5. Rabaptin-5 interacts with both active Rab4 and Rab5 (Vitale et al., 1998), suggesting an important link between the endocytic and recycling pathways. In addition to Rabex-5, Rab5, and Rab4, a number of Rabaptin-5–interacting partners have been documented. Rabaptin-5 has been found to interact with tuberin (Xiao et al., 1997), GAP-43 (Neve et al., 1998), γ adaptins (Hirst et al., 2000), Rabphillin3 (Ohya et al., 1998), and the MAK-V protein kinase (Korobko et al., 2000). Finally, a Rabaptin-5 homologue (neurocrescin) involved in cone growth and exocytosis has been reported (Nishimune et al., 1997). Taken together, this suggests that the Rabaptin-5/Rabex-5 complex may play an important role in linking different components of the endocytic pathway and/or coupling different pathways. It will therefore be important to clarify the nature of these interactions and putative roles to fully understand the function of Rabaptin-5.
The Rab5 FYVE effector proteins EEA1 and Rabenosyn-5 are present on the early endosomes but absent from the plasma membrane (Mu et al., 1995; Nielsen et al., 2000; Wilson et al., 2000). This distribution is consistent with the asymmetric localization between CCV and early endosomes of the phosphatidylinositol 3-kinase hVPS34-p150 and its product phosphatidylinositol 3-phosphate (Christoforidis et al., 1999b; Gillooly et al., 2000). Furthermore, we were unable to detect large patches of Rab5 on the plasma membrane in comparison with the early endosomes (Sonnichsen et al., 2000). These observations suggest that the participation of the Rabaptin-5/Rabex-5 complex in the formation of a Rab5 membrane domain together with the other Rab5 effectors may be restricted to the early endosomes. This mechanism may be desirable to ensure directionality of vesicular transport to the latter compartment.
The activation of Rab5 necessary for clathrin-coated vesicle formation may require the Rabaptin-5/Rabex-5 complex without the formation of large patches of Rab5 on the plasma membrane. However, we have here uncovered the ability of Rabex-5 to be recruited as a free molecule independently of both Rabaptin-5 and Rab5 (Figure 3A). This suggests that a Rabex-5 receptor distinct from Rab5 might be present on endosomes and CCV derived from the plasma membrane. Interestingly, this is similar to the largely Sec4p-independent targeting of the Sec2p to vesicles and to the bud site in yeast (Elkind et al., 2000). This also suggests a potential dissociation of the complex on the membrane, because Rabex-5 appears to be entirely complexed to Rabaptin-5 in cytosol. These observations raise several questions. What is the putative Rabex-5 receptor? How is the dissociation of the complex regulated? Is there a functional difference between free and complexed Rabex-5 on the membrane? Is free Rabex-5 required to activate Rab5 for the formation of CCV at the plasma membrane (McLauchlan et al., 1998) or for the motility of early endosomes (Nielsen et al., 1999)? It will now become important to investigate the mechanism of the Rabex-5 recruitment, by identifying the putative receptor and determining its distribution.
ACKNOWLEDGMENTS
We are grateful to Savvas Christoforidis and Heidi McBride for valuable discussions and critical reading of the manuscript. R.L. and M.M. were recipients of a Max-Planck and Human Frontier Science Program Fellowships respectively. This work was supported by the Max Planck Gesellschaft and by grants from the Human Frontier Science Program (RG-432/96), EU TMR (ERB-CT96–0020), and Biomed (BMH4-97-2410) (M.Z.).
REFERENCES
- Alexandrov K, Horiuchi H, Steele-Mortimer O, Seabra MC, Zerial M. Rab escort protein-1 is a multifunctional protein that accompanies newly prenylated rab proteins to their target membranes. EMBO J. 1994;13:5262–5273. doi: 10.1002/j.1460-2075.1994.tb06860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock JB, Klumperman J, Davanger S, Scheller R. Syntaxin 6 functions in trans-Golgi network vesicle trafficking. Mol Biol Cell. 1997;8:1261–1271. doi: 10.1091/mbc.8.7.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- Burd C, Mustol PA, Schu PV, Emr SD. A yeast protein related to a mammalian Ras-binding protein, Vps9p, is required for localization of vacuolar proteins. Mol Cell Biol. 1996;16:2369–2377. doi: 10.1128/mcb.16.5.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Peterson M, Cowles CR, Emr SD. A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Mol Biol Cell. 1997;8:1089–1104. doi: 10.1091/mbc.8.6.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoforidis S, McBride HM, Burgoyne RD, Zerial M. The Rab5 effector EEA1 is a core component of endosome docking. Nature. 1999a;397:621–625. doi: 10.1038/17618. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999b;1:249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- Christoforidis S, Zerial M. An affinity chromatography approach leading to the purification and identification of novel Rab effectors. Methods. 1999;20:403–410. doi: 10.1006/meth.2000.0953. [DOI] [PubMed] [Google Scholar]
- Cosulich SC, Horiuchi H, Zerial M, Clarke PR, Woodman PG. Cleavage of rabaptin-5 blocks endosome fusion during apoptosis. EMBO J. 1997;16:6182–6191. doi: 10.1093/emboj/16.20.6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers FP, Armstrong SA, Seabra MC, Brown MS, Goldstein JL. REP-2, a Rab escort protein encoded by the choroideremia-like gene. J Biol Chem. 1994;269:2111–2117. [PubMed] [Google Scholar]
- Elkind NB, Walch-Solimena C, Novick PJ. The role of the COOH terminus of Sec2p in the transport of post-Golgi vesicles. J Cell Biol. 2000;149:95–110. doi: 10.1083/jcb.149.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvel J-P, Chavrier P, Zerial M, Gruenberg J. Rab5 controls early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- Gournier H, Stenmark H, Rybin V, Lippe R, Zerial M. Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. EMBO J. 1998;17:1930–1940. doi: 10.1093/emboj/17.7.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Roth D, Walch-Solimena, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicle to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst J, Lui WW, Bright NA, Totty N, Seaman MN, Robinson MS. A family of proteins with gamma-adaptin and VHS domains that facilitate trafficking between the trans-Golgi network and the vacuole/lysosome. J Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi H, Lippe R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, Zerial M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90:1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- Jones S, Newman C, Liu F, Segev N. The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol Biol Cell. 2000;11:4403–4411. doi: 10.1091/mbc.11.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. Adaptors for clathrin-mediated traffic. Annu Rev Cell Dev Biol. 1999;15:705–732. doi: 10.1146/annurev.cellbio.15.1.705. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T. Clathrin. Annu Rev Biochem. 2000;69:699–727. doi: 10.1146/annurev.biochem.69.1.699. [DOI] [PubMed] [Google Scholar]
- Korobko IV, Korobko EV, Kiselev SL. The MAK-V protein kinase regulates endocytosis in mouse. Mol Gen Genet. 2000;264:411–418. doi: 10.1007/s004380000293. [DOI] [PubMed] [Google Scholar]
- Li G, Barbieri MA, Colombo MI, Stahl PD. Structure features of the GTP-binding defective rab5 mutants required for their inhibitory activity on endosome fusion. J Biol Chem. 1994;269:14631–14635. [PubMed] [Google Scholar]
- Lippe R, Horiuchi H, Zerial M. Regulators and effectors of small GTPases: expression, purification and properties of the Rab4/5 specific Rabex/Rabaptin complex. Methods Enzymol. 2001;329:132–145. doi: 10.1016/s0076-6879(01)29074-0. [DOI] [PubMed] [Google Scholar]
- Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- McBride HM, Rybin V, Murphy C, Giner A, Teasdale R, Zerial M. Oligomeric complexes link Rab5 effectors with NSF and drive membrane fusion via interactions between EEA1 and syntaxin 13. Cell. 1999;98:377–386. doi: 10.1016/s0092-8674(00)81966-2. [DOI] [PubMed] [Google Scholar]
- McLauchlan H, Newell J, Morrice N, Osborne A, West M, Smythe E. A novel role for Rab5-GDI in ligand sequestration into clathrin-coated pits. Curr Biol. 1998;8:34–45. doi: 10.1016/s0960-9822(98)70018-1. [DOI] [PubMed] [Google Scholar]
- Mills IG, Jones AT, Clague MJ. Involvement of the endosomal autoantigen EEA1 in homotypic fusion of early endosomes. Curr Biol. 1998;8:881–884. doi: 10.1016/s0960-9822(07)00351-x. [DOI] [PubMed] [Google Scholar]
- Mu F, Callaghan JM, Steele-Mortimer O, Stenmark H, Parton RG, Campbell PL, McCluskey J, Yeo JP, Tock EPC, Toh BH. EEA1, an early endosome-associated protein. J Biol Chem. 1995;270:13503–13511. doi: 10.1074/jbc.270.22.13503. [DOI] [PubMed] [Google Scholar]
- Nair J, Müller H, Peterson M, Novick P. Sec2 protein contains a coiled-coil domain essential for vesicular transport and a dispensable carboxy terminal domain. J Cell Biol. 1990;110:1897–1909. doi: 10.1083/jcb.110.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neve RL, Coopersmith R, McPhie DL, Santeufemio C, Pratt KG, Murphy CJ, Lynn SD. The neuronal growth-associated protein GAP-43 interacts with rabaptin-5 and participates in endocytosis. J Neurosci. 1998;18:7757–7767. doi: 10.1523/JNEUROSCI.18-19-07757.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- Nishimune H, Uyeda A, Nogawa M, Fujimori K, Taguchi T. Neurocrescin: a novel neurite-outgrowth factor secreted by muscle after denervation. Neuroreport. 1997;8:3649–3654. doi: 10.1097/00001756-199711100-00045. [DOI] [PubMed] [Google Scholar]
- Ohya T, Sasaki T, Kato M, Takai Y. Involvement of rabphilin3 in endocytosis through interaction with Rabaptin5. J Biol Chem. 1998;273:613–617. doi: 10.1074/jbc.273.1.613. [DOI] [PubMed] [Google Scholar]
- Ridley AJ, Self AJ, Kasmi F, Paterson HF, Hall A, Marshall CJ, Ellis C. rho family GTPase activating proteins p190, bcr and rhoGAP show distinct specificities in vitro and in vivo. EMBO J. 1993;12:5151–5160. doi: 10.1002/j.1460-2075.1993.tb06210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Barbieri MA, Pryse KM, Chua M, Morisaki JH, Stahl PD. Endosome fusion in living cells overexpressing GFP-rab5. J Cell Sci. 1999;112:3667–3675. doi: 10.1242/jcs.112.21.3667. [DOI] [PubMed] [Google Scholar]
- Rybin V, Ullrich O, Rubino M, Alexandrov K, Simon I, Seabra C, Goody R, Zerial M. GTPase activity of Rab5 acts as a timer for endocytic membrane fusion. Nature. 1996;383:266–269. doi: 10.1038/383266a0. [DOI] [PubMed] [Google Scholar]
- Schmid SL. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- Seals DF, Eitzen G, Margolis N, Wickner WT, Price A. A Ypt/Rab effector complex containing the Sec1 homolog Vps33p is required for homotypic vacuole fusion. Proc Natl Acad Sci USA. 2000;97:9402–9407. doi: 10.1073/pnas.97.17.9402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A, Gaullier JM, D'Arrigo A, Stenmark H. The Rab5 effector EEA1 interacts directly with syntaxin-6. J Biol Chem. 1999;274:28857–28860. doi: 10.1074/jbc.274.41.28857. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Peak-Chew SY, Pelham HR. Ric1p and Rgp1p form a complex that catalyzes nucleotide exchange on Ypt6p. EMBO J. 2000;19:4885–4894. doi: 10.1093/emboj/19.18.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–914. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, Vitale G, Ullrich O, Zerial M. Rabaptin-5 is a direct effector of the small GTPase Rab5 in endocytic membrane fusion. Cell. 1995;83:423–432. doi: 10.1016/0092-8674(95)90120-5. [DOI] [PubMed] [Google Scholar]
- Swanton E, Bishop N, Woodman P. Human rabaptin-5 is selectively cleaved by caspase-3 during apoptosis. J Biol Chem. 1999;274:37583–37590. doi: 10.1074/jbc.274.53.37583. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Horiuchi H, Bucci C, Zerial M. Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature. 1994;368:157–160. doi: 10.1038/368157a0. [DOI] [PubMed] [Google Scholar]
- Vitale G, Rybin V, Christoforidis S, Thornqvist P, McCaffrey M, Stenmark H, Zerial M. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J. 1998;17:1941–1951. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, Novick P. Sec2 mediates nucleotide exchange on Sec4 and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Sacher M, Ferro-Novick S. TRAPP stimulates guanine nucleotide exchange on Ypt1p. J Cell Biol. 2000;151:289–296. doi: 10.1083/jcb.151.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JM, de Hoop M, Zorzi N, Toh BH, Dotti CG, Parton RG. EEA1, a tethering protein of the early sorting endosome, shows a polarized distribution in hippocampal neurons, epithelial cells, and fibroblasts. Mol Biol Cell. 2000;11:2657–2671. doi: 10.1091/mbc.11.8.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser AE, Sato TK, Emr SD. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J Cell Biol. 2000;151:551–562. doi: 10.1083/jcb.151.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao GH, Shoarinejad F, Jin F, Golemis EA, Yeung RS. The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. J Biol Chem. 1997;272:6097–6100. doi: 10.1074/jbc.272.10.6097. [DOI] [PubMed] [Google Scholar]