Progranulin (PGRN) protein encoded by the granulin (GRN) gene has been recently implicated in several neurodegenerative diseases [2,5]. While haploinsufficiency of PGRN leads to frontotemporal lobar degeneration (FTLD) [2,5], the most prevalent form of early onset dementia after Alzheimer’s disease (AD), complete loss of PGRN is known to cause neuronal ceroid lipofuscinosis (NCL) [1,13], a group of lysosomal storage diseases. PGRN is a secreted glycoprotein of 7.5 granulin repeats [2,5]. However, within the cell, PGRN is localized to lysosomes through two independent trafficking pathways [8,17]. Furthermore, GRN is transcriptionally co-regulated with a number of essential lysosomal genes by the transcriptional factor TFEB [3]. While all this evidence suggests an essential role of PGRN in regulating lysosomal function, how PGRN does so is still unclear.
Cathepsin D (CTSD) is a lysosomal aspartic-type protease involved in many neurodegenerative diseases [14]. Mutations in the cathepsin D gene (CTSD) result in NCL in humans [9]. Interestingly, mice deficient in CTSD also develop TDP-43 aggregates (Supplementary Fig. 1) [7], a hallmark of FTLD with GRN mutations. FTLD patients with GRN mutations exhibit typical pathological features of NCL [7]. These data support that lysosomal dysfunction might serve as a common mechanism for FTLD and NCL and suggest that PGRN and CTSD might function together to regulate lysosomal activities. In support of this hypothesis, granulin motifs are found in cathepsin-like cysteine proteases in plants [11,16]. These lines of evidence led us to postulate that an interaction between PGRN and CTSD may exist.
To test the physical interaction between PGRN and CTSD, FLAG-tagged CTSD was co-transfected with untagged PGRN in HEK293T cells. Cell lysates were immunoprecipitated with anti-FLAG antibodies. PGRN signal is detected in anti-FLAG CTSD immunoprecipitates but not in the controls (Fig. 1a), suggesting a physical interaction between PGRN and CTSD. Since CTSD is known to interact with prosaposin (PSAP) [6,10], which we previously showed to bind to PGRN as well [17], it is possible that the PGRN and CTSD interaction might be bridged by endogenous PSAP in HEK293T cells. To rule out this possibility, we compared the interaction between PGRN and CTSD in control N2a cells or N2a cells with PSAP expression depleted using the CRISPR/Cas9 system [17]. PSAP ablation has no effect on PGRN-CTSD binding in the co-immunoprecipitation assay (Fig. 1b), indicating that the interaction between PGRN and CTSD is not mediated by PSAP. Co-immunoprecipitation studies between individual granulins and CTSD suggest that multiple granulin motifs interact with CTSD (Supplemental Fig. 2a and 2b).
Fig. 1.
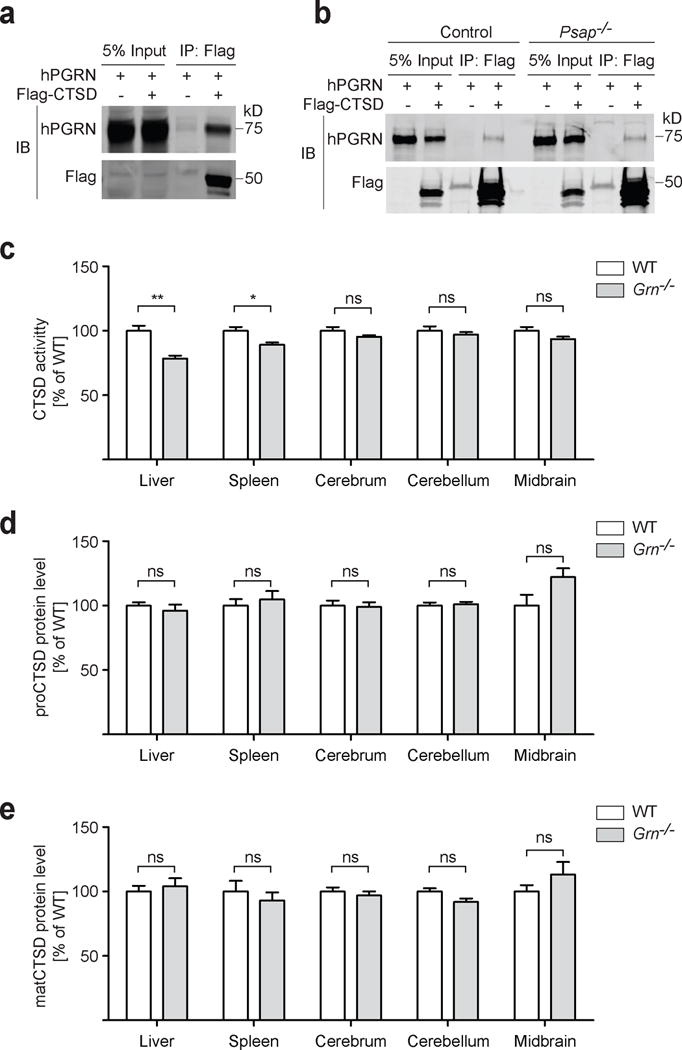
PGRN binds to CTSD and regulates its activity. a HEK293T cells transfected with indicated constructs were lysed and immunoprecipitated with anti-FLAG antibodies. b Control N2a cells or Psap−/−N2a cells generated using Cas9/CRIPSR were transfected with indicated constructs, lysed and immunoprecipitated with anti-FLAG antibodies. c, CTSD activities in tissue lysates of 2 month old WT or Grn−/− mice as indicated. d, e, The levels of both the pro (d) and mature (e) forms of cathepsin D in the tissue lysates of WT or Grn−/− mice are quantified and normalized to GAPDH. n=5–6, +/− SEM, *p-value <0.05, **p-value <0.01, ns, not significant, Student’s t-test.
With the physical interaction between PGRN and CTSD confirmed, next we wanted to investigate its functional relevance. Since CTSD deficiency results in much more severe lysosomal phenotypes than PGRN deficiency [4], we hypothesized that PGRN might regulate CTSD activities. Therefore, we measured CTSD activities in 2-month-old wild type (WT) and PGRN deficient mice before the appearance of any obvious lysosomal abnormalities or glial activation. Indeed, liver and spleen lysates from PGRN-deficient mice showed significantly lower CTSD activities compared to those from WT mice (Fig. 1c), without any changes in CTSD protein levels or maturation status (Fig. 1d, 1e, Supplementary Fig. 3). Lysates from cerebrum and cerebellum also show a trend of lower CTSD activities in Grn−/− mice (Fig. 1c). Notably, in Grn−/− midbrain, although the protein levels of CTSD are slightly increased, CTSD activities are slightly lower than WT, indicating that the midbrain is one of earliest affected brain regions in Grn−/− mice (Fig. 1c–e). It should be noted that Grn−/− mice do not develop TDP-43 pathology and neurodegeneration as seen in FTLD patients [12]. Thus it is possible that PGRN more strongly regulates CTSD activity in humans than in mice. Indeed, a recent study demonstrated reduced cathepsin D activity in fibroblasts derived from FTLD patients with heterozygous GRN mutations [15].
In summary, we demonstrate that PGRN interacts with the lysosomal protease CTSD and maintains its proper activity in vivo. CTSD mediated proteolysis is essential to neuronal cell homeostasis through the degradation of aggregates delivered to lysosomes via autophagy or endocytosis [14]. Therefore by regulating CTSD activity, PGRN may modulate protein homeostasis. This could potentially explain the TDP-43 aggregation observed in FTLD with GRN mutations. Although the mechanism by which PGRN regulates CTSD activity remains to be determined, our data argues that reduced CTSD activities are a disease mechanism for FTLD with GRN mutations.
Supplementary Material
Acknowledgments
We thank Dr. Paul Saftig and Dr. Jianhua Zhang for Ctsd+/− mice, Dr. Haiyuan Yu for his kind gift of Cathepsin D cDNA and Mrs. Xiaochun Wu for technical assistance. This work is supported by funding to F.H. from Weill Institute for Cell and Molecular Biology and NINDS (R01NS088448) and by funding to X. Z. from the Weill Institute Fleming Postdoctoral Fellowship.
Footnotes
Conflict of interest: The authors declare no competing conflict of interest.
References
- 1.Almeida MR, Macario MC, Ramos L, Baldeiras I, Ribeiro MH, Santana I. Portuguese family with the co-occurrence of frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis phenotypes due to progranulin gene mutation. Neurobiol Aging. 2016;41:200 e201–205. doi: 10.1016/j.neurobiolaging.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 2.Bateman A, Bennett HP. The granulin gene family: from cancer to dementia. Bioessays. 2009;31:1245–1254. doi: 10.1002/bies.200900086. [DOI] [PubMed] [Google Scholar]
- 3.Belcastro V, Siciliano V, Gregoretti F, Mithbaokar P, Dharmalingam G, Berlingieri S, Iorio F, Oliva G, Polishchuck R, Brunetti-Pierri N, di Bernardo D. Transcriptional gene network inference from a massive dataset elucidates transcriptome organization and gene function. Nucleic Acids Res. 2011;39:8677–8688. doi: 10.1093/nar/gkr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carcel-Trullols J, Kovacs AD, Pearce DA. Cell biology of the NCL proteins: What they do and don’t do. Biochim Biophys Acta. 2015;1852:2242–2255. doi: 10.1016/j.bbadis.2015.04.027. [DOI] [PubMed] [Google Scholar]
- 5.Cenik B, Sephton CF, Kutluk Cenik B, Herz J, Yu G. Progranulin: a proteolytically processed protein at the crossroads of inflammation and neurodegeneration. J Biol Chem. 2012;287:32298–32306. doi: 10.1074/jbc.R112.399170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gopalakrishnan MM, Grosch HW, Locatelli-Hoops S, Werth N, Smolenova E, Nettersheim M, Sandhoff K, Hasilik A. Purified recombinant human prosaposin forms oligomers that bind procathepsin D and affect its autoactivation. Biochem J. 2004;383:507–515. doi: 10.1042/BJ20040175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gotzl JK, Mori K, Damme M, Fellerer K, Tahirovic S, Kleinberger G, Janssens J, van der Zee J, Lang CM, Kremmer E, Martin JJ, Engelborghs S, Kretzschmar HA, Arzberger T, Van Broeckhoven C, Haass C, Capell A. Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol. 2014;127:845–860. doi: 10.1007/s00401-014-1262-6. [DOI] [PubMed] [Google Scholar]
- 8.Hu F, Padukkavidana T, Vaegter CB, Brady OA, Zheng Y, Mackenzie IR, Feldman HH, Nykjaer A, Strittmatter SM. Sortilin-mediated endocytosis determines levels of the frontotemporal dementia protein, progranulin. Neuron. 2010;68:654–667. doi: 10.1016/j.neuron.2010.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ketterer S, Gomez-Auli A, Hillebrand LE, Petrera A, Ketscher A, Reinheckel T. Inherited diseases caused by mutations in cathepsin protease genes. FEBS J. 2016 doi: 10.1111/febs.13980. [DOI] [PubMed] [Google Scholar]
- 10.Laurent-Matha V, Lucas A, Huttler S, Sandhoff K, Garcia M, Rochefort H. Procathepsin D interacts with prosaposin in cancer cells but its internalization is not mediated by LDL receptor-related protein. Exp Cell Res. 2002;277:210–219. doi: 10.1006/excr.2002.5556. [DOI] [PubMed] [Google Scholar]
- 11.Palfree RG, Bennett HP, Bateman A. The Evolution of the Secreted Regulatory Protein Progranulin. PLoS ONE. 2015;10:e0133749. doi: 10.1371/journal.pone.0133749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberson ED. Mouse models of frontotemporal dementia. Ann Neurol. 2012;72:837–849. doi: 10.1002/ana.23722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith KR, Damiano J, Franceschetti S, Carpenter S, Canafoglia L, Morbin M, Rossi G, Pareyson D, Mole SE, Staropoli JF, Sims KB, Lewis J, Lin WL, Dickson DW, Dahl HH, Bahlo M, Berkovic SF. Strikingly different clinicopathological phenotypes determined by progranulin-mutation dosage. Am J Hum Genet. 2012;90:1102–1107. doi: 10.1016/j.ajhg.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vidoni C, Follo C, Savino M, Melone MA, Isidoro C. The Role of Cathepsin D in the Pathogenesis of Human Neurodegenerative Disorders. Med Res Rev. 2016;36:845–870. doi: 10.1002/med.21394. [DOI] [PubMed] [Google Scholar]
- 15.Ward ME, Chen R, Huang HY, Ludwig C, Telpoukhovskaia M, Taubes A, Boudin H, Minami SS, Reichert M, Albrecht P, Gelfand JM, Cruz-Herranz A, Cordano C, Alavi MV, Leslie S, Seeley WW, Miller BL, Bigio E, Mesulam MM, Bogyo MS, Mackenzie IR, Staropoli JF, Cotman SL, Huang EJ, Gan L, Green AJ. Individuals with progranulin haploinsufficiency exhibit features of neuronal ceroid lipofuscinosis. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aah5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada K, Matsushima R, Nishimura M, Hara-Nishimura I. A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 2001;127:1626–1634. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou X, Sun L, Bastos de Oliveira F, Qi X, Brown WJ, Smolka MB, Sun Y, Hu F. Prosaposin facilitates sortilin-independent lysosomal trafficking of progranulin. J Cell Biol. 2015;210:991–1002. doi: 10.1083/jcb.201502029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


