Abstract
Biological informational polymers such as nucleic acids have the potential to provide unambiguous evidence of life beyond Earth. To this end, we are developing an automated in situ life-detection instrument that integrates nucleic acid extraction and nanopore sequencing: the Search for Extra-Terrestrial Genomes (SETG) instrument. Our goal is to isolate and determine the sequence of nucleic acids from extant or preserved life on Mars, if, for example, there is common ancestry to life on Mars and Earth. As is true of metagenomic analysis of terrestrial environmental samples, the SETG instrument must isolate nucleic acids from crude samples and then determine the DNA sequence of the unknown nucleic acids. Our initial DNA extraction experiments resulted in low to undetectable amounts of DNA due to soil chemistry–dependent soil-DNA interactions, namely adsorption to mineral surfaces, binding to divalent/trivalent cations, destruction by iron redox cycling, and acidic conditions. Subsequently, we developed soil-specific extraction protocols that increase DNA yields through a combination of desalting, utilization of competitive binders, and promotion of anaerobic conditions. Our results suggest that a combination of desalting and utilizing competitive binders may establish a “universal” nucleic acid extraction protocol suitable for analyzing samples from diverse soils on Mars. Key Words: Life-detection instruments—Nucleic acids—Mars—Panspermia. Astrobiology 17, 747–760.
1. Introduction
Widespread synthesis of the known building blocks of life, that is, amino acids and nucleobases, within the early planetary nebula (Nuevo et al., 2009, 2012; Ciesla and Sandford, 2012) may have biased life as we know it toward the utilization of nucleic acids as the basic medium for information storage and transference (e.g., deoxyribonucleic acid—DNA—and ribonucleic acid—RNA). Recent work also demonstrates synthesis of ribose and related sugars under conditions similar to those of the late solar nebula (Meinert et al., 2016). Life on Mars, if it developed independently, may have adopted a similar medium of information storage and readout transfer (DNA, RNA), although this assumes that there is a unitary chemical solution to the problem of genetic transmission. We propose that, if life has transferred between Earth and Mars, perhaps billions of years ago, the highly evolved tools of modern genomics and metagenomics could be applied to the search for extant or preserved life on Mars (Ruvkun et al., 2002, Isenbarger et al., 2008).
Common ancestry of life on Mars with life on Earth could have been mediated by the estimated billion tons of rock transferred between Mars and Earth during the Late Heavy Bombardment period (Gladman and Burns, 1996; Gladman et al., 1996). In this scenario, viable microbes may have been transported between planets within meteoroids, some of which experienced nonsterilizing transfers in space (Weiss et al., 2000; Fritz et al., 2005; Shuster and Weiss, 2005) and tolerable shock pressures and heating upon planetary entry (Ruvkun et al., 2002, Horneck et al., 2008, Isenbarger et al., 2008). In such a case, comparing genetic sequencing data from martian “life” with conserved (Makarova et al., 1999; Harris et al., 2003) genetic sequences from known Earth life (i.e., the ribosomal RNA) (Woese et al., 1975) could discriminate forward contamination from true life detection and establish a possible common ancestry (Ruvkun et al., 2002; Isenbarger et al., 2008).
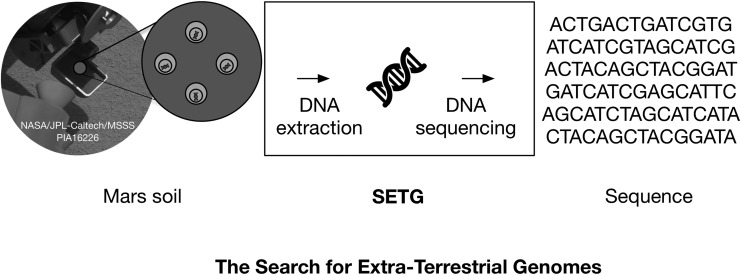
We are developing an automated nucleic acid extraction and sequencing instrument (Lui et al., 2011; Carr et al., 2013a, 2013b, 2016), Search for Extra-Terrestrial Genomes (SETG), that is capable of analyzing a variety of environmental samples relevant to the search for evidence of life (related or otherwise to life on Earth) on Mars as well as on other habitable bodies in the Solar System (Fig. 1). On Earth, metagenomic sequencing of DNA extracted from environmental samples has been an invaluable tool for identifying new and unique “unculturable” microorganisms (Schloss and Handelsman, 2005). However, complex soil matrices like those containing iron oxides, for example, have strong inhibitory side effects toward nucleic acid extraction due to problems of competitive binding (Jiang et al., 2012; Hurt et al., 2014) and the production of destructive hydroxyl radicals from Fe3+ and Fe2+ redox cycling (Imlay and Linn, 1988; Imlay et al., 1988). Additional inhibition may emerge in the presence of silicates (Volossiouk et al., 1995; Melzak et al., 1996; Trevors, 1996), clay minerals (Greaves and Wilson, 1969), high concentrations of salt minerals, and acidic conditions (Henneberger et al., 2006). These soil-DNA interactions are especially problematic in low-biomass terrestrial environments (Barton et al., 2006; Azua-Bustos et al., 2012, 2015) and pose similar challenges for the extraction of nucleic acids from Mars analog soils (Navarro-Gonzalez et al., 2003; Mojarro et al., 2016).
FIG. 1.
Overview of the SETG instrument. Soils collected by a sample-handling system will be processed; any extracted nucleic acids will then be conditioned and sequenced in situ. Any resulting sequences can be compared to all known Earth sequences to determine if such sequences represent forward contamination or putative martian life. Image adapted from PIA16226 (NASA/JPL-Caltech/MSSS).
Extensive literature exists on how to mitigate soil-DNA interactions and increase DNA extraction yields from a variety of soil species (e.g., Purdy et al., 1996; Takada-Hoshino and Matsumoto, 2004; Barton et al., 2006; Henneberger et al., 2006; Herrera and Cockell, 2007; Direito et al., 2012; Lever et al., 2015). The use of competitive binders (skim milk, RNA, poly-dIdC) (Volossiouk et al., 1995; Takada-Hoshino and Matsumoto, 2004; Barton et al., 2006), sample dialysis (Barton et al., 2006), and chelation of calcium and iron inhibitors through the use of ethylenediaminetetraacetic acid (EDTA) or ethylene glycol tetraacetic acid (EGTA) (Wade and Garcia-Pichel, 2003; Barton et al., 2006) in combination with traditional soil nucleic acid extraction kits (Yakimov et al., 2002; Wade and Garcia-Pichel, 2003; Takada-Hoshino and Matsumoto, 2004; Walker et al., 2005; Barton et al., 2006; Brown and Wolfe, 2006; Henneberger et al., 2006) is an effective method of extracting PCR-quality DNA from various complex terrestrial environments. These solutions, however, require involved steps, additional equipment, and large volumes of reagents that would increase the complexity and size of an in situ SETG instrument on Mars.
Our goal for SETG is to generate a versatile nucleic acid extraction protocol capable of processing samples from diverse environments on Mars (Fig. 1). A typical benchtop DNA extraction protocol requires bead beating, centrifugation, and frequent spin-column nucleic acid purification steps, while archaic protocols require freezing samples in liquid nitrogen, manual grinding with a mortar and pestle, phenol/chloroform nucleic acid isolation, and several alcohol purification steps. Although the aforementioned extraction methods have been proven effective, we wish to develop a system that employs minimal extraction steps and reagent volumes, requires low power, and is compact for variable spaceflight applications involving rover/lander or human operations. In this study, we utilize PureLyse Bacterial gDNA Kits (ClaremontBio, 01.351.06), which are miniature (dime-sized), low-power (6 V), rapid (3 min protocol) solid-phase nucleic acid extraction devices, as the precursor to a “mature” SETG extraction module. Additionally, PureLyse-related hardware has been validated in orbit as part of NASA's WetLab-2 platform aboard the International Space Station to extract and purify RNA from cells (Robert Doebler, personal communication, 2016).
To characterize PureLyse for SETG's application, we challenge DNA extraction performance with spores of Bacillus subtilis ATCC 6633 (Fig. 2), a hardy microorganism with spaceflight heritage known to survive exposure in orbit (Horneck et al., 2012) and in synthetic Mars analog soils under martian temperature, pressure, and radiation (Schuerger et al., 2012). We first test the recovery of Escherichia coli DNA and the extraction of B. subtilis spore DNA from synthetic Mars analog soils in order to characterize soil-DNA interactions that may adversely affect DNA extraction performance on Mars. We then develop modified DNA extraction protocols to mitigate the identified soil-DNA interactions. Modified extraction protocols were based on 50 mg soil samples, the upward sample loading capacity of PureLyse. Finally, we determine the purity of extracted DNA via quantitative real-time polymerase chain reaction (qPCR) as a proxy for sequencing, which would be used on the full SETG instrument, to verify that no compounds were coextracted from the soil that would inhibit DNA sequencing.
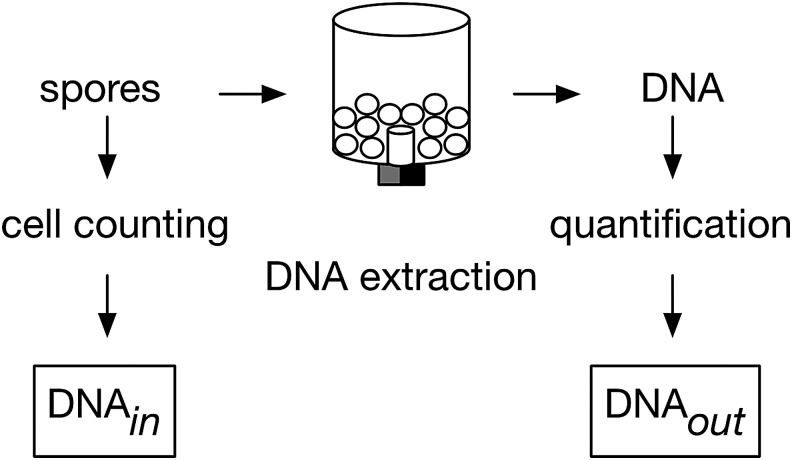
FIG. 2.
Simplified experimental overview of extraction of spore DNA. Counted DNase-treated spores of Bacillus subtilis ATCC 6633 (DNAin) are loaded into PureLyse, processed, and yield DNA quantified by a dsDNA-specific fluorometric assay (DNAout).
2. Materials and Methods
2.1. Approach
Here, we show that a PureLyse-like system may be adapted to extract nucleic acids from martian or Mars-like soils. Our two primary experiments are focused on “recovery of free DNA” and “extraction of spore DNA” from synthetic Mars analog soils. The purpose of the “recovery of free DNA” experiment is to establish interactions that may occur between free DNA, defined as extracellular DNA able to freely interact in a solvent, and soil. In contrast, the purpose of the “extraction of spore DNA” experiments is to determine PureLyse lysis and extraction performance of DNA insulated by an endospore coat (i.e., B. subtilis spores) in soils. These experiments culminate in the generation of extraction modifications that mitigate soil-DNA interactions and maximize DNA yields from spore DNA in soil. We consider a DNA extraction yield of 5% or greater to be adequate for subsequent sequencing; this would allow us to achieve, at a minimum, our sensitivity target of 1 ppb, which corresponds to ∼104 B. subtilis spores in a 50 mg sample (Carr et al., 2016) as we approach a mature SETG instrument. Here, we focus on higher cell counts and leave it to future work to demonstrate that a similar extraction yield can be achieved at lower cell counts.
Finally, the purpose of the “soil carryover” experiment is to evaluate the purity of extracted DNA for the presence of soil contaminants or “carryover” that may inhibit downstream sequencing; as a proxy, we utilized qPCR, which relies on polymerase and serves as a cost-effective proxy for current nanopore sequencing technologies, which use polymerase (Fuller et al., 2016) or helicase (Derrington et al., 2016) enzymes. Of note, qPCR is the standard method used to evaluate the quality of DNA molecules prior to traditional next-generation (e.g., Illumina, Ion Torrent) sequencing.
2.2. Synthetic Mars analog soils
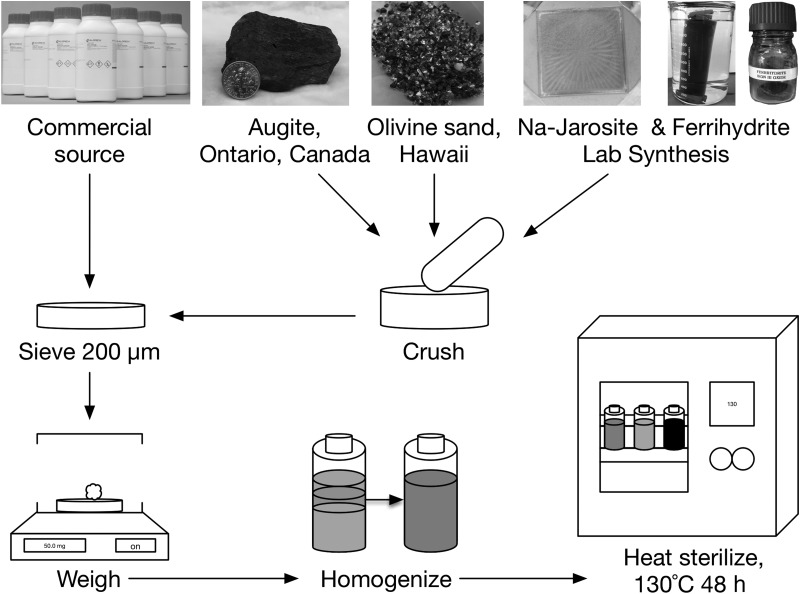
Six synthetic Mars analog soils and one lunar analog soil, representing unaltered martian basalt, were used. Five of the synthetic Mars analog soils were prepared in accordance with in situ measurements (e.g., X-ray diffraction) from rover and lander surface sites as described by Schuerger et al. (2012). The five synthetic Mars analogues represent the salt-rich soils of Gusev Crater (Ming et al., 2006; Morris et al., 2008), jarosite-containing acidic soils of Meridiani Planum (Klingelhöfer et al., 2004; Morris et al., 2006), carbonate-rich alkaline soils of Chryse Planitia (Clark et al., 1982; Wänke et al., 2001), weakly alkaline and perchlorate-rich soils of Vastitas Borealis (Ming et al., 2008), and the pervasive global aeolian soils (Bell et al., 2000). These synthetic Mars analog soils are henceforth referred to as salt, acid, alkaline, perchlorate, and aeolian, respectively. In addition, a commercially available spectral analogue of Mars (Orbitec, JSC Mars-1A) (Allen et al., 2000) and lunar analogue (Orbitec, JSC-1A) (McKay et al., 1994), designated JSC and basalt, were also used.
All minerals and salts were commercially sourced from Sigma-Aldrich (calcium carbonate—481807, calcium phosphate—04231, ferric sulfate—307718, gypsum—237132, halite—S9888, hematite—310050, kieserite—434183, magnesium chloride—208337, Ti-magnetite—310069, magnesite—63062, sodium carbonate—451614, sodium perchlorate—310514, sodium sulfate—204447) with the exception of pyroxene, olivine, anhydrite, ferrihydrite, and natrojarosite (Fig. 3). Pyroxene was purchased from Ward's Science (Catalog #466474). Olivine sand was collected from Papalōlea beach, near South Point, Hawaii, in accordance with the work of Schuerger et al. (2012) and cleaned with a 0.2 M sodium acetate solution (Doug Ming, personal communication, 2015). Anhydrite was produced by dehydrating gypsum (Sigma-Aldrich, 237132) at 300°C overnight (Atoji, 1959), and 2-line ferrihydrite was chemically synthesized from reagent-grade iron (III) nitrate (Sigma-Aldrich, 216828) (Schwertmann and Cornell, 2008). Lastly, natrojarosite was synthesized from reagent-grade iron (III) sulfate (Sigma-Aldrich, 307718) and sodium sulfate (Sigma-Aldrich, S9627) (Drouet and Navrotsky, 2003).
FIG. 3.
Overview of synthetic Mars analog synthesis process, adapted from Schuerger et al. (2012). Synthetic samples are controlled, reproducible, and simulate various martian environments.
Individual minerals, salts, and commercial analog soils were hand-crushed with a Coorstek alumina mortar and pestle and passed through a 200 μm stainless steel sieve (Fig. 3) (Schuerger et al., 2012). Mineral and salt powders were then individually weighed and then mixed and homogenized in 100 g batches to represent the acid, aeolian, alkaline, perchlorate, and salt synthetic analog soils (Schuerger et al., 2012) in addition to the JSC and basalt soils (Fig. 3). All soils were heat-sterilized at 130°C for 48 h prior to any experimentation in a Fisher Science Isotemp 282A vacuum oven (Fig. 3), cooled overnight, and stored in air-tight borosilicate bottles (Corning, 1395-100) at room temperature (Schuerger et al., 2012).
2.3. Bacillus subtilis spores
Spore suspensions of Bacillus subtilis (ATCC 6633) were purchased from NAMSA (Part# SBS-08), and spore titer was verified by plating on lysogeny broth agar media followed by counting colonies after incubation at 37°C for 24 h. Spores were treated with DNAse I (New England Biolabs, M0303L) prior to any extractions.
2.4. DNA yield quantitation
We define DNA yield from our experiments as
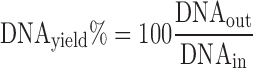 |
We assume a single genome copy (3,987,576 base pairs) per DNAse I-treated spore of B. subtilis for calculating DNAin. DNAout was quantified using a double-stranded, DNA-specific fluorometric assay (Invitrogen, Qubit dsDNA HS Assay Kit, Q32854) and a Qubit 2.0 fluorometer (Invitrogen, Q32866, limit of detection of 0.1 ng/mL, dsDNA).
2.5. PureLyse
The PureLyse Bacterial gDNA Extraction Kit is a Claremont Biosolutions OmniLyse mechanical cell disruption module (30,000 rpm bead beating), coupled with proprietary binding and elution buffers, that enables rapid solid-phase extraction of nucleic acids (including RNA). The standard off-the-shelf protocol advises 2 min lysis with 1× binding buffer solution (100 μL 8× binding buffer and 700 μL water), 45 s wash with 3 mL of 0.25× binding buffer wash (∼100 μL 8× binding buffer and 2.9 mL water), and a 1 min elution with 200 μL 1× elution buffer.
2.6. Recovery of free DNA from synthetic Mars analog soils
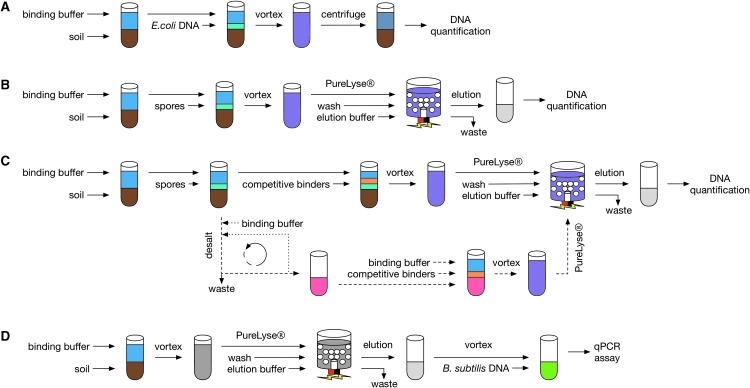
Purified Escherichia coli DNA (USB Corporation, #14380, 1859 ng) was added to each set of microcentrifuge tubes containing 0–50 mg (0, 10, 20, 30, 40, 50 mg, n = 3) of synthetic Mars analog soil and 800 μL of 1× PureLyse binding buffer solution. The microcentrifuge tube mixtures were homogenized on a digital vortex mixer (VWR, #14005-824) at 1000 rpm for 1 min and immediately centrifuged (Eppendorf, Centrifuge 5418) for 5 min at 10,000 rpm. The amount of recoverable DNA from the supernatant was then measured with a fluorometric assay (Fig. 4A).
FIG. 4.
Experiment overview. (A) Recovery of free DNA from synthetic Mars analog soils. (B) Baseline extractions of spore DNA from synthetic Mars analog soils using the standard PureLyse protocol. (C) Modified extractions of spore DNA from synthetic Mars analog soils. (D) Soil carryover qPCR assay.
2.7. Baseline extractions of spore DNA from synthetic Mars analog soils
An estimated 2.2 × 108 spores (about 1 μg of DNA) of B. subtilis were processed with 0–50 mg (water control and 10, 20, 30, 40, 50 mg, n = 3) of synthetic Mars analog soil following the standard DNA extraction protocol (Fig. 4B). DNA yields were calculated from two combined elutions (200 μL each) with a fluorometric assay.
2.8. Modified extractions of spore DNA from synthetic Mars analog soils
As above, equal amounts of B. subtilis spores (2.2 × 108, ∼1 μg DNA) were combined with 50 mg of each synthetic Mars analog soil as well as a water control (n = 3) and processed with modified DNA extraction protocols developed per analog soil.
Water (control) samples were processed with 40 μL of phenol solution (Sigma, P4557) added to 760 μL of binding buffer solution (100 μL 8× binding buffer and 660 μL water) to create a final 1× binding buffer concentration and 5% phenol solution.
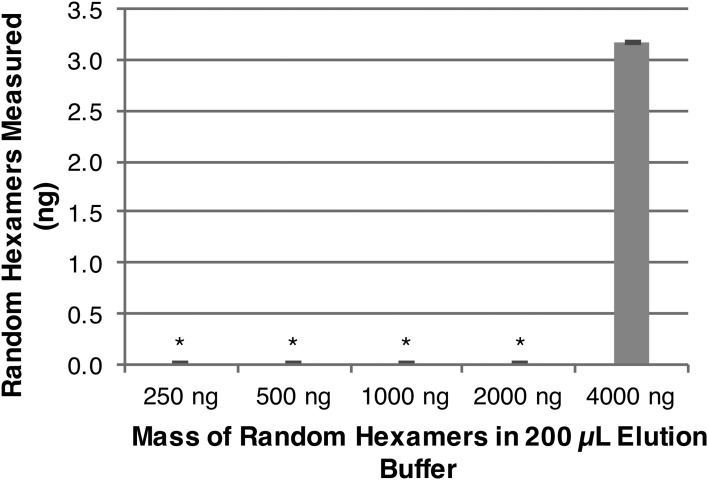
Basalt samples were processed with 150 ng of random hexamer primers (Promega, Random C1181). The soil and primers were homogenized for 1 min at 1000 rpm, then processed following the standard DNA extraction protocol (Fig. 4C). Random hexamer primers used here were removed from the elutions (Mojarro et al., 2016) prior to measuring DNA yield with an extra cleaning step (Zymo, Genomic Clean & Concentrator). Subsequent testing demonstrated that random hexamer primer concentrations up to 20 ng/μL are not measurable using the dsDNA-specific fluorometric assay (Fig. 5).
FIG. 5.
Random hexamer primer detection by a dsDNA-specific fluorometric assay. Random hexamers (250, 500, 1000, 2000, 4000 ng) were added to 200 μL of the PureLyse elution buffer, homogenized, then quantified on a Qubit 2.0 fluorometer (standard error shown, n = 3). Random hexamer loads were not detectable until 20 ng/μL (4000 ng in 200 μL), likely due to increased hybridization. These results suggest that using random hexamer primers (less than 20 ng/μL) as competitive binders will not bias DNA yield results with this dsDNA-specific fluorometric assay. *At or below 0.01 ng limit of detection.
Alkaline soil samples were processed with a modified DNA extraction protocol containing 4× binding buffer solution (400 μL 8× binding buffer and 400 μL water) and 1× wash solution (Fig. 4B).
JSC soil samples were processed inside an anaerobic chamber (Sigma, Atmosphere Bag, Z530212): after flushing the chamber with nitrogen gas, reagents were degassed for 1 h (Butler et al., 1994) in the presence of oxygen scavengers (Thermo Scientific, AnaeroGen, 2.5L) before processing via the standard DNA extraction protocol (Fig. 4B).
Salt soil samples were suspended in 800 μL of 8× binding buffer solution and desalted on a 100K Amicon Ultra column (Z740183) prior to extraction. Samples were then processed with 4× binding buffer solution and 1× wash solution (1.5 mL 8× binding buffer and 1.5 mL water) (Fig. 4C).
Perchlorate and aeolian soil samples were prepared in accordance with the salt soil protocol. Furthermore, perchlorate soil samples included 300 ng of random hexamer primers, while the aeolian soil samples included 500 ng (Fig. 4C) when processed with the 4× binding buffer solution.
Lastly, acid soil samples were also prepared following the salt protocol, but with the following modifications: samples were resuspended in multiple volumes of 8× binding buffer within the 100K Amicon Ultra column and desalted until the color of the waste became clear. Acid soil samples were then extracted using a 4× binding buffer solution that included 2 mg of skim milk (40 mg of skim milk per 1 g of soil, Takada-Hoshino and Matsumoto, 2004) and 2× wash solution (Fig. 4C).
Table 1 compiles the modified DNA extraction protocols developed in this study.
Table 1.
Summary of Methods for Extracting Nucleic Acids from Mars Analog Soils (50 mg Soil)
| Synthetic sample | Model geographic location on Mars | Soil characteristics | Proposed DNA interaction(s) | Modified DNA extraction | Baseline yield (%) | Modified yield (%) | Notes |
|---|---|---|---|---|---|---|---|
| Water | — | — | SASPs inhibit DNA extraction | 5% phenol extraction | 14.7% | 43.0% | — |
| Basalt | Subsurface | Unaltered basalt, ∼50% SiO2 | Adsorption to silica | 150 ng of random hexamer primer | 12.2% | 13.7% | JSC-1A lunar regolith |
| Alkaline | Chryse Planitia | Carbonate-rich (Mg, Ca) | Adsorption to silica Binding to Ca2+ |
4× binding buffer | 6.62% | 11.2% | — |
| JSC | Global | Highly oxidized | Hydroxyl radical generation DNA adsorption to silica DNA binding to Fe3+ |
Degas reagents/Anaerobic extraction | 3.04% | 6.39% | JSC Mars-1A spectral analogue |
| Salt | Gusev Crater | Salt-rich, highly oxidized | High salts DNA adsorption to silica DNA binding to Ca2+ DNA binding to Fe3+ |
Desalt soil suspended in 8× binding buffer in 100 kDa column prior to extraction. Resuspend in 4× binding buffer and 1× wash. | 1.57% | 6.64% | — |
| Perchlorate | Vastitas Borealis | Weakly alkaline, perchlorate-rich, highly oxidized | High salts DNA adsorption to silica DNA binding to Ca2+ DNA binding to Fe3+ Hydroxyl radical generation |
Desalt soil suspended in 4× binding buffer in 100 kDa column. Re-suspend in 4× binding buffer, 300 ng random primer hexamer, and 1× wash. | 0.58% | 6.49% | — |
| Aeolian | Global | Highly oxidized | High salts DNA adsorption to silica DNA binding to Ca2+ DNA binding to Fe3+,2+ Hydroxyl radical generation |
Desalt soil suspended in 4× binding buffer in 100 kDa column. Resuspend in 4× binding buffer, 500 ng random primer hexamer, and 1× wash. |
0.23% | 5.03% | — |
| Acid | Meridiani Planum | Highly oxidized, acidic pH 1.42, contains jarosite | DNA destruction by low pH Hydroxyl radical generation DNA adsorption to silica DNA binding to Ca2+ DNA binding to Fe3+ |
Desalt soil in 100 kDa column in multiple 8× binding buffer solutions. Re-suspend in 4× binding solution, 2 mg skim milk, and 2× wash. | 0% | 6.61% | Samples were desalted in continuous 8× binding buffer volumes until the waste color became clear |
2.9. Soil carryover
A total of 50 mg of synthetic Mars analog soil was processed following the standard DNA extraction protocol (n = 3) without B. subtilis spores. Bacillus subtilis DNA, purified from vegetative cells, was then added to the “blank” elution to achieve a final concentration of 0.2 ng DNA/μL (Fig. 4D). A total of 5 μL of the simulated extract, containing 1 ng of DNA, was then amplified in a 20 μL (total) reaction volume of PerfeCta SYBR Green FastMix and Integrated DNA Technologies Readymade 16s rRNA primers (Universal). Our qPCR mix and 96-well plate were prepared inside an AirClean 600 PCR Workstation by an Andrew Alliance liquid handling robot. Thermocycling conditions were as follows: (1) 94°C for 3 min, (2) 94°C for 45 s, (3) 50°C for 1 min, (4) 72°C for 90 s, (5) plate read (6) repeat steps 2–5 40 times.
3. Results
3.1. Recovery of free DNA from synthetic Mars analog soil
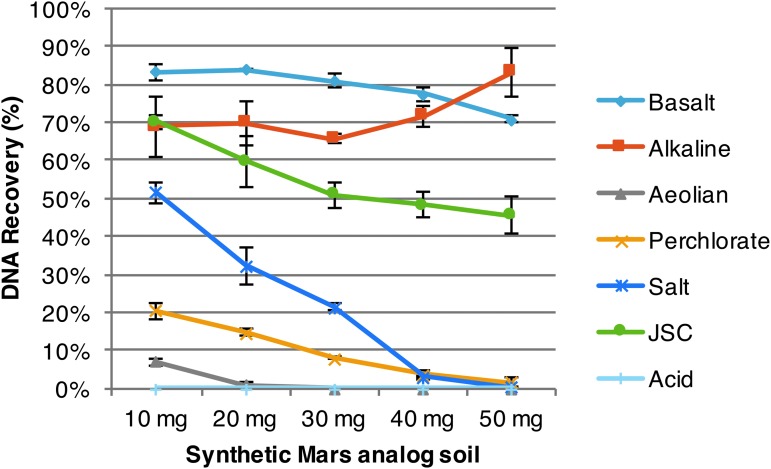
Approximately 80% of DNA added to the sample was recoverable in the basalt and alkaline, for all quantities of soil tested (Fig. 6). Otherwise, the aeolian, perchlorate, salt, and JSC soils exhibited an inferred linear relationship, where increasing soil mass decreases DNA recovery (Fig. 6). No measurable DNA (lower limit of detection 0.1 ng/mL) was recovered from the acid soils (10–50 mg) (Fig. 6), which may be due to the destruction of DNA through depurination and hydrolysis in low pH (Schroeder et al., 2006; Bernhardt and Tate, 2012).
FIG. 6.
Recovery of free DNA from synthetic Mars analog soils. Purified Escherichia coli DNA, ∼2 μg, was added to synthetic Mars analog soils (0–50 mg); DNA recovery profiles identify general soil-DNA interactions, namely competitive adsorption to minerals, binding to free cations, and presumed destruction (standard error shown, n = 3).
3.2. Baseline and modified extractions of spore DNA from water
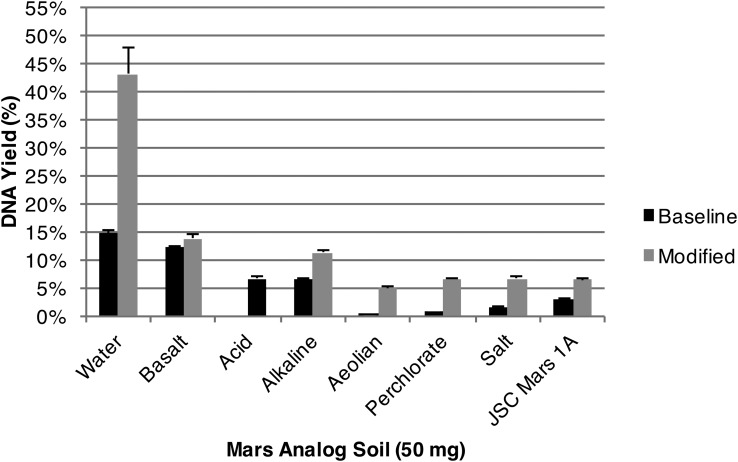
Baseline DNA extractions of B. subtilis spores from our water (control) sample produced 14.7% DNA yield (Fig. 7). This result is significantly lower than manufacturer studies of PureLyse that have demonstrated ∼90% DNA extraction yields from Escherichia coli (ClaremontBio Solutions, 2011) with multiple elutions. PureLyse kits operate by adsorbing nucleic acids onto zirconium-silicate microbeads via salt bridges that link the phosphate backbone under high salt and moderately acidic (∼3.4 pH) conditions. Our low DNA extraction yields from B. subtilis spores may be caused by small acid-soluble proteins (SASPs) that interfere with the extraction process (Setlow and Setlow, 1993; Moeller et al., 2008). If the DNA backbone is in fact bound by these acidic proteins, it will disregard the salt bridges, and DNA will be discarded in the waste. To test this model, “on the fly” protein separation was facilitated with phenol. A 5% final solution (Chomczynski and Sacchi, 1987; Volossiouk et al., 1995) resulted in significantly increased DNA yields from 14.7% to 43% (Fig. 8). We consider the final phenol concentration limiting, as solutions containing greater than 5% phenol dissolved the OmniLyse module.
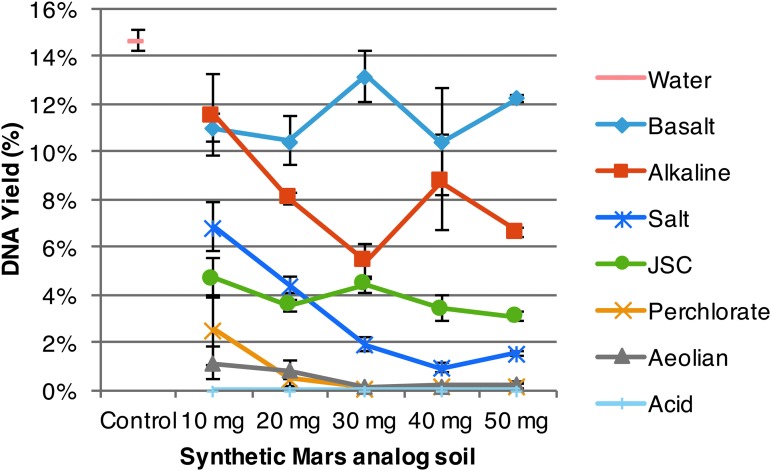
FIG. 7.
Baseline extraction yields of spore DNA from synthetic Mars analog soils and water. Bacillus subtilis spores, 2.23 × 108 (∼1 μg DNA), were added to water (control) and 10–50 mg of basalt, alkaline, aeolian, acid, perchlorate, salt, and JSC (standard error shown, n = 3). All soils adversely affect DNA extraction performance; low DNA yield from water is likely due to SASPs that interfere with solid-phase extraction.
FIG. 8.
Modified and baseline extraction yields of spore DNA from synthetic Mars analog soils and water. DNA yields from 50 mg of soil (including water control) and 2.23 × 108 Bacillus subtilis spores (∼1 μg DNA) were increased with soil-specific modified extraction protocols (standard error shown, n = 3). Our results suggest that a combination of desalting and competitive binders may establish a “universal” Mars protocol.
3.3. Baseline extractions of spore DNA from synthetic Mars analog soils
The baseline DNA extractions of B. subtilis spores from synthetic analog soils (10–50 mg) showed a similar trend as the free DNA soil recovery assay (Figs. 6 and 7), resulting in decreasing DNA yields with increasing soil mass (Fig. 7). At 50 mg, basalt and alkaline soils moderately affected yields compared to baseline water (control) extractions (Fig. 7) with about 12% DNA yield. Meanwhile, JSC, perchlorate, aeolian, salt, and acid dramatically decreased yields (Fig. 7), less than 3% as observed earlier (Fig. 6).
3.4. Modified extractions of spore DNA from synthetic Mars analog soils
Basalt soil sample DNA yields were increased from 12.2% to 13.7% (Fig. 8) to nearly replicate water control yields by employing random hexamer primers as competitive binders to prime DNA adsorption sites prior to cell lysis. Decreased yields in basalt soil samples are likely associated with competitive DNA adsorption to silicates (Melzak et al., 1996; Trevors, 1996; Zhou et al., 1996).
Alkaline soil sample DNA yields were increased from 6.6% to 11.2% (Fig. 8), comparable to unmodified basalt soil DNA yields, by increasing binding buffer concentration. Although nearly identical to basalt, alkaline samples contain 4 wt % (Ca, Mg) carbonate that may bind DNA. Previous studies that focused on carbonate environments (Barton et al., 2006) have utilized EGTA, a calcium-specific chelator, to remove DNA-binding calcium cations (Barton et al., 2006). Although EDTA has a lower calcium affinity (which the binding buffer presumably contains), we increased the binding buffer concentration. At a 4× binding buffer concentration, chelator specificity became negligible.
JSC soil sample DNA yields were increased from 3.0% to 6.4% (Fig. 8). Analysis of the JSC analog soil by Allen et al. (2000) revealed a strong Fe3+:Fe2+ couple, present in a 3:1 ratio, that may lead to the production of destructive hydroxyl radicals (Imlay and Linn, 1988; Imlay et al., 1988) under aqueous conditions. Thereby presumed DNA destruction was prevented by removing dissolved oxygen from our reagents through degassing in an anaerobic chamber.
Salt soil sample DNA yields were increased from 1.6% to 6.6% (Fig. 8) by desalting and without any further treatment with competitive binders. The salt soil is composed of 13.9 wt % salt minerals (calcite, gypsum, kierserite, magnesium chloride) that bind DNA and inhibit elution (occurs under low salt). Successful dialysis techniques utilized by Barton et al. (2006) with carbonate-rich samples informed a pre-extraction desalting step.
For aeolian and perchlorate soil samples, the combination of desalting and competitive binders (random hexamer primers) increased DNA extraction yields from 0.2% to 5.0% (Fig. 8) in the aeolian soil and 0.6% to 6.5% in the perchlorate soil samples (Fig. 8). Extraction of DNA from aeolian and perchlorate soil samples when using the unmodified protocol resulted in greatly reduced DNA yields due to presumed production of hydroxyl radicals that destroy DNA (Imlay and Linn, 1988; Imlay et al., 1988) and high concentrations of DNA-binding iron cations (Fe3+,2+); (Volossiouk et al., 1995; Takada-Hoshino and Matsumoto, 2004; Barton et al., 2006). Before implementing our optimized protocol, we initially addressed hydroxyl radicals by degassing reagents inside an anaerobic chamber; however, yields minimally increased when compared to JSC soil samples. Comparable yields to an anaerobic treatment resulted when both soils were desalted prior to extraction. We conclude that the majority of DNA is lost due to strong competitive adsorption to iron-bearing minerals (ferrihydrite, hematite, Ti-magnetite) once hydroxyls and free cations have been addressed.
Acid sample DNA yields were increased to 6.6% (Fig. 8) by employing a combination of desalting and skim milk (Takada-Hoshino and Matsumoto, 2004). Initial extractions from the acid soils yielded no detectable DNA due to presumed destruction by high acidity (pH of 1.42, Figs. 7 and 8). Buffering pH prior to extractions resulted in measurable DNA yields, <0.7%, similar to the untreated aeolian or perchlorate soils (Figs. 7 and 8). Desalting mildly improved yields, though high iron concentrations from 27 wt % ferric sulfate, 1 wt % jarosite, and 2.5 wt % hematite resulted in lower extraction yields in comparison to aeolian or perchlorate soils. A combination of desalting and competitive binders increased yields, but again the increase was minimal, and we found that random hexamer primer loads greater than 5 μg began to inhibit DNA extraction.
Table 1 compiles the proposed soil-DNA interactions identified in this study.
3.5. Soil carryover
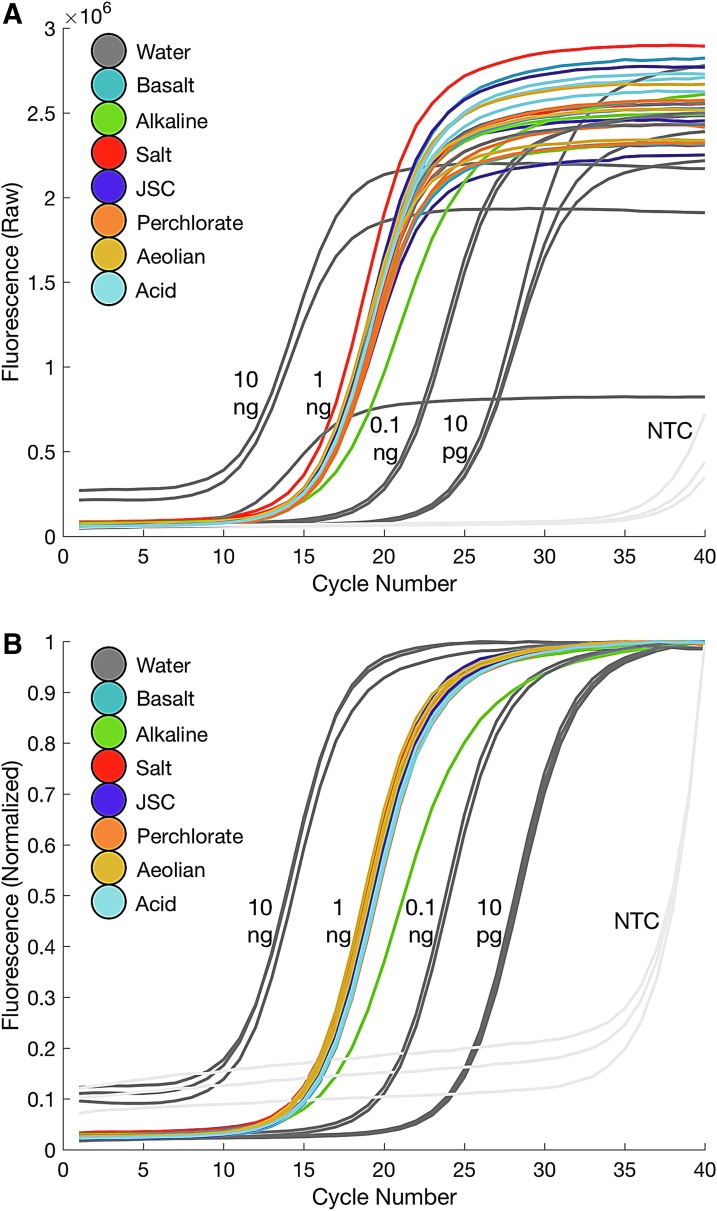
We observed successful qPCR amplifications from all simulated extractions containing 1 ng of B. subtilis DNA (added post-extraction) (Fig. 9), suggesting high DNA elution purity from PureLyse.
FIG. 9.
Quantitative PCR (qPCR) amplification curves reveal successful amplification from all synthetic Mars analog soils (perchlorate, alkaline, aeolian, acid, salt, JSC, and basalt) minus one alkaline sample (likely due to pipetting variability) suggest high PureLyse elution purity (1 ng of B. subtilis DNA, 10 ng to 10 pg and NTC standards, n = 3). (A) Raw fluorescence (B) Normalized fluorescence.
4. Discussion
4.1. Extraction and recovery of DNA from synthetic Mars analog soils
Here, we demonstrate that, with minimal soil-specific modifications, PureLyse Bacterial gDNA Extraction Kits are capable of extracting PCR-quality DNA from synthetic Mars analog soils representing a variety of martian environments (Figs. 8 and 9). We confirm that DNA extraction difficulties primarily derive from binding by free cations, adsorption to minerals, and destruction due to hydroxyl radicals and high acidity. Our results suggest that desalting soils prior to extraction is an effective approach for purging highly disruptive cations such as Fe3+,2+ (participate in redox cycling, bind DNA) and Ca2+ (bind DNA) along with additional inhibitory ions (e.g., Cl-, Na+, ClO4-, CO3+, SO4-, HSO4-). In addition, coating mineral adsorption sites with competitive binders also increases DNA yields. Random hexamer primers and skim milk were both effective competitors; however, we recognize that skim milk exhibits a higher affinity toward soils, while random hexamer primers appear nonselective and may actually inhibit DNA extraction if hexamer quantities are greater than the combined soil-PureLyse carrying capacity. Many of our experiments here focus on addressing single soil-DNA interactions, such as the basalt (adsorption) and alkaline (Ca2+ binding) soils. Further optimization focused on soils containing multiple soil-DNA interactions like the aeolian, perchlorate, and acid soil may further increase yields. We conclude that a combination of desalting and employing competitive binders is the most effective method for extracting DNA, of the conditions tested, and may be a suitable starting point with which to approach the development of a universal martian soil DNA extraction protocol.
4.2. Environmental analogues and limits of detection
Our current experiments rely on appreciable amounts of DNA that establish resolvable soil-DNA interaction profiles (Figs. 6 and 7). However, we must note that the large spore populations used in this study (2.2 × 108 spores per 50 mg soil, ∼1 μg DNA) are unlikely to be observed in any terrestrial Mars analog environment or on Mars itself. Studies of microbial communities in the Atacama Desert and Antarctic McMurdo Dry Valleys (MDV) have reported detectable microbial populations as low as 500–5000 spores per 50 mg of soil (1 × 103 cfu per gram of soil), although about 104 to 106 cells per gram of soil is more typical (Navarro-Gonzalez et al., 2003; Goordial et al., 2016). Current environmental analog testing is focused on optimizing extraction protocols with Atacama/MDV-like microbial populations in the synthetic Mars analog soils and environmental samples from Mars-relevant sites. The OmniLyse cell-lysis component of the PureLyse Bacterial gDNA Extraction Kits has previously demonstrated the ability to lyse down to 50 spores of B. subtilis and has been characterized as the “best yield” extraction method by the United States Department of Agriculture in comparison to 11 other extraction methods (Irwin et al., 2014). Prior validation of OmniLyse suggests that DNA extraction of low spore populations analogous to Mars-like environments is feasible.
4.3. Small acid-soluble proteins and Mars
Small acid-soluble proteins (SASPs) within spores of B. subtilis bind to DNA and furnish protection from heat, high salts, desiccation, and UV radiation (Moeller et al., 2009, 2012). SASPs may contribute to the resilience of B. subtilis spores, and if life exists on Mars today, it is reasonable to consider that some martian organisms may have inherited or evolved homologous or analogous sporulation-protective programs that could also affect simple DNA isolation protocols. Thus, we must carefully review this possibility as we mature our Mars extraction protocols. To mitigate the effect of SASPs, we also developed an “on the fly” protein separation protocol involving a 5% phenol solution that increases DNA yields from upward of 14.7% to 43% (Fig. 8). It may be possible to further mitigate protein interactions and increase DNA yields from B. subtilis with a phenol-compatible DNA extraction kit, although phenol may not be acceptable aboard a Mars payload due to its potential risk for organic contamination to other instruments. Phenol modifications were purposely excluded from our spore DNA extraction from soils due to human health concerns with large-n experiments. However, it may yet be possible to develop an alternative solution that uses the same principle to separate DNA and protein. In this case, significant increases in extraction yield may be possible for the synthetic Mars analog soils. However, the ability to form spores is not common in bacterial phylogeny; it is mainly found in Gram-positive Firmicute bacteria (Traag et al., 2013).
4.4. DNA elution purity and sequencing
Recovering DNA does not always mean success (Barton et al., 2006). Soil contaminants (e.g., metal ions) can coelute with DNA and inhibit downstream analysis (e.g., PCR, sequencing) without preventative or additional purification steps. We determined that the PureLyse elution is of high purity (Fig. 9) and may not contain any compounds that inhibit certain nanopore sequencing technologies that rely on PCR-participating enzymes. While in this study we employed universal 16s rRNA primers, future studies, including those with low spore populations, can utilize single-copy B. subtilis specific primers in order to resolve discrete spore DNA extraction yields at low cell counts, even in the presence of exogenous contamination. In addition, Takada-Hoshino and Matsumoto (2004) and Barton et al. (2006) showed that samples treated with synthetic DNA (poly-dIdC) or skim milk, competitive binders used to increase DNA yields, do not inhibit amplification and thus may not inhibit nanopore sequencing either.
5. Conclusions
Life on Mars, if it exists or existed in the not-so-distant past, may potentially be detected via in situ nucleic acid extraction and sequencing aboard a future Mars rover/lander. Martian metagenomics could conceivably identify extraterrestrial genomes and discern conserved regions that may also suggest a Mars-Earth shared ancestry (Ruvkun et al., 2002; Isenbarger et al., 2008). Here, we have developed methodologies that enable such downstream nucleic acid detection or analysis technologies for future Mars exploration. The results indicate soil chemistry must be considered when extracting nucleic acids from Mars analogues or future Mars samples. Substantial challenges remain to be addressed, mainly engineering of an automated system and a Mars-compatible lysis and extraction module that incorporates all key steps for successful DNA extraction (desalting, competitive binding, DNA-protein separation), and utilizing biological reagents in the context of planetary protection regulations. Our work suggests that nucleic acid–based life detection on Mars may be feasible in the context of extant or recently dead life-intact cells; however, extracellular DNA may be bound to minerals or destroyed once hydrated. Additional work is required to understand the precise conditions under which DNA bound to minerals can be displaced to facilitate detection. Similar challenges apply to preservation and extraction of ancient DNA on Earth, with the added challenge of warm temperatures and their associated high hydrolysis rates.
Abbreviations Used
- EDTA
ethylenediaminetetraacetic acid
- EGTA
ethylene glycol tetraacetic acid
- MDV
McMurdo Dry Valleys
- qPCR
quantitative real-time polymerase chain reaction
- SASPs
small acid-soluble proteins
- SETG
Search for Extra-Terrestrial Genomes
Acknowledgments
This work was supported by NASA MATTISSE award NNX15AF85G to M.T.Z. We also thank members of the SETG team and Robert Doebler, Mark Brown, Doug Ming, and Carl Fuller.
Author Disclosure Statement
No competing financial interests exist.
References
- Allen C.C., Griffin C., Steele A. and Wainwright N. (2000) JSC Mars-1: martian regolith simulant [abstract 1797]. In 28th Lunar and Planetary Science Conference, Lunar and Planetary Institute, Houston [Google Scholar]
- Atoji M. (1959) Anhydrite obtained by the dehydration of gypsum. J Chem Phys 30:341–342 [Google Scholar]
- Azua-Bustos A., Urrejola C., and Vicuña R. (2012) Life at the dry edge: microorganisms of the Atacama Desert. FEBS Lett 586:2939–2945 [DOI] [PubMed] [Google Scholar]
- Azua-Bustos A., Caro-Lara L., and Vicuña R. (2015) Discovery and microbial content of the driest site of the hyperarid Atacama Desert, Chile. Environ Microbiol Rep 7:388–394 [DOI] [PubMed] [Google Scholar]
- Barton H.A., Taylor N.M., Lubbers B.R., and Pemberton A.C. (2006) DNA extraction from low-biomass carbonate rock: an improved method with reduced contamination and the low-biomass contaminant database. J Microbiol Methods 66:21–31 [DOI] [PubMed] [Google Scholar]
- Bell J.F., McSween H.Y., Crisp J.A., Morris R.V., Murchie S.L., Bridges N.T., Johnson J.R., Britt D.T., Golombek M.P., Moore H.J., Ghosh A., Bishop J.L., Anderson R.C., Brückner J., Economou T., Greenwood J.P., Gunnlaugsson H.P., Hargraves R.M., Hviid S., Knudsen J.M., Madsen M.B., Reid R., Rieder R., and Soderblom L. (2000) Mineralogic and compositional properties of martian soil and dust: results from Mars Pathfinder. J Geophys Res: Planets 105:1721–1755 [Google Scholar]
- Bernhardt H.S. and Tate W.P. (2012) Primordial soup or vinaigrette: did the RNA world evolve at acidic pH? Biol Direct 7, doi: 10.1186/1745-6150-7-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.B. and Wolfe G.V. (2006) Protist genetic diversity in the acidic hydrothermal environments of Lassen Volcanic National Park, USA. J Eukaryot Microbiol 53:420–431 [DOI] [PubMed] [Google Scholar]
- Butler I.B., Schoonen M., and Rickard D.T. (1994) Removal of dissolved oxygen from water: a comparison of four common techniques. Talanta 41:211–215 [DOI] [PubMed] [Google Scholar]
- Carr C.E., Rowedder H., Lui C.S., Zlatkovsky I., Papalias C.W., Bolander J., Myers J.W., Bustillo J., Rothberg J.M., Zuber M.T., and Ruvkun G. (2013a) Radiation resistance of sequencing chips for in situ life detection. Astrobiology 13:560–569 [DOI] [PubMed] [Google Scholar]
- Carr C.E., Rowedder H., Vafadari C., Lui C.S., Cascio E., Zuber M.T., and Ruvkun G. (2013b) Radiation resistance of biological reagents for in situ life detection. Astrobiology 13:68–78 [DOI] [PubMed] [Google Scholar]
- Carr C.E., Mojarro A., Tani J., Bhattaru S.A., Zuber M.T., Fuller C.W., Finney M., Church G., and Ruvkun G. (2016) Advancing the search for extra-terrestrial genomes. In 2016 IEEE Aerospace Conference, IEEE, Piscataway, NJ, doi: 10.1109/AERO.2016.7500859 [DOI] [Google Scholar]
- Chomczynski P. and Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159 [DOI] [PubMed] [Google Scholar]
- Ciesla F.J. and Sandford S.A. (2012) Organic synthesis via irradiation and warming of ice grains in the solar nebula. Science 336:452–454 [DOI] [PubMed] [Google Scholar]
- ClaremontBio Solutions. (2011) PureLyse and OmniLyse Data Sheet, Claremont Biosolutions, Upland, CA, pp 1–4 [Google Scholar]
- Clark B.C., Baird A.K., Weldon R.J., Tsusaki D.M., Schnabel L., and Candelaria M.P. (1982) Chemical composition of martian fines. J Geophys Res: Planets 87:10059–10067 [Google Scholar]
- Derrington I.M., Craig J.M., Stava E., Laszlo A.H., Ross B.C., Brinkerhoff H., Nova I.C., Doering K., Tickman B.I., Ronaghi M., Mandell J.G., Gunderson K.L. and Gundlach J.H. (2015) Subangstrom single-molecule measurements of motor proteins using a nanopore. Nature Biotechnology 33: 1073–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Direito S.O.L., Marees A., and Röling W.F.M. (2012) Sensitive life detection strategies for low-biomass environments: optimizing extraction of nucleic acids adsorbing to terrestrial and Mars analogue minerals. FEMS Microbiol Ecol 81:111–123 [DOI] [PubMed] [Google Scholar]
- Drouet C. and Navrotsky A. (2003) Synthesis, characterization, and thermochemistry of K-Na-H3O jarosites. Geochim Cosmochim Acta 67:2063–2076 [Google Scholar]
- Fritz J., Artemieva N., and Greshake A. (2005) Ejection of martian meteorites. Meteorit Planet Sci 40:1393–1411 [Google Scholar]
- Fuller C.W., Kumar S., Porel M., Chien M., Bibillo A., Stranges P.B., Dorwart M., Tao C., Li Z., Guo W., Shi S., Korenblum D., Trans A., Aguirre A., Liu E., Harada E.T., Pollard J., Bhat A., Cech C., Yang A., Arnold C., Palla M., Hovis J., Chen R., Morozova I., Kalachikov S., Russo J.J., Kasianowicz J.J., Davis R., Roever S., Church G.M. and Ju J. (2016) Real-time single-molecule electronic DNA sequencing by synthesis using polymer-tagged nucleotides on a nanopore array. Proc Nat Acad Sci USA 113: 5233–5238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladman B.J. and Burns J.A. (1996) Mars meteorite transfer: simulation. Science 274:161–162 [DOI] [PubMed] [Google Scholar]
- Gladman B.J., Burns J.A., Duncan M., and Lee P. (1996) The exchange of impact ejecta between terrestrial planets. Science 271:1387–1392 [Google Scholar]
- Goordial J., Davila A., Lacelle D., Pollard W., Marinova M.M., Greer C.W., DiRuggiero J., McKay C.P., and Whyte L.G. (2016) Nearing the cold-arid limits of microbial life in permafrost of an upper dry valley, Antarctica. ISME J 10:1613–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves M.P. and Wilson M.J. (1969) The adsorption of nucleic acids by montmorillonite. Soil Biol Biochem 1:317–323 [Google Scholar]
- Harris J.K., Harris J.K., Kelley S.T., Spiegelman G.B., and Pace N.R. (2003) The Genetic Core of the Universal Ancestor. Genome Research 13:407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneberger R.M., Walter M.R., and Anitori R.P. (2006) Extraction of DNA from acidic, hydrothermally modified volcanic soils. Environmental Chemistry 3:100–105 [Google Scholar]
- Herrera A. and Cockell C.S. (2007) Exploring microbial diversity in volcanic environments: a review of methods in DNA extraction. J Microbiol Methods 70:1–12 [DOI] [PubMed] [Google Scholar]
- Horneck G., Stöffler D., Ott S., Hornemann U., Cockell C.S., Moeller R., Meyer C., de Vera J.-P., Fritz J., Schade S., and Artemieva N.A. (2008) Microbial rock inhabitants survive hypervelocity impacts on Mars-like host planets: first phase of lithopanspermia experimentally tested. Astrobiology 8:17–44 [DOI] [PubMed] [Google Scholar]
- Horneck G., Moeller R., Cadet J., Douki T., Mancinelli R.L., Nicholson W.L., Panitz C., Rabbow E., Rettberg P., Spry A., Stackebrandt E., Vaishampayan P., and Venkateswaran K.J. (2012) Resistance of bacterial endospores to outer space for planetary protection purposes—experiment PROTECT of the EXPOSE-E mission. Astrobiology 12:445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt R.A., Robeson M.S., Shakya M., Moberly J.G., Vishnivetskaya T.A., Gu B., and Elias D.A. (2014) Improved yield of high molecular weight DNA coincides with increased microbial diversity access from iron oxide cemented sub-surface clay environments. PLoS One 9, doi: 10.1371/journal.pone.0102826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J.A. and Linn S. (1988) DNA damage and oxygen radical toxicity. Science 240:1302–1309 [DOI] [PubMed] [Google Scholar]
- Imlay J.A., Chin S.M., and Linn S. (1988) Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640–642 [DOI] [PubMed] [Google Scholar]
- Irwin P., Nguyen L., He Y., Paoli G., Gehring A., and Chen C.-Y. (2014) The near-quantitative sampling of genomic DNA from various food-borne Eubacteria. BMC Microbiol 14:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenbarger T.A., Carr C.E., Johnson S.S., Finney M., Church G.M., Gilbert W., Zuber M.T., and Ruvkun G. (2008) The most conserved genome segments for life detection on Earth and other planets. Orig Life Evol Biosph 38:517–533 [DOI] [PubMed] [Google Scholar]
- Jiang S., Zhuang J., Wang C., Li J., and Yang W. (2012) Highly efficient adsorption of DNA on Fe 3+–iminodiacetic acid modified silica particles. Colloids Surf A Physicochem Eng Asp 409:143–148 [Google Scholar]
- Klingelhöfer G., Morris R.V., Bernhardt B., and Schröder C. (2004) Jarosite and hematite at Meridiani Planum from Opportunity's Mössbauer spectrometer. Science 306:1740–1745 [DOI] [PubMed] [Google Scholar]
- Lever M.A., Torti A., Eickenbusch P., Michaud A.B., Šantl-Temkiv T., and Jørgensen B.B. (2015) A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front Microbiol 6:1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui C., Carr C.E., Rowedder H., Ruvkun G., and Zuber M.T. (2011) SETG: an instrument for detection of life on Mars ancestrally related to life on Earth. In 2011 IEEE Aerospace Conference, IEEE, Piscataway, NJ, doi: 10.1109/AERO.2011.5747299 [DOI] [Google Scholar]
- Makarova K.S., Aravind L., Galperin M.Y., Grishin N.V., Tatusov R.L., Wolf Y.I. and Koonin E.V. (1999) Comparative Genomics of the Archaea (Euryarchaeota): Evolution of Conserved Protein Families, the Stable Core, and the Variable Shell. Genome Research 9:608–628 [PubMed] [Google Scholar]
- McKay D.S., Carter J.L., and Boles W.W. (1994) JSC-1: a new lunar soil simulant [abstract 963]. In 24th Lunar and Planetary Science Conference, Lunar and Planetary Institute, Houston [Google Scholar]
- Meinert C., Myrgorodska I., De Marcellus, P., Buhse T., Nahon L., Hoffmann S.V., d'Hendecourt L.L.S., and Meierhenrich U.J. (2016) Ribose and related sugars from ultraviolet irradiation of interstellar ice analogs. Science 352:208–212 [DOI] [PubMed] [Google Scholar]
- Melzak K.A., Sherwood C.S., and Turner R. (1996) Driving forces for DNA adsorption to silica in perchlorate solutions. J Colloid Interface Sci 181:635–644 [Google Scholar]
- Ming D.W., Mittlefehldt D.W., Morris R.V., Golden D.C., Gellert R., Yen A., Clark B.C., Squyres S.W., Farrand W.H., Ruff S.W., Arvidson R.E., Klingelhöfer G., McSween H.Y., Rodionov D.S., Schröder C., de Souza P.A., Jr., and Wang A. (2006) Geochemical and mineralogical indicators for aqueous processes in the Columbia Hills of Gusev Crater, Mars. J Geophys Res 111, doi: 10.1029/2005JE002560 [DOI] [Google Scholar]
- Ming D.W., Morris R.V., Woida R., Sutter B., Lauer H.V., Shinohara C., Golden D.C., Boynton W.V., Arvidson R.E., Stewart R.L., Tamppari L.K., Gross M., and Smith P. (2008) Mars 2007 Phoenix Scout mission Organic Free Blank: method to distinguish Mars organics from terrestrial organics. J Geophys Res 113, doi: 10.1029/2007JE003061 [DOI] [Google Scholar]
- Moeller R., Setlow P., Horneck G., Berger T., Reitz G., Rettberg P., Doherty A.J., Okayasu R., and Nicholson W.L. (2008) Roles of the major, small, acid-soluble spore proteins and spore-specific and universal DNA repair mechanisms in resistance of Bacillus subtilis spores to ionizing radiation from X rays and high-energy charged-particle bombardment. J Bacteriol 190:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller R., Setlow P., Reitz G., and Nicholson W.L. (2009) Roles of small, acid-soluble spore proteins and core water content in survival of Bacillus subtilis spores exposed to environmental solar UV radiation. Appl Environ Microbiol 75:5202–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller R., Schuerger A.C., Reitz G., and Nicholson W.L. (2012) Protective role of spore structural components in determining Bacillus subtilis spore resistance to simulated Mars surface conditions. Appl Environ Microbiol 78:8849–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojarro A., Ruvkun G., and Zuber M.T. (2016) Methods for extracting nucleic acids from Mars analog regolith [abstract 1643]. In 47th Lunar and Planetary Science Conference, Lunar and Planetary Institute, Houston [Google Scholar]
- Morris R.V., Klingelhöfer G., Schröder C., Rodionov D.S., Yen A., Ming D.W., de Souza P.A., Wdowiak T., Fleischer I., Gellert R., Bernhardt B., Bonnes U., Cohen B.A., Evlanov E.N., Foh J., Gütlich P., Kankeleit E., McCoy T., Mittlefehldt D.W., Renz F., Schmidt M.E., Zubkov B., Squyres S.W., and Arvidson R.E. (2006) Mössbauer mineralogy of rock, soil, and dust at Meridiani Planum, Mars: Opportunity's journey across sulfate‐rich outcrop, basaltic sand and dust, and hematite lag deposits. J Geophys Res: Planets 111, doi: 10.1029/2006JE002791 [DOI] [Google Scholar]
- Morris R.V., Klingelhöfer G., Schröder C., Fleischer I., Ming D.W., Yen A.S., Gellert R., Arvidson R.E., Rodionov D.S., Crumpler L.S., Clark B.C., Cohen B.A., McCoy T.J., Mittlefehldt D.W., Schmidt M.E., de Souza P.A., and Squyres S.W. (2008) Iron mineralogy and aqueous alteration from Husband Hill through Home Plate at Gusev Crater, Mars: results from the Mössbauer instrument on the Spirit Mars Exploration Rover. J Geophys Res: Planets 113, doi: 10.1029/2008JE003201 [DOI] [Google Scholar]
- Navarro-Gonzalez R., Rainey F.A., and Molina P. (2003) Mars-like soils in the Atacama Desert, Chile, and the dry limit of microbial life. Science 302:1018–1021 [DOI] [PubMed] [Google Scholar]
- Nuevo M., Milam S.N., Sandford S.A., Elsila J.E., and Dworkin J.P. (2009) Formation of uracil from the ultraviolet photo-irradiation of pyrimidine in pure H2O ices. Astrobiology 9:683–695 [DOI] [PubMed] [Google Scholar]
- Nuevo M., Milam S.N., and Sandford S.A. (2012) Nucleobases and prebiotic molecules in organic residues produced from the ultraviolet photo-irradiation of pyrimidine in NH3 and H2O+NH3 ices. Astrobiology 12:295–314 [DOI] [PubMed] [Google Scholar]
- Purdy K.J., Embley T.M., Takii S., and Nedwell D.B. (1996) Rapid extraction of DNA and rRNA from sediments by a novel hydroxyapatite spin-column method. Appl Environ Microbiol 62:3905–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G., Finney M., Church G., Zuber M., and Gilbert W. (2002) A robotic-PCR detector for DNA-based life on other planets. In Signs of Life: A Report Based on the April 2000 Workshop on Life Detection Techniques, National Academies Press, Washington, DC, pp 137–140 [Google Scholar]
- Schloss P.D. and Handelsman J. (2005) Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol 6, doi: 10.1186/gb-2005-6-8-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder G.K., Lad C., Wyman P., Williams N.H., and Wolfenden R. (2006) The time required for water attack at the phosphorus atom of simple phosphodiesters and of DNA. Proc Natl Acad Sci USA 103:4052–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuerger A.C., Golden D.C., and Ming D.W. (2012) Biotoxicity of Mars soils 1. Dry deposition of analog soils on microbial colonies and survival under martian conditions. Planet Space Sci 72:91–101 [Google Scholar]
- Schwertmann U. and Cornell R.M. (2008) Iron Oxides in the Laboratory: Preparation and Characterization, Wiley-VCH, Weinheim [Google Scholar]
- Setlow B. and Setlow P. (1993) Binding of small, acid-soluble spore proteins to DNA plays a significant role in the resistance of Bacillus subtilis spores to hydrogen peroxide. Appl Environ Microbiol 59:3418–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster D.L. and Weiss B.P. (2005) Martian surface paleotemperatures from thermochronology of meteorites. Science 309:594–600 [DOI] [PubMed] [Google Scholar]
- Takada-Hoshino Y. and Matsumoto N. (2004) An improved DNA extraction method using skim milk from soils that strongly adsorb DNA. Microbes Environ 19:13–19 [Google Scholar]
- Traag B.A., Pugliese A., Eisen. J.A., and Losick R. (2013) Gene conservation among endospore-forming bacteria reveals additional sporulation genes in Bacillus subtilis. J Bacteriol 190:253–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevors J.T. (1996) DNA in soil: adsorption, genetic transformation, molecular evolution and genetic microchip. Antonie Van Leeuwenhoek 70:1–10 [DOI] [PubMed] [Google Scholar]
- Volossiouk T., Robb E.J., and Nazar R.N. (1995) Direct DNA extraction for PCR-mediated assays of soil organisms. Appl Environ Microbiol 61:3972–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade B.D. and Garcia-Pichel F. (2003) Evaluation of DNA extraction methods for molecular analyses of microbial communities in modern calcareous microbialites. Geomicrobiol J 20:549–561 [Google Scholar]
- Walker J.J., Spear J.R., and Pace N.R. (2005) Geobiology of a microbial endolithic community in the Yellowstone geothermal environment. Nature 434:1011–1014 [DOI] [PubMed] [Google Scholar]
- Wänke H., Brückner J., Dreibus G., Rieder R., and Ryabchikov I. (2001) Chemical composition of rocks and soils at the Pathfinder site. Space Sci Rev 96:317–330 [Google Scholar]
- Weiss B.P., Kirschvink J.L., Baudenbacher F.J., Vali H., Peters N.T., Macdonald F.A., and Wikswo J.P. (2000) A low temperature transfer of ALH84001 from Mars to Earth. Science 290:791–795 [DOI] [PubMed] [Google Scholar]
- Woese C.R., Fox G.E., Zablen L., Uchida T., Bonen L., Pechman K., Lewis B.J. and Stahl D. (1975) Conservation of primary structure in 16S ribosomal RNA. Nature 254:83–86 [DOI] [PubMed] [Google Scholar]
- Yakimov M.M., Giuliano L., Crisafi E., Chernikova T.N., Timmis K.N., and Golyshin P.N. (2002) Microbial community of a saline mud volcano at San Biagio-Belpasso, Mt. Etna (Italy). Environ Microbiol 4:249–256 [DOI] [PubMed] [Google Scholar]
- Zhou J., Bruns M.A., and Tiedje J.M. (1996) DNA recovery from soils of diverse composition. Appl Environ Microbiol 62:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]