Abstract
Precise targeting of genetic lesions alone has been insufficient to extend brain tumor patient survival. Brain cancer cells are diverse in their genetic, metabolic, and microenvironmental compositions, accounting for their phenotypic heterogeneity and disparate responses to therapy. These factors converge at the level of the epigenome, representing a unified node that can be disrupted by pharmacologic inhibition. Aberrant epigenomes define many childhood and adult brain cancers, as demonstrated by widespread changes to DNA methylation patterns, redistribution of histone marks, and disruption of chromatin structure. In this review, we describe the convergence of genetic, metabolic, and micro-environmental factors upon mechanisms of epigenetic deregulation in brain cancer. We discuss how aberrant epigenetic pathways identified in brain tumors affect cell identity, cell state, and neoplastic transformation, in addition to the potential to exploit these alterations as novel therapeutic strategies for the treatment of brain cancer.
INTRODUCTION
Brain tumors encompass a wide spectrum of over 120 histologically, demographically, clinically and molecularly distinct diseases1, and are one of the most common causes of cancer-related death in children and adults. Genome-sequencing studies have uncovered the landscape of genetic alterations present in many pediatric and adult cancer types2, and highlights a convergence on deregulated epigenomes in the form of aberrant DNA methylation signatures, histone modification patterns, and disorganized chromatin architecture3–7. In adult glioblastoma (GBM, World Health Organization grade IV glioma), the most aggressive and prevalent adult primary intrinsic brain cancer, nearly 46% of patients harbor at least one mutation of an epigenetic regulator amidst a diversity of oncogenic pathway mutations8. Equally striking is the pediatric counterpart of glioblastoma where one highly prevalent mutation occurs in a histone protein9. Somatic mutations and structural variations that target regulators of epigenetic modifications and functional regulatory elements have been reported across several aggressive pediatric and adult brain cancers such as glioblastoma 5, 8–10, medulloblastoma 6, 11–18, ependymoma19, atypical teratoid rhabdoid tumors (ATRT)20, 21, diffuse intrinsic pontine gliomas (DIPG) 22–27, and embryonal tumors with multilayered rosettes (ETMR) 28. The function of these epigenetic alterations is likely context dependent, but ultimately influences cell identity and cell state transitions during neoplastic transformation (Figure 1). Brain cancer cells are not only heterogeneous in their genetic composition, but also reside in varying microenvironments and interact with different cell types. Therefore, factors such as altered cellular metabolism and the microenvironment may critically define the neoplastic effects of epigenetic programs in the process of brain tumor development7, 29–41. In this review, we will detail the collective genetic, metabolic, and microenvironmental alterations present during brain tumorigenesis, and discuss the impact these changes have upon epigenetic programs important for cell state transition or maintenance. Further, we will highlight the therapeutic potential of targeting brain tumor cell state by modulation of epigenetic signatures.
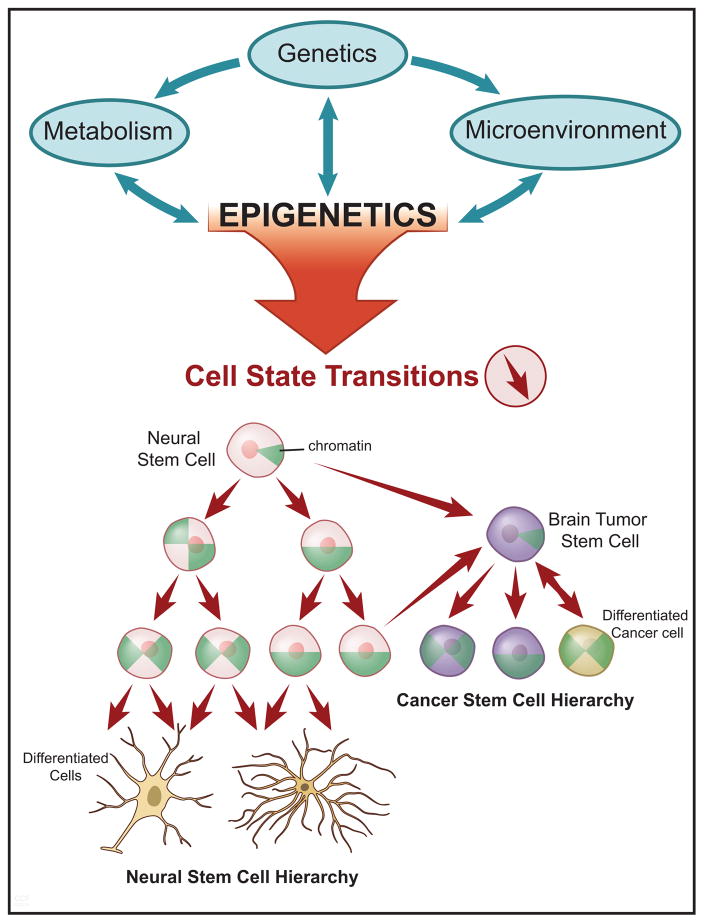
Figure 1. The Epigenetic Gateway to Cell Identity and Neoplastic Transformation.
A schematic depicting the genetic, metabolic, and microenvironmental interactions (green arrows) with epigenetic programs in cancer (top panel). In the lower panel, a diagram illustrating the cell state transitions (red arrows) influenced by altered epigenetic landscapes and their relevance to both normal neural stem cell, and cancer stem cell hierarchies (lower panel). Within the cells are green pie-shaped triangles, which represent the restructuring of chromatin architecture and progression towards closed chromatin in the most differentiated cell state.
The Epigenetic Gateway to Cell Identity and Neoplastic Transformation
Cancer cells are characterized by a state of uncontrolled proliferation and replicative immortality42. The epigenetic landscape defines cell state, supporting epigenetic control as an essential node of transformation. It is now clear based on Nobel prize-winning work of Shinya Yamanaka43 and many others, that the state of a cell is dynamic and more plastic than previously thought. Various studies demonstrating direct cell conversion to specific lineages, including multiple types of neural progenitors that are the putative cell of origin of many brain tumors highlight the ability of cells to transform their state with the introduction of only a few transcription factors44–46. Cancer cells capitalize on this cellular plasticity to acquire developmental programs that endow upon the cell limitless self-renewal capacity, similar to that of reprogrammed induced pluripotent stem cells (iPSCs) and neural stem cells. In fact, there are close parallels between cellular reprogramming and oncogenic transformation. Yamanaka transcription factors, including SOX2 and MYC 47–49, and many of the epigenetic modifier genes that are necessary for cellular reprogramming have an oncogenic role in cancer (reviewed in 50). Suva et al. demonstrated, similar to direct conversion of non-transformed cells that they could reprogram a differentiated cancer cell into a tumor-propagating cell (i.e. satisfying a key functional criterion for glioma brain tumor stem cells (BTSCs)) with four master transcription factors (POU3F2, SOX2, SALL2 and OLIG2)48. Restoring, at least in part, the epigenome of a native BTSC was necessary to regain tumorigenic potential, supporting the concept that epigenomic programs define the cancer cell state. Resetting the epigenetic landscape of BTSCs using a method similar to iPSC reprogramming, and given external cues to set up an epigenetic program distinct from brain tissue attenuates tumor formation51, 52. While these studies and others demonstrate in a laboratory setting that epigenetic regulation can drive or inhibit cancer growth, human tumors are not formed from the exogenous introduction of transcription factors. Tumors, in particular brain tumors, are heterogeneous at the single cell level and organized in a hierarchical structure composed of cells with varying cell states53, 54. Genetic alterations, signaling alterations, metabolic alterations and microenvironmental conditions converge to dictate the epigenetic landscape of individual cells (Figure 1). This landscape, in turn, defines cell state and influences cell signaling, metabolism, the microenvironment and even the genetic landscape15, 55–58. Molecular alterations within cancer cells promote cancer growth, but multiple deregulated pathways may converge to create an oncologic epigenome: the altered epigenome may lock cells in a stem-like state, inhibiting normal differentiation19, 53, 59–61. In concert, tumor epigenomes inhibit tumor suppressor gene expression, drive oncogenic activation, and further render the cell of origin susceptible to neoplastic transformation2, 55–57, 62.
Convergence on Chromatin Architecture
Characterization of histone modifications and their role in normal cellular function has provided insight into the potential mechanisms of epigenetic de-regulation in brain cancer63, 64. Octamers of histone proteins are responsible for wrapping 147 base pair units of double-stranded DNA into compacted subunits called nucleosomes. Post-translational modification of histones by methylation, acetylation, phosphorylation, sumoylation, and ubiquitylation etc., instruct states of euchromatin and heterochromatin (As reviewed in 65). Histone modifications further define distinct regions of the epigenome such as enhancers, promoters, and gene bodies (Figure 2). Modifications of histone amino acid residues are mediated by enzymes (‘epigenetic writers’), such as histone methyltransferases (i.e. Enhancer of Zeste Homolog 2, EZH2) and acetyltransferases (i.e. P300/CREB-binding protein, CBP), which catalyze the addition of methyl or acetyl groups, and histone demethylases (i.e. Jumonji Domain Containing 3, JMJD3) and deacetylases (i.e. histone deacetylases, HDACs), which facilitate their removal (‘epigenetic erasers’). Proteins that recognize histone modifications, known as histone ‘readers’, recruit additional proteins and protein complexes that facilitate transcriptional regulation. The organization of larger-scale chromatin structure is regulated by chromatin remodelers and chromatin associated proteins. In brain cancer, mutations have been identified at nearly all levels of chromatin regulation from mutations of histones, to enzymes that catalyze histone modification, to proteins that facilitate larger-order chromatin structure (Figure 2).
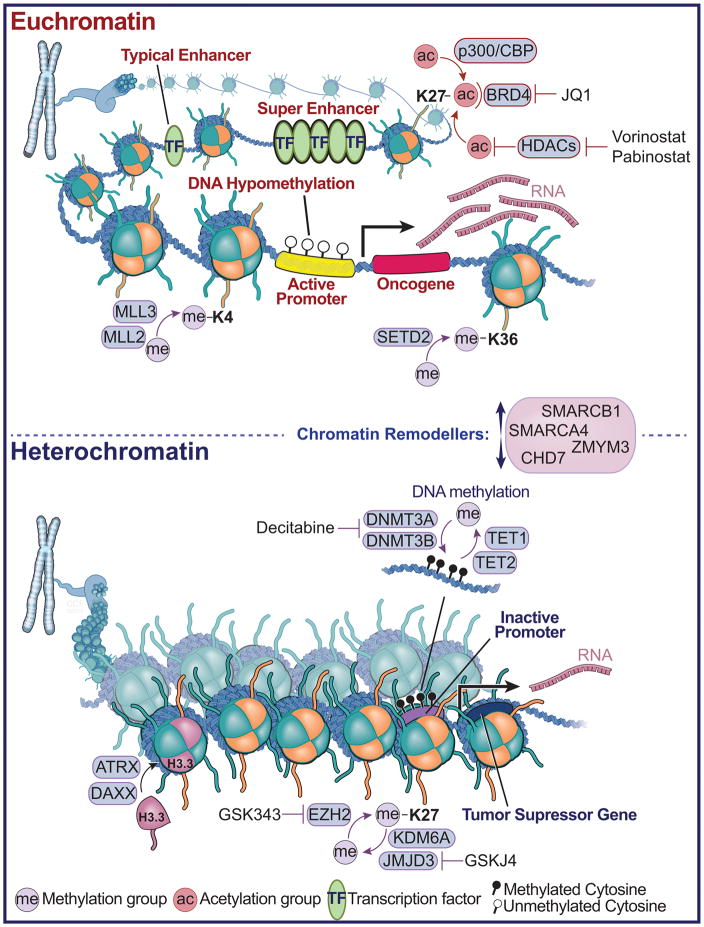
Figure 2. Brain Tumors Converge on Chromatin Architecture.
A diagram depicting euchromatin and histone modifications that mediate ‘active’ transcription in cancer cells (top panel). Shown are various histone modifications and enzymes, which catalyze the addition of post-translational modifications such as histone methylation and acetylation, or bind to these modifications, such as the BRD4, which binds acetylated lysine residues on histones. The green ovals represent transcription factor binding sites and locations of enhancers, or clusters of enhancers, termed super-enhancers. Also shown are drug compounds, which inhibit the removal (Vorinostat and Pabinostat - Histone deacetylase (HDAC) inhibitor) or detection of acetylation (JQ1). Shown in the middle are the chromatin remodelers, which facilitate the landscape of higher order chromatin structure towards euchromatin or heterochromatin. In the lower panel, heterochromatin is depicted and the associated modifications that mediate tumor suppressor gene silencing. These include the DNA methyltransferase family of enzymes, which catalyze the addition of methyl groups to cytosine - guanine di-nuclueotides, and TET enzymes, which facilitate DNA de-methylation through 5-methyl cytosine hydroxylation. Also shown is EZH2, which methylates at histone H3 at the 27th position, and the associated histone H3K27 de-methylases KDM6A and JMJD3. Shown are chemical inhibitors that reverse the methylation marks deposited or removed by these methyltransferase and demethylase enzymes related to heterochromatin (Decitabine, GSK343, GSKJ4). ATRX and DAXX are depicted which function to incorporate the histone H3.3 variant, and which are frequently mutated in pediatric high-grade glioma.
Childhood Tumors Highlight Epigenetic Dependencies in Brain Cancer
An emerging theme in brain cancer sequencing studies is the relative decreased prevalence of mutations observed in childhood versus adult brain tumors5, 6, 8, 16, 17, 19, 20, 66–71. This holds true for other pediatric cancers such as infant leukemia, neuroblastoma, and retinoblastoma, which exhibit lower mutation rates as compared to highly mutated adult tumors, such as melanoma and lung cancer17, 19, 66–68, 70, 71. Of the few recurrent mutations identified in brain cancer genomes, many target chromatin associated proteins or histone proteins themselves, termed ‘landscaping genes’, due to their potential widespread effects on transcriptional programs4–9, 12, 13, 15, 17, 19, 28, 69. ATRTs harbor remarkably silent genomes, yet exhibit recurrent mutations or deletions of the SMARCB1 gene (SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily B) 20, 21, 72 (Figure 2). SMARCB1 encodes a subunit of the SWI/SNF chromatin-remodeling complex, which functions as a tumor suppressor protein that is highly mutated in several cancers73. Homozygous deletion of SMARCB1 in mice leads to embryonic lethality, while heterozygous loss leads to aggressive tumors that recapitulate human rhabdoid tumors74–76. It is important to note that SMARCB1 loss is deleterious to a vast majority of cells, and mutation within an exclusive cellular and developmental context leads to neoplastic transformation77. As shown in Drosophila neuroblasts, proper lineage specification by the SWI/SNF component Osa (ARID1) prevents tumorigenesis, by restricting self-renewal and inhibiting de-differentiation78. Two groups recently described the genetic landscape of another aggressive pediatric brain tumor, ependymoma, in which hindbrain tumors exhibit no recurrent mutations in coding space and no evidence of recurrent gene fusions or focal somatic copy number alterations19, 79, 80. This was in contrast to its direct adult ependymoma counterpart, which harbored widespread genomic instability81. The DNA methylome of infant hindbrain ependymoma displays aberrant DNA hypermethylation at CpG islands, described as a CpG Island Methylator Phenotype (CIMP). Importantly, hypermethylated genes converged upon embryonic stem cell (ESC) targets regulated by the Polycomb Repressor Complex 2 (PRC2), suggesting that epigenomic alterations could be disrupting cell state and differentiation programs important to ependymoma development. A link between ESC programs and cancer is further demonstrated in the embryonal brain tumor ETMR, which harbors a fusion between a highly amplified microRNA cluster (C19MC) and TTYH1 (Tweety family member 1)28, 82, 83. A downstream consequence of the fusion is aberrant overexpression of a novel DNA methyltransferase 3B (DNMT3B) isoform normally and exclusively expressed in the first weeks of neural tube development. Observations in ATRT, ependymoma, and ETMR, alongside several other cancers, suggest that neoplastic transformation is a process dependent on proper maintenance of stem cell programs through tight chromatin regulation. While these aberrant epigenetic events have been observed through genome-wide approaches, future validation will be needed to model these alterations during the initiation and progression processes of brain tumorigenesis.
Mutations of Histone Proteins
Recurrent genetic lesions linking epigenomic programs to brain tumor formation is perhaps best exemplified in pediatric glioblastoma and DIPG, which harbor frequent mutation of H3F3A, encoding the H3.3 histone variant, and to a lesser extent HISTH1B and HISTH1C, encoding the H3.1 variant9, 22, 23, 26, 84–87. These mutations target the histone H3 lysine 27th position (K27M), a direct site important for epigenetic post-translational modifications, and the neighboring residue G34R or G34V, which is thought to affect a nearby lysine residue at the 36th position (H3K36)10 (Figure 2). The H3.3 K27M mutation affects a site of post-translational modification and is associated with global decreased K27 methylation and increased K27 acetylation88. Further, the K27M mutant results in aberrant redistribution of residual patterns of H3K27me3 within the tumor epigenome85, 86. ESC-derived neural precursor cells (NPCs) can be transformed with a combination of H3.3-K27M over-expression, shRNA knockdown of TP53, and over-expression of PDGFRA (platelet-derived growth factor receptor A)89. Importantly, ESCs and terminally differentiated cells are resistant to transformation, suggesting that the effect of the K27M mutation is highly restricted to a cell type occurring within a defined NPC population during embryonic development. The temporally and anatomically distinct tumors defined by K27M and G34R/V mutations, suggest unique cells of origin and/or cell states that are required for tumor initiation87. Mutations have also been reported in the proteins that facilitate histone H3.3 incorporation, such as alpha thalassemia/mental retardation syndrome X-linked (ATRX) and death-domain associated protein (DAXX)9, 90. The significance and functional characterization of these mutations in the setting of epigenomic reprogramming remains an area of active and future investigation.
Mutations of Histone Modifiers
Enzymes that catalyze the addition or removal of modifications are recurrently mutated, amplified, or deleted in brain cancer genomes. These include MLL2 and MLL3 (mixed-lineage leukemia 2/3 in medulloblastoma and adult glioblastoma)6, 16, 17, 69, SMARCB1 (ATRT)20, 21, SMARCA4 (glioblastoma, medulloblastoma, ATRT)5, 8, 9, 14, 16, 17, 69, 87, 91, and SETD2 (SET domain containing 2 in both pediatric and adult glioblastoma)91 mutations occurring in a diverse set of adult and pediatric brain tumors (Figure 2). Whole-exome and whole-genome sequencing studies of medulloblastoma have revealed the most commonly mutated chromatin modifier to be MLL2, which mediates histone H3 lysine 4 (H3K4me3) tri-methylation, a mark of active transcription6, 11, 14, 16, 17. Further, the histone lysine 27 demethylase, KDM6A, is recurrently mutated, and associated with increased H3K27me3 levels in a group of medulloblastomas with a poor prognosis (Group 4), which also overexpress EZH2. Poor prognosis medulloblastomas (Groups 3 and 4, which are not driven by sonic hedgehog and wnt signaling) also harbor subgroup-associated mutations in CHD7 (chromodomain helicase DNA binding protein 7) and ZMYM3 (zinc finger, MYM-type 3), which converge on regulation of gene expression by H3K4me3. Given the role of H3K27me3 in repressing lineage specific genes in stem cells, it is hypothesized that Group 3 and 4 medulloblastomas retain stem-like signatures through accumulation of H3K27me3 and abrogation of H3K4me3 mediated transcription. Interestingly, these alterations are in contrast to the global loss of H3K27me3 levels in pediatric glioblastoma, and perhaps suggest that perturbation of a global balance and/or distribution of H3K27me3 and H3K4me3 patterns may reflect cell state specific dependencies in neoplastic transformation. A major effort moving forward will be functional characterization of these epigenetic alterations and identification of specific developmental cell types where their epigenetic deregulation promotes tumor formation.
Genomic Regulatory Elements of Brain Tumors
The convergence on histone modifications and chromatin regulation highlights the importance of understanding and mapping these modifications in brain tumors. In tumors, such as pediatric glioblastoma and ependymoma, histone modification mapping by chromatin immunoprecipitation followed by high density sequencing (ChIP-seq) has demonstrated aberrant epigenetic patterns of histone H3K27 tri-methylation10, 19, 85. The linkage between epigenetic modifications and cell identity and lineage specification underscores the importance of understanding the epigenetic landscape in brain cancer. Recent studies have highlighted the importance of clusters of enhancer elements, termed ‘Super-Enhancers’, which both identify and regulate genes involved in cell identity and disease92 (Figure 2). These epigenomic features can be co-opted in cancer by mutations and structural variations93. In Group 3 medulloblastoma, Super-Enhancers are hijacked by structural variations, which lead to aberrant activation of GFI1 and GFI1b (growth factor independent 1 transcription repressor) oncogenes12. In several brain tumors, non-coding mutations have been observed in the promoter regions of TERT (telomerase reverse transcriptase, which encodes the catalytic subunit of the enzyme telomerase), which are enriched in tumors characterized by low-rates of self-renewal94, 95. The consequence of these mutations in glioblastoma is the aberrant recruitment of the GABP (GA Binding Protein) transcription factor96. Future in depth sequencing of non-coding regions and integration with histone modification and transcription factor maps may uncover crucial genes that maintain cell state and the factors that govern their expression.
Altered DNA Methylation Patterns in Brain Cancer
Changes in DNA methylation patterns have been widely reported in cancer in the form of DNA hyper-methylation and silencing of tumor suppressor genes, and loss of methylation of oncogenes and repetitive elements97. To date, genome-wide studies focusing largely on promoter regions and CpG islands, have revealed novel mechanisms of oncogenic and tumor suppressor gene regulation in cancer. Examples include widespread accumulation of DNA methylation in IDH1 (Isocitrate dehydrogenase 1) mutated gliomas (see metabolism section below)39, 98, and the establishment of CIMP phenotypes in other tumors, such as ependymoma (Figure 2). Further, an important application of DNA methylation profiling is to identify signatures associated with genetic lesions, and the use of DNA methylation as a method for robust molecular stratification8, 19, 21, 87. It is also posited that DNA methylation patterns may reflect the specific cellular states and/or cells of origin present during transformation. Advances in our understanding of the epigenomic landscapes of normal human and murine neural stem cells and cellular hierarchies may shed light upon the potential cell identity and cell state transitions that occur in the early stages of brain tumor initiation. Technological advances have also allowed for genome-wide characterization of brain tumor DNA methylomes using whole-genome bisulfite sequencing (WGBS). Early WGBS studies have revealed novel mechanisms of transcriptional regulation in medulloblastoma and ependymoma, and have provided an integrated view of DNA methylation and histone modification landscapes in brain tumors15, 19.
Epigenetic Perturbation of Genetic Landscapes
In addition to influences on cell state, epigenetic alterations have been shown to have widespread effects on the genetic landscape of tumor cells. For example, methylated cytosine bases are highly prone to mutation by spontaneous deamination to thymine, thus creating opportunities for deregulation of tumor suppressor genes and oncogenes in the absence of intact DNA repair mechanisms99. Furthermore, it has been shown that hypomethylation of transposable elements have been observed widely in cancer, and may contribute significantly to genomic instability through aberrant translocation of DNA sequences100. At the chromatin level a direct association between histone modifications and genetic alteration is evidenced in tumors that overexpress the H3K9/36me3 lysine demethylase KDM4A/JMJD2A, which leads to regional DNA copy gain in the absence of global chromosomal instability58. This illustrates a scenario in which aberrant chromatin modulator expression could establish somatic copy number changes during neoplastic transformation58. From cancer genome sequencing studies, evidence is emerging that links regional mutation density with the degree of heterochromatin as marked by H3K9me356. These findings demonstrate that somatic mutations are not distributed uniformly across the human genome, and are associated with epigenomic topographies derived from the most likely cell type of origin and cell state during malignant transformation55.
Cellular Microenvironment Influences Epigenetic State of Brain Tumor Cells
Brain tumor cells do not exist in isolation, but are part of a dynamic and spatially distributed system, interacting with a wide-diversity of environments and cell types. For example, active neuronal activity promotes mitosis of the putative cells of origin in high-grade glioma through NLGN3 (Neuroligin 3) secretion101. Brain tumor stem cells (BTSCs), in particular, exhibit a complex relationship with their microenvironment: they can actively modify and shape their own environment but are also regulated, supported, and directed by microenvironmental signals (Figure 3). This intricate crosstalk is crucial to maintain a stem cell state and occurs within a localized, supportive microenvironment around the stem cells called a niche. There are a multitude of factors within the stem cell niche that affect the cellular state of brain tumor cells including nutrient availability, hypoxia, pH, and cell-cell interactions. In other systems, stem cell state maintenance and cell state change or differentiation are governed epigenetically.102 So far little is known at the mechanistic level as to how niche cues regulate brain tumor epigenetics. However, a number of studies have revealed how external environmental cues functionally change brain tumor cell state through unexplored epigenetic mechanisms.
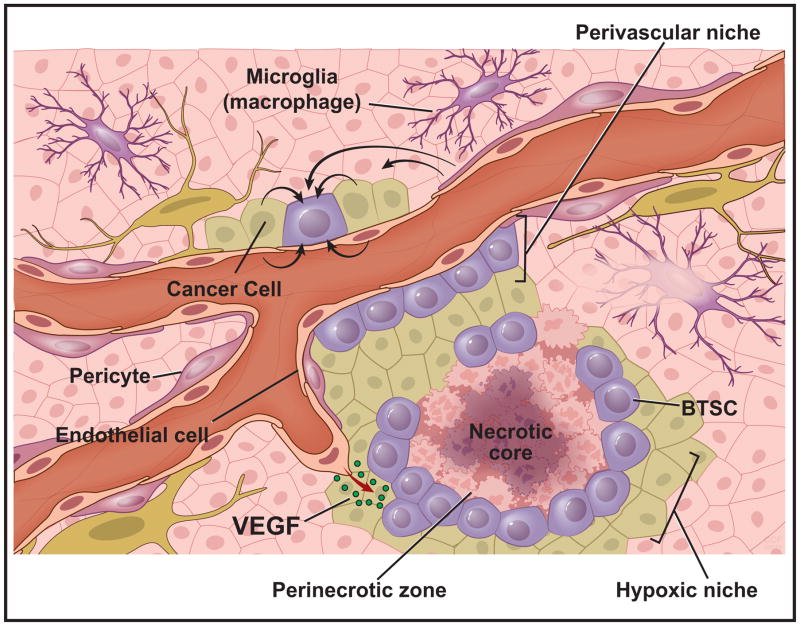
Figure 3. Cellular Microenvironment Influences Epigenetic State of Brain Tumor Cells.
An illustration of the brain tumor microenvironment highlighting the perivascular and hypoxic niches, which dictate interacting cell types and nutrient availabilities. Both cancer cells (light green) and brain tumor cells (purple-round) exist in dynamic microenvironments containing exogenous signals from surrounding microglia (purple), pericytes (dark pink), endothelial cells (light pink), and other neoplastic cells. These interactions occur in the presence of variable growth factor gradients (ie. VEGF), oxygen availability, and nutrient levels (i.e. glucose, acetate, glutamine etc.).
The Hypoxic Niche
Areas of hypoxia and necrosis can be a diagnostic feature of many malignant tumors, including glioblastoma. Historically, this has been hypothesized as the expected occurrence when a tumor’s growth outpaces its blood supply, leaving behind starved and/or dying cells, but recent studies have revealed that micro- and macro- cellular relationships within a tumor’s hypoxic niche are far more complex. Many normal adult stem cell niches as well many steps of embryonic development are naturally hypoxic.103 Hypoxic niche support of stem cells may be a conserved feature of development, normal tissue maintenance, and cancer. Although cells in nutrient-rich environments have the resources to facilitate rapid proliferation and tumor growth, it may be the cells within the hypoxic niche that actually drive tumor progression and recurrence due to the stem-like transcriptional and epigenetic adaptations they undergo in this environment (Figure 3).
The direct molecular responses of brain tumor cells to hypoxia are principally mediated by the hypoxia-inducible factor (HIF) family of transcription factors, especially HIF1α and HIF2α.104 In glioblastoma biopsies, BTSCs are enriched in peri-necrotic regions in the context of HIF activation.105 A number of studies have demonstrated that hypoxia directly mediates expansion of the BTSC pool and that this is dependent on HIF1 and HIF2. 31, 106 However, whereas HIF1α appears generally necessary for glioma survival in hypoxia, HIF2α is specifically necessary to sustain BTSC.31 This may be mediated through HIF2 enhancement of MYC transcriptional activity30 that is required for BTSC maintenance and proliferation.107
Little is known about the direct epigenetic consequences of hypoxia and HIF activation in brain cancer, but exploration of this field is beginning. In NPCs of the developing brain, HIF1α interacts with Notch signaling and can affect cell fate decisions through epigenetic alteration.41 In glioblastoma, the histone methyltransferase mixed-lineage leukemia 1 (MLL1) is induced by hypoxia; and loss of MLL1 reduces the expression of HIF transcripts and HIF2α protein,108 indicating a potential feedback loop sustaining hypoxic response. Depletion of MLL1 inhibited the expression of HIF2α and target genes, including vascular endothelial growth factor (VEGF), and reduced BTSC self-renewal, growth, and tumorigenicity.108
In other cancers, HIF-independent hypoxia mediated epigenetic silencing of tumor suppressor genes has been described. Specifically, the BRCA1 and RAD51 promoters have been shown to be repressed by local chromatin restructuring via H3K4 demethylation, H3K9 methylation, and H3K9 deacetylation109. It is important to note that a growing number of epigenetic modifiers, which are deregulated in numerous cancer types, are dependent upon proper oxygen maintenance (see metabolism section below). As one example, various cancer cell lines grown in vitro versus the hypoxic conditions they experience in an in vivo xenograft setting, result in a global induction of DNA hypomethylation110.
For a wide variety of cancers, extracellular solid tumor pH has been determined to be significantly more acidic than in normal tissues111 Tumor hypoxia in particular can induce a metabolic shift that causes acidosis,112 although these two microenvironmental components can also occur independently.32 Importantly, acidic conditions promote the expression of BTSC markers, self-renewal and tumor growth through augmentation of HIF2 transcriptional responses.35 In response to an acidic environment, and decreasing intracellular pH (pH(i)), cancer cells have been shown to respond and attempt to regulate pH(i), by global de-acetylation, which is accompanied by extensive redistribution of acetylation across the genome113. This suggests that exposure to low pH, either derived extrinsically from the niche or created autonomously by cellular alteration of the niche, promotes malignancy through the induction of distinct cellular phenotypes (i.e. BTSC), and is a process tightly associated with epigenetic alterations.
The Perivascular Niche
A hallmark of glioblastoma is the development of histologic regions of microvascular proliferation, often displaying highly disorganized angiogenic vessels and overall high vascularity (Figure 3). Angiogenesis is essential for tumor survival and is the canonical downstream effect of HIF transcriptional activity. Medulloblastoma cells and BTSCs both consistently secrete elevated VEGF levels40, 114. This effect is markedly enhanced by hypoxia and serves to increase endothelial migration, motility and vasculogenesis114. This suggests initial epigenetic state changes within endothelial cells as BTSCs recruit blood vessels through VEGF secretion, followed by epigenetic adaptation of the BTSCs as they adopt a new cell state to complement their changing niche. The regions around these blood vessels are high in oxygen and nutrients and harbor an increased number of stem cells115. Cells within glioblastoma, medulloblastoma, ependymoma and oligodendroglioma are located in close proximity to tumor capillaries. Within this perivascular niche, soluble factors released from the endothelial cells can promote self-renewal and proliferation of BTSCs29. In medulloblastoma, perivascular stem cells are resistant to radiation and likely give rise to tumor recurrence,116 echoing similar findings in glioblastoma117.
Infiltration and enrichment of tumor-associated macrophages (TAMs) is a common feature of glioblastoma, where TAMs are preferentially located in the perivascular niche (Figure 3)118. Their mutual enrichment and proximity has suggested a relationship between TAMs and BTSCs. Although activated M2 TAMs have well known pro-tumor effects119 including in glioma,120 the mechanisms of the potential BTSC-TAM relationship have been largely undefined. Recently, Zhou et al. al. demonstrated that BTSCs preferentially secrete the cytokine periostin (POSTN), which attracts TAMs. POSTN repression resulted in a striking reduction in TAM density, inhibition of tumor growth, and improved survival of tumor-bearing mice121. TAMs or microglia in the glioblastoma microenvironment may also play a significant role in TGFβ and NF-κB–dependent mesenchymal differentiation, enabling glioblastoma cells to switch subtypes to a more radio-resistant cell state.122 This may further be governed through aberrant activation of the STAT3 (Signal transducer and activator of transcription 3) pathway in glioma by frequent loss or repression of the tumor-suppressor phosphatase PTPRD (protein tyrosine phosphatase, receptor type, D)123. For these state-change events to be lasting and maintained by the niche, BTSCs must adopt a stem-like chromatin state.
Epigenetic regulation of tumors by endothelial cell signals
Beyond being a good place for a stem cell to grow due to the abundance of oxygen, nutrients, and growth factors, cells of the perivascular niche directly interact and bidirectionally communicate with brain tumor stem cells (Figure 3). The molecular mechanisms through which the perivascular niche controls brain tumor stem cell state are beginning to be discovered. BTSCs express Notch receptors while endothelial cells of the niche express Notch-activating ligands124. Whereas co-xenograft of brain tumor cells and endothelial cells increases tumor initiation and growth, knockdown of Notch ligands in the co-injected endothelial cells reduces tumor growth29, 124. BTSCs in the hypoxic niche secrete VEGF,114 which in turn can both recruit new blood vessel formation and stimulate endothelial cells to secrete Notch ligand,125 which can then stimulate Notch signaling in BTSCs. This feed-forward loop may be yet another example of microenvironmental modification initiated by BTSCs to promote maintenance of their own cell state.
Cancer cell dormancy is a potential mechanism to explain many detrimental clinical findings including resistance to chemotherapy, tumor recurrence, and metastasis.126 Entry and exit from cancer dormancy is mediated by epigenetic alterations, signaling pathways, and transcriptional circuits that are also known to drive stem cell reprogramming and maintenance126. The key coagulation mediator Tissue Factor (F3) expressed by vascular endothelial cells and is linked with malignant breast cancer127, where protein secretion by endothelial cells during neovascularization my trigger an exit from dormancy and cancer proliferation.128 In glioma, cancer cell dormancy may be governed by F3. F3 activity enabled glioma cells to form a microenvironment containing angiogenic and inflammatory cells. Strikingly, glioma cells lacking F3 remain viable but dormant unless they are supplemented with exogenous F3.129 This work suggests that microenvironmental changes triggering exit from dormancy are accompanied by more permanent epigenetic, genetic, and phenotypic changes in the glioma cells resulting in tumorigenesis.
Influences from the microenvironment can affect, promote, preserve, and even dictate brain tumor cell states. These findings could have vast clinical implications and suggest therapeutic targets greatly needed in this disease. However, we currently lack the basic mechanistic understandings of how these phenotypic changes in brain tumor cell states are affected and maintained at the chromatin level. Advancing technologies that allow epigenetic analysis at high fidelity with lower numbers of cells may enable such studies to be performed in the near future.
Cellular Metabolism Influences Brain Cancer Epigenetic State
The metabolic state of brain tumor cells is highly influenced by alterations in tumor microenvironment, and linked directly to changes in global epigenetic patterns (Figure 4). Microenvironmental alterations dictate fuel sources available to brain tumor cells, such as glucose130, 131, acetate33, 132 and glutamine133, which limit or alter the distribution of substrates required for post-translational epigenetic modifications33, 132. Mutations of metabolic pathways have been observed in several cancers, in addition to brain tumors, as a means of disrupting epigenetic and cellular state134, 135. In glioma, one of the most common recurrent mutations occurs in IDH1, resulting in the accumulation of an oncometabolite (R-2-hydroxyglutarate, R-2-HG), which functions to inhibit the activity of multiple α-ketoglutarate (α-KG) dependent dixoygenases. Through competitive inhibition, R-2-HG impairs the activity a wide-variety of histone and DNA demethylases such as the Jmjc domain-containing histone demethylases (KDMs), RNA demethylases, and the TET (ten-eleven translocation) family of DNA hydroxylases that facilitate DNA de-methylation. (Figure 4). These enzymes comprise a family of 2-oxoglutarate-dependent dioxygenases that depend upon iron and oxygen for their function, further linking, metabolic and hypoxic regulation with epigenetic programs. However, these widespread effects also increase the difficulty of deciphering the functional consequence(s) of the IDH mutations, specifically whether some of these effects are a mere product of increased R-2-HG production. One of several consequences from IDH mutations is aberrant methylation of histones at several lysine residues, and acquisition of a CpG island methylator phenotype through DNA hypermethylation37–39, 98. While the function of IDH1 mutations in glioblastoma remains to be fully characterized, the result of increased histone methylation prevents lineage-specific progenitors from differentiating into terminally differentiated cells38. Furthermore, chemical inhibition of IDH1 has been shown to promote glioma differentiation136. Like pediatric glioblastomas, which harbor K27M mutations, the convergence on epigenetic programs elicited by metabolic state changes, suggests that these types of mutations may function to activate a stem- or progenitor- cell states required for tumorigenesis.
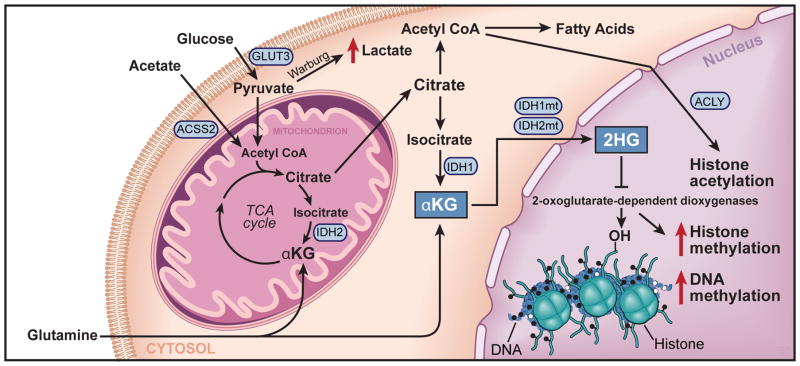
Figure 4. Cellular Metabolism Influences Brain Cancer Epigenetic State.
A schematic of metabolic pathways present within a bran tumor cell with emphasis on transport proteins (GLUT3), and enzymatic effectors (IDH1/2 mutations, ACSS2, and ACLY (ATP citrate lyase)), which alter tumor metabolism and ultimately epigenetic programs. The result of the IDH1 mutation is emphasized, which results in the accumulation of 2-hydroxyglutarate, a metabolite that inhibits the function of iron, oxygen, and α-ketoglutarate dependent demethylase enzymes, thus leading to aberrant accumulation of both DNA and histone methylation.
Like all cancers, brain cancers display the Warburg effect, a preferential utilization of aerobic glycolysis for energy supplies and macromolecule synthesis. This is especially true within the hypoxic niche where both oxygen and nutrients supplied by distant blood vessels are scarce (Figure 3). One method utilized by BTSCs to meet their metabolic needs is to co-opt expression of the high affinity glucose transported, GLUT3, to efficiently scavenge glucose from their environment 137. (Figures 3 and 4) More strikingly, non-stem glioblastoma cells grown in restricted glucose exhibited increased levels of the ESC master transcription factors and showed significant functional enrichment for stem-like cells, indicating adaptation and reprogramming to a more stem-like state 137. The exact epigenetic mechanisms underlying these adaptations are so far unknown. However, the genomic locus of GLUT3 is part of a conserved, 200-kb gene cluster that is highly enriched for genes associated with pluripotency, including the master ESC transcription factor NANOG 138. This region is under control of another the master ESC transcription factor, OCT4 (Octamer-binding transcription factor 4) 139. It is possible that cancer cells can gain GLUT3 expression and stem cell properties simultaneously by epigenetically de-repressing this region of chromatin during stem cell reprogramming.
Another mechanism utilized by brain tumors and brain metastases to meet tumor metabolic demands is the utilization of acetate by the enzyme ACSS2 (Acetyl-coenzyme A synthetase, cytoplasmic).33, 132 Acetate and co-enzyme A are oxidized by ACSS2 to form the central metabolite Acetyl-CoA necessary for a wide variety of cellular processes including epigenetic modulation through histone acetylation.140 Histone acetylation has a very short half-life in tumor cells creating an abundant supply of intracellular acetate to be utilized by ACSS2,33 and also necessitating a continued active upkeep of histone modifications to maintain cell state. Indeed, acetate is utilized by ACSS2 in both brain tumor models and in brain tumor patients and its expression correlates with tumor aggressiveness in a variety of cancers including brain tumors.132
Brain Tumor Therapy by Disrupting Epigenetic Regulators of Cell State
The convergence of genetic, metabolic, and microenvironmental alterations on cell state, and the dependence of cell state on epigenomic programs, suggests that targeting epigenetic mechanisms could be a valuable strategy for treatment of brain tumors. Numerous preclinical studies have shown that brain tumors are sensitive to a variety of inhibitors of epigenetic modifications, several of which are FDA approved (Figure 2)141–143. These include DNA methylation and HDAC inhibitors, such as decitabine and vorinostat, respectively. Targeted epigenetic modulation has already shown promise in numerous pre-clinical models of brain tumors characterized by aberrant epigenetic programs. In the case of DIPGs that harbor the H3.3 K27M mutant, and global loss of H3K27 tri-methylation, a H3K27 demethylase (JMJD3) inhibitor (GSK-J4) has been shown to be effective for reducing tumor growth by elevating H3K27 tri-methylation144. Furthermore, GSK-J4 exhibits synergistic activity with the HDAC inhibitor, pabinostat145. In brain tumors such as glioblastoma, ATRT and ependymoma, characterized by aberrant H3K27me3 patterns, highly specific EZH2 inhibitors (i.e. GSK343) have been shown to be effective at restricting tumor growth in pre-clinical models19, 146, 147. A novel avenue of targeting histone modification is inhibiting the readers of acetylation (i.e. BRD4, bromodomain containing 4), which mark active enhancers and super-enhancers, using inhibitors of bromodomain containing proteins, such as JQ1148. JQ1 treatment has been shown to be effective in both MYC and SHH (sonic hedgehog)-driven medulloblastoma by targeting cancer dependency genes driven by super-enhancers149, 150. This represents a collection of early studies examining an emerging concept of reversing epigenetic signatures in brain tumors using novel small molecule epigenetic inhibitors. Understanding the function and potential requirements of specific epigenetic marks in brain tumors, alongside development of specific epigenetic drugs, may reveal new opportunities for rationale and targeted therapeutics. Targeting cellular state through manipulation of epigenetic regulators represents an alternative or complementary approach to ‘drugging’ specific genetic lesions.
Moving Forward
Genomic sequencing of several types of brain tumors -- astrocytomas, oligodendrogliomas, medulloblastomas, ependymomas, meningiomas, ATRT -- have yielded remarkably granular genomic landscapes. Glioblastoma and other brain tumors harbor mutations that are infrequent in isolation but disrupt normal function of a limited cohort of pathways (p53, retinoblastoma, receptor tyrosine kinase signaling, and chromatin-associated molecules). Sadly, this avalanche of information has made relatively modest impact on the clinical practice of neuro-oncology. Therapeutic trials against driving genetic abnormalities amenable to therapeutic targeting – like EGFR – have been largely negative in most brain cancers. Standard-of-care for most brain tumors remains focused on maximal surgical resection, radiotherapy, and chemotherapy. Indirect targeting of the tumor through anti-angiogenics (e.g. bevacizumab) and immunotherapies (vaccines, adoptive therapies, immune checkpoint inhibitors, and oncolytic viruses) have demonstrated preclinical activity but mixed efficacy in clinical trials. The convergence of genomic alterations, microenvironmental conditions, and metabolic reprogramming to create an epigenetic landscape that promotes aberrant activation and maintenance of stem cell-like transcriptional programs may offer a coherent strategy to improve diagnosis, prediction of prognosis, and therapies. Global chromatin reprogramming may be detectable at the earliest stage of transformation empowering early detection and prognosis. Circulating DNA and tumor cells have proven informative of tumor development and progression, suggesting that simultaneous assessment of tumor genetics and epigenetics may better inform the status of tumors. Currently unclear is the landscape, prevalence and importance of non-coding mutations and structural variations, which will be revealed as future brain tumor whole-genomes are sequenced to greater depth. Delineating the functional consequences of non-coding alterations will benefit from comprehensive and integrated mapping of histone modifications and chromatin structure in brain tumors. Epigenomic mapping, such as enhancer profiles, may also reveal the master transcription factors important for maintaining cancer cell state in addition to the mechanisms that lead to oncogenic transformation. The multiple influences upon epigenetic mechanisms including both intrinsic factors (i.e. mutations) and extrinsic factors (i.e. microenvironment) may complicate epigenomic mapping of brain tumors. However, identifying pathways of convergence and dependencies on epigenetic programs may reveal important insights into the molecular biology of brain tumors, and new avenues for cell state therapies. While targeting epigenetic regulators in tumor cells – e.g. inhibitors of IDH1 or BRD4 – may offer benefit, sustained tumor control will be most likely achieved with combinatorial targeting strategies with conventional or targeted therapies. Potentially, inhibitors of chromatin-associated proteins could induce synthetic lethality with other treatments, disrupt the growth of heterogeneous tumor populations, and attenuate mechanisms of progression. Transforming neuro-oncology care will require more complex modeling of tumor biology through the integration of epigenetics and the multi-dimensional interactions with genetics, metabolism, and the microenvironment. Caution must be exercised as each new advance in oncology has been hailed as a potential cure, but reprogramming tumor cells towards a differentiated phenotype could reverse therapeutic resistance, immune escape, invasion, and angiogenesis.
Acknowledgments
We sincerely apologize to those investigators whose work we were unable to cite due to space limitations. We thank David Schumick (Center of Medical Art and Photography, Cleveland Clinic) for assistance with figure preparation. This work was supported by The Banting Fellowship (SCM),, James S. McDonnell Foundation (JNR) and NIH grants: F32 CA189647 (CGH), F30 CA183510 (TEM), T32 GM007250 MSTP (TEM),, CA154130 (JNR), R01 CA169117 (JNR), R01 CA171652 (JNR), R01 NS087913 (JNR), R01 NS089272 (JNR). MDT is supported by a CIHR Clinician Scientist Phase II award, funds from the Garron Family Chair in Childhood Cancer Research at The Hospital for Sick Children and The University of Toronto, and operating funds from the Canadian Institutes of Health Research, the National Institutes of Health (R01CA159859 and R01CA148699) and the Pediatric Brain Tumor Foundation.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta neuropathologica. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettegowda C, et al. Exomic sequencing of four rare central nervous system tumor types. Oncotarget. 2013;4:572–583. doi: 10.18632/oncotarget.964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiao Y, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget. 2012;3:709–722. doi: 10.18632/oncotarget.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan H, et al. IDH1 and IDH2 mutations in gliomas. The New England journal of medicine. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartzentruber J, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 10.Bjerke L, et al. Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer discovery. 2013;3:512–519. doi: 10.1158/2159-8290.CD-12-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubuc AM, et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta neuropathologica. 2013;125:373–384. doi: 10.1007/s00401-012-1070-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Northcott PA, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature. 2014;511:428–434. doi: 10.1038/nature13379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Northcott PA, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nature genetics. 2009;41:465–472. doi: 10.1038/ng.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Northcott PA, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hovestadt V, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature. 2014;510:537–541. doi: 10.1038/nature13268. [DOI] [PubMed] [Google Scholar]
- 16.Jones DT, et al. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pugh TJ, et al. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mack SC, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee RS, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. The Journal of clinical investigation. 2012;122:2983–2988. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torchia J, et al. Molecular subgroups of atypical teratoid rhabdoid tumours in children: an integrated genomic and clinicopathological analysis. The lancet oncology. 2015;16:569–582. doi: 10.1016/S1470-2045(15)70114-2. [DOI] [PubMed] [Google Scholar]
- 22.Buczkowicz P, et al. Genomic analysis of diffuse intrinsic pontine gliomas identifies three molecular subgroups and recurrent activating ACVR1 mutations. Nature genetics. 2014;46:451–456. doi: 10.1038/ng.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontebasso AM, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. 2014;46:462–466. doi: 10.1038/ng.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malkin H, et al. Nature genetics [Google Scholar]
- 25.Taylor KR, et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nature genetics. 2014;46:457–461. doi: 10.1038/ng.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu G, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature genetics. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu G, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nature genetics. 2014;46:444–450. doi: 10.1038/ng.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleinman CL, et al. Fusion of TTYH1 with the C19MC microRNA cluster drives expression of a brain-specific DNMT3B isoform in the embryonal brain tumor ETMR. Nature genetics. 2014;46:39–44. doi: 10.1038/ng.2849. [DOI] [PubMed] [Google Scholar]
- 29.Calabrese C, et al. A perivascular niche for brain tumor stem cells. Cancer cell. 2007;11:69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 30.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fukumura D, et al. Hypoxia and acidosis independently up-regulate vascular endothelial growth factor transcription in brain tumors in vivo. Cancer research. 2001;61:6020–6024. [PubMed] [Google Scholar]
- 33.Comerford SA, et al. Acetate dependence of tumors. Cell. 2014;159:1591–1602. doi: 10.1016/j.cell.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eyler CE, et al. Glioma stem cell proliferation and tumor growth are promoted by nitric oxide synthase-2. Cell. 2011;146:53–66. doi: 10.1016/j.cell.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hjelmeland AB, et al. Acidic stress promotes a glioma stem cell phenotype. Cell death and differentiation. 2011;18:829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charles N, et al. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell stem cell. 2010;6:141–152. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koivunen P, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slongo ML, et al. Functional VEGF and VEGF receptors are expressed in human medulloblastomas. Neuro-oncology. 2007;9:384–392. doi: 10.1215/15228517-2007-032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mutoh T, Sanosaka T, Ito K, Nakashima K. Oxygen levels epigenetically regulate fate switching of neural precursor cells via hypoxia-inducible factor 1alpha-notch signal interaction in the developing brain. Stem cells. 2012;30:561–569. doi: 10.1002/stem.1019. [DOI] [PubMed] [Google Scholar]
- 42.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han DW, et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell stem cell. 2012;10:465–472. doi: 10.1016/j.stem.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 46.Najm FJ, et al. Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nature biotechnology. 2013;31:426–433. doi: 10.1038/nbt.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gangemi RM, et al. SOX2 silencing in glioblastoma tumor-initiating cells causes stop of proliferation and loss of tumorigenicity. Stem cells. 2009;27:40–48. doi: 10.1634/stemcells.2008-0493. [DOI] [PubMed] [Google Scholar]
- 48.Suva ML, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkar A, Hochedlinger K. The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell stem cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suva ML, Riggi N, Bernstein BE. Epigenetic reprogramming in cancer. Science. 2013;339:1567–1570. doi: 10.1126/science.1230184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stricker SH, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes & development. 2013;27:654–669. doi: 10.1101/gad.212662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Friedmann-Morvinski D, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel AP, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer M, et al. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:851–856. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Polak P, et al. Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature. 2015;518:360–364. doi: 10.1038/nature14221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–507. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 57.Liu L, De S, Michor F. DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes. Nature communications. 2013;4:1502. doi: 10.1038/ncomms2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Black JC, et al. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154:541–555. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ben-Porath I, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nature genetics. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Easwaran H, et al. A DNA hypermethylation module for the stem/progenitor cell signature of cancer. Genome research. 2012;22:837–849. doi: 10.1101/gr.131169.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143:313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tessarz P, Kouzarides T. Histone core modifications regulating nucleosome structure and dynamics. Nature reviews Molecular cell biology. 2014;15:703–708. doi: 10.1038/nrm3890. [DOI] [PubMed] [Google Scholar]
- 66.Hodis E, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pugh TJ, et al. The genetic landscape of high-risk neuroblastoma. Nature genetics. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson G, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature. 2012;488:43–48. doi: 10.1038/nature11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang J, et al. A novel retinoblastoma therapy from genomic and epigenetic analyses. Nature. 2012;481:329–334. doi: 10.1038/nature10733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang J, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hasselblatt M, et al. High-resolution genomic analysis suggests the absence of recurrent genomic alterations other than SMARCB1 aberrations in atypical teratoid/rhabdoid tumors. Genes, chromosomes & cancer. 2013;52:185–190. doi: 10.1002/gcc.22018. [DOI] [PubMed] [Google Scholar]
- 73.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:5187–5191. doi: 10.1073/pnas.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roberts CW, Galusha SA, McMenamin ME, Fletcher CD, Orkin SH. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumors in mice. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13796–13800. doi: 10.1073/pnas.250492697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Klochendler-Yeivin A, et al. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO reports. 2000;1:500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guidi CJ, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Molecular and cellular biology. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- 78.Eroglu E, et al. SWI/SNF complex prevents lineage reversion and induces temporal patterning in neural stem cells. Cell. 2014;156:1259–1273. doi: 10.1016/j.cell.2014.01.053. [DOI] [PubMed] [Google Scholar]
- 79.Parker M, et al. C11orf95-RELA fusions drive oncogenic NF-kappaB signalling in ependymoma. Nature. 2014;506:451–455. doi: 10.1038/nature13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson RA, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Witt H, et al. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer cell. 2011;20:143–157. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li M, et al. Frequent amplification of a chr19q13.41 microRNA polycistron in aggressive primitive neuroectodermal brain tumors. Cancer cell. 2009;16:533–546. doi: 10.1016/j.ccr.2009.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sin-Chan P, Huang A. DNMTs as potential therapeutic targets in high-risk pediatric embryonal brain tumors. Expert opinion on therapeutic targets. 2014;18:1103–1107. doi: 10.1517/14728222.2014.938052. [DOI] [PubMed] [Google Scholar]
- 84.Khuong-Quang DA, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta neuropathologica. 2012;124:439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 86.Chan KM, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes & development. 2013;27:985–990. doi: 10.1101/gad.217778.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sturm D, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer cell. 2012;22:425–437. doi: 10.1016/j.ccr.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 88.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Funato K, Major T, Lewis PW, Allis CD, Tabar V. Use of human embryonic stem cells to model pediatric gliomas with H3.3K27M histone mutation. Science. 2014;346:1529–1533. doi: 10.1126/science.1253799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heaphy CM, et al. Altered telomeres in tumors with ATRX and DAXX mutations. Science. 2011;333:425. doi: 10.1126/science.1207313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fontebasso AM, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta neuropathologica. 2013;125:659–669. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Killela PJ, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:6021–6026. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Remke M, et al. TERT promoter mutations are highly recurrent in SHH subgroup medulloblastoma. Acta neuropathologica. 2013;126:917–929. doi: 10.1007/s00401-013-1198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bell RJ, et al. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science. 2015 doi: 10.1126/science.aab0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zardo G, et al. Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nature genetics. 2002;32:453–458. doi: 10.1038/ng1007. [DOI] [PubMed] [Google Scholar]
- 98.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rideout WM, 3rd, Coetzee GA, Olumi AF, Jones PA. 5-Methylcytosine as an endogenous mutagen in the human LDL receptor and p53 genes. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 100.Tubio JM, et al. Mobile DNA in cancer. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345:1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Venkatesh HS, et al. Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell. 2015;161:803–816. doi: 10.1016/j.cell.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell stem cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 104.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nature reviews Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Seidel S, et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2 alpha. Brain : a journal of neurology. 2010;133:983–995. doi: 10.1093/brain/awq042. [DOI] [PubMed] [Google Scholar]
- 106.Soeda A, et al. Hypoxia promotes expansion of the CD133-positive glioma stem cells through activation of HIF-1alpha. Oncogene. 2009;28:3949–3959. doi: 10.1038/onc.2009.252. [DOI] [PubMed] [Google Scholar]
- 107.Wang J, et al. c-Myc is required for maintenance of glioma cancer stem cells. PloS one. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Heddleston JM, et al. Hypoxia-induced mixed-lineage leukemia 1 regulates glioma stem cell tumorigenic potential. Cell death and differentiation. 2012;19:428–439. doi: 10.1038/cdd.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lu Y, Chu A, Turker MS, Glazer PM. Hypoxia-induced epigenetic regulation and silencing of the BRCA1 promoter. Molecular and cellular biology. 2011;31:3339–3350. doi: 10.1128/MCB.01121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shahrzad S, Bertrand K, Minhas K, Coomber BL. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics. 2007;2:119–125. doi: 10.4161/epi.2.2.4613. [DOI] [PubMed] [Google Scholar]
- 111.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature reviews Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 112.Chiche J, Brahimi-Horn MC, Pouyssegur J. Tumour hypoxia induces a metabolic shift causing acidosis: a common feature in cancer. Journal of cellular and molecular medicine. 2010;14:771–794. doi: 10.1111/j.1582-4934.2009.00994.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.McBrian MA, et al. Histone acetylation regulates intracellular pH. Molecular cell. 2013;49:310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bao S, et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer research. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 115.Christensen K, Schroder HD, Kristensen BW. CD133+ niches and single cells in glioblastoma have different phenotypes. Journal of neuro-oncology. 2011;104:129–143. doi: 10.1007/s11060-010-0488-y. [DOI] [PubMed] [Google Scholar]
- 116.Hambardzumyan D, et al. PI3K pathway regulates survival of cancer stem cells residing in the perivascular niche following radiation in medulloblastoma in vivo. Genes & development. 2008;22:436–448. doi: 10.1101/gad.1627008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 118.Pietras A, et al. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell stem cell. 2014;14:357–369. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 120.Pyonteck SM, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nature medicine. 2013;19:1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhou W, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nature cell biology. 2015;17:170–182. doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bhat KP, et al. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ortiz B, et al. Loss of the tyrosine phosphatase PTPRD leads to aberrant STAT3 activation and promotes gliomagenesis. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:8149–8154. doi: 10.1073/pnas.1401952111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu TS, et al. Endothelial cells create a stem cell niche in glioblastoma by providing NOTCH ligands that nurture self-renewal of cancer stem-like cells. Cancer research. 2011;71:6061–6072. doi: 10.1158/0008-5472.CAN-10-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lobov IB, et al. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155:750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Contrino J, Hair G, Kreutzer DL, Rickles FR. In situ detection of tissue factor in vascular endothelial cells: correlation with the malignant phenotype of human breast disease. Nature medicine. 1996;2:209–215. doi: 10.1038/nm0296-209. [DOI] [PubMed] [Google Scholar]
- 128.Ghajar CM, et al. The perivascular niche regulates breast tumour dormancy. Nature cell biology. 2013;15:807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Magnus N, et al. Tissue factor expression provokes escape from tumor dormancy and leads to genomic alterations. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:3544–3549. doi: 10.1073/pnas.1314118111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee JV, et al. Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell metabolism. 2014;20:306–319. doi: 10.1016/j.cmet.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wellen KE, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mashimo T, et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cascon A, et al. Whole-exome sequencing identifies MDH2 as a new familial paraganglioma gene. Journal of the National Cancer Institute. 2015;107 doi: 10.1093/jnci/djv053. [DOI] [PubMed] [Google Scholar]
- 135.Hao HX, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Flavahan WA, et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nature neuroscience. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Korkola JE, et al. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer research. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
- 139.Levasseur DN, Wang J, Dorschner MO, Stamatoyannopoulos JA, Orkin SH. Oct4 dependence of chromatin structure within the extended Nanog locus in ES cells. Genes & development. 2008;22:575–580. doi: 10.1101/gad.1606308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ecke I, et al. Antitumor effects of a combined 5-aza-2’deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer research. 2009;69:887–895. doi: 10.1158/0008-5472.CAN-08-0946. [DOI] [PubMed] [Google Scholar]
- 142.Milde T, et al. HD-MB03 is a novel Group 3 medulloblastoma model demonstrating sensitivity to histone deacetylase inhibitor treatment. Journal of neuro-oncology. 2012;110:335–348. doi: 10.1007/s11060-012-0978-1. [DOI] [PubMed] [Google Scholar]
- 143.Spiller SE, Ditzler SH, Pullar BJ, Olson JM. Response of preclinical medulloblastoma models to combination therapy with 13-cis retinoic acid and suberoylanilide hydroxamic acid (SAHA) Journal of neuro-oncology. 2008;87:133–141. doi: 10.1007/s11060-007-9505-1. [DOI] [PubMed] [Google Scholar]
- 144.Pal R, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nature medicine. 2014;20:1394–1396. doi: 10.1038/nm.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Grasso CS, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. 2015 doi: 10.1038/nm0715-827a. [DOI] [PubMed] [Google Scholar]
- 146.Alimova I, et al. Inhibition of EZH2 suppresses self-renewal and induces radiation sensitivity in atypical rhabdoid teratoid tumor cells. Neuro-oncology. 2013;15:149–160. doi: 10.1093/neuonc/nos285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kim E, et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Henssen A, et al. BET bromodomain protein inhibition is a therapeutic option for medulloblastoma. Oncotarget. 2013;4:2080–2095. doi: 10.18632/oncotarget.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Link BA, et al. Inhibition of BRD4 attenuates tumor cell self-renewal and suppresses stem cell signaling in MYC driven medulloblastoma. Nature medicine. 2014;5:2355–2371. doi: 10.18632/oncotarget.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]