Abstract
Objective
Scleroderma is a genetically complex autoimmune disease with substantial phenotypic heterogeneity. Previous genome-wide association studies have identified common genetic variants associated with disease risk, but these studies are not designed to capture rare or potential causal variants. Our goal was to identify rare, as well as common genetic variants in patients with diffuse cutaneous systemic sclerosis (dcSSc) through whole exome sequencing (WES) in order to identify potential causal variants.
Methods
We generated WES data for 32 dcSSc patients with or without interstitial lung disease (ILD) and 17 healthy “in-house” controls. Variants were annotated and filtered by quality, minor allele frequency, and deleteriousness on gene function. We applied a gene burden test to identify novel dcSSc and dcSSc-associated ILD candidate genes that were enriched with deleterious variants in cases compared to in-house controls as well as controls from the 1000 Genomes Project (n=130).
Results
We identified 70 genes that were enriched with deleterious variants in dcSSc patients. Two of them (BANK1 and TERT) are in pathways previously implicated in SSc or ILD pathogenesis or known susceptibility loci. Newly-identified genes are significantly enriched in the extracellular matrix-related pathway (COL4A3, COL4A4, COL5A2, COL13A1, and COL22A1), which is relevant to the fibrotic features of dcSSc, and the DNA repair pathway (XRCC4).
Conclusion
This study demonstrates the value of WES for the identification of novel gene variants and pathways that may contribute to scleroderma risk and/or severity. The candidate genes we discovered are potential targets for in-depth functional studies.
Introduction
Systemic sclerosis (SSc) is a complex disease characterized by autoimmunity, vasculopathy and fibrosis. The prevalence of SSc in the US is estimated to be 250 to 300 cases per million, with a female to male ratio of 4.6 to 1 (1). SSc is a heterogeneous disorder with phenotypic variation in the extent of skin disease, presence of specific autoantibodies, internal organ involvement (2), and disease outcome. For example, mortality rates for diffuse cutaneous SSc (dcSSc) are greater than for limited cutaneous SSc. This is likely due to the increased rates of organ involvement, such as interstitial lung disease (ILD), in dcSSc (2).
Several genome-wide association studies (GWAS) have been performed to date, revealing a number of loci associated with increased risk of SSc. These studies also suggest that genetic variation may explain, at least in part, the observed phenotypic heterogeneity of the disease (3). However, GWAS are designed to capture only common genetic variation, and to date these risk loci collectively explain a relatively small proportion of SSc heritability.
Targeted sequencing of the protein-coding part of the genome (whole exome sequencing, WES) has become a popular approach for identifying rare variants that affect protein function and may contribute to disease pathogenesis. For the current study, we performed WES to identify both rare and common variants predicted to be deleterious, and characterized their aggregate effects based on enrichment in specific genes and pathways. Using this strategy, we have identified potential causal variants in established SSc risk loci as well as novel candidate genes and pathways involved in dcSSc and dcSSc-associated ILD.
Patients and Methods
Patients
We studied 32 patients diagnosed with dcSSc at the University of California, San Francisco (UCSF) using established criteria for the disease and major disease subsets (4, 5) (Table 1). Patient-reported ethnicity was verified by ancestry composition analysis (Supp. Methods). dcSSc patients were classified as having ILD based on evidence of reticulation and architectural distortion on high resolution computed tomography of the chest (6) or restrictive lung disease on pulmonary function tests. Twenty-six of the patients were female, 19 had ILD, and 23 had >90% European ancestry and were defined as EUR dcSSc samples (Supp. Figures 1 and 2).
Table 1.
Thirty-two patients with diffuse cutaneous systemic sclerosis.
| Women | 26 (81.3%) |
| Men | 6 (18.8%) |
|
| |
| Age at diagnosis (years) | |
|
| |
| All | 45.2 ± 12.2 |
| Women | 44.7 ± 11.9 |
| Men | 47.2 ± 14.5 |
|
| |
| Clinical features | |
|
| |
| ILD | 19 (59.4%) (N=32) |
| Raynaud's | 31 (96.9%) (N=32) |
| Digital ulcers | 16 (57.1%) (N=28) |
| SRC | 3 (9.4%) (N=32) |
N: number of subjects for which information was available
Abbreviations: ILD, interstitial lung disease; SRC, scleroderma renal crisis
In-house exome control data
WES data for 17 healthy controls were generated in the same sequencing center and analyzed using the same protocols as the dcSSc patients.
DNA extraction
Genomic DNA was extracted from whole blood using QIAGEN Puregene Blood Kits (Valencia, CA) according to the manufacturer’s protocol.
Whole-exome sequencing
Library preparation was performed using the NuGEN Ovation Ultralow Library Systems or Nextera DNA Sample Preparation Kit (San Carlos, CA). Exome capture was performed using the Nimblegen SeqCap EZ Human Exome Library v3.0 Kit (Madison, WI) according to the manufacturer’s protocols. Sequencing was performed on an Illumina HiSeq 2000 sequencer with a paired-end read length of 100 bp in the Genomics Core Facility at UCSF. Exome data generated in this project was registered in the NCBI BioProject (BioProject ID PRJNA316441). NCBI BioSample Accessions for all dcSSc samples are listed in Supp. Table 1.
Next generation sequencing data analysis
Variant analysis and joint genotyping (analyzing the entire set of samples for genotype calls) were performed on dcSSc and in-house controls according to procedures recommended by the GATK Best Practices (Supp. Methods).
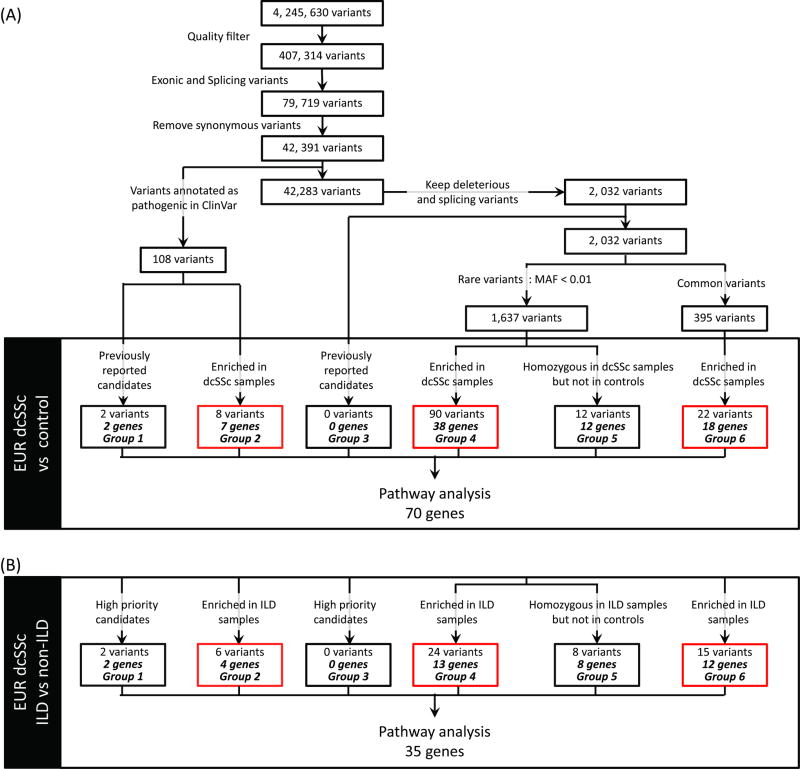
The joint genotyped variant list was annotated and further filtered using Variant Tools (7). The following filters were applied to generate a preliminary variant list: (i) remove false positive variants (Variant Quality Score Recalibration tranche sensitivity < 99.00) and low quality variants that had low depth of coverage (DP < 10) and poor genotype quality (GP < 20); (ii) keep exonic or splicing variants based on UCSC hg19 known gene annotation; and (iii) remove synonymous variants. This preliminary variant list was further filtered by previously reported candidate genes, deleteriousness and allele frequency (Figure 1 and Supp. Methods). Case-control gene burden analysis (see below) was performed on both the rare and common deleterious variants to identify genes with enrichment of deleterious variants in dcSSc samples. Genes with rare variants that were homozygous in dcSSc cases but not present in control samples were also identified.
Figure 1.
Variant filtration and gene burden analysis to identify dcSSc and dcSSc-associated ILD susceptibility genes.
(A) Variant filtration for all variants identified from 32 dcSSc samples. Gene burden analysis was performed using EUR dcSSc samples as cases and EUR in-house and public samples as controls. The variant list for all groups can be found in Supp. Table 5. (B) Gene burden analysis was carried out by comparing EUR dcSSc-associated ILD patients (cases) to dcSSc non-ILD patients (controls). The variant list for all groups can be found in Supp. Table 6. Quality filters applied were VQSR tranche sensitivity < 99.00, DP > 10 and GQ > 20 in which DP is read depth and GQ is the genotyping quality in phred scale. MAF, minor allele frequency in 1000 Genomes European (phase 1) or ESP6500 European American population; Variants enriched in dcSSc/ILD samples (red box), variants that satisfied the criteria described in the case-control gene burden analysis; Pathway analysis, pathway analysis performed using g:Profiler.
Case-control gene burden analysis
To minimize false positive associations due to population substructure, we performed case-control gene burden analysis on EUR samples only (n=23). Two types of controls were used: (i) 130 European samples from the 1000 Genomes Project phase 3 study; and (ii) 11 in-house EUR controls. For each gene, the number of alleles from the retained deleterious variants was counted (see Figure 1 for the different groups of retained variants). All available genotype calls in the EUR samples regardless of DP and GQ contributed to the allele count. The gene-level allele frequency was calculated for the dcSSc and control groups separately. The gene burden ratio was calculated by dividing the allele frequency in cases by the allele frequency in controls.
An enrichment of deleterious variants of a gene in the dcSSc group was identified if (i) the gene burden ratio was > 1.5 fold with both types of controls; or (ii) there were at least 3 case samples having deleterious alleles in the gene when it was not possible to calculate a gene burden ratio due to zero allele frequency in the controls. The gene burden ratio was also calculated using European dcSSc samples with and without ILD as cases and controls, respectively.
Pathway analysis
Pathway analysis was performed using g:Profiler with the hierarchical filtering of “best per parent group (strong)” and a Bonferroni corrected p < 0.05 as the significance threshold (http://biit.cs.ut.ee/gprofiler, accessed on January 30, 2016).
Results
Summary of WES data
WES data were generated from 32 dcSSc patients with a mean coverage of 51X on targeted exome regions (Supp. Table 1). An average of 81.3% of the targeted regions was covered by at least 10 reads. The in-house controls had a mean coverage of 82X on targeted exome regions and an average of 93.5% of those regions were covered by at least 10 reads (Supp. Table 2).
Deleterious variants in SSc or ILD-associated genes
A total (union) of 4,245,630 variants were identified from the 32 dcSSc samples (Figure 1). After applying the quality filters, we found 42,391 exonic, splicing and nonsynonymous variants, including a small number of stop-gain, stop-loss, frameshift, and non-frameshift indels (Supp. Table 3). Of these, 108 variants were identified as deleterious based on the “pathogenic” annotation in ClinVar, of which two are in previously reported candidate genes (group 1, Supp. Table 4, Supp. Figure 3). These include the SSc-related gene BANK1 and the ILD-related gene, TERT. An additional 2,032 variants predicted to be deleterious were identified based on the ensemble LR score. Not surprisingly, the deleterious variant in the ILD-associated gene, TERT, was found in a dcSSc patient who also had ILD (Supp. Figure 3B). For the SSc-associated BANK1 gene, the ILD patients were predominantly homozygous for the deleterious allele.
Novel SSc candidate genes enriched with deleterious variants
In order to identify novel genes and pathways that could increase our understanding of dcSSc pathogenesis, we performed a gene burden analysis to identify genes for which deleterious variants were enriched in our European dcSSc samples compared to in-house and public control samples. Seven such genes were identified (group 2 in Figure 1A and Supp. Table 5).
Rare variants are more likely than common ones to affect protein function and result in clinically-relevant consequences (8). Thus, for variants that were predicted to be deleterious and did not overlap with previously reported candidates, we grouped them into rare (MAF <1%) and common variants. We then performed a gene burden analysis for variants within each of these groups. We identified 38 genes (group 4 in Figure 1A and Supp. table 7) with rare, deleterious variants enriched in the European dcSSc samples compared to in-house and public control samples.
We also identified 18 genes with common, deleterious variants (group 6 in Figure 1A and Supp. Table 5). Further, since the functional impact of rare, deleterious variants is likely to be greater when present as two copies, we identified rare, deleterious variants that were homozygous in the European dcSSc samples and absent in the controls (group 5 in Figure 1A and Supp. Table 5). In total, we identified 2 known and 68 novel candidate genes that have not been previously associated with SSc or ILD.
We also performed a gene burden analysis on the 20 top genes in a recently published WES study of 78 scleroderma patients (9) and observed enrichment of variants for three of these genes, ASB10, KRTAP17-1 and GDF2 (Supp. Methods).
Pathway discovery
Using our methodology (Figure 1), we identified a total of 70 genes as candidates for increased risk of dcSSc (Supp. Table 5). In order to identify the associated biological pathways, we performed pathway analysis using g:Profiler and identified the extracellular-matrix related “collagen biosynthesis and modifying enzymes” pathway as significantly overrepresented (Table 2), which is consistent with the fibrotic nature of dcSSc.
Table 2.
Pathway analysis for dcSSc and ILD candidate genes.
| Pathway | p-value* | Genes |
|---|---|---|
| Based on genes identified in dcSSc and control comparison | ||
| Collagen biosynthesis and modifying enzymes | 2.38E-03 | COL4A3, COL4A4, COL5A2, COL13A1 COL22A1 |
| Based on genes only identified in ILD vs. non-ILD comparison | ||
| 2-LTR circle formation | 5.00E-02 | XRCC4 |
p values corrected for multiple testing using Bonferroni correction
Genes and pathways enriched in ILD
In order to identify variants that might predispose dcSSc patients to ILD, we repeated the variant filtration and gene burden analyses comparing European dcSSc samples with and without ILD (Figure 1B). A total of 35 genes were identified (Supp. Table 6), compared to the 70 genes identified in the case-control comparison. They formed a union of 87 genes, 18 of which were shared between the dcSSc and ILD groups while 52 and 17 genes were unique to the dcSSc and ILD groups, respectively (Supp. Table 7). Pathway analysis was performed on the 17 genes unique to the ILD list, which identified the XRCC4 DNA repair gene within the “2-LTR circle formation” pathway as significantly overrepresented (Table 2).
Discussion
Our goal for this study was to perform WES to identify potentially causal variants for dcSSc. Since ILD is a major cause of mortality in these patients, we also sought to identify variants that might contribute to ILD pathogenesis.
We first focused on identifying variants in genes previously associated with SSc and ILD (group 1 and 3 in Figure 1). For example, variants in the TERT gene have been previously associated with ILD in a GWAS (10). We identified a variant (rs34094720) in a dcSSc patient with ILD that is classified as pathogenic in ClinVar and has been associated with the autosomal recessive form of dyskeratosis congenita, which is characterized by increased risk of pulmonary fibrosis (OMIM 613989).
Consistent with previous GWAS of dcSSc (10, 11), we identified the BANK1 variant, rs10516487, as a dcSSc candidate (Figure 1A, group 1). rs10516487 was previously associated with dcSSc and systemic lupus erythematosus (10–12). This variant was found in 11 dcSSc patients and 7 in-house controls. Of interest, more dcSSc patients with ILD were homozygous for this variant (4 dcSSc-ILD patients compared to 1 control). Previous work indicates that this variant alters the binding affinity of BANK1 for the calcium channel IP3R, resulting in B-cell hyperactivity (12).
We also sought to identify novel genes or biological pathways that may increase the risk of dcSSc, including both rare, and potentially private variants, as well as common variants. To account for additive effects of multiple pathogenic variants that affect the same gene or pathway, we assessed aggregate effects of variants using gene burden and pathway analyses. Together these analyses identified 70 genes that may contribute directly to dcSSc, with or without ILD, including two genes previously implicated in SSc or ILD, and 68 novel candidates.
We also identified enrichment in variants in three of the top genes (ASB10, KRTAP17-1 and GDF2) identified in a recently published WES of SSc (9). Inclusion criteria for that study were much broader than the current study, which focused on dcSSc. For example, they included patients with limited systemic sclerosis and focused on pulmonary arterial hypertension as a severity measure. Given the heterogeneity of the disease and these differences in inclusion criteria, it is not surprising that there was not more overlap in the variants identified through WES.
Biologic pathways previously implicated in SSc pathogenesis include genes from large-scale GWAS (Supp. Table 8), interferon response or regulatory factors (IRF5 and IRF8), TGF-β, Wnt and PPAR-γ pathways (3). We identified an additional pathway based on our WES data (Table 2) that has not been implicated in the broader group of immune/autoimmune related conditions and may therefore be specific to dcSSc. Indeed, our observed enrichment of deleterious variants in genes within the extracellular matrix-related pathway in dcSSc patients is consistent with the extensive fibrosis that characterizes this disease.
We also sought to identify potential causal variants for the development of ILD among SSc patients. There is growing evidence that in spite of the overlap of some clinical features, SSc-associated ILD and idiopathic pulmonary fibrosis have different genetic contributions (14). Our pathway analyses that focused on variants enriched among dcSSc patients with ILD highlighted the XRCC4 DNA repair gene in the HIV viral infection-related “2-LTR circle formation” pathway (Table 2). Of interest, previous work implicates XRCC4 in DNA damage repair and the development of autoantibodies (15).
Although our sample size was smaller than optimal for identification of potential causal variants for SSc, we sought to limit genetic heterogeneity and increase our statistical power by focusing on dcSSc. We also applied joint genotype calling according to GATK Best Practices to increase the power of our study instead of applying the single sample calling approach that is standard for studies of this size. The main advantage of joint calling is that it resolves genotype call at every site where any individual in the cohort has evidence for a variant and therefore makes a clear distinction between a homozygous reference call versus “no call” due to insufficient data. One reported drawback of joint calling is the potential to miss a small fraction of private variants (those unique to individual samples) in low-coverage/confidence positions. We decided to take the advantage of the benefits of joint genotyping to generate unambiguous genotype calls that allowed us to produce reliable allele frequency statistics for gene burden analysis. Further, to balance specificity and sensitivity of the gene burden analysis, we first identified a relatively reliable preliminary variant list by applying a quality filter of VQSR tranche sensitivity < 99.00, DP > 10 and GQ > 20 at an early stage of variant filtering and then maximized the number of samples contributing to the allele frequency calculation.
In summary, we have performed WES to identify genetic variants that may be involved in the development of dcSSc, and/or concurrent ILD. The variants highlighted include previously implicated genes as well as novel genes and pathways. This approach significantly extends the work of GWAS and provides insight into potential disease-specific mechanisms. While additional work is required to validate these findings and define the underlying functional mechanisms, these results can help guide future efforts to elucidate the pathogenesis of SSc through additional genetic and functional studies.
Supplementary Material
Acknowledgments
The authors wish to thank the UCSF Genomics Core Facility for generating the exome sequencing data for this project, Dr. Anne Slavotinek for sharing her exome data as in-house control samples and Dr. Marquitta J. White for statistical advice.
Grants and Financial Supporters:
ACY Mak was supported by NIH training grant T32 AR007175. This project was funded by a UCSF Institute for Human Genetics pilot grant and the Scleroderma Research Foundation.
References
- 1.Mayes MD, Lacey JV, Jr, Beebe-Dimmer J, Gillespie BW, Cooper B, Laing TJ, et al. Prevalence, incidence, survival, and disease characteristics of systemic sclerosis in a large US population. Arthritis Rheum. 2003 Aug;48(8):2246–55. doi: 10.1002/art.11073. [DOI] [PubMed] [Google Scholar]
- 2.Hachulla E, Launay D. Diagnosis and classification of systemic sclerosis. Clin Rev Allergy Immunol. 2011 Apr;40(2):78–83. doi: 10.1007/s12016-010-8198-y. [DOI] [PubMed] [Google Scholar]
- 3.Korman BD, Criswell LA. Recent advances in the genetics of systemic sclerosis: toward biological and clinical significance. Curr Rheumatol Rep. 2015 Mar;17(3) doi: 10.1007/s11926-014-0484-x. 21,014-0484-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van den Hoogen F, Khanna D, Fransen J, Johnson SR, Baron M, Tyndall A, et al. 2013 classification criteria for systemic sclerosis: an American College of Rheumatology/European League against Rheumatism collaborative initiative. Arthritis Rheum. 2013 Nov;65(11):2737–47. doi: 10.1002/art.38098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, Jr, et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol. 1988 Feb;15(2):202–5. [PubMed] [Google Scholar]
- 6.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, et al. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.San Lucas FA, Wang G, Scheet P, Peng B. Integrated annotation and analysis of genetic variants from next-generation sequencing studies with variant tools. Bioinformatics. 2012 Feb 1;28(3):421–2. doi: 10.1093/bioinformatics/btr667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012 Jul 6;337(6090):100–4. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao L, Emond MJ, Louie T, Cheadle C, Berger AE, Rafaels N, et al. Whole-exome sequencing identifies rare variants in ATP8B4 as a risk factor for systemic sclerosis. Arthritis Rheumatol. 2016 Jan;68(1):191–200. doi: 10.1002/art.39449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dieude P, Wipff J, Guedj M, Ruiz B, Melchers I, Hachulla E, et al. BANK1 is a genetic risk factor for diffuse cutaneous systemic sclerosis and has additive effects with IRF5 and STAT4. Arthritis Rheum. 2009 Nov;60(11):3447–54. doi: 10.1002/art.24885. [DOI] [PubMed] [Google Scholar]
- 11.Rueda B, Gourh P, Broen J, Agarwal SK, Simeon C, Ortego-Centeno N, et al. BANK1 functional variants are associated with susceptibility to diffuse systemic sclerosis in Caucasians. Ann Rheum Dis. 2010 Apr;69(4):700–5. doi: 10.1136/ard.2009.118174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008 Feb;40(2):211–6. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 13.Rossi S, Testa F, Attanasio M, Orrico A, de Benedictis A, Corte MD, et al. Subretinal Fibrosis in Stargardt's Disease with Fundus Flavimaculatus and ABCA4 Gene Mutation. Case Rep Ophthalmol. 2012 Sep;3(3):410–7. doi: 10.1159/000345415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herzog EL, Mathur A, Tager AM, Feghali-Bostwick C, Schneider F, Varga J. Review: interstitial lung disease associated with systemic sclerosis and idiopathic pulmonary fibrosis: how similar and distinct? Arthritis Rheumatol. 2014 Aug;66(8):1967–78. doi: 10.1002/art.38702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palomino GM, Bassi CL, Wastowski IJ, Xavier DJ, Lucisano-Valim YM, Crispim JC, et al. Patients with systemic sclerosis present increased DNA damage differentially associated with DNA repair gene polymorphisms. J Rheumatol. 2014 Mar;41(3):458–65. doi: 10.3899/jrheum.130376. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.