Summary
Female meiosis provides an opportunity for selfish genetic elements to violate Mendel’s Law of Segregation by increasing the chance of segregating to the egg [1]. Centromeres and other repetitive sequences can drive in meiosis by cheating the segregation process [2], but the underlying mechanisms are unknown. Here we show that centromeres with more satellite repeats house more nucleosomes that confer centromere identity, containing the histone H3 variant CENP-A, and bias their segregation to the egg relative to centromeres with fewer repeats. CENP-A nucleosomes predominantly occupy a single site within the repeating unit that becomes limiting for centromere assembly on smaller centromeres. We propose that amplified repetitive sequences act as selfish elements by promoting expansion of CENP-A chromatin and increased transmission through the female germline.
Results and Discussion
Mendelian inheritance assumes that alleles are transmitted through the germline with equal probability to gametes. Selfish elements on chromosomes can violate Mendel’s Law of Segregation, however, to increase their transmission. The asymmetry of female meiosis, which produces a single egg from two rounds of cell division, provides an opportunity to cheat by increasing the chance of segregating to the egg. Such female meiotic drive depends on three components [1]. The first is asymmetry in meiotic cell division and cell fate, which is explained by having half of the genome eliminated in polar bodies. The second is asymmetry in the meiotic spindle (unpublished results, T.A. & M.A.L.) that can be exploited by selfish elements if they preferentially attach to the side that will keep them in the egg. The third component is a difference between the homologous chromosomes that influences their segregation, such as the presence of a selfish element. Putative selfish elements in various plants and animals include those that implicate centromeres [3–5], the chromosomal locus that typically directs chromosome inheritance by nucleating the kinetochore at cell division [6,7], and those that involve non-centromeric loci [8,9]. In all cases, the underlying mechanisms by which any of these selfish elements violate Mendelian segregation are unknown. For centromeres, since there is a strong epigenetic component to specifying their location at a single location on the chromosome, both epigenetic and genetic features must be considered. In principle, a selfish element could consist of an alteration in local epigenetic information, a special DNA sequence, and/or copy number variation of a functional repetitive centromere element. Here, using a mouse model system to study differences between centromeres, we test these models for meiotic drive in the mammalian female germline.
We first focused our attention on the specialized nucleosomes containing the histone variant, CENP-A, which defines the location of kinetochore formation [10–13]. Since these nucleosomes contain both the epigenetic mark and the functional centromeric DNA, they represent the primary candidate to be altered between centromeres that are unequally segregated in female meiosis. In our model system to examine differences between centromeres, we previously measured abundance of a microtubule-binding kinetochore protein (HEC1) at centromeres from three mouse strains: CF-1, C57BL/6J, and CHPO [5]. We refer to CF-1 and C57BL/6J centromeres as “stronger” because they have increased HEC1 levels relative to “weaker” CHPO centromeres. CF-1 and C57BL/6J have complementary advantages as stronger centromere strains: C57BL/6J is an inbred strain with its reference genome available, and CF-1 is an outbred strain that efficiently produces hybrid offspring with CHPO. We first considered the possibility that centromere strength relates to sequence changes in the CENP-A protein, itself, which is rapidly evolving relative to core histones [14]. CENP-A is extremely long-lived at centromeres [13,15,16], and changes in CENP-A levels are heritable on some Drosophila centromeres from one generation to the next [17]. The CENP-A polypeptide is 100% identical in all three strains (as we found by sequencing the ORF from each strain; corresponding to GenBank: NP_031707), however, indicating that the molecular determinants of centromere strength are likely in the DNA.
To isolate centromeric DNA from stronger and weaker centromere strains, we enriched CENP-A nucleosomes from C57BL/6J and CHPO, respectively, by native chromatin immunoprecipitation (ChIP). High-throughput sequencing and quantitative PCR (qPCR) showed enrichment of repetitive minor satellite DNA (hereafter referred to as minor satellite) in isolations from each strain (Figure 1A,B; Figure S1A; Figure S1E), as expected because minor satellite is the major site of CENP-A nucleosome assembly in mouse strains studied to date [18]. Other than canonical centromere satellites, we find no enrichment in CENP-A nucleosome ChIP of repetitive elements in either CHPO or C57BL/6J that could have formed an alternative centromere assembly site, as proposed in M. Caroli [19] (Table S1). Mouse CENP-A nucleosomes protect a shorter length of DNA from micrococcal nuclease (MNase) digestion relative to the total (input) nucleosome population (Figure S1F-G), similar to humans and rice [20–23]. Furthermore, high-throughput sequencing of the isolated DNA revealed a striking specificity of CENP-A nucleosomes for a single major position within minor satellite (Figure 1C,E), whereas the input nucleosomes are present at multiple highly populated positions throughout the monomer repeat sequence (Figure 1D,F). This strong and specific phasing of CENP-A nucleosomes on both CHPO and C57BL/6J centromeres highlights the extraordinarily close relationship between the typical centromeric DNA sequence and the centromere-specifying CENP-A nucleosomes in mouse. The major site occupied by CENP-A nucleosomes is centered – the location of its presumed dyad axis of symmetry – within 10 bp of the 3′ end of the CENP-B box, the 17 bp recognition motif for CENP-B [24] (Figure 1C,E). Thus, the relationship between CENP-A nucleosomes and minor satellite could be intrinsic to the local DNA sequence or mediated via CENP-B.
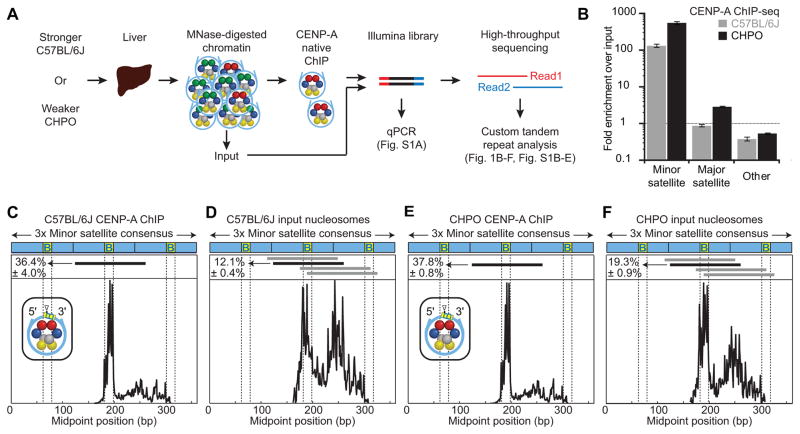
Figure 1. CENP-A nucleosomes occupy a larger proportion of minor satellite DNA in weaker centromeres and are highly phased.
(A) Schematic for CENP-A ChIP analysis of nuclease-protected DNA fragments from stronger (C57BL/6J) or weaker (CHPO) centromere strains. See also Table S1.
(B) CENP-A ChIP sequencing results. Fold-enrichment was calculated as the fraction of reads that are minor or major satellite in the ChIP sample divided by the fraction in the input sample (mean±SEM, n=3 independent experiments). See Figure S1E.
(C–F) Midpoint position of CENP-A ChIP (C, E) or input (D, F) reads (size 100–160 bp) along the trimer minor satellite consensus sequence. Vertical lines indicate the 17-bp CENP-B box. The major CENP-A nucleosome position (identified in the CENP-A ChIP samples) is indicated by a horizontal black line above the respective midpoint values and schematized (inset) for CENP-A ChIP with a triangle representing the dyad position. The same nucleosome position is indicated in the input samples. Numbers to the left of the positions indicate the percentage of reads (mean±SEM, n=3 independent experiments) where the midpoint spans the 10 bp at the 3′ end of the CENP-B box (yellow, labeled B). Horizontal gray lines indicate other major nucleosome positions in the input samples. See Figure S1H for multiple sequence alignment. Similar results were obtained from a CENP-A ChIP-seq analysis in another stronger centromere strain (CF-1, Figure S1B–D).
Given the close association of CENP-A nucleosomes with minor satellite sequences, two other results from our ChIP-seq experiments suggest how stronger and weaker centromeres might differ from each other. First, the enrichment of minor satellite in CENP-A ChIP from CHPO (weaker) centromeres, relative to input, was much greater (300–400-fold) than the enrichment from C57BL/6J (75–100-fold) (Figure 1B; Figure S1A; Table S1). This difference raises the possibility that there is less minor satellite overall in CHPO and therefore fewer of these sequences in the input. Second, minor satellite sequences associated with the input nucleosomes reveal the signature CENP-A phasing as the dominant minor satellite nucleosome site in CHPO, whereas it is only one of several similarly populated sites in C57BL/6J (Figure 1F,D, respectively). The analysis of minor satellite from C57BL/6J is consistent with centromeres from human cells, where CENP-A nucleosomes are a minor component of centromeric chromatin (<10% of total nucleosomes) [25]. Minor satellite from CHPO, however, exhibits more of the preferred nucleosome positions characteristic of those containing CENP-A, which indicates less minor satellite occupied by other nucleosomes. Together, these two results suggest the hypothesis that weaker (CHPO) centromeres have a restricted number of minor satellite repeats that can act as functional assembly sites for CENP-A nucleosomes, which would restrict the amount of centromeric chromatin. Additionally, major satellite DNA, which typically forms constitutive heterochromatin and not CENP-A nucleosomes [18,26], is slightly enriched in CHPO isolations but not in C57BL/6J (Figure 1B; Figure S1A; Table S1). This finding suggests that although CENP-A nucleosomes are predominantly found on minor satellite, they extend partially into major satellite because of the limited availability of minor satellite in CHPO.
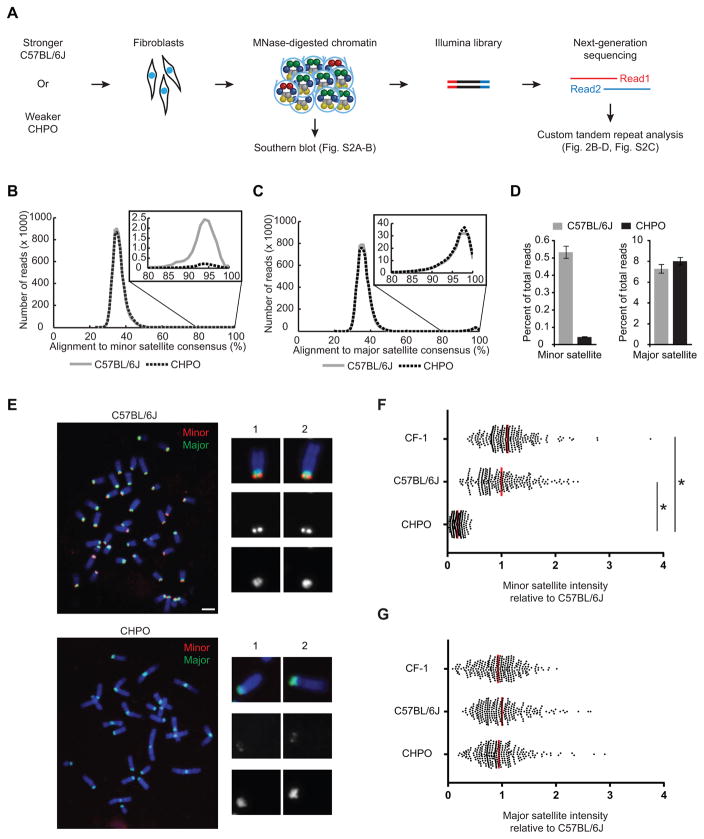
To test our hypothesis that weaker centromeres are limited for CENP-A assembly sites, we determined the abundance, nucleosomal DNA length distributions, and sequence composition of minor satellite in weaker and stronger centromere strains. We performed MNase digestion coupled to high-throughput sequencing (MNase-seq) to analyze total mononucleosome populations from primary cell cultures from each strain (Figure 2A). We first confirmed that the MNase-digested material that we sequenced included most detectable minor and major satellite DNA from both species (Figure S2A–B). Our sequencing revealed that the fraction of the genome aligning to major satellite was similar for the two strains (7–8%) and consistent with a previous report [27]. In contrast, only 0.05% of the CHPO genome aligned to minor satellite, compared to 0.5% for C57BL/6J (Figure 2B–D). Notably, there was a 2.4±0.4-fold higher proportion of short nucleosomal DNA lengths (100–119 bp) for minor satellite in CHPO relative to C57BL/6J, while major satellite nucleosome lengths were similar between the strains (Figure S2D–F). This finding is consistent with the notion that CENP-A nucleosomes occupy a very high proportion of minor satellite DNA at weaker (CHPO) centromeres. Importantly, the sequences of the minor satellite monomers were similar between the two strains (Figure S2G), which allowed us to use fluorescence in situ hybridization (FISH) as a second approach to probe the amount of minor satellite at each centromere. FISH analysis of metaphase chromosome spreads from CF-1, C57BL/6J and CHPO primary embryonic fibroblasts shows a 6-fold increase in minor satellite at CF-1 and C57BL/6J centromeres compared to CHPO centromeres, whereas the amount of major satellite DNA is similar among these strains (Figure 2E–G; Figure S3). Together, these results show, based on two complementary approaches, that stronger centromeres from CF-1 or C57BL/6J contain 6 to 10-fold more minor satellite at centromeres than weaker centromeres from CHPO. Indeed, accounting for the size of the genome and the percentage that is minor satellite repeats, and assuming a similar number of CENP-A nucleosomes as measured for human centromeres [25], CENP-A nucleosomes would account for 2–8% in C57BL/6J versus 13–52% in CHPO of the total number of nucleosomes assembled on minor satellite DNA (see Table S2).
Figure 2. Stronger centromeres contain more minor satellite than weaker centromeres.
(A–D) MNase-seq of C57BL/6J (stronger) and CHPO (weaker), as shown in the schematic (A). Reads were aligned to a trimer of minor satellite or a dimer of major satellite consensus sequences (see Figure S1E). Histograms show distribution of reads aligning to minor (B) or major (C) satellite, with 80–100% range expanded in insets. The percent of reads that aligned with ≥ 80% identity was calculated (D, mean±SEM, n=3 independent experiments). See also Figure S2 and Table S2.
(E–G) FISH analysis of minor (red) and major (green) satellite on metaphase chromosomes of C57BL/6J and CHPO (see also Figure S3). Representative images are shown (E), with single chromosomes magnified in insets. Minor (F) and major (G) satellite signals were quantified for CF-1, C57BL/6J and CHPO. Each dot represents one centromere; red bar, mean; n≥260 in each case; * p < 0.0001; scale bars 5 μm.
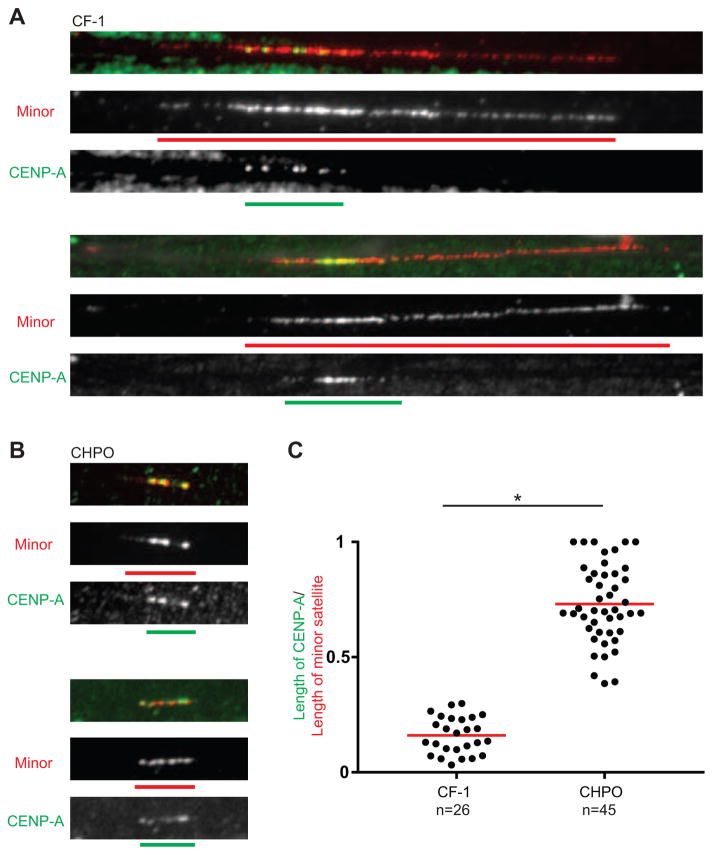
To determine how centromere chromatin is organized at stronger and weaker centromeres, we simultaneously visualized CENP-A by immunofluorescence and minor satellite by FISH on stretched chromatin fibers. The length of minor satellite is ~6-fold greater at stronger (CF-1) centromeres compared to weaker (CHPO) (30±13 vs. 4.7±1.6 μm), as expected from our sequencing and metaphase chromosome FISH analyses. We detect CENP-A only on minor satellite in both cases, but it occupies only 20% of the total minor satellite region, on average, at stronger centromeres compared to 70% at weaker centromeres (Figure 3). These results suggest a model in which CENP-A nucleosomes are restricted to minor satellite, filling the length of available minor satellite at weaker centromeres but only a small fraction of the minor satellite at stronger centromeres. Current models of centromeric chromatin include an important role for interspersed nucleosomes containing histone H3 [28–30], to spatially separate kinetochore and inner centromere functions and/or provide sites for nascent CENP-A nucleosome assembly that doubles the total number of CENP-A nucleosomes at centromeres upon exit from cell division [6,7]. Taking these H3 nucleosomes into account, our centromere stretching (Figure 3) and sequencing (Figure 2) experiments support the notion that weak centromeres are at or near the lower limit for the remaining nucleosome assembly sites available for CENP-A.
Figure 3. CENP-A nucleosomes fill the minor satellite region at weaker centromeres.
(A and B) Representative images of CENP-A (green) and minor satellite (red) localized by immunofluorescence and FISH, respectively, on extended chromatin fibers from stronger (A, CF-1) or weaker (B, CHPO) centromere strains. Green and red bars show the length of CENP-A and minor satellite signals, respectively. Scale bar, 5 μm.
(C) Ratio of the length of CENP-A to the length of minor satellite. Each dot represents one centromere; red bar, mean; * p < 0.0001.
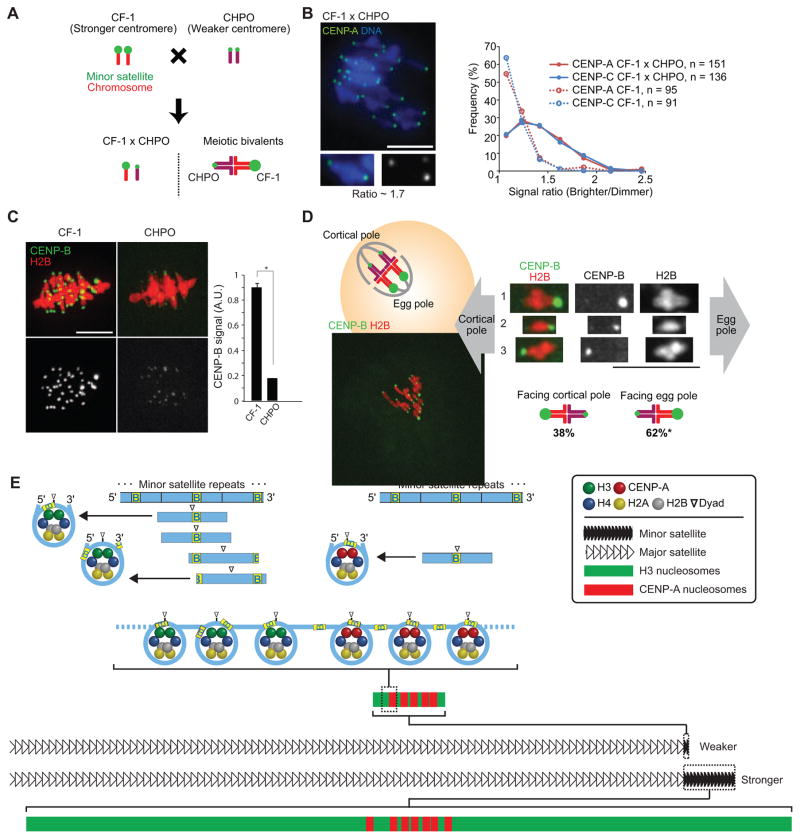
To test the functional significance of observed differences in centromere DNA sequence and chromatin organization, we crossed CF-1 and C57BL/6J to CHPO to examine bivalents with the homologous chromosomes paired in meiosis I (Figure 4A). Both stronger and weaker centromeres are present on the same bivalent, so any differences must be intrinsic to the centromeres. We examined the centromere proteins CENP-A, which specifies the sites of kinetochore assembly, and CENP-C, which directly interacts with CENP-A nucleosomes and recruits other kinetochore proteins [6,13]. We find differences in both CENP-A and CENP-C levels across the bivalents in CF-1 × CHPO and C57BL/6J × CHPO hybrid oocytes, with a median ratio of 1.3 in each case (Figure 4B; Figure S4A–C). We did not see such differences across bivalents from the parental CF-1 strain, in which centromeres of homologous chromosomes should be the same. In contrast, an epigenetic mark at pericentric heterochromatin (H3K9me3) is similar across bivalents in CF-1 × CHPO hybrid oocytes and CF-1 oocytes (Figure S4D). Consistent with our results in oocytes, we find more CENP-A at CF-1 centromeres compared to CHPO centromeres in CF-1 × CHPO hybrid embryonic fibroblast cells, using minor satellite FISH to distinguish the two (Figure S4E–G). These results indicate that greater abundance of minor satellites is associated with increased levels of proteins that bind the centromere DNA in both oocytes and somatic cells. Centromeric chromatin is likely similar, therefore, in these cell types, although we cannot exclude the possibility that the organization in oocytes might be different from what we observe in somatic cells (Figure 3).
Figure 4. Stronger centromeres bind more CENP-A and CENP-C and orient preferentially to the egg in meiosis I.
(A) Progeny of a CF-1 × CHPO cross have meiotic bivalents with both weaker (CHPO) and stronger (CF-1) centromeres.
(B) CF-1 × CHPO oocytes were stained for CENP-A and CENP-C at metaphase I (see also Figure S4A–C). Image shows CENP-A staining, with a single bivalent magnified in the inset (arrows indicate paired centromeres). Graph is a histogram of CENP-A (red) and CENP-C (blue) intensity ratios, calculated as the brighter divided by the dimmer signal for each bivalent. CF-1 oocytes (dashed lines) are shown as controls.
(C) CF-1 or CHPO oocytes expressing CENP-B-mCherry and H2B-EGFP were imaged live at metaphase I. Graph shows quantification of CENP-B signals (mean±SEM, n≥340 centromeres from ≥27 oocytes in each case, pooled from three independent experiments). * p < 0.001; A.U., arbitrary units.
(D) Schematic shows bivalents in CF-1 × CHPO oocytes, with CF-1 centromeres facing the egg. Image shows a CF-1 × CHPO oocyte expressing CENP-B-EGFP and H2B-mCherry, shortly before anaphase onset; dashed white lines show cortex and spindle outline. The orientation of each bivalent was determined using CENP-B-EGFP intensity to distinguish CF-1 (brighter) and CHPO (dimmer) centromeres. * Significantly different from 50% (n=133, p<0.005). Images (B–D) are maximal intensity z-projections; insets are optical slices showing single bivalents. Scale bars, 10 μm.
(E) Model of our proposal that the amount of minor satellite determines centromere strength by constraining the spreading of CENP-A nucleosomes. Top: CENP-A nucleosomes (right) but not bulk nucleosomes (left) are strongly positioned on the minor satellite consensus, with the digestion-protected fragment centered on the last 10 bp of the 17-bp CENP-B box (yellow, labeled B). Major nucleosome positions are indicated by horizontal lines below the minor satellite consensus. Bottom: stronger centromeres contain more minor satellite DNA and centromeric proteins (e.g., CENP-A) than weaker centromeres. CENP-A nucleosomes localize to a small fraction of the minor satellite of stronger centromeres but occupy the length of the minor satellite DNA of weaker centromeres. CENP-A nucleosomes are shown with the dyad consistently positioned on the CENP-B box, whereas the position is variable for H3 nucleosomes (note that linkers between nucleosomes are drawn without CENP-B boxes for simplicity, and the linker length is drawn arbitrarily).
We previously found that bivalents in CF-1 × CHPO hybrid oocytes are positioned off-center on the spindle at metaphase I, with differences in HEC1 levels consistent with what we show here for CENP-A and CENP-C [5]. Taken together, these findings lead to a prediction of functional differences in microtubule interactions between weaker and stronger centromeres, which could bias meiotic segregation. To test this prediction, we examined the orientation of bivalents labeled with fluorescently-tagged CENP-B in CF-1 × CHPO hybrid oocytes. Because minor satellite contains CENP-B boxes, intensity of the CENP-B signal should allow us to distinguish between CF-1 and CHPO centromeres due to the massive difference in minor satellite abundance. Indeed, we find a 5-fold difference in CENP-B intensity at centromeres in CF-1 oocytes vs. CHPO oocytes (Figure 4C). In CF-1 × CHPO hybrid oocytes, each bivalent contained one bright and one dim CENP-B signal, corresponding to stronger and weaker centromeres. We followed these bivalents live and measured their orientation within 30 min of anaphase I onset (Figure 4D). In 62% of bivalents the weaker centromere oriented towards the cortex, indicating that it would segregate to the polar body while the stronger centromere remains in the egg (Figure 4D).
Based on these data, we propose a model for how centromere function depends on the underlying DNA sequence (Figure 4E). CENP-A nucleosomes are predominantly found on minor satellite and are highly phased, suggesting co-evolution of specialized histone and sequence contributing to the function and strength of centromere chromatin. Furthermore, the amount of minor satellite at a centromere constrains the spreading of CENP-A nucleosomes. Even when minor satellite is severely limited, as at weaker centromeres, we find only slight enrichment of major satellite associated with CENP-A. At stronger centromeres, in contrast, CENP-A chromatin can expand because it occupies only a fraction of the available minor satellite. CENP-A expansion would lead to formation of a larger kinetochore, preferential orientation on an asymmetric spindle, and retention in the egg in female meiosis. Our findings indicate that the number of CENP-A nucleosomes is not simply determined by the amount of minor satellite, as the differences in CENP-A levels between weaker and stronger centromeres are not as large as the differences in minor satellite abundance. Indeed, there is a well-established epigenetic pathway for assembly of new CENP-A chromatin in each cell cycle [6,7] that may limit its expansion. The relationship between satellite DNA abundance and CENP-A chromatin likely has important consequences in many contexts in biology. For example, competition between neighboring repetitive α-satellite DNA arrays has been proposed to underlie the preferential epigenetic silencing of the smaller array in humans [31].
An alternative to our model is that differences in CENP-A levels between weaker and stronger centromeres are maintained solely by an epigenetic pathway, independent of DNA sequence. In the Drosophila male germline, for example, pre-existing CENP-A in spermatocytes influences the amount of CENP-A in the offspring [17]. In support of our model, as opposed to a purely epigenetic model, the phasing of CENP-A nucleosomes and the restriction of CENP-A nucleosomes to minor satellite argues that these sequences are significant. A dependence on DNA sequence is also the most straightforward explanation for how differences between weaker and stronger centromeres are maintained over many mitotic cell cycles and through the germline in our experimental system. Overall, our findings explain how amplified repetitive sequences can act as selfish elements and provide a molecular basis for centromere drive. Furthermore, they reconcile the epigenetic pathway for centromere specification with the typical accumulation of repetitive sequences in centromere evolution.
STAR methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ben E. Black (blackbe@mail.med.upenn.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Mouse strains were purchased from the Jackson Laboratory (ZALENDE/EiJ, stock #001392 corresponds to CHPO; C57BL/6J, stock# 000664) or Envigo (CF-1, # 033). All mice used in this study were female at 7–14 week old unless otherwise indicated. All animal experiments were approved by the Institutional Animal Care and Use Committee and were consistent with the National Institutes of Health guidelines.
Primary cell lines
Mouse lung fibroblasts
Lung fibroblast cells were derived from female adult mice of C57BL/6J and CHPO strains and cultured into primary cell lines in growth medium: Dulbecco’s Modified Eagle’s medium (DMEM) with 10% FBS, penicillin–streptomycin at 37°C in a humidified atmosphere with 5% CO2 [32].
Mouse embryonic fibroblasts
Primary mouse embryonic fibroblasts (MEFs) were isolated from E12.5 embryos of both female and male mice of CF-1, C57BL/6J, and CHPO strains [33] and cultured in growth medium: Dulbecco’s Modified Eagle’s medium (DMEM) with 10% FBS, penicillin–streptomycin and GlutaMAX (Gibco) at 37°C in a humidified atmosphere with 5% CO2.
Cell lines
CF-1xCHPO hybrid mouse embryonic fibroblasts
Primary mouse embryonic fibroblasts (MEFs) were isolated from a female CF-1 × CHPO hybrid mouse at E12.5 and immortalized by transfection of SV40 large T antigen [34] (a gift from Dr. Brad Johnson at the University of Pennsylvania) using TransIT-X2R Dynamic Delivery System (Mirus) to obtain immortalized MEFs.
METHOD DETAILS
Sequencing of CENP-A cDNA
RNA was extracted from spleen of CF-1, C57BL/6J and CHPO using TRIzol (Invitrogen), and cDNA was synthesized using SuperScript™ II Reverse Transcriptase (Invitrogen). CENP-A was amplified using PCR with the two sets of primers (FW: 5′-CTCGCGTTCGGTTTCGGCAGCAGGACC-3′, RV: 5′-CTCCTCCGACACCACTGTCAAGC-3′) and (FW: 5′-ACTGCTGGCGACCGAGTTCTGG-3′, RV: 5′-ACAGTTTTAAGGGCACCGTGTAGCC-3′). The alignment of CENP-A protein was obtained in Lasergene (DNASTAR).
Chromatin immunoprecipitation
Nuclei were isolated from 2 flash-frozen mouse livers (CHPO, C57BL/6J, or CF-1) per CENP-A ChIP. Livers were homogenized in 4 ml ice-cold Buffer I (0.32 M sucrose, 60 mM KCl, 15 mM NaCl, 15 mM Tris-Cl, pH 7.5, 5 mM MgCl2, 0.1 mM EGTA, 0.5 mM DTT, 0.1 mM PMSF, 1 mM leupeptin/pepstatin, 1 mM aprotinin) per g of tissue by Dounce homogenization. Homogenate was filtered through 100 μm cell strainer (Falcon) and centrifuged at 6000 × g for 10 min at 4°C. The pellet was resuspended in the same volume Buffer I. An equivalent volume ice-cold Buffer I supplemented with 0.2% IGEPAL was added for a final concentration of 0.1%, and samples were incubated on ice for 10 min to release nuclei. 4 ml nuclei were gently layered on top of 8 ml ice-cold Buffer III (1.2 M sucrose, 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.1 mM EGTA, 15 mM Tris, pH 7.5, 0.5 mM DTT, 0.1 mM PMSF, 1 mM leupeptin/pepstatin, 1 mM aprotinin) and centrifuged at 10,000 × g for 20 min at 4°C with no brake. Pelleted nuclei were resuspended in Buffer A (0.34 M sucrose, 15 mM HEPES, pH 7.4, 15 mM NaCl, 60 mM KCl, 4 mM MgCl2, 1 mM DTT, 0.1 mM PMSF, 1 mM leupeptin/pepstatin, 1 mM aprotinin) to 400 ng/μl, flash-frozen in liquid nitrogen, and stored at −80°C. Nuclei were digested with MNase (Affymetrix), using 0.05–0.1 U/μg chromatin in Buffer A supplemented with 3 mM CaCl2 for 10 min at 37°C. The reaction was quenched with 10 mM EGTA on ice for 5 min, and an equal volume of 2× Post-MNase Buffer (40 mM Tris, pH 8, 220 mM NaCl, 4 mM EDTA, 2% Triton X-100, 0.5 mM DTT, 0.5 mM PMSF, 1 mM leupeptin/pepstatin, 1 mM aprotinin) was added prior to centrifugation at 18,800 × g for 15 min at 4°C. The supernatant containing the MNase-digested chromatin was pre-cleared with 100 μl 50% Protein G Sepharose bead (GE Healthcare) slurry in 1× Post-MNase Buffer for ~ 2 h at 4°C with rotation. Beads were blocked in NET Buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 1 mM EDTA, 0.1% IGEPAL, 0.25% gelatin, and 0.03% NaN3) and washed prior to use. Pre-cleared supernatant was divided so that an estimated 250 μg chromatin was used for CENP-A ChIP (10 μg α-mouse-specific CENP-A antibody [35]) and 12.5 μg was saved as input for further processing. The CENP-A ChIP was rotated at 4°C for 2 h. Immunocomplexes were recovered by addition of 100 μl 50% NET-blocked protein G Sepharose bead slurry followed by overnight rotation at 4°C. The beads were washed three times with Wash Buffer 1 (150 mM NaCl, 20 mM Tris-HCl, pH 8, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), once with High Salt Wash Buffer (500 mM NaCl, 20 mM Tris-HCl, pH 8, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), and the chromatin was eluted 2× each with 200 μl Elution Buffer (50 mM NaHCO3, 0.32 mM sucrose, 50 mM Tris, pH 8, 1 mM EDTA, 1% SDS) at 65°C for 10 min at 1500 rpm. The input sample was adjusted to a final volume of 400 μl with Elution Buffer. To each 400 μl input and ChIP sample, 16.8 μl of 5 M NaCl and 1 μl of RNAse A (10 mg/ml) was added. After 1 h at 37°C, 4 μl of 0.5M EDTA and 12 μl Proteinase K (2.5 mg/ml, Roche) were added, and samples were incubated for another 2 h at 42°C. The resulting Proteinase K-treated samples were subjected to a phenol-chloroform extraction followed by purification with a QiaQuick PCR Purification column (Qiagen).
Chromatin preparation for MNase-seq
For each mouse strain, 1–2 million primary lung fibroblast cells were grown in 10 cm plates, trypsinized, pelleted (330 × g, 5 min, 4°C) and resuspended in 2 ml ice-cold Buffer I. 2 ml of ice-cold Buffer II (Buffer I + 0.1% IGEPAL) was added to release nuclei, cells were mixed by inversion and placed on ice for 10 min. 4 ml nuclei were gently layered on top of 8 ml ice-cold Buffer III and centrifuged at 10,000 × g for 20 min at 4°C with no brake. Pelleted nuclei were resuspended by gentle shaking in Buffer A to 400 ng/μl, flash-frozen in liquid nitrogen, and stored at −80°C. 5–20 μg of chromatin in Buffer A was supplemented with 3 mM CaCl2 and digested with 0.05 U/μg of MNase (Affymetrix) for 10–15 min at 37°C to obtain a predominant mononucleosome population. Reactions were quenched with 10 mM EGTA and DNA was purified using a QIAquick PCR purification kit (Qiagen).
Southern blotting
200 ng of MNase-digested chromatin (see section Chromatin preparation for MNase-seq) was run on a 1% agarose gel for 12–16 h at 45 V. The gel was post-stained with ethidium bromide, imaged, and transferred to membrane (Amersham) overnight. After UV crosslinking, the membrane was prehybridized for 30 min at 65°C in Church Buffer (0.5 M sodium phosphate, 1 mM EDTA, 7% SDS, 1% BSA) in a hybridization bottle. DNA probes were made by radiolabeling minor and major satellite templates (detailed below in section “Probes of minor satellites and major satellites for Southern blotting and FISH”) with [α-32P]dCTP using a Rediprime II DNA labeling kit (GE) and cleaned up with MicroSpin G-25 columns (GE). Probes were denatured for 10 min at 95°C, chilled for 2 min on ice and then added directly to the hybridization bottle and incubated overnight at 65°C in a rotating oven. The following day, the membrane was washed 1× with 1.5× SSC/0.1% SDS then incubated for 30 min at 65°C in 225 mM NaCl, 22.5 mM sodium citrate, followed by 1× wash with 1× SSC/0.1% SDS and another incubation for 30 min at 65°C in 150 mM NaCl, 15 mM sodium citrate). The membrane was then transferred to plastic wrap and placed in a phosphoimager cassette and exposed to a phosphoscreen for 4–24 h. Screens were imaged on a Typhoon 9200 scanner (GE).
High-throughput sequencing
Unamplified nucleosomes from MNase-seq, input, or ChIP DNA (see sections Chromatin preparation for MNase-seq and Chromatin immunoprecipitation) was first analyzed using an Agilent 2100 Bioanalyzer High Sensitivity Kit in order to determine the quantity of DNA on the basis of fluorescence intensity. DNA libraries were prepared for multiplexed sequencing according to Illumina recommendations as described [20] with minor modifications using NEB enzymes. Libraries of the input nucleosomes sequences or MNase-seq preparations were used to assess the degree of nucleosome digestion for each experiment to avoid using over-digested samples in our analysis. Briefly, 50 ng of MNase-digested DNA (MNase-seq) or 5 ng input or ChIP DNA (ChIP-seq) was end-repaired and A-tailed. Illumina TruSeq adaptors were ligated, libraries were size-selected to exclude polynucleosomes, and adapter-modified DNA fragments were enriched by PCR using KAPA polymerase. Libraries were submitted for 150-bp, paired-end Illumina sequencing on a NextSeq 500 instrument.
Paired-end sequencing analysis
Paired-end reads, resulting from either ChIP-seq or MNase-seq, were converted to a name-sorted SAM file using picard-tools and samtools [36], then joined in MATLAB using the ‘localalign’ function to determine the overlapping region between the paired-end reads (requiring ≥95% overlap identity; see [20]), and adapter sequences were removed if present. For analysis of minor and major satellite DNA, we used a custom tandem repeat analysis as described [20] with the following modifications. Joined reads were aligned to a trimerized mouse minor satellite consensus (GenBank: X14464.1) [37], to a dimerized mouse major satellite consensus (GenBank: V00846.1) [38], or to the reverse complement of those tandem consensus sequences. Those joined reads aligning with ≥ 80% identity were chosen for further analysis, and the midpoints of the joined reads along the consensus sequences were used to generate nucleosome midpoint position plots (Figure 1C–F). Nucleosome occupancy maps (Figure S2E–F) were generated from the positions of joined reads along the consensus sequence. Normalization was performed by dividing the read count at each bin (at 1-bp increments) by the sum of counts over all bins over the three length classes. To calculate the percent of total reads, the number of joined reads aligning to the consensus sequence in either the forward or reverse complement orientation (without double-counting any joined read) was divided by the total number of joined reads. ChIP fold-enrichment was calculated as the fraction of reads mapping to minor (or major) satellite from the CENP-A ChIP divided by the fraction of reads mapping to minor (or major) satellite in the input.
Multiple sequence alignment analysis
200 minor satellite joined MNase-seq reads were randomly selected and aligned using ClustalW [39]. For CENP-A ChIP, 200 minor satellite joined reads with midpoints centered on the CENP-B box were randomly selected and aligned using Clustal Omega [40]. The multiple alignments were manually edited using Jalview [41], and sequence logos were created using the WebLogo server (Figure S1H; Figure S2G) [42].
Alternative repeat analysis using RepeatMasker
10,000 joined reads were randomly selected from CENP-A ChIP or input datasets for both C57BL/6J and CHPO and submitted to RepeatMasker [43]. For uniform annotation of elements across all data sets, FASTAs from all data sets were merged prior to submission, and the output was sorted according to original data sets before analysis. Number of elements was extracted, and fold enrichment was calculated for each repeat by dividing the number of elements in the CENP-A ChIP by the number in the input. For each repeat with an average fold-enrichment greater than 1 for either C57BL6/J or CHPO, a one-tailed t-test was performed comparing the number of elements of CENP-A ChIP and input replicates (Table S1).
Quantitative real-time PCR
Quantitative real-time PCR (qPCR) was performed as previously described [20] with modifications. Briefly, each reaction was assembled in a 10 μl volume using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) at 1x, 1 μM each primer, and input or ChIP DNA at 0.125 ng per reaction, as determined by Bioanalyzer. Triplicate reactions were loaded into a 384-well plate. Thermal cycling was performed in a LightCycler 480 (Roche) using the following cycling parameters: initial denaturation at 95°C for 2 min, 40 cycles of 94°C for 15 s, 58°C for 1 min. Cycle threshold (Ct) values were determined using LightCycler 480 Software. A standard curve was obtained for each primer pair. Fold enrichment (FE) was calculated from the amplification factor (AF) determined from the standard curve and the difference in cycle threshold (ΔCt) between input DNA and CENP-A ChIP DNA for each primer pair (FE = AFΔCt). Average fold-enrichment was calculated from three biologic replicates (independent ChIPs from different animals). The following primer sequences were used for qPCR [44]: major satellite (FW: 5′-GACGACTTGAAAAATGACGAAATC-3′, RV: 5′-CATATTCCAGGTCCTTCAGTGTGC-3′) and minor satellite (FW: 5′-CATGGAAAATGATAAAAACC-3′, RV: 5′-CATCTAATATGTTCTACAGTGTGG-3′).
Probes of minor satellite and major satellite for Southern blotting and FISH
Plasmids containing minor or major satellite sequences were provided by Dr. Rachel O’Neill (University of Connecticut) as templates for Southern blotting and FISH probes: First minor satellite template (~3.5 monomers)
5′-GGAAACATGATAAAAACCACAGTGTAGAACAGATTAGATGGGTGAGTTACACTGAAAAACACAT TCTTTGGAAACAGGATTTGTAGAACAGTGTATATCAATGAATTACAATGAGAAACATGGAAAAT GATAAAAACCACACTGTACAACAGATTAGATGAGTGAGTTACACTGAAAAACACATTCCTTGGA AACGGGATTTGTAGAATAGTGTATATCAATGAGTTACAATGAGAAACATGGAAAATGATAAAAA CCACACTGTAGAACAGATTAGATGAGTGAGTTACACTGAAAAACACATTCGTTGGAAACGGGAT TTGTAGAACAGTGTATATCAATGAGTTACAATGAAAAACATGGAAAATGATGAAAACCACACTG TAGAACATATTAGATGAGTGAGTTACACTGAAAAACACATTCGTTGGAAACGGGATTTGTAGAA CAGTGTATATCAATG-3′
Major satellite template (~4 monomers)
5′-GACCTGGATATGGCGAGTAAACTGAAAATTACGGAAAATGAGAAATACACACTTTAGGACGTGA AATATGGCGAGGAAAACTGAAAAAGGTGGAAAATTTAGAAATGTCCACTGTAGGACGTGGAATA TGGCAAGAAAACTGAAAATCATGGAAAATGAGAAACATCCACTTGACGACTTGAAAAATGACGA AATCACTAAAAAAAGTGAAAAATGAGAAATGCACACTGAAGGACCTGGAATATGGCGAGAAAAC TGAAAATCACGGAAAATGAGAAATACACACTGTAGGACGTGAAATATGGCGAGGAAAACTGAAA AAGGTGGAAAATTTAGAAATGTCCACTGTAGGAAGTGGAATATGGCAAGAAAACTGAAAATCAT GGAAAATGAGAAACATCCACTTGACGACTTGAAAAATGACGATATCACTAAAAAACGTGAAAAA TGAGAAATGCACAGTGAAGGACCTGGAATATGGCGAGAAAACTGAAAATCACTGAAAATGAGAA ATACACACTTTAGGACGTGAAATATGGCGAGGAAAACTGAAAAAGGTGGAAAATTTAGAAATGT CCACTGTAGGACGTGGAATATGGCAAGAAAACTGAAAATCATGGAAAATGAGAAACATCCACTT GACGACTTGAAAAATGACGAAATCACTAAAAAAAGTGAAAAATGAAAATGCACACTGTAGGACC TGGAATATGGCGAGAAAACTGAAAATCACGGAAAATGAGAAATACACACTTTAGGACGTGAAAT ATGGCGAGGAATACTGAAAAAGGTGGAAAATTTAGAAATGTCCACTGTAGGACGTGATATGGCA AGAAAACTGAAATCATGGAAATGAGAACATCCACTTGACGACTTGAAATGACGAATCACTAAAA AAGTGAAAATGAGAAATGCACACTGA-3′
Because minor satellite monomers from C57BL/6J and CHPO exhibit nucleotide variation, we made an additional minor satellite template. We cloned minor satellite repeats by amplification of genomic DNA of C57BL/6J using PCR with the following primers: 5′-GGAAAATGATAAAAACCACACTG -3′ and 5′-TCATTGATATACACTGTTCTACA - 3′. Amplified PCR products were gel-purified and ligated into a TOPO TA cloning vector, transformed into the E. coli strain DH5α (Invitrogen) and a single clone was used as a second minor satellite template. The insert was confirmed to have minor satellite monomer variants different from the one shown above.
Second minor satellite template (~2.5 monomers):
5′-GGAAAATGATAAAAACCACACTGTAGAACATTTTAGATGAGTGAGTTACACTGAAAAACACATA CGTTGGAAACCGGCATTGTAGAACAGTGTATATCAATGAGTTACAATGAAAAACATGGAAAATG ATAAAAACCACACTGTAGAACATTTTAGATGAGTGAGTTACACTGAAAAACACATACGTTGGAA ACCGGCATTGTAGAACAGTGTATATCAATGAGTTACAATGAGAAACATGGAAAATGATAAAAAC CACACTGTAGAACATATTAGATGAGTGAGTTACACTGAAAAACACATACGTTGGAAACCTGCAT TGTAGAACAGTGTATATCAATGA-3′
To make FISH probes, plasmids containing minor or major satellite templates were nick translated with biotin dUTP (Roche) or digoxigenin dUTP (Roche) using a Nick Translation Kit (Roche). The two minor satellite probes were used together.
FISH of metaphase chromosome spreads
Mitotic cells of MEFs from CF-1, C57BL/6J and CHPO strains were collected after cells were incubated with nocodazole (100 ng/ml) for 3 h, and metaphase chromosome spreads were prepared using 3:1 methanol:acetic acid fixation. After dehydration in ethanol series (70%, 90%, 100%; each 5 min), metaphase chromosomes were denatured in 70% formamide/2× SSC at 77°C for 3 min, and dehydrated in ethanol series at –20C°. Probes were denatured separately in hybridization buffer (50% formamide, 10% Dextran Sulfate, 2× SSC, 10 μg sheared single-stranded salmon DNA) at 77°C for 15 min, and hybridized to chromosomes at 37°C overnight in a humidified chamber. Slides were washed in 50% formamide/2× SSC for 10 min at 42°C and 2× SSC for 5 min at RT, and incubated with either Streptavidin AF 594 (1:100, Life technologies) or Fluorescein anti-digoxigenin (1:100, Roche) for 30 min at 37°C. After slides were washed three times in 2× SSC for 5 min each, chromosomes were counterstained with DAPI.
Images were acquired with a 63× objective on an inverted fluorescence microscope (DMI6000B; Leica) equipped with a charge-coupled device camera (ORCA AG; Hamamatsu Photonics), using μManager software [45]. For each sample, images were collected at 0.2 μm z-sections and a maximal intensity z projection was used for analysis. The mean intensities of FISH signals at each centromere were calculated in ellipses drawn around centromeres after background subtraction using Fiji/ImageJ [46,47]. FISH signals were obtained from two independent cell lines obtained from each strain, and two experiments were conducted for each cell line. Statistical differences of minor satellites between stronger (CF-1 and C57BL/6J) and weaker (CHPO) centromeres were determined by an unpaired t-test.
IF-FISH on chromatin fibers
Spleens obtained from CF-1 or CHPO mice were homogenized in a pre-chilled glass homogenizer with 1 ml of ice-cold PBS, and cells were pelleted at 600 × g for 10 min. Chromatin fibers were then obtained using two different protocols [48,49]. For the first protocol, cell pellets were resuspended in nuclear isolation buffer (150 mM NaCl, 10 mM HEPES, pH 7.4, 1.5 mM MgCl2, 10 mM KCl, 0.5% NP-40, 0.5 mM DTT, 1mM PMSF, 1 μg/ml pepstatin, 1 μg/ml aprotinin/leupeptin) and 10 μl of nuclei suspension was put on positively charged slides (Superfrost Plus Microscope Slides; Fisher). Slides were placed in lysis buffer (25 mM Tris–HCl pH 7.5, 0.5 M NaCl, 1% Triton X-100, 0.5 M Urea) for 10 min, and then fixed with 4% paraformaldehyde for 10 min [48]. For the second protocol, after centrifugation the cell pellet was resuspended to 6–7×104 cells/ml in hypotonic buffer (0.8% Sodium Citrate) for 2 min [49]. The cell suspension was spun onto positively charged slides (Superfrost Plus Microscope Slides; Fisher) using Cytospin 4 (Thermo) at 800 rpm for 4 min, and the slides were placed in lysis buffer for 15 min, permeabilized in PBS with 0.1% Triton X-100 for 15 min, and then place in lysis buffer for 15 min. Slides were fixed with 4% paraformaldehyde.
For CENP-A staining, slides were placed in blocking solution (1% BSA, 0.5% Triton X-100, 0.02% Sodium Azide in PBS) for 1 h and stained with rabbit anti-CENP-A (1:200, Cell Signaling #2048S) for 1 h, followed by Alexa Flour 488 donkey anti-rabbit IgG (1:200, Life Technologies) for 1 h. Slides were washed with PBS, and fibers were fixed with 4% paraformaldehyde for 10 min at RT and then stored in PBS at 4°C until FISH labeling. Minor satellite FISH was performed as described above. Images were captured with a 100 × 1.4 NA objective on an inverted fluorescence microscope (DMI6000; Leica) equipped with a charge-coupled device camera (QuantEM 512SC, Photometrics) and a SPECTRA × Light Engine (Lumencor), controlled by MetaMorph software (Molecular Devices).
We used the following two criteria to choose chromatin fibers for analysis: (1) the minor satellite signals can be traced for its entire length, and (2) dot-like signals of CENP-A and minor satellite indicate the fiber is not bundled with other fibers. Segmented lines were drawn along CENP-A or minor satellite signals, the lengths of the lines were measured using Fiji/Image J, and the ratio of the length of CENP-A to the length of minor satellite was calculated for each fiber. Results were pooled from five different independent experiments, and statistical differences between stronger (CF-1) and weaker (CHPO) centromeres were determined by an unpaired t-test.
IF-FISH on intact mitotic cells
Immortalized CF-1×CHPO hybrid MEFs were grown on coverslips and incubated with nocodazole (100 ng/ml) for 2 h to enrich mitotic cells, followed by incubation with a kinesin-5 inhibitor, STLC (Sigma) for 1 h, to generate monopolar spindles. IF-FISH on intact cells was conducted as previously described [50]. Cells were fixed in 2% paraformaldehyde for 10 min at RT, permeabilized in 0.5% Triton X-100 for 5 min at 4°C, then incubated with IF block (2% FBS, 2% BSA, 0.1% Tween, 0.02% Sodium Azide in PBS). Cells were then incubated with λ-phosphatase (80U/μl, New England Biolabs) for 2 h at 30°C to remove phosphate groups blocking the CENP-A epitope, and stained with rabbit anti-CENP-A (1:200) for 1 h, followed by Alexa Fluor 488 donkey anti-rabbit IgG (1:200) for 1 h. Cells were then washed with PBS, fixed with 4% paraformaldehyde for 10 min at RT, permeabilized in 0.7% Triton X-100/0.1M HCl for 10 min at 4°C. Chromosomes were denatured at 77°C for 30 min, and then cells were incubated overnight at 37°C with 40 ng of minor satellite probes that were denatured separately at 77°C for 15 min in hybridization buffer (50% formamide, 10% Dextran Sulfate, 2× SSC, 10 μg sheared single-stranded salmon DNA). After three washes each with 50% formamide/2× SSC and 2× SSC at 37°C, 5 min each, samples were incubated with Alexa Flour 647 streptavidin (1:100, ThermoFisher).
Confocal images of mitotic cells were captured with a 63× NA 1.4 objective using a Leica STED 3× Super Resolution laser scanning confocal system on an inverted fluorescence microscope (DMI6000, Leica) equipped with an XY Piezo-Z stage (Applied Scientific Instrumentation), controlled by LAS × Software (Leica). Images were collected as z-stacks at 0.3 μm intervals and analyzed using Fiji/ImageJ.
CF-1 and CHPO centromeres were distinguished within an individual cell based on bright or dim minor satellite signals, respectively. Ellipses were drawn around the centromeres, and CENP-A intensity was integrated over each ellipse in optical slices containing the centromere, after subtracting background. Eleven cells were analyzed from three independent experiments. Statistical differences between stronger (CF-1) and weaker (CHPO) centromeres were determined by an unpaired t-test.
Oocyte collection and culture
Female mice (7–14 wk-old) were hormonally primed with 5U of Pregnant Mare Serum Gonadotropin (PMSG, Calbiochem) 48 h prior to oocyte collection. Germinal vesicle (GV)-intact oocytes were collected in bicarbonate-free minimal essential medium with polyvinylpyrrolidone and Hepes (MEM-PVP) [51], denuded from cumulus cells, and cultured in Chatot-Ziomek-Bavister (CZB) [52] medium with 2.5 μM milrinone covered with mineral oil (Sigma) in a humidified atmosphere of 5% CO2 at 37°C. To induce meiotic entry, oocytes were washed through several drops of CZB media without milrinone.
Immunostaining of MI oocytes
7–8 h after milrinone washout, oocytes were fixed with 2% paraformaldehyde in PBS (+0.1% Triton X-100) for 20 min. For CENP-A staining, oocytes were treated with λ-phosphatase (80U/μl, New England Biolabs) for 2 h at 30°C to remove phosphate groups blocking the CENP-A epitope. Oocytes were then permeabilized for 15 min at RT (room temperature; 0.1% Triton X-100 in PBS) and blocked for 15 min at RT (0.1% BSA, 0.01% Tween-20 in 1× PBS) prior to staining. Oocytes were labelled with rabbit anti-CENP-A (1:200, Cell Signaling #2048S), rabbit anti-CENP-C (1:2000, a gift from Dr. Yoshinori Watanabe, University of Tokyo [53]), rabbit anti-H3K9me3 (1:500, Abcam ab8898) or CREST Human Autoantibody Against Centromere (1:50, Immunovision) for 1 h, followed by fluorescently-labelled goat or donkey anti-rabbit IgG (1:500, Life Technologies) or anti-human IgG (1:100, Life Technologies) for 1 h. Following each staining step, oocytes were washed in blocking buffer three times (10–15 min each wash). Finally, oocytes were mounted in Vectashield (Vector) containing DAPI.
CF-1 × CHPO and C57BL/6J × CHPO hybrid oocytes contain six bivalents of homologous telocentrics and seven trivalents, in which seven CHPO metacentric fusions pair with homologous CF-1 telocentrics [5]. We focused only on bivalents, which can be distinguished from trivalents based on chromosomal morphology, for measurement of CENP-A and CENP-C signal intensity. Using Fiji/ImageJ, optical slices containing centromeres from the same bivalent were added to produce a sum projection. Ellipses were drawn around the centromeres, and signal intensity was integrated over each ellipse after subtracting background, obtained by measuring the average intensity of a region near the centromeres. Ratios were obtained for each bivalent by dividing the intensity of the brighter centromere by that of the dimmer centromere.
Oocyte microinjection and live imaging
CF-1, CHPO, and CF-1 × CHPO oocytes at GV stage were subjected to inter-cytoplasmic microinjection in MEM-PVP medium at RT with a micromanipulator (Narishige) and a picoinjector (Medical Systems Corp). Each oocyte was injected with 2 pl of cRNA, then incubated for 16–18 h at 37°C in a humidified atmosphere of 5% CO2 to allow protein expression. The following cRNAs were used for microinjection: H2B-EGFP (human histone H2B with EGFP at the C-terminus) at a final concentration of 600 ng/μl, CENP-B-mCherry (mouse CENP-B with mCherry at the C-terminus) at 1300 ng/μl, H2B-mCherry (human histone H2B with mCherry at the C-terminus) at 400 ng/μl, CENP-B-EGFP (mouse CENP-B with EGFP at the C-terminus) at 1040 ng/μl. The cRNAs were synthesized using the Transcript Aid T7 High Yield Transcription kit (Thermo Fisher Scientific) and purified by phenol-chloroform extraction.
Live imaging of injected oocytes was performed after meiotic resumption induced by milrinone washout. Oocytes were placed into 3.5 μl drops of CZB media covered with mineral oil in a 35 mm tissue culture dish (FluoroDish FD35–100) in a heated environmental chamber with a stage top incubator (Incubator BL and Heating Insert P; PeCon GmBH) to maintain 5% CO2 in air and 37°C. Confocal images were collected with a microscope (DMI4000 B; Leica) equipped with a 63× 1.3 NA glycerol-immersion objective lens, an xy piezo Z stage (Applied Scientific Instrumentation), a spinning disk confocal scanner (Yokogawa Electric Corporation), an electron multiplier charge-coupled device camera (ImageEM C9100-13; Hamamatsu Photonics), and an LMM5 laser merge module with 488- and 593-nm diode lasers (Spectral Applied Research) controlled by MetaMorph software (Molecular Devices). For analysis of CENP-B at centromeres, oocytes expressing CENP-B-mCherry and H2B-EGFP were imaged as z-stacks with 0.4 μm spacing at a single time point 7 h after GVBD. CENP-B signal was quantified in regions drawn manually around individual centromeres (n > 10 per oocyte, n > 10 oocytes, 3 independent experiments) in the z-slice with the brightest signal after subtracting background.
For the biased orientation assay, GV oocytes from CF-1 × CHPO were collected and microinjected for CENP-B-EGFP and H2B-mCherry. Oocytes were induced for meiotic resumption by washing out milrinone and imaged live as described above 7–12 h after GVBD to capture the timing just before anaphase onset. Images were taken every 30 min. Spindle position was confirmed by DIC images, and the fraction of bivalents with the stronger centromere oriented towards the egg was quantified (n=133 bivalents from 26 oocytes, pooled from four independent experiments). Chi square analysis was used to determine statistical significance of a difference from 50:50 ratio in Figure 4D.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details of the experiments can be found in the figure legends, table footnotes, and the Method Details section of the STAR Methods. Statistical details include exact value of n, what n represents, definition of center, and dispersion and precision measures. Statistical test were performed as described in the Method Details section using either GraphPad Prism or MATLAB, and a p value of less than 0.05 was judged as statistically significant.
DATA AND SOFTWARE AVAILABILITY
Data Resources
Raw data files for the DNA sequencing analysis have been deposited in the NCBI Sequence Read Archive (SRA) under project accession number SRA: SRP109959.
Custom Scripts
Custom scripts used for tandem repeat analysis of paired-end sequencing reads are available at SourceForge, see Key Resource Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Alexa Fluor® 488 Goat Anti-Human IgG (H+L) | Life technologies | A11013 |
| Alexa Flour 594 donkey anti-rabbit IgG | Life technologies | A11008 |
| Alexa Flour 594 donkey anti-rabbit IgG | Life technologies | A21207 |
| Fluorescein antidigoxigenin | Roche | 11207741910 |
| CREST Human Autoantibody Against Centromere | Immunovision | HCT-0100 |
| Anti-histone H3K9me3 antibody | Abcam | ab8898 |
| CENP-A (C51A7) Rabbit antibody for immunostaining | Cell signaling | 2048 |
| Rabbit α-mouse-specific CENP-A antibody for ChIP | Beth Sullivan, Duke University [35] | N/A |
| Rabbit α-mouse-specific CENP-C antibody | Yoshinori Watanabe, University of Tokyo [53] | N/A |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| SuperScript™ II Reverse Transcriptase | invitrogen | 18064014 |
| Rediprime II DNA labeling kit | GE | RPN1633 |
| MicroSpin G-25 columns | GE | 27-5325-01 |
| Nocodazole | sigma | M1404 |
| Dextran sulfate | Amresco | E516 |
| Sheared single-stranded salmon DNA | Sigma | D7656 |
| TransIT-X2® Dynamic Delivery System | Mirus | MIR 6004 |
| λ-phosphatase | New England Biolabs | P0753S |
| STLC | Sigma | 164739 |
| Pregnant Mare Serum Gonadotropin (PMSG) | Calbiochem | 367222 |
| mineral oil | Sigma | M5310 |
| milrinone | Sigma | M4659 |
| Transcript Aid T7 High Yield Transcription kit | ThermoFisher | K0441 |
| Digoxigenin-11-dUTP, alkali-stable | Roche | 11093088910 |
| Anti-Digoxigenin-Fluorescein, Fab fragments | Roche | 11207741910 |
| Nick Translation Kit | Roche | 10976776001 |
| Biotin-16-dUTP | Roche | 11093070910 |
| Streptavidin AF 594 | Life technologies | S11227 |
| Alexa Flour 647 streptavidin | ThermoFisher | S21374 |
| Micrococcal Nuclease | Affymetrix | 70196Y; CAS: 9013-53-0 |
| Protein G Sepharose 4 Fast Flow | GE | 17061801 |
| KAPA HiFi DNA Polymerase with dNTPs | Kapa Biosystems | KK2102 |
| 10× T4 DNA Ligase Buffer with 10 mM ATP | New England Biolabs | B0202S |
| T4 DNA Polymerase | New England Biolabs | M0203S |
| Klenow DNA Polymerase | New England Biolabs | M0210S |
| T4 Polynucleotide Kinase (PNK) | New England Biolabs | M0201S |
| Klenow Fragment (3′ to 5′ exo minus) | New England Biolabs | M0212S |
| Quick DNA Ligation Kit | New England Biolabs | M2200S |
| QIAquick PCR Purification Kit | Qiagen | 28104 |
| MinElute PCR Purification Kit | Qiagen | 28004 |
| Critical Commercial Assays | ||
| SYBR Green JumpStart Taq ReadyMix | Sigma | S4438 |
| Agilent High Sensitivity DNA Kit | Agilent | 5067-4626 |
| NextSeq 500/550 Mid Output Kit v2 (300 cycles) | Illumina | FC-404-2003 |
| Deposited Data | ||
| Raw data | This paper | SRA: SRP109959 |
| Experimental Models: Cell Lines | ||
| Primary mouse embryonic fibroblasts of C57BL/6J | This paper | N/A |
| Primary mouse embryonic fibroblasts of CF-1 | This paper | N/A |
| Primary mouse embryonic fibroblasts of CHPO | This paper | N/A |
| Immortalized mouse embryonic fibroblasts of CF-1 × CHPO hybrid mice | This paper | N/A |
| Primary lung fibroblasts of C57BL/6J | This paper | N/A |
| Primary lung fibroblasts of CHPO | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | 000664 |
| Mouse: ZALENDE/EiJ (CHPO) | The Jackson Laboratory | 001392 |
| Mouse: NSA (CF-1) | Envigo | 033 |
| Oligonucleotides | ||
| CENP-A cDNA sequencing primer1: (FW 5′-CTCGCGTTCGGTTTCGGCAGCAGGACC-3′, RV: 5′-CTCCTCCGACACCACTGTCAAGC-3′) | This paper | N/A |
| CENP-A cDNA sequencing primer2: (FW 5′-ACTGCTGGCGACCGAGTTCTGG-3′, RV: 5′-ACAGTTTTAAGGGCACCGTGTAGCC-3′) | This paper | N/A |
| Primers for cloning of minor satellite repeats: FW: 5′-GGAAAATGATAAAAACCACACTG-3′, RV: 5′-TCATTGATATACACTGTTCTACA-3′ | This paper | N/A |
| Major satellite primers for qPCR: (FW: 5′-GACGACTTGAAAAATGACGAAATC-3′, RV: 5′-CATATTCCAGGTCCTTCAGTGTGC-3′) | [44] | N/A |
| Minor satellite primers for qPCR: (FW: 5′-CATGGAAAATGATAAAAACC-3′, RV: 5′-CATCTAATATGTTCTACAGTGTGG-3′) | [44] | N/A |
| Recombinant DNA | ||
| Plasmid containing minor satellite repeats | Rachel O’Neill, University of Connecticut | N/A |
| Plasmid containing major satellite repeats | Rachel O’Neill | N/A |
| Plasmid containing SV40 large T antigen | Brad Johnson, University of Pennsylvania [34] | N/A |
| Software and Algorithms | ||
| GraphPad Prizm | GraphPad | http://www.graphpad.com/ |
| Lasergene | DNASTAR | https://www.dnastar.com/ |
| samtools-0.1.19 | [36] | http://samtools.sourceforge.net/ |
| ClustalW | [39] | http://www.ebi.ac.uk/Tools/msa/clustalw2/ |
| Clustal Omega | [40] | http://www.ebi.ac.uk/Tools/msa/clustalo/ |
| Jalview | [41] | http://www.jalview.org/ |
| WebLogo | [42] | http://weblogo.berkeley.edu/logo.cgi |
| RepeatMasker open-4.0.5 and open-4.0.6 | [43] | http://www.repeatmasker.org |
| FIJI/ImageJ | [46,47] | https://fiji.sc/ |
| picard-tools-2.2.4 | N/A | http://broadinstitute.github.io/picard/ |
| MATLAB R2013b | https://www.mathworks.com/ | |
| Custom scripts for tandem repeat analysis of paired-end sequencing reads | This paper | https://sourceforge.net/p/blacklab-mouse-seq/code |
| Other | ||
Supplementary Material
Acknowledgments
We thank M. Riegman for assistance with cell culture, G. Logsdon and T. Christopher for help with Illumina library preparation and sequencing, respectively, P. Heun for experimental advice, R. O’Neill for minor and major satellite plasmids and discussions, F.B. Johnson for the SV40 large T antigen plasmid, and Y. Watanabe for rabbit anti-CENP-C antibody. The research was supported by NIH grants GM107086 (M.A.L. and R.M.S) and GM082989 (B.E.B), by an NIH postdoctoral fellowship award GM108360 (J.M.D.) and by JSPS postdoctoral fellowship for research abroad (T.A.)
Footnotes
Author Contributions
A.I.O., J.M.D., T.A., S.J.F., R.M.S., M.A.L., and B.E.B. designed experiments. A.I.O., J.M.D., T.A., S.J.F., L.C., and K.Y. performed experiments. B.A.S. provided essential reagents and methodologies. A.I.O., J.M.D., M.A.L., and B.E.B. wrote the manuscript. All authors edited the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pardo-Manuel De Villena F, Sapienza C. Nonrandom segregation during meiosis: The unfairness of females. Mamm Genome. 2001;12:331–339. doi: 10.1007/s003350040003. [DOI] [PubMed] [Google Scholar]
- 2.Chmatal L, Schultz RM, Black BE, Lampson MA. Cell biology of cheating—transmission of centromeres and other selfish elements through asymmetric meiosis. Prog Mol Subcell Biol. 2017 doi: 10.1007/978-3-319-58592-5_16. in press. [DOI] [PubMed] [Google Scholar]
- 3.Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- 4.Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 5.Chmatal L, Gabriel SI, Mitsainas GP, Martinez-Vargas J, Ventura J, Searle JB, Schultz RM, Lampson MA. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Curr Biol. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westhorpe FG, Straight AF. The centromere: Epigenetic control of chromosome segregation during mitosis. Cold Spring Harb Perspect Biol. 2015;7:a015818. doi: 10.1101/cshperspect.a015818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawe RK. Handbook of Maize: Genetics and Genomics. New York, NY: Springer New York; 2009. Maize centromeres and knobs (neocentromeres) pp. 239–250. [Google Scholar]
- 9.Didion JP, Morgan AP, Clayshulte AMF, Mcmullan RC, Yadgary L, Petkov PM, Bell TA, Gatti DM, Crowley JJ, Hua K, et al. A multi-megabase copy number gain causes maternal transmission ratio distortion on mouse chromosome 2. PLoS Genet. 2015;11:e1004850. doi: 10.1371/journal.pgen.1004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–21. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 11.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A–H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tachiwana H, Kagawa W, Shiga T, Osakabe A, Miya Y, Saito K, Hayashi-Takanaka Y, Oda T, Sato M, Park SY, et al. Crystal structure of the human centromeric nucleosome containing CENP-A. Nature. 2011;476:232–235. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 13.Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LET, Van Duyne GD, Vinogradov SA, et al. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348:699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 15.Bodor DL, Valente LP, Mata JF, Black BE, Jansen LET. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell. 2013;24:923–32. doi: 10.1091/mbc.E13-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smoak EM, Stein P, Schultz RM, Lampson MA, Black BE. Long-term retention of CENP-A nucleosomes in mammalian oocytes underpins transgenerational inheritance of centromere identity. Curr Biol. 2016;26:1110–1116. doi: 10.1016/j.cub.2016.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raychaudhuri N, Dubruille R, Orsi GA, Bagheri HC, Loppin B, Lehner CF. Transgenerational propagation and quantitative maintenance of paternal centromeres depends on Cid/Cenp-A presence in Drosophila sperm. PLoS Biol. 2012;10:e1001434. doi: 10.1371/journal.pbio.1001434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guenatri M, Bailly D, Maison C, Almouzni G. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J Cell Biol. 2004;166:493–505. doi: 10.1083/jcb.200403109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kipling D, Mitchell AR, Masumoto H, Wilson HE, Nicol L, Cooke HJ. CENP-B binds a novel centromeric sequence in the Asian mouse Mus caroli. Mol Cell Biol. 1995;15:4009–20. doi: 10.1128/mcb.15.8.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasson D, Panchenko T, Salimian KJ, Salman MU, Sekulic N, Alonso A, Warburton PE, Black BE. The octamer is the major form of CENP-A nucleosomes at human centromeres. Nat Struct Mol Biol. 2013;20:687–95. doi: 10.1038/nsmb.2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacoste N, Woolfe A, Tachiwana H, Garea AV, Barth T, Cantaloube S, Kurumizaka H, Imhof A, Almouzni G. Mislocalization of the Centromeric Histone Variant CenH3/CENP-A in Human Cells Depends on the Chaperone DAXX. Mol Cell. 2014;53:631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 22.Zhang T, Talbert PB, Zhang W, Wu Y, Yang Z, Henikoff JG, Henikoff S, Jiang J. The CentO satellite confers translational and rotational phasing on cenH3 nucleosomes in rice centromeres. Proc Natl Acad Sci. 2013;110:E4875–E4883. doi: 10.1073/pnas.1319548110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nechemia-Arbely Y, Fachinetti D, Miga KH, Sekulic N, Soni GV, Kim DH, Wong AK, Lee AYK, Nguyen C, Dekker B, et al. Human centromeric CENP-A chromatin is a homotypic, octameric nucleosome at all cell cycle points. J Cell Biol. 2017 doi: 10.1083/jcb.201608083. jcb.201608083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. J Cell Biol. 1989;109:1963–73. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. The quantitative architecture of centromeric chromatin. Elife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peters AHFM, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 27.Kit S. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J Mol Biol. 1961;3:711–716. doi: 10.1016/s0022-2836(61)80075-2. [DOI] [PubMed] [Google Scholar]
- 28.Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: A repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol. 2004;11:1076–83. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stimpson KM, Song IY, Jauch A, Holtgreve-Grez H, Hayden KE, Bridger JM, Sullivan BA. Telomere Disruption Results in Non-Random Formation of De Novo Dicentric Chromosomes Involving Acrocentric Human Chromosomes. PLoS Genet. 2010;6:e1001061. doi: 10.1371/journal.pgen.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seluanov A, Vaidya A, Gorbunova V. Establishing primary adult fibroblast cultures from rodents. J Vis Exp. 2010:8–11. doi: 10.3791/2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conner DA. Mouse embryo fibroblast (MEF) feeder cell preparation. Curr Protoc Mol Biol. 2001;Chapter 23(Unit 23.2) doi: 10.1002/0471142727.mb2302s51. [DOI] [PubMed] [Google Scholar]
- 34.Yu Y, Alwine JC. Human cytomegalovirus major immediate-early proteins and simian virus 40 large T antigen can inhibit apoptosis through activation of the phosphatidylinositide 3′-OH kinase pathway and the cellular kinase Akt. J Virol. 2002;76:3731–8. doi: 10.1128/JVI.76.8.3731-3738.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maloney KA, Sullivan LL, Matheny JE, Strome ED, Merrett SL, Ferris A, Sullivan BA. Functional epialleles at an endogenous human centromere. Proc Natl Acad Sci. 2012;109:13704–13709. doi: 10.1073/pnas.1203126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. Genome Project Data Processing Subgroup, 1000 Genome Project Data Processing (2009) The Sequence Alignment/Map format and SAMtools Bioinformatics. 1000;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wong AKC, Rattner JB. Sequence organization and cytological localization of the minor satellite of mouse. Nucleic Acids Res. 1988;16:11645–11661. doi: 10.1093/nar/16.24.11645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horz W, Altenburgerl W. Nucleotide sequence of mouse satellite DNA. Nucleic Acids Res. 1981;9:683–696. doi: 10.1093/nar/9.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2014;7:539–539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smit A, Hubley R, Green P. RepeatMasker Open-4.0. 2013–2015 < http://www.repeatmasker.org>.
- 44.Martens JHA, O’Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–12. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using μManager software. J Biol methods. 2014;1:10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem. 2006;281:12760–12766. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan BA. Methods in molecular biology. Clifton, N.J: 2010. Optical Mapping of Protein–DNA Complexes on Chromatin Fibers; pp. 99–115. [DOI] [PubMed] [Google Scholar]
- 50.Chaumeil J, Micsinai M, Skok JA. Combined immunofluorescence and DNA FISH on 3D-preserved interphase nuclei to study changes in 3D nuclear organization. J Vis Exp. 2013:e50087. doi: 10.3791/50087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stein P, Schindler K. Mouse oocyte microinjection, maturation and ploidy assessment. J Vis Exp, e2851. 2011 doi: 10.3791/2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil. 1989;86:679–88. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
- 53.Kim J, Ishiguro K, Nambu A, Akiyoshi B, Yokobayashi S, Kagami A, Ishiguro T, Pendas AM, Takeda N, Sakakibara Y, et al. Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature. 2014;517:466–471. doi: 10.1038/nature14097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.