Abstract
Objective
This first pilot study of genome-wide expression as predictor of antidepressant response in late-life depression examined genome-wide transcriptional profiles in a randomized placebo-controlled trial of combined methylphenidate and citalopram.
Methods
Genome-wide transcriptional profiles were examined in peripheral blood leukocytes sampled at baseline and 16 weeks from 35 older adults with major depression, who were randomized to methylphenidate + citalopram, citalopram + placebo, or methylphenidate + placebo. Methylphenidate doses ranged between 10 and 40mg/day, and citalopram doses ranged between 20 and 60mg/day. Remission was defined as Hamilton Depression Rating Scale score of 6 or below. Early remission was achieved in the first 4 weeks of treatment. We hypothesized that differential gene expression at baseline can predict antidepressant response.
Results
We analyzed gene expression in 24 remitters and 11 non-remitters. At baseline, we found three genes showing higher expression in all remitters versus non-remitters that satisfied the established level of significance: a fold change of 2 and p-value of 0.05 that included HLA-DRB5, SELENBP1, and LOC388588. Two gene transcripts showed higher expression in early remitters at baseline compared with non-remitters. The first gene was CA1 carbonic anhydrase gene, on chromosome 8 involved in respiratory function (fold change 2.54; p=0.03). The second gene was the SNCA-α-synuclein gene, implicated, which binds to dopamine transporter (fold change 2.1; p=0.03).
Conclusions
Remission to antidepressants in geriatric depression may be associated with a particular gene expression profile in monoaminergic and metabolic pathways and needs to be replicated in a larger sample.
Keywords: geriatric depression, gene expression, antidepressant response, remission, citalopram, methylphenidate
Introduction
Late-life depression (LLD) is a major health issue and has a rising prevalence given population aging (Ferrari et al., 2013; UN, 2013). The Global Burden of Disease Study 2010 suggests depressive disorders were the second leading cause of years lived with disability in 2010 (Ferrari et al., 2013). In addition to the high and rising burden of LLD, conventional antidepressant therapies are limited by modest efficacy. In LLD, a recent meta-analysis of clinical trials suggests a response rate of 48% and a remission rate of 33.7%, both very similar to response and remission rates found in adult patients (Kok et al., 2012).
It is important to understand the underlying biological mechanisms of treatment resistance with non-remission to medications. Response to psychiatric medications in LLD is highly heterogeneous and complex; however, there is a paucity of research into genomic and transcriptional predictors of treatment response, especially in older adults (Shiroma et al., 2010). Because of the increased burden of disease, accelerated response and remission are particularly important (Lavretsky et al., 2015). Typical targets of examination in pharmacogenetic depression studies include candidate genes from the monoaminergic neurotransmitter pathways related to the putative mechanisms of action (i.e., serotonin, adrenalin, and dopamine), neuroplasticity (e.g., BDNF, CREB1) (Alexopoulos et al., 2010; Taylor et al., 2011; Murphy et al., 2013), treatment resistance (ABCB1 (MDR1)) (Sarginson et al., 2010), or immune modulation (Kalman et al., 2005). The search for a genetic mechanism of treatment response in depression still continues with only a few confirmed results.
Methylphenidate is a promising adjunctive therapy in LLD, which may be useful in optimizing the treatment outcomes (Lavretsky et al., 2003; Lavretsky et al., 2006; Lavretsky et al., 2008). Methylphenidate is a dopamine reuptake inhibitor and, as a single agent, has been shown to be effective and safe in a few open and controlled studies in the older people (Satel and Nelson, 1989; Murray and Cassem, 1998; Volkow et al., 1998; Lavretsky and Kumar, 2001; Lavretsky et al., 2003; Lavretsky et al., 2006). Two large series of medically ill mixed-age patients (Woods et al., 1986; Masand et al., 1991) suggested the value of adjunctive dextroamphetamine (range: 2.5–30 mg) and methylphenidate (range: 5–30 mg/day) in relieving depression with the rapid onset of response within 48 h. The use of dopaminergic agents like methylphenidate in LLD could be particularly useful in the treatment of LLD given the observed reduction in dopamine neurotransmission with aging (Volkow et al., 1998; Lavretsky et al., 2003; Lavretsky et al., 2006). Recently, our group conducted the first randomized placebo-controlled trial aimed to test the clinical efficacy of methylphenidate (MPH) in combination with citalopram (CIT) used to improve antidepressant response in LLD (Lavretsky et al., 2015). This study demonstrated a faster and improved response in the combined treatment group compared with either drug with placebo. The rate of improvement in the citalopram plus methylphenidate group was significantly higher than that in the citalopram plus placebo group in the first 4 weeks of the trial.
Peripheral blood gene-expression studies have shown to provide a good proxy for what may take place in the brain of psychiatric patients and can assist in predicting response to treatment (Mamdani et al., 2014). In our pilot controlled trial (Lavretsky et al., 2008) of methylphenidate in 15 older outpatients with major depression, those with DAT VNTR 10/10 genotype had greater cognitive executive dysfunction at baseline but responded preferentially to methylphenidate added to citalopram with a greater reduction in depression severity over time compared with other subjects. Only one prior study has explored gene expression biomarkers of response to antidepressants at a genome-wide level. We are not aware of any studies in late-life populations that explore gene expression biomarkers of response at a genome-wide level (Pitychoutis et al., 2013).
The present pilot study examined leukocyte gene transcriptional alterations in remitters versus non-remitters with antidepressant treatment in a subsample of 35 participants from the parent study (Lavretsky et al., 2015). Genome-wide transcriptional profiling was carried out in the peripheral blood mononuclear cell (PBMC) samples obtained at baseline and post-intervention. We hypothesized that genome-wide transcriptional analyses will reveal potential predictors of remission in the dopaminergic pathways based on our pilot data.
Methods
Study procedures
From August 2008 to September 2012, 510 individuals were screened according to eligibility criteria by phone, yielding 203 individuals for a diagnostic interview. After describing the details of the study to interested and eligible subjects, written informed consent was obtained in accordance with the procedures set by the UCLA Institutional Review Board.
Inclusion and exclusion criteria
Inclusion criteria were (i) current episode of unipolar major depressive disorder according to DSM-IVTR criteria; (ii) Hamilton Depression Rating Scale (HDRS-24) score≥16 (Hamilton, 1959); and (iii) Mini-mental state examination (MMSE) (Folstein et al., 1975) score≥ 26. Exclusion criteria were (i) history of any other psychiatric disorders (other than unipolar major depressive disorder with or without comorbid anxiety symptoms); (ii) severe or acute unstable medical illness, including the presence of either atrial or ventricular arrhythmia, or acute ischemic changes on the baseline electrocardiogram; (iii) acute suicidal or violent behavior or history of suicide attempt within the last year; or (iv) any other central nervous system diseases. Patients were free of psychotropic medications for at least 2 weeks before starting the trial.
Primary outcomes
Remission versus non-remission was the primary outcome of the trial. Remission was defined as HDRS-24 score of 6 or below. Early remission was determined by remission by week 4, while delayed response was response at any time point afterwards (between 4 and 16 weeks). We measured comorbid symptoms of anxiety, apathy, medical and vascular risk factors, health-related quality of life, and cognitive performance.
Randomization procedures
Randomization was performed using a computer-generated schedule. Because there were three groups, we used block randomization to maintain balance over the course of the study with a random mix of block lengths of three and six to help further preserve the blind. Allocation concealment was implemented using sealed, sequentially numbered boxes that were identical in appearance for the three treatment groups. In order to monitor the internal validity of the randomization and blinding in the trial, we created a guessing scale for the study staff in the first year of the trial, and the accuracy of our guessing assignment in two independent trials was 35%.
Intervention procedures
Participants were seen in person weekly for the first 4 weeks, while methylphenidate dose was titrated for evaluation of safety and detection of accelerated response and every 2 weeks thereafter for the remainder of 16 weeks. Treatment with both drugs was initiated simultaneously after the baseline assessment in order to track accelerated response. Participants were given a weekly supply of the given study medications prepared and dispensed by the UCLA pharmacy in matching capsules: 20 mg/day citalopram and methylphenidate 2.5mg (or one cap) twice a day recommended at 9AM and 3PM, or the matching number of capsules of placebo as a starting dose. We used a 5- to 40-mg flexible dose of methylphenidate that was increased based on the response and tolerability assessment during each weekly visit in the first 4 weeks of treatment. The dose range was established in two of our pilot studies that were dedicated to the dose finding and safety evaluation of the optimal methylphenidate dose in older adults (Lavretsky and Kumar, 2001; Lavretsky et al., 2003) The dose of the methylphenidate was increased at each visit if subjects had the Clinical Global Impression (CGI)-Improvement (Guy, 1976) scores of 3 and greater, and they showed no serious adverse effects. The increment increase of methylphenidate occurred in the first 4 weeks of the trial by 2.5mg twice a day every 4 days between days 4 and 28 of treatment or until subjects were able to achieve CGI score of 1 or 2. After day 28 of methylphenidate titration, subjects remained on the same dose through the end of the trial. If subjects showed minimal improvement with CGI improvement score of 3 or greater by day 28 of treatment, citalopram dose was increased to 40mg and continued to the end of the trial in the majority of subjects, with the exception of 13 subjects, who received another increase in citalopram dose to 60mg at weeks 7–8 of the trial owing to insufficient response. The allowed dose adjustment for methylphenidate was decreasing by two pills, to a minimum of 5mg a day, and decreasing citalopram dose to 20 mg. If subjects could not tolerate the minimum allowed dose, they were discontinued from the trial. The use of concomitant rescue medications during the treatment trial was restricted to the use of lorazepam up to 1 mg/day.
Assessment instruments
Subjects were evaluated using validated assessment instruments that included the Hamilton Depression Rating Scale (HDRS-24), Montgomery–Åsberg Depression Rating Scale (Montgomery and Åsberg, 1979), and CGI (Guy, 1976) to measure depression severity and change over time. We measured anxiety symptoms using the Hamilton Anxiety (HAM-A) Rating Scale (Hamilton, 1960). Cerebrovascular Risk Factor Prediction Chart (Truelsen et al., 1994) and Cumulative Illness Rating Scale-Geriatrics (Miller et al., 1992) assessed medical comorbidity. Other instruments, and the Medical Outcomes Study Short Form 36-Item Health Survey. The primary outcome measures were administered at all visits. The rest of the clinical measures were administered at baseline and the end of the study by two raters (H. L. and N. S. C.).
Safety and adherence assessments
Vital signs and weight were measured at baseline and at each visit in addition to a 12-lead electrocardiogram performed at baseline, and at weeks 3 and 16, if any cardiac complaints were present. A physical examination was administered at baseline and week 16, or upon early termination. Side effects were assessed at all visits by the Udvalg for Kliniske Undersogelser Side Effect Rating Scale (Lingjaerde et al., 1987). Treatment compliance was assessed by employing indirect measures of adherence including questioning of the patients, returned pill count, and drug level measures at weeks 3, 8, and 16. Plasma levels of citalopram and metabolite, as well as ritalin and ritalinic acid levels, were obtained.
Leukocyte analysis
Biological genomic analyses were conducted on a randomly selected subsection of the larger clinical trial. RNA samples were quality checked using an Agilent 2200 (Agilent Technologies, Santa Clara, CA, USA) tapestation and quantitated with ribogreen. Total RNA of 100–200 ng was used for amplification/labeling using the Ambion total prep 96 kit. Amplified and labeled samples were hybridized to Illumina expression chips according to the standard Illumina protocol (Cole et al., 2011). Post-hybridization washing and staining were carried out using a SciGene Little Dipper robotic processing platform. Chips were scanned on our Illumina iScan confocal scanner.
Gene expression profiling and analysis
Genome-wide transcriptional profiles were collected from peripheral blood leukocytes sampled at baseline and 16 weeks from 35 older adults with major depression who were randomly selected for testing and were randomized to MPH + CIT, CIT + placebo, or MPH + placebo. Peripheral blood mononuclear cells were isolated by Ficoll density gradient centrifugation of antecubital venipuncture samples drawn between 10 AM and 11 AM at baseline and 16 weeks later following the completion of intervention procedures. Genome-wide transcriptional profiling was carried out with Illumina single color Human BeadChip HT12 v4, according to manufacture’s protocols. Remission and non-remission were determined across the treatment groups.
Statistical analysis
All data were entered into the database at the time of their collection and analyzed after completion of the trial. Raw cell files were loaded into GeneSpring GX 10.0.2 software (Agilent Technologies) for analysis. Quantile normalization was used, and samples were compared with a fold change and t-test analysis. A fold change of greater than 2 and a p-value of less than 0.05 were considered significant. Significant genes were further submitted to Database for Annotation, Visualization and Integrated Discovery (DAVID) analyses (Huang da et al., 2009a; 2009b), to identify biological meaning. Owing to the small number of non-remitters, non-parametric analyses of variance were performed to compare remitters and non-remitters on demographic and clinical variables.
Results
Table 1 presents the baseline demographic and clinical characteristics of the intervention in the 35 intervention completers. Of the 35 study participants, 24 responded to the treatments by meeting the criteria for clinical remission. Thirteen subjects remitted early, within the first 4 weeks of treatment. Remitters and non-remitters did not differ statistically at baseline on demographic measures (sex and education) except for age. Non-remitters were older than remitters (R = 67.2 (5.7) vs. NR = 73.5 (9.3); Wilcoxon statistic = 254.5, p = 0.05). Non-remitters also had higher anxiety scores (R = 7.8 vs. NR = 11.2; Wilcoxon statistic = 265.5, p = 0.02) and lower MMSE scores than responders (R = 28.8 vs. NR = 27.5; Wilcoxon statistic = 133, p = 0.02). Controlling for the group difference in age, the differences for MMSE and HAM-A were no longer significant. Remitters and non-remitters were not significantly different in their drug doses or plasma drug levels (although remitters were borderline different from non-remitters (p = 0.08) in their citalopram levels).
Table 1.
Comparison of clinical and demographic factors at baseline
| Responder (n = 24)
|
Non-responder (n = 11)
|
||||
|---|---|---|---|---|---|
| Variables | Mean (SD) | N (%) | Mean (SD) | N (%) | p-value |
| Age (years) | 67.2 (5.7) | 73.5 (9.3) | 0.05 | ||
| Sex (female) | 15 (68.6) | 4 (31.5) | 0.3 | ||
| Education (years) | 15.5 (1.9) | 14.3 (4.7) | 0.7 | ||
| Age of first episode (years) | 39.6 (21.72) | 41.4 (29.3) | 0.9 | ||
| Medical burden | |||||
| CIRS | 4.9 (3.7) | 5.5 (3.2) | 0.6 | ||
| CVRF | 11.0 (6.2) | 10.7 (4.6) | 0.7 | ||
| Mood | |||||
| HAM-D (screen) | 18.2 (2.3) | 20.3 (3.6) | 0.11 | ||
| HAM-D (16 weeks) | 4.5 (4.4) | 12.6 (3.9) | 0.0006 | ||
| MADRS (screen) | 17.0 (2.4) | 20.1 (4.0) | 0.07 | ||
| MADRS (16 weeks) | 5.5 (5.3) | 10.8 (4.2) | 0.008 | ||
| HAM-A* (screen) | 7.8 (2.3) | 11.2 (4.7) | 0.02 | ||
| HAM-A (16 weeks) | 2.7 (2.2) | 6.0 (3.3) | 0.01 | ||
| Cognition | |||||
| MMSE* | 28.8 (1.2) | 27.5 (1.6) | 0.02 | ||
| Drug doses (ng/ml) | |||||
| Citalopram | 35.00 (20.6) | 16.36 (23.3) | 0.06 | ||
| Methylphenidate | 14.38 (12.4) | 11.82 (9.5) | 0.55 | ||
| Drug levels (ng/ml) | |||||
| Citalopram | 91.01 (75.1) | 34.82 (53.9) | 0.08 | ||
| Dexmethylphenidate | 34.20 (34.3) | 13.38 (20.7) | 0.15 | ||
| Ritalin | 1.64 (2.3) | 0.56 (0.7) | 0.48 | ||
CIRS, Cumulative Illness Rating Scale; CVRF, Cerebrovascular Risk Factor Prediction Chart; HAM-D, Hamilton Depression Rating Scale; MADRS, Montgomery–Åsberg Depression Rating Scale; HAM-A, Hamilton Anxiety Rating Scale; MMSE, Mini-mental state examination.
Significance is reduced to p = 0.07 for HAM-A and p = 0.08 for MMSE, when controlling for age.
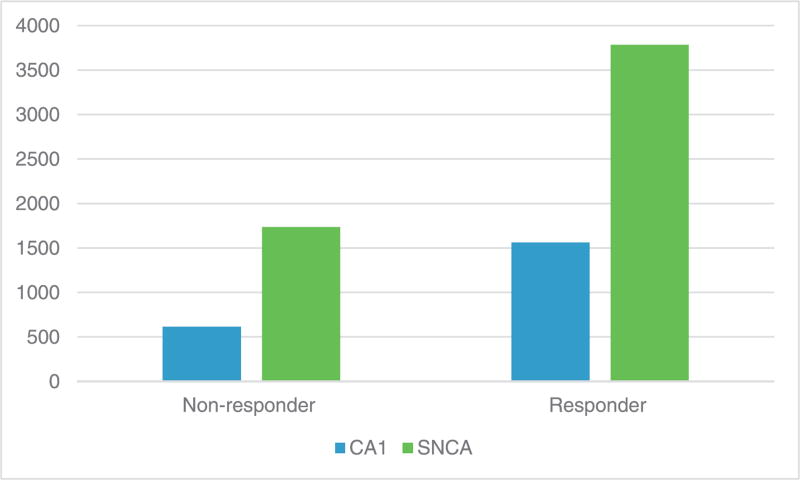
At baseline, 18 genes had higher expression in the early remitters versus non-remitter group with a p-value of less than 0.05 and a fold change of greater than 2 (see Supporting Information Supplement 1 for full list). When considering biological significance of these 18 genes, only two were considered relevant to the mechanism of action of the utilized antidepressants in this study. These genes included CA1 carbonic anhydrase gene on chromosome 8 involved in reversible hydration of CO2 and respiratory function (fold change 2.54; p=0.03) and SNCA-α-synuclein gene implicated in Parkinson’s disease that binds to dopamine transporter (fold change 2.1; p = 0.03). Figure 1 outlines a diagrammatic representation of the fold change in gene expression for each gene. There were no significant correlations between remission and non-remission with final drug doses of CIT and MPH, nor plasma drug levels of CIT, dexmethylphenidate and ritalin, or CA1 and SNCA levels. There were no significant relationships between remitters and non-remitters and genomic data at follow-up.
Figure 1.
Change in gene expression in early remitters versus non-remitters. NB: Y-axis denotes gene expression. The p-value between early remission and non-remission for CA-1 is 0.03; for SNCA, 0.03.
We ran DAVID analyses to look at biologically significant functions among the 18 differentially expressed genes in the early remitter response comparison. A total of 28 Gene Ontology groups were found with a p-value of <0.1 (Supporting Information Supplement 2). Of interest, cytosol and protein stabilization were found significant at p-value of 0.015 and 0.025, respectively. Additionally, ion and cellular homeostasis were also significant (p = 0.005 and p = 0.04, respectively). Finally, immune system development was found to be significant (p = 0.023).
When comparing all remitters (i.e., both early and delayed; n=24) with non-remitters (N = 11), three genes had a higher expression in remitters versus non-remitters. These genes satisfied a fold change of greater than 2 and a p-value of less than 0.05. These genes included major histocompatibility complex, class II, DR beta 5 (HLA-DRB5; fold change 6.53, p = 0.02), selenium binding protein 1 (SELENBP1; fold change 2.02, p = 0.04), and LOC388588 (fold change 2.39, p = 0.03). An additional gene, ALAS2, satisfied a fold change of greater than 2 and a p-value of less than 0.1 (Supporting Information Supplement 1). In this analysis, CA1 and SNCA were not significantly affected.
Discussion
Our preliminary report is the first to suggest a unique transcriptional signature in older remitters and non-remitters to antidepressant treatment in the monoaminergic and metabolic pathways important for neuroplasticity and brain aging.
We are not aware of any other study in LLD exploring gene expression biomarkers of antidepressant response at a genome-wide level (Pitychoutis et al., 2013); however, a single study investigated a defined number of probesets predicting treatment response. Specifically, in a study of 77 adult outpatients, Mamdani et al. (2014) reported differences in baseline peripheral gene expression between remitters and non-remitters in an 8-week citalopram treatment trial. A total of 434 probesets displayed significant correlation to change in score (HAM-D 21), and 33 probesets were differentially expressed between eventual remitters and non-remitters. Probesets for SMAD 7 (SMA-related and MAD-related protein 7) and SIGLECP3 (sialic acid-binding immunoglobulin-like lectin, pseudogene 3) were the most significant differentially expressed genes following FDR correction, and both were down-regulated at baseline in individuals who responded to treatment. Our pilot study does not show significance for these gene expression markers.
In our study, it is important to understand the mechanisms and possible relevance of the genes related to early remission and standard remission. In early remission, the first gene found was CA1 carbonic anhydrase gene located on chromosome 8. This gene is associated with reversible hydration of CO2, respiratory function, water and ion transport, and pH regulation. One or more of various genetically distinct isoforms is expressed nearly ubiquitously in most living cells (Schneider et al., 2013). Some isoforms are located in the cytosol, while others express their activity at the extracellular membrane (Schneider et al., 2013). Owing to the expression of multiple isoforms at different intracellular and extracellular locations, it is difficult to discriminate the contribution of these isoforms (Schneider et al., 2013). Extracellular CA activity was found in hippocampal slices, where the enzyme was implicated in the regulation of excitatory transmission (Fedirko et al., 2007). Functional activity was found again in rodent in vitro hippocampal neurons, where the anion exchanger AE3 activity was enhanced by enzymatic activity (Svichar et al., 2009), and for H+ buffering (Chen and Chesler, 1992). The DAVID analysis revealed ion and cellular homeostasis were significantly affected in the responder group, which is consistent with the aforementioned details about CA1. Taken together, the exact effect of CA in treatment response to antidepressants is not clear; however, it may have a role in modulating excitatory transmission in the brain. The second gene we found correlated to early remission was SNCA-α-synuclein. The function of this gene has been implicated in Parkinson’s disease and is associated with dopamine transporter function (Nuber et al., 2013). The α-synuclein protein is involved in deposition as insoluble fibrils, proteinase K resistant and is modified by truncation, phosphorylation, oxidation, nitrosylation, and ubiquitination (Nuber et al., 2013). Growing evidence points towards modulation of synaptic plasticity of newborn neurons in olfaction by α-synuclein protein (Lazarini and Lledo, 2011). Similarly, in depression, α-synuclein has been shown to decrease olfactory bulb neurogenesis (Winner et al., 2004; Marxreiter et al., 2009; Lelan et al., 2011), whereas treatment with antidepressants resulted in an increased neurogenesis in α-synuclein transgenic mice (expressing human α-synuclein under the oligodendrocyte-specific myelin basic protein promoter (MBP1-hαsyn tg mice)) (Kohl et al., 2012; Ubhi et al., 2012). In these same transgenic mice, antidepressants might be of interest as anti-inflammatory and α-synuclein-reducing agents for multiple system atrophy and other related α-synucleinopathies {Valera, 2014 #285}. Multiple system atrophy is a neurodegenerative disease characterized by the pathological accumulation of α-synuclein within oligodendroglial cells {Valera, 2014 #285}. This accumulation is accompanied by neuroinflammation, thereby leading to neuronal death {Valera, 2014 #285}. Evidence from other rodent studies suggests a high vulnerability of rat dopaminergic synapses to conversion of transgenic human α-synuclein into insoluble neurotoxic conformers (Nuber et al., 2013). The DAVID analysis revealed that cytosol and protein stabilization, and immune system development were significantly affected in the responder group, and this is consistent with the aforementioned details on SNCA.
Three genes were involved in standard remission (HLA-DRB5, SELENBP1, and LOC388588); however, their role in enhanced treatment response is large unknown. HLA-DRB5 is associated with multiple sclerosis, Parkinson’s disease, immunocompetence, and histocompatibility {Lincoln, 2009 #278} {Cano, 2007 #279} {Handel, 2010 #280}. We are not aware of any studies exploring this gene in depression. Intuitively, increased HLA-DRB5 expression in remitters suggests immune function is involved in the treatment response. SELENBP1 is involved with selenium metabolism {Amar, 2010 #281}, and we are not aware of any studies of this gene in depression. Selenium is implicated in neuroprotection; therefore, modulation of SELENBP1 may impact selenium and therefore the neuroprotective mechanisms {Yeo, 2007 #282} {Saito, 1998 #283}. The role of LOC388588 gene has not been explored in depression and neuroscience, and hence, it is unclear what mechanistic role it has in enhancing treatment remission in LLD.
We chose not to correct the correlations between remitters and non-remitters and genomic data for multiple comparisons, as this is the first study of this kind exploring genome-wide data on treatment response in LLD. This study is intended as a hypothesis-generating study where it is more important not to miss possibly important findings rather than to prematurely discard potentially useful observations because of type 2 errors caused by corrections for multiplicity {Rothman, 1990 #228}. The statistical analyses used in this study did not control for age, gender, or treatment group. This is a limitation that should be addressed in future research as these factors may influence results.
Despite the preliminary nature and small sample, our results are of interest, suggesting a unique transcriptional signature of early remission and standard remission in antidepressant treatment with citalopram and/or methylphenidate in the monoaminergic and metabolic pathways important for neuroplasticity during brain aging. Our identified biomarkers of response may be specific to our population and the study drugs, and hence, these results will require future replications in a large sample.
Supplementary Material
Key points.
This is the first genome-wide expression biomarker study of remission to antidepressants in late-life depression (LLD).
We explored the response of subjects with LLD to either methylphenidate and citalopram, citalopram, and placebo or methylphenidate and placebo.
We found three genes showing increased expression in all remitters versus non-remitters at baseline that satisfied the established level of significance (a fold change of 2 and p-value of 0.05) including HLA-DRB5, SELENBP1, and LOC388588.
In those who showed early remission within the first 4 weeks of treatment, two gene transcripts showed increased expression at baseline versus non-remitters, CA1 carbonic anhydrase gene, and SNCA-α-synuclein gene.
Remission to antidepressants in LLD may be associated with a particular gene expression profile in monoaminergic, neuroplastic, and metabolic pathways.
Acknowledgments
This work was supported by NIH grants MH077650 and MH086481 to Dr. Lavretsky.
Footnotes
Conflict of interest
None declared.
Clinical trial registration
NCT00602290. Effectiveness of Methylphenidate in Improving Cognition and Function in Older Adults with Depression.
Additional supporting information may be found in the online version of this article at the publisher’s website.
References
- Alexopoulos GS, Glatt CE, Hoptman MJ, et al. BDNF val66met polymorphism, white matter abnormalities and remission of geriatric depression. J Affect Disord. 2010;125:262–268. doi: 10.1016/j.jad.2010.02.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JC, Chesler M. pH transients evoked by excitatory synaptic transmission are increased by inhibition of extracellular carbonic anhydrase. Proc Natl Acad Sci USA. 1992;89:7786–7790. doi: 10.1073/pnas.89.16.7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Cacioppo JT. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci USA. 2011;108:3080–3085. doi: 10.1073/pnas.1014218108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedirko N, Avshalumov M, Rice ME, Chesler M. Regulation of postsynaptic Ca2+ influx in hippocampal CA1 pyramidal neurons via extracellular carbonic anhydrase. J Neurosci. 2007;27:1167–1175. doi: 10.1523/JNEUROSCI.3535-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari AJ, Charlson FJ, Norman RE, et al. Burden of depressive disorders by country, sex, age, and year: findings from the global burden of disease study 2010. PLoS Med. 2013;10:e1001547. doi: 10.1371/journal.pmed.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Guy W. Clinical Global Impressions (CGI) In: Services UDoHaH., editor. ECDEU Assessment Manual for Psychopharmacology. Alcohol Drug Abuse and Mental Health Administration, NIMPH Psychopharmacology Research Branch; Rockville: 1976. pp. 218–222. [Google Scholar]
- Hamilton M. The assessment of anxiety states by rating. Br J Med Psychol. 1959;32:50–55. doi: 10.1111/j.2044-8341.1959.tb00467.x. [DOI] [PubMed] [Google Scholar]
- Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23:56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009a;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009b;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman J, Palotas A, Juhasz A, et al. Impact of venlafaxine on gene expression profile in lymphocytes of the elderly with major depression—evolution of antidepressants and the role of the “neuro-immune” system. Neurochem Res. 2005;30:1429–1438. doi: 10.1007/s11064-005-8513-9. [DOI] [PubMed] [Google Scholar]
- Kohl Z, Winner B, Ubhi K, et al. Fluoxetine rescues impaired hippocampal neurogenesis in a transgenic A53T synuclein mouse model. Eur J Neurosci. 2012;35:10–19. doi: 10.1111/j.1460-9568.2011.07933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok RM, Nolen WA, Heeren TJ. Efficacy of treatment in older depressed patients: a systematic review and meta-analysis of double-blind randomized controlled trials with antidepressants. J Affect Disord. 2012;141:103–115. doi: 10.1016/j.jad.2012.02.036. [DOI] [PubMed] [Google Scholar]
- Lavretsky H, Kim MD, Kumar A, Reynolds CF., 3rd Combined treatment with methylphenidate and citalopram for accelerated response in the elderly: an open trial. J Clin Psychiatry. 2003;64:1410–1414. doi: 10.4088/jcp.v64n1202. [DOI] [PubMed] [Google Scholar]
- Lavretsky H, Kumar A. Methylphenidate augmentation of citalopram in elderly depressed patients. Am J Geriatr Psychiatry. 2001;9:298–303. [PubMed] [Google Scholar]
- Lavretsky H, Park S, Siddarth P, Kumar A, Reynolds CF., 3rd Methylphenidate-enhanced antidepressant response to citalopram in the elderly: a double-blind, placebo-controlled pilot trial. Am J Geriatr Psychiatry Off J A Assoc Geriatr Psychiatry. 2006;14:181–185. doi: 10.1097/01.JGP.0000192503.10692.9f. [DOI] [PubMed] [Google Scholar]
- Lavretsky H, Reinlieb M, St Cyr N, et al. Citalopram, methylphenidate, or their combination in geriatric depression: a randomized, double-blind, placebo-controlled trial. Am J Psychiatry. 2015;172:561–569. doi: 10.1176/appi.ajp.2014.14070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavretsky H, Siddarth P, Kumar A, Reynolds CF., 3rd The effects of the dopamine and serotonin transporter polymorphisms on clinical features and treatment response in geriatric depression: a pilot study. Int J Geriatr Psychiatry. 2008;23:55–59. doi: 10.1002/gps.1837. [DOI] [PubMed] [Google Scholar]
- Lazarini F, Lledo PM. Is adult neurogenesis essential for olfaction? Trends Neurosci. 2011;34:20–30. doi: 10.1016/j.tins.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Lelan F, Boyer C, Thinard R, et al. Effects of human alpha-synuclein A53T-A30P mutations on SVZ and local olfactory bulb cell proliferation in a transgenic rat model of Parkinson disease. Parkinson’s Dis. 2011;2011:987084. doi: 10.4061/2011/987084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingjaerde O, Ahlfors UG, Bech P, Dencker SJ, Elgen K. The UKU side effect rating scale. A new comprehensive rating scale for psychotropic drugs and a cross-sectional study of side effects in neuroleptic-treated patients. Acta Psychiatr Scand Suppl. 1987;334:1–100. doi: 10.1111/j.1600-0447.1987.tb10566.x. [DOI] [PubMed] [Google Scholar]
- Mamdani F, Berlim MT, Beaulieu MM, Turecki G. Pharmacogenomic predictors of citalopram treatment outcome in major depressive disorder. World J Biol Psychiatry Off J World Fed Soc Biol Psychiatry. 2014;15:135–144. doi: 10.3109/15622975.2013.766762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marxreiter F, Nuber S, Kandasamy M, et al. Changes in adult olfactory bulb neurogenesis in mice expressing the A30P mutant form of alpha-synuclein. Eur J Neurosci. 2009;29:879–890. doi: 10.1111/j.1460-9568.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- Masand P, Pickett P, Murray GB. Psychostimulants for secondary depression in medical illness. Psychosomatics. 1991;32:203–208. doi: 10.1016/S0033-3182(91)72093-8. [DOI] [PubMed] [Google Scholar]
- Miller MD, Paradis CF, Houck PR, et al. Rating chronic medical illness burden in geropsychiatric practice and research: application of the Cumulative Illness Rating Scale. Psychiatry Res. 1992;41:237–248. doi: 10.1016/0165-1781(92)90005-n. [DOI] [PubMed] [Google Scholar]
- Montgomery SA, Åsberg M. A new depression scale designed to be sensitive to change. Br J Psychiatry. 1979;134:382–389. doi: 10.1192/bjp.134.4.382. [DOI] [PubMed] [Google Scholar]
- Murphy GM, Jr, Sarginson JE, Ryan HS, et al. BDNF and CREB1 genetic variants interact to affect antidepressant treatment outcomes in geriatric depression. Pharmacogenet Genomics. 2013;23:301–313. doi: 10.1097/FPC.0b013e328360b175. [DOI] [PubMed] [Google Scholar]
- Murray GB, Cassem E. Use of stimulants in depressed patients with medical illness. In: Nelson JG, editor. Geriatric Psychopharmacology. Marcel Decker, Inc; New York-Basel-Hong Kong: 1998. pp. 245–258. [Google Scholar]
- Nuber S, Harmuth F, Kohl Z, et al. A progressive dopaminergic phenotype associated with neurotoxic conversion of alpha-synuclein in BAC-transgenic rats. Brain J Neurol. 2013;136:412–432. doi: 10.1093/brain/aws358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitychoutis PM, Kokras N, Sanoudou D, Dalla C, Papadopoulou-Daifoti Z. Pharmacogenetic considerations for late life depression therapy. Expert Opin Drug Metab Toxicol. 2013;9:989–999. doi: 10.1517/17425255.2013.794786. [DOI] [PubMed] [Google Scholar]
- Sarginson JE, Lazzeroni LC, Ryan HS, et al. ABCB1 (MDR1) polymorphisms and antidepressant response in geriatric depression. Pharmacogenet Genomics. 2010;20:467–475. doi: 10.1097/FPC.0b013e32833b593a. [DOI] [PubMed] [Google Scholar]
- Satel SL, Nelson JC. Stimulants in the treatment of depression: a critical overview. J Clin Psychiatry. 1989;50:241–249. [PubMed] [Google Scholar]
- Schneider HP, Alt MD, Klier M, et al. GPI-anchored carbonic anhydrase IV displays both intra- and extracellular activity in cRNA-injected oocytes and in mouse neurons. Proc Natl Acad Sci U S A. 2013;110:1494–1499. doi: 10.1073/pnas.1221213110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiroma PR, Geda YE, Mrazek DA. Pharmacogenomic implications of variants of monoaminergic-related genes in geriatric psychiatry. Pharmacogenomics. 2010;11:1305–1330. doi: 10.2217/pgs.10.118. [DOI] [PubMed] [Google Scholar]
- Svichar N, Waheed A, Sly WS, et al. Carbonic anhydrases CA4 and CA14 both enhance AE3-mediated Cl-HCO3- exchange in hippocampal neurons. J Neurosci. 2009;29:3252–3258. doi: 10.1523/JNEUROSCI.0036-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WD, McQuoid DR, Ashley-Koch A, et al. BDNF val66met genotype and 6-month remission rates in late-life depression. Pharmacogenomics J. 2011;11:146–154. doi: 10.1038/tpj.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truelsen T, Lindenstrom E, Boysen G. Comparison of probability of stroke between the Copenhagen City Heart Study and the Framingham Study. Stroke. 1994;25:802–807. doi: 10.1161/01.str.25.4.802. [DOI] [PubMed] [Google Scholar]
- Ubhi K, Inglis C, Mante M, et al. Fluoxetine ameliorates behavioral and neuropathological deficits in a transgenic model mouse of alpha-synucleinopathy. Exp Neurol. 2012;234:405–416. doi: 10.1016/j.expneurol.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Population Division DoEaSA., editor. UN. World Population Ageing 2013. 2013. [Google Scholar]
- Volkow ND, Gur RC, Wang GJ, et al. Association between decline in brain dopamine activity with age and cognitive and motor impairment in healthy individuals. Am J Psychiatry. 1998;155:344–349. doi: 10.1176/ajp.155.3.344. [DOI] [PubMed] [Google Scholar]
- Winner B, Lie DC, Rockenstein E, et al. Human wild-type alpha-synuclein impairs neurogenesis. J Neuropathol Exp Neurol. 2004;63:1155–1166. doi: 10.1093/jnen/63.11.1155. [DOI] [PubMed] [Google Scholar]
- Woods SW, Tesar GE, Murray GB, Cassem NH. Psychostimulant treatment of depressive disorders secondary to medical illness. J Clin Psychiatry. 1986;47:12–15. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.