Abstract
Even though gene repression is a powerful approach to exogenously regulate cellular behavior, developing a platform to effectively repress targeted genes, especially for stem cell applications, remains elusive. Herein, we introduce a nanomaterial-based platform that is capable of mimicking the function of transcription repressor proteins to downregulate gene expression at the transcriptional level for enhancing stem cell differentiation. We developed the NanoScript platform by integrating multiple gene repression molecules with a nanoparticle. First, we show a proof-of-concept demonstration using a GFP-specific NanoScript to knockdown GFP expression in neural stem cells (NSCs-GFP). Then, we show that a Sox9-specific NanoScript can repress Sox9 expression to initiate enhanced differentiation of NSCs into functional neurons. Overall, the tunable properties and gene knockdown capabilities of NanoScript enables its utilization for gene-repression applications in stem cell biology.
Keywords: gene regulation, gene knockdown, nanoparticles, neuronal differentiation, transcription repressor proteins
TOC image
It Will Knock You Down: A NanoScript-based platform is designed to knock down transcriptional gene expression in stem cells. The tunable and non-viral NanoScript platform, which is comprised of a nanoparticle functionalized with specific small molecules, has been demonstrated to both effectively knock down GFP in GFP-labeled neural stem cells (NSCs), and repress Sox9 expression in NSCs to induce differentiation into functional neurons.
Stem cell differentiation and cellular reprogramming is fundamentally regulated through a process known as gene regulation.[1] Gene regulation is an inherent cellular mechanism through which gene expression is either increased or decreased, and this has a direct impact on cellular behavior such as proliferation, migration, and differentiation.[2] There are two types of gene regulation: 1) gene activation, which refers to an increase in the expression levels of a targeted gene, and 2) gene repression, which refers to a decrease in the expression levels of targeted genes.[3]
The complex process of gene regulation is intrinsically regulated by transcription factors (TFs), which function by binding to specific gene sequences, thereby controlling the initiation of transcription of genetic information.[4] Once TFs bind to their target gene, the gene can either be activated or repressed, depending on which domains are present on the TFs. A typical TF contains three fundamental domains: 1) a DNA-Binding Domain (DBD) which is sequence-specific and binds to target sequences, 2) a nuclear localization domain to enable the TF proteins entry inside the nucleus, and 3) either an activation domain or a repression domain (RD). If an activation domain is present on the TF, then the targeted gene will be transcribed and gene expression will be upregulated;[5] and if a RD is present on the TF, then the target gene will be repressed and gene expression will be downregulated.[6]
We recently developed a nanomaterial-based platform called NanoScript, which was designed to mimic the fundamental structure and function of TF activator proteins.[7] NanoScript was designed by attaching specific small molecules on a nanoparticle. These small molecules emulate the function of individual domains on TF proteins, and when multiple small molecules are assembled together on a single nanoparticle, the resulting NanoScript platform can mimic the function and structure of natural TF proteins. NanoScript is a platform with interchangeable components that can be modified depending on the desired application. Our previous NanoScript included a series of small molecules to mimic a subset of TF activator proteins. However, even though there are reports of small molecule-based approaches for transcriptional gene knockdown in in vitro systems,[8] there have been no previous reports to either demonstrate NanoScript’s capability to mimic TF repressor proteins or to develop a nanomaterial-based platform that can effectively repress genes at the transcriptional level. We speculate that by modifying our NanoScript platform with repression-specific small molecules, we can design NanoScript to mimic transcriptional repressor proteins for effectively downregulating genes to induce stem cell differentiation.
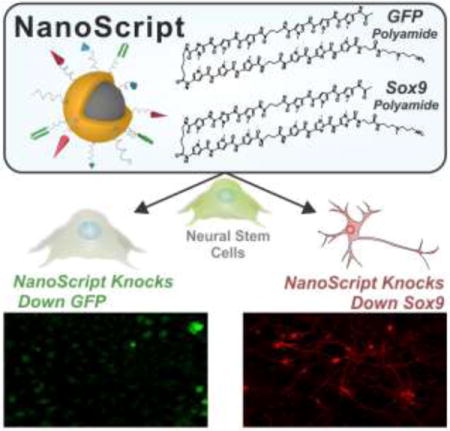
Herein, we developed the NanoScript platform to effectively mimic the fundamental structure and gene-silencing function of TF repressor proteins. In order to emulate the function of each domain on natural TF repressor proteins, NanoScript was constructed by assembling multiple gene repression molecules, which function to inhibit and block the recruitment of factors to the DNA binding site to prevent gene expression, together on a multifunctional nanoparticle (Figure 1a, b). We performed a proof-of-concept experiment by successfully repressing endogenous expression of green fluorescence protein (GFP) in neural stem cells. Moreover, NanoScript was utilized to repress the neuro-specific gene Sox9 in neural stem cells which induced their differentiation into neurons (Figure 1c). The primary advantage of our multifunctional NanoScript platform over conventional approaches is its ability to tether multiple repressor molecules, which function through different mechanisms, on a single nanoparticle to synergistically repress gene expression.
Figure 1. Schematic Representation of NanoScript-based Gene Repression.
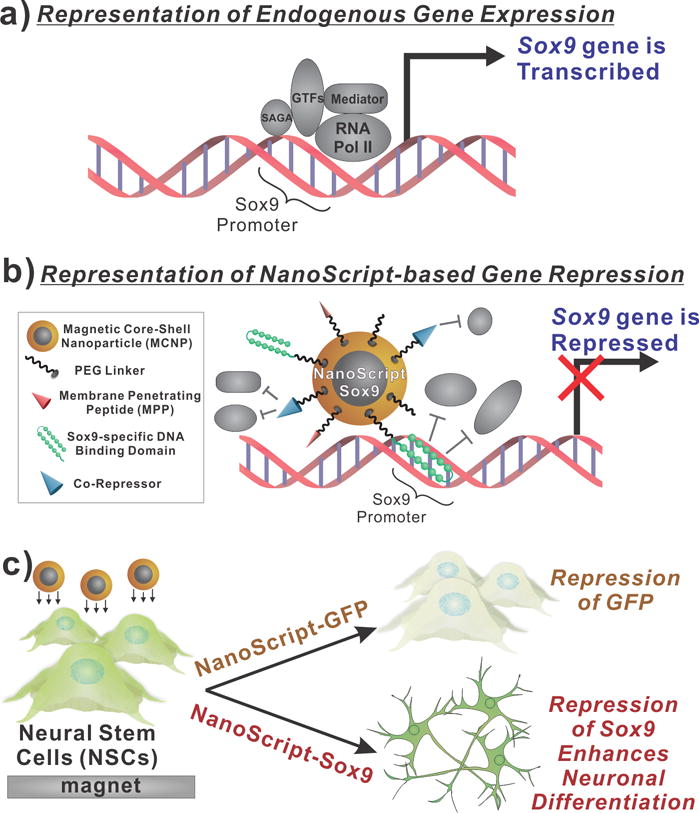
(a) When components of the transcriptional basal complex assemble on a target DNA sequence, such as the Sox9 promoter sequence, the corresponding gene is transcribed. (b) NanoScript-based gene expression is based on the synergistic effect of the DNA Binding Domain molecule for steric hindrance and the co-repressor molecule to disrupt the formation of the transcriptional basal complex on the target DNA sequence. (c) To demonstrate NanoScript-based repression in neural stem cells (NSCs), a GFP-specific NanoScript silences expression of GFP and a Sox9-specific NanoScript represses Sox9 to induce neuronal differentiation.
The NanoScript platform was functionalized with multiple molecules in order to emulate the function and structure TF repressor proteins. The first component of NanoScript is the hairpin polyamide molecule, specific for the GFP and Sox9 genes. The hairpin polyamide is a small molecule comprised of the pyrrole (Py) and imidazole (Im) groups which binds to A-T and G-C base pairs on the DNA respectively with nanomolar affinity.[9] The binding of the hairpin polyamide to the DNA sterically hinder the attachment of enzymes like RNA Polymerase II to the binding site, which in turn, prevents the gene from being transcribed.[8b] The GFP promoter sequence was obtained from the company from which GFP-labeled rat neural stem cells (rNSCs) were purchased (Figure S1). A hairpin polyamide with a sequence of PyPyPy-β-PyPyIm-γ-PyPyPy-β-PyImPy-β-Dp-NH2 (γ is γ-aminobutyric acid, β is β-alanine, and Dp is dimethylaminopropylamide) that targets the GFP promoter was synthesized using a previously established solid-phase synthesis protocol (Figure S2a).[7a] An in-vitro binding assay study was performed using surface plasmon resonance (SPR) and revealed a high nanomolar binding affinity (Figure S2b). Moreover, we synthesized a Sox9-specific hairpin polyamide with a sequence of PyPyPy-β-PyImPy-γ-PyPyPy-β-PyImIm-β-Dp-NH2 that also showed nanomolar binding affinity to its target sequence. (Figure S3).[10]
The second molecule is the corepressor peptide with a sequence of WRPW. The WRPW peptide was specifically chosen because: 1) it has been demonstrated to induce gene repression by preventing the formation of the basal transcriptional machinery at the binding site, 2) it induces repression of genes via the Groucho family proteins, which have been demonstrated to play a role in neurogenesis, and 3) it is a short tetrapeptide with only 4 amino acids, and hence it is readily soluble in physiological environments.[11] The third molecule is the membrane penetrating peptide (MPP) which has been previously demonstrated to effectively shuttle nanoparticles across the plasma and nuclear membrane.[12]
These small molecules (hairpin polyamide, WRPW peptide, and MPP) were conjugated to PEG-based linker molecules to enhance their solubility, with the PEG linker having with a thiol terminus to enable functionalization onto the magnetic core-shell nanoparticles (MCNPs).[13] MCNPs were chosen because of their high biocompatibility, inert properties, ability to induce magnetofection by placing a magnet underneath the culture plate to attract MCNP onto the cell surface, and multifunctional gold surface which enable attachment of multiple molecules on a single nanoparticle.[14] After the nanoparticles were functionalized with the PEG-terminated small molecules (refer to methods in SI for details), the resulting platform was termed NanoScript (Figure 2a). Using a combination of dynamic light scattering and transmission electron microscopy, we found the size of the MCNP to be 17.3 nm (Figure S4), and after functionalization, the size of both NanoScript-GFP and NanoScript-Sox9 was found to be about 45 nm (Figure 2b). Through UV-vis absorption spectroscopy, we observed a shift in the plasmon resonance which is indicative of surface functionalization (Figure S5). Based on previously demonstrated studies which show that there are approximately 4.3 ligands/nm2, we predict that there are approximately 3,902 ligands on the nanoparticle.[15] Moreover, the monodispersity of NanoScript was confirmed through transmission electron microscopy, wherein we visualized well-rounded and monodispersed sizes (Figure 2c). Furthermore, we tested if NanoScript can localize within the nucleus by labeling NanoScript with an Alexa Flour 568 dye and transfecting them into rat neural stem cells (rNSCs). After 24 hours, we performed fluorescence imaging and NanoScript was detected within the nucleus (Figure 2d, Figure S6). We also performed transmission electron microscopy on cellular cross-sections and found that NanoScript was distributed in the nucleus and cytoplasm (Figure S7). Moreover, we performed inductively coupled plasma optical emission spectrometry (ICP-OES) and observed that NanoScript was uptaken inside the cells (Figure S8).
Figure 2. Construction and Characterization of NanoScript.
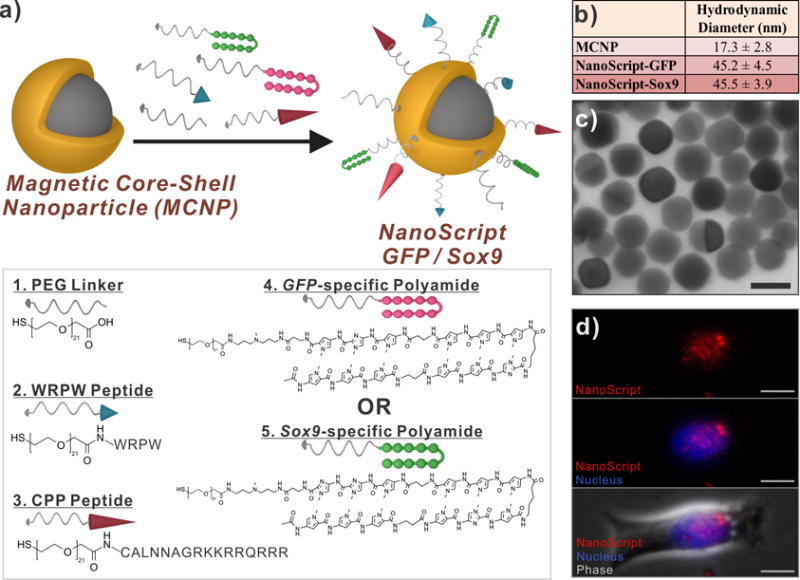
(a) The magnetic core-shell nanoparticle (MCNP) was functionalized with PEG-terminated biomolecules, through thiol-gold interactions, to develop the NanoScript platform specific for either GFP or Sox9. (b) The hydrodynamic diameter and (c) transmission electron micrographs of NanoScript (scale bar = 20 nm). (d) A dye-labeled NanoScript (red) was transfected into rat NSCs and NanoScript was detected within the nucleus (blue) (scale bar = 20 μm).
To test if NanoScript can repress gene expression, we performed a proof-of-concept demonstration using a GFP-specific NanoScript (termed NanoScript-GFP) on GFP-labeled rNSCs. These GFP-labeled rNSCs intrinsically express GFP, and its expression can easily be detected through fluorescence imaging.[16] Hence, we can evaluate the gene repression capability of NanoScript-GFP by observing GFP knockdown in rNSCs (Figure 3a). The rNSCs were transfected with NanoScript-GFP and expression of GFP was evaluated at different time points using fluorescence imaging. During the transfection, a magnet was placed underneath the culture plate for 15 min to induce magnetically-facilitated delivery, which is a technique to attract NanoScript onto the cellular surface, in a similar manner as our previous paper.[14b] After 4 days, the GFP expression was significantly repressed as compared to the control; and control conditions which included unconjugated GFP polyamide, unconjugated WRPW peptide, nanoparticle with WRPW (MCNP-WRPW), and nanoparticle with GFP polyamide only (MCNP-GFP), showed decreased GFP knockdown (Figure 3b). In the control conditions, attachment of either WRPW or GFP polyamide to the nanoparticle increased GFP knockdown as compared to unconjugated WRPW or GFP polyamide. The same nanoparticle core was used for each condition. Moreover, the MPP was shown to have almost no direct influence on GFP knockdown (Figure S9). The intensity of GFP fluorescence expression in the images of all the conditions was quantified, and these results not only confirmed the trend observed in the fluorescence images, but revealed a time-dependent increase of GFP knockdown (Figure 3c). Finally, we tested GFP mRNA levels using qPCR and observed a similar trend of decreasing GFP expression using NanoScript (Figure S10). Collectively, these results suggest that NanoScript-GFP can not only repress endogenous expression of GFP, but also that the cooperative function of the polyamide and WRPW on the same nanoparticle enhances gene knockdown.
Figure 3. NanoScript-GFP Effectively Silences GFP Expression.
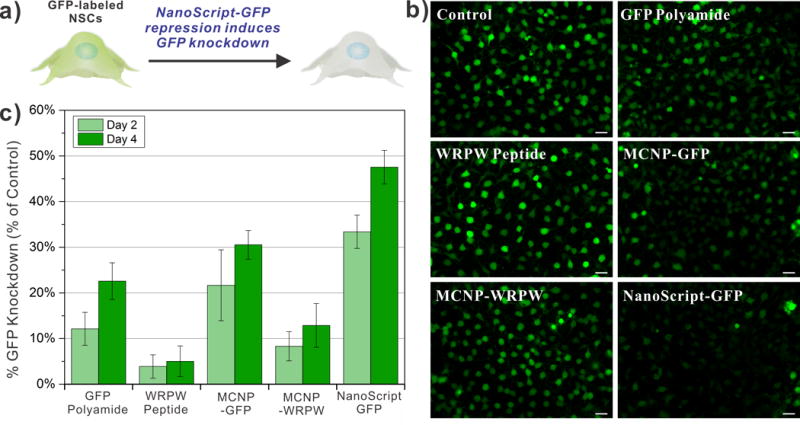
(a) Schematic representation of NanoScript-GFP downregulating GFP expression (i.e. reduction in green fluorescence from the cell) in GFP-labeled rat NSCs. (b) Fluorescence images of GFP-labeled rNSCs 4 days post transfection showing a maximal decrease in GFP expression (green) when NanoScript-GFP is transfected (scale bar = 20 μm). A 1 nM concentration of MCNPs was applied during the transfection. (c) Quantification of GFP expression in the fluorescence images corroborates the trend in the images and reveals that NanoScript-GFP has the highest GFP knockdown as compared to the controls. Quantification of GFP knockdown is an average from 6 images and standard error is from three independent trials.
Although this proof-of-concept demonstration indicates that NanoScript can repress gene expression, the real challenge and central goal is to translate the NanoScript-based gene repression approach for stem cell differentiation applications. To this end, the Sox9 gene has been identified as a critical gene to regulate neuronal differentiation in stem cells. Studies have shown that repression of Sox9 in neural stem cells initiates a pathway to guide their differentiation into neurons.[16–17] Hence, we developed a Sox9-specific NanoScript (termed NanoScript-Sox9), and we predict that if NanoScript-Sox9 can effectively repress Sox9 in human neural stem cells (hNSCs), enhanced differentiation into neurons can be observed (Figure 4a).
Figure 4. NanoScript-Sox9 Represses Sox9 to Induce Functional Neuronal Differentiation.
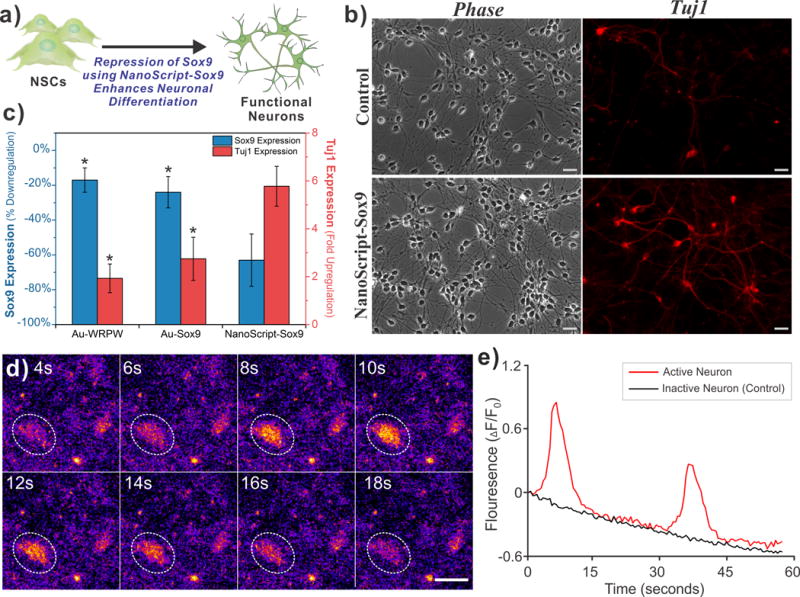
(a) Schematic representation of Sox9 repression in human NSCs by NanoScript-Sox9 induces enhanced neuronal differentiation. (b) Fluorescence images of hNSC stained with Tuj1 5 days post-transfection shows greater Tuj1 expression (red) when NanoScript-Sox9 is transfected. (Scale bar = 20 μm). (c) Gene expression analysis using qPCR in hNSCs reveals that repression of Sox9 correlates with an upregualtion of Tuj1. (Percent down-regulation of Sox9 and fold up-regulation of Tuj1 was calculated by normalizing to the housekeep gene, GAPDH, from the control) Standard error is from three independent trials (* = P < 0.05). (d) Spontaneous calcium fluctuations via Fluo4 fluorescence (orange/yellow color) for an active neuron (white circle) during 18 seconds of imaging (scale bar = 20 μm). (e) Traces for the normalized fluorescence change (ΔF/F0) representing spontaneous calcium ion influx for an active neuron (red line) and an inactive neuron (black line). Decreasing trend of the fluorescence is due to mild photobleaching.
To test this, we transfected NanoScript-Sox9 into hNSCs (refer to methods in SI for protocol) and then we evaluated the expression of neuronal markers through qPCR and immunocytochemistry. Specifically, the expression of Tuj1 was evaluated because it is a prominent marker for neurons.[18] We predict that the suppression of Sox9 by NanoScript-Sox9 should lead to enhanced neuronal differentiation, and hence, an increase in Tuj1 expression. We fixed and stained for Tuj1 on Day 5, and the resulting fluorescence images indicated a greater expression of Tuj1 as compared to the control (Figure 4b). This was further confirmed by testing gene expression through qPCR, wherein the expression levels induced by NanoScript-Sox9 showed a decrease of Sox9 expression by 63% and a 5.7-fold increase in Tuj1 expression as compared to the control (Figure 4c). The expression levels of other control conditions (nanoparticle with WPRW and nanoparticle with Sox9 polyamide) were also able to induce Sox9 repression and Tuj1 expression, but not as strongly as compared to the NanoScript-Sox9 conditions (Figure 4c). The expression levels of additional control experiments including unconjugated Sox9 polyamide and WRPW showed minimal changes as compared to the control (Figure S11). Expression of Sox9 protein levels was further evaluated using immunostaining which revealed a similar decreasing trend in the NanoScript-Sox9 condition (Figure S12). Moreover, by performing scanning electron microscopy (SEM), we were able to visualize neurons in high resolution (Figure S13). High cell survival was confirmed with a cell viability assay (Figure S14).
To evaluate if the induced neurons have spontaneous neuronal activity, we monitored changes in intracellular calcium levels. Functionally active neurons are known to spontaneously fire action potentials that allow influx of cations including calcium.[19] Using a commercially available calcium indicator dye, Fluo4, changes in intracellular calcium concentrations were visualized and its fluorescence intensity was quantified. After 7 days post-transfection, we performed calcium imaging using Fluo4 and observed changes in fluorescence levels in the induced neurons (Figure 4d, Video S1). Furthermore, we quantified the fluorescence changes and observed spontaneous fluctuations of calcium ions in the active neuron over a 60 second period while the control inactive neuron did not show any changes in fluorescence (Figure 4e). These results suggest that the induced neurons show functional activity.
In summary, the overall goal of introducing a tunable and efficient platform that can mimic TF repressor proteins for effectively repressing genes to induce stem cell differentiation was achieved. As a result, this is the first-ever demonstration of utilizing a nanomaterial-based platform for emulating the function of TF repressor proteins to downregulate gene expression at the transcriptional level for inducing stem cell differentiation. We developed the NanoScript platform by functionalizing a nanoparticle with multiple gene repression molecules such as gene-specific polyamides and the WRPW peptide. We first show a proof-of-concept demonstration that utilizes a GFP-specific NanoScript to knockdown GFP expression in GFP-labeled rNSCs. Then we show that a Sox9-specific NanoScript can repress Sox9 expression in hNSCs to initiate enhanced differentiation into functional neurons. The only difference between these two demonstrations is the gene-specific polyamide, thus highlighting the versatility and tunability of the NanoScript platform.
Furthermore, the results from both demonstrations (GFP knockdown and Sox9 repression) suggested that the synergistic effect of the polyamide and WRPW peptide on the NanoScript is needed for enhanced gene repression. One hypothesis for this result is that the two molecules contribute to gene repression through two different mechanisms. Previously reported mechanistic studies have shown that the binding of the polyamide to the target DNA sequence sterically occlude factors like RNA polymerase II for assembling on the DNA;[8b] and the WRPW peptide is known to initiate the Groucho family proteins which are well-established corepressor factors that prevents the formation of the transcriptional basal complex.[11b] By assembling both molecules on the NanoScript, we not only synergistically enhance gene repression, but enable NanoScript to more closely mimic the structure of TF repressor proteins.
While NanoScript does not completely knockdown GFP, its knockdown level is comparable to other nanomaterial-based methods that regulate GFP knockdown at the translational level.[16] Other methods for inducing neuronal differentiation such as viral vectors, small molecules, and nanomaterial-based platforms have been developed,[16, 20] but because of NanoScript unique features including its non-viral gene regulation and interchangeable components, we are further investigating to optimize and evaluate NanoScript against current methods. Furthermore, previous studies have shown that the extra cellular matrix plays a role in inducing neuronal differentiation,[21] and so, the effect of ECM on assisting in NanoScript-based differentiation may require further investigation. Thus far, the current NanoScript platform is primarily applicable for gene regulation in adherent cells, but when NanoScript was evaluated for gene regulation in hNSCs cultured in suspension, we observed knockdown of the Sox9 gene (Figure S15); however, we are further optimizing and investigating the potential of applying NanoScript for other cell types, such as those in suspension.
In conclusion, the introduction of NanoScript platform as an approach to repress gene expression will significant impact the field of stem cell biology. First, because NanoScript regulates gene repression in a non-viral manner, it can be a candidate for stem cell-based research and potential therapies. Second, the high cell viability of NanoScript-transfected rat and human NSCs ensures the potential applicability of NanoScript for other stem cells lines. Third, by simply redesigning the polyamide sequence to complement a targeted gene, it is possibly to modify NanoScript to target and repress almost any gene of interest. We are confident that the versatility, effectiveness, and tunable properties of NanoScript will give scientists a new tool for gene-regulating applications such as stem cell biology and cellular reprogramming.
Supplementary Material
Acknowledgments
This work was financially supported the NIH Director’s Innovator Award (1DP20D006462-01), NIH R21 (1R21NS085569-01), New Jersey Commission on Spinal Cord (CSR13ERG005), and NSF CHE-1429062. We express gratitude to Prof. Gene Hall (Rutgers University) for assisting in ICP-OES measurements.
Contributor Information
Sahishnu Patel, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA).
Sy-Tsong Dean Chueng, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA).
Perry T. Yin, Department of Biomedical Engineering, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA)
Kholud Dardir, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA).
Zhichao Song, Department of Cell Biology & Neuroscience, W.M. Keck Center for Collaborative Neuroscience, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA).
Nicholas Pasquale, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA).
Kelvin Kwan, Department of Cell Biology & Neuroscience, W.M. Keck Center for Collaborative Neuroscience, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA).
Hiroshi Sugiyama, Department of Chemistry, Graduate School of Science, Kyoto University, Kyoto, 606-8501 (Japan).
Ki-Bum Lee, Department of Chemistry and Chemical Biology, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA); Department of Biomedical Engineering, Rutgers, The State University of New Jersey, Piscataway, NJ 08854 (USA).
References
- 1.Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA. Cell Stem Cell. 2010;7:249–257. doi: 10.1016/j.stem.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rockman MV, Kruglyak L. Nat Rev Genet. 2006;7:862–872. doi: 10.1038/nrg1964. [DOI] [PubMed] [Google Scholar]
- 3.Reik W. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 4.Spitz F, Furlong EE. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 5.a) Weake VM, Workman JL. Nat Rev Genet. 2010;11:426–437. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]; b) Mapp AK, Ansari AZ, Ptashne M, Dervan PB. Proc Natl Acad Sci U S A. 2000;97:3930–3935. doi: 10.1073/pnas.97.8.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds N, O’Shaughnessy A, Hendrich B. Development (Cambridge, England) 2013;140:505–512. doi: 10.1242/dev.083105. [DOI] [PubMed] [Google Scholar]
- 7.a) Patel S, Jung D, Yin PT, Carlton P, Yamamoto M, Bando T, Sugiyama H, Lee KB. ACS Nano. 2014;8:8959–8967. doi: 10.1021/nn501589f. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Patel S, Pongkulapa T, Yin P, Pandian GN, Rathnam C, Bando T, Vaijayanthi T, Sugiyama H, Lee KB. Journal of the American Chemical Society. 2015 doi: 10.1021/ja511298n. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Patel S, Yin PT, Sugiyama H, Lee KB. ACS Nano. 2015;9:6909–6917. doi: 10.1021/acsnano.5b00709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Dickinson LA, Gulizia RJ, Trauger JW, Baird EE, Mosier DE, Gottesfeld JM, Dervan PB. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12890–12895. doi: 10.1073/pnas.95.22.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Leung CH, Chan DS, Ma VP, Ma DL. Medicinal research reviews. 2013;33:823–846. doi: 10.1002/med.21266. [DOI] [PubMed] [Google Scholar]
- 9.a) Gottesfeld JM, Neely L, Trauger JW, Baird EE, Dervan PB. Nature. 1997;387:202–205. doi: 10.1038/387202a0. [DOI] [PubMed] [Google Scholar]; b) Dervan PB, Edelson BS. Curr Opin Struct Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 10.Mertin S, McDowall SG, Harley VR. Nucleic Acids Res. 1999;27:1359–1364. doi: 10.1093/nar/27.5.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Fisher AL, Ohsako S, Caudy M. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Courey AJ, Jia S. Genes Dev. 2001;15:2786–2796. doi: 10.1101/gad.939601. [DOI] [PubMed] [Google Scholar]; c) Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 12.Krpetić Ze, Saleemi S, Prior IA, Sée V, Qureshi R, Brust M. ACS Nano. 2011;5:5195–5201. doi: 10.1021/nn201369k. [DOI] [PubMed] [Google Scholar]
- 13.Jokerst JV, Lobovkina T, Zare RN, Gambhir SS. Nanomedicine. 2011;6:715–728. doi: 10.2217/nnm.11.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.a) Colombo M, Carregal-Romero S, Casula MF, Gutierrez L, Morales MP, Bohm IB, Heverhagen JT, Prosperi D, Parak WJ. Chemical Society Reviews. 2012;41:4306–4334. doi: 10.1039/c2cs15337h. [DOI] [PubMed] [Google Scholar]; b) Shah B, Yin PT, Ghoshal S, Lee KB. Angewandte Chemie International Edition. 2013;52:6190–6195. doi: 10.1002/anie.201302245. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Scherer F, Anton M, Schillinger U, Henke J, Bergemann C, Kruger A, Gansbacher B, Plank C. Gene Ther. 2002;9:102–109. doi: 10.1038/sj.gt.3301624. [DOI] [PubMed] [Google Scholar]
- 15.Hinterwirth H, Kappel S, Waitz T, Prohaska T, Lindner W, Lämmerhofer M. ACS Nano. 2013;7:1129–1136. doi: 10.1021/nn306024a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Solanki A, Shah S, Yin PT, Lee KB. Sci Rep. 2013;3 doi: 10.1038/srep01553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grienberger C, Konnerth A. Neuron. 2012;73:862–885. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 20.a) Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Shah S, Solanki A, Sasmal PK, Lee KB. Journal of the American Chemical Society. 2013;135:15682–15685. doi: 10.1021/ja4071738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Liu M, Yan Y, Yang ST. World Journal of Stem Cells. 2014;6:11–23. doi: 10.4252/wjsc.v6.i1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


