SUMMARY
Select humans and animals control persistent viral infections via adaptive immune responses that include production of neutralizing antibodies. The precise genetic basis for the control remains enigmatic. Here, we report positional cloning of the gene responsible for production of retrovirus-neutralizing antibodies in mice of the I/LnJ strain. It encodes the beta subunit of the non-classical Major Histocompatibility Complex class II (MHC-II)-like molecule H2-O, a negative regulator of antigen presentation. The recessive and functionally null I/LnJ H2-Ob allele supported the production of virus-neutralizing antibodies independently of the classical MHC haplotype. Subsequent bioinformatics and functional analyses of the human H2-Ob homologue, HLA-DOB revealed both loss- and gain-of-function alleles, which could affect the ability of their carriers to control infections with Human Hepatitis B (HBV) and C (HCV) viruses. Thus, understanding of the previously unappreciated role of H2-O (HLA-DO) in immunity to infections may suggest new approaches in achieving neutralizing immunity to viruses.
INTRODUCTION
Resistance and sensitivity to viral infections depend on the genetic make-up of the host. Genome-wide association studies (GWASs), which test DNA sequence variation across the human genome for linkage with genetic traits (e.g. resistance to infections), have been used to uncover associated genetic loci. Outcomes of infections with persistent viruses, such as Human Immunodeficiency Virus (HIV), Human Hepatitis C Virus (HCV) and Human Hepatitis B virus (HBV) have been linked to the Major Histocompatibility Complex (MHC). The ability to control HIV in some ‘elite controllers’ has been linked to specific MHC Class I (MHC-I) alleles (Fellay et al., 2007; Kosmrlj et al., 2010; Kulkarni et al., 2011; Miura et al., 2009; Pereyra et al., 2010) and persistence of HCV and HBV have been linked to MHC Class II (MHC-II) alleles (Chang et al., 2014; Duggal et al., 2013; Li et al., 2016). MHC involvement in virus clearance was expected since MHC-I and MHC-II genes control CD8+ T cell responses and CD4+ T cell-dependent antibody (Ab) responses, respectively. Since elite controllers are capable of producing virus-specific cytotoxic T-cell mediated immune responses (Walker and Yu, 2013), alleles of HLA-C and HLA-B were proposed as candidates for HIV control (Kosmrlj et al., 2010; Kulkarni et al., 2011; Miura et al., 2009b). Similarly, as spontaneous HCV clearance correlates with the early appearance of virus-neutralizing antibodies (Abs) (Osburn et al., 2014; Pestka et al., 2007) several MHC-II alleles of HLA-DQ were considered as candidates mediating efficient, CD4+ T cell-dependent humoral responses to this virus (Duggal et al., 2013). Further, the production of neutralizing Abs that play a key role in the recovery from infection with HBV (Ciupe et al., 2014; Huang et al., 2006) were also linked to specific HLA-DQ alleles (Chang et al., 2014; Li et al., 2016).
GWAS is a powerful tool, but its predictive capacity relies on the frequency of recombination, which varies from locus to locus. This is especially important when considering critical regions within the MHC locus, one of the most gene-rich regions in the human genome where many of the linked genes play important roles in immune system regulation. Therefore, additional approaches to resolve involvement of specific MHC genes in immunity to infection are required to test the possibility that other closely linked genes might actually determine the phenotype. In contrast to humans, where genetic manipulations are impossible, animal models serve as powerful tools that can be exploited to discover underlying mechanisms that alleviate viral diseases. Mice are resistant to human viruses such as HCV or HIV; however, they have their own persistent viruses, such as Mouse Mammary Tumor Virus (MMTV, a betaretrovirus) and Murine Leukemia Virus (MuLV, a gammaretrovirus) that can be used as models to discover genes responsible for the outcome of infection. Mice from genetically distinct strains exhibit selective susceptibility to MMTV or MuLV infections and the mechanisms controlling these viruses are linked to adaptive immune responses [reviewed in (Dudley et al., 2016) (Miyazawa et al., 2008)]. The anti-retroviral Ab response in C57BL/6 (B6) mice is mapped to a single dominant gene, ‘recovery from Friend virus 3’ (Rfv3), located on chromosome 15 (Hasenkrug et al., 1995). Rfv3 mediates the Ab response to MuLV, but not to MMTV. In contrast, mice of the I/LnJ strain are unique in their ability to control both MMTV and MuLV via virus-neutralizing Ab responses (Case et al., 2008; Purdy et al., 2003). I/LnJ mice become infected with either retrovirus, but neutralizing Abs they produce not only render the viruses noninfectious, but also prevent the emergence of immune escape variants (Case et al., 2008; Case et al., 2005; Purdy et al., 2003). A single recessive locus, virus infectivity controller 1 (vic1), controls resistance to these retroviruses in I/LnJ mice (Purdy et al., 2003). I/LnJ mice carry a resistant allele of vic1 (vic1R), whereas MMTV-susceptible C3H/HeN and B6, as well as MMTV- and MuLV-susceptible BALB/cJ mice carry the susceptible alleles (vic1S) (Case et al., 2008; Golovkina, 2000; Kane et al., 2011; Purdy et al., 2003). The vic1 gene has been preliminary mapped to the MHC locus (Kane et al., 2011). However, the unusual recessive nature of resistance conferred by vic1R suggested that its dominant counterpart vic1S was a negative regulator of the retrovirus-specific immune response.
Here, using positional cloning approach, we identify vic1 as H2-Ob which encodes the beta subunit (Oβ in mice and DOβ in humans) of the non-classical MHC-II-like molecule H2-O (HLA-DO or DO in humans). The recessive I/LnJ H2-Ob allele resulted in a functionally null H2-O protein. Thus, the common functional H2-Ob allele is responsible for the lack of Ab responses in virus-susceptible mice. Computational and subsequent functional analyses of human HLA-DOB alleles revealed genetic variants with both loss- and gain-of-function mutations. GWASs have previously linked the outcome (persistence or clearance) of both HCV and HBV infections to the HLA-DQ locus (Chang et al., 2014; Li et al., 2016). We have mapped HLA-DOB within the same linkage disequilibrium (LD) block as the HLA-II DQ locus, thus suggesting that HLA-DOB could contribute to the control of both viruses as much as the HLA-II DQ genes. Finally, at least one gain-of-function DOB variant (G77V) was linked to the inability to clear HCV infection. These findings reveal the previously unknown role of H2-O (HLA-DO) in the control of persistent viral infections.
RESULTS
Mapping of the vic1 gene to the Major Histocompatibility Complex locus
In order to determine the genetic mechanism by which I/LnJ mice mount a highly potent anti-retroviral immune response, a positional cloning approach was taken. The vic1 locus was previously mapped to a ~54.4-Mb region on Chromosome 17 between 8.3 Mb and 63 Mb (Case et al., 2008; Kane et al., 2011). To confirm the role of this locus in the resistance to retroviruses, the locus was transferred from the I/LnJ background to the MMTV-susceptible C3H/HeN and B6 backgrounds, as well as the MMTV- and MuLV-susceptible BALB/cJ background. As a result, congenic C3H/HeNvic1i/i, B6vic1i/i, and BALB/cJvic1i/i lines were produced (Figure 1A). Upon infection with retroviruses, all individual congenic strains generated viral neutralizing immune responses (Figure 1, shown as line A), recapitulating the phenotype of the I/LnJ strain (Case et al., 2008; Kane et al., 2011). The perfect correlation between the vic1R genotype and the viral resistance phenotype for each of the congenic strains clearly indicated that the production of the anti-viral immune response was controlled by the I/LnJ vic1 locus.
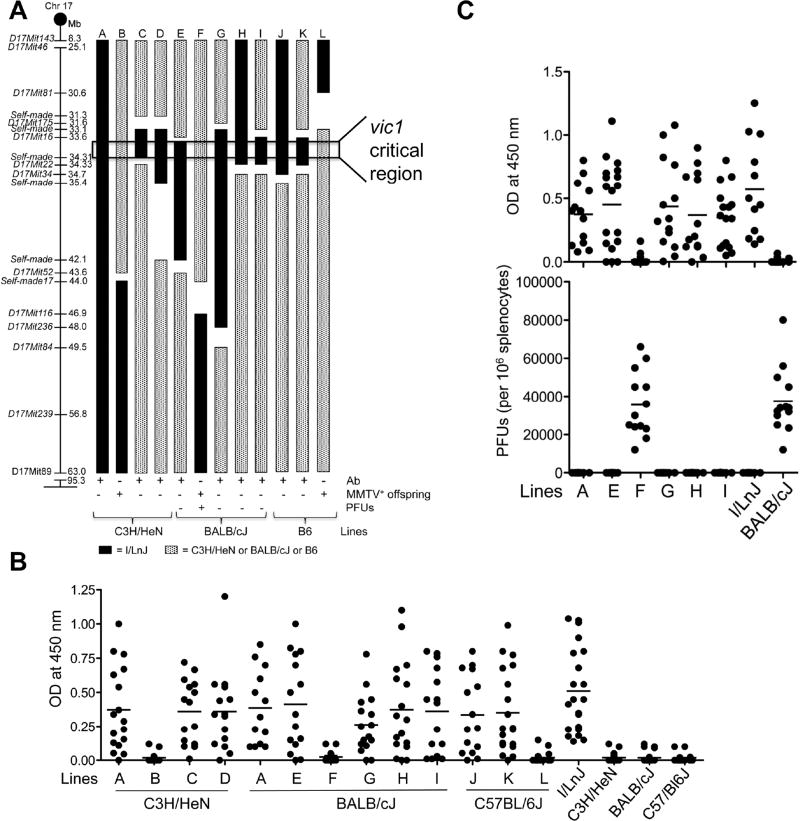
Figure 1. Mapping the retroviral resistance gene.
(A) Reduction the vic1 critical region to ~0.71 Mb via a congenic approach. Diagram of Chromosome 17, with microsatellite markers and their positions (in Mbs). A-L; schematic maps of congenic mouse strains used to fine-map the vic1 region. Solid bars represent the resistant (I/LnJ) chromosome and patterned bars represent chromosomal segments from susceptible mouse strains (C3H/HeN, BALB/cJ or B6). Gaps indicate regions of undefined genotypes. Line A, which contains the originally mapped vic1 region, was transferred to the C3H/HeN background (this Figure), the BALB/cJ background (Figures 1B and 1C) and to the B6 background (Kane et al., 2011). C3H/HeN-, BALB/cJ- and B6- based strains were infected with MMTV, while BALB/cJ-based strains were also infected with MuLV. Relevant anti-viral response phenotypes (anti-MMTV and anti-MuLV IgG2a/2c–specific Abs and MuLV plaque forming units, PFUs, three months post infection of 6–8 week old mice) are shown below. MMTV-neutralizing capacity of Abs was tested in the offspring produced by injected females (2–3 litters from each line). Females from lines A, C-E, G-K produced uninfected offspring, whereas mice from lines B, F and L transmitted infectious virus (MMTV+ offspring).
(B) Anti-MMTV response in mice from congenic lines. Mice shown in Figure 1A were infected with MMTV at 6–8 weeks of age and screened for anti-virus IgG2a/2c–specific Abs by ELISA three months later. Backgrounds obtained from incubation with secondary Abs alone were subtracted. Data are presented as mean of >15 mice per line and were combined from >15 independent experiments.
(C) Anti-virus Abs and PFUs in MuLV-infected mice from BALB/cJvic1i/i congenic lines. Mice were injected with MuLV at 6–8 weeks and were screened for anti-virus Abs as well as PFUs three months later. Means are combined from 8 independent experiments with >13 mice per mouse line used.
To further reduce the size of the 54.7 Mb vic1 critical region, a series of congenic strains containing smaller overlapping regions within I/LnJ vic1 locus were generated and mice with new recombination points within the vic1 locus were made homozygous for the I/LnJ-derived locus (required because the resistance mechanism was recessive) and subsequently analyzed for a virus-neutralizing immune response. MMTV-infected mice from lines A (positive control), C, D, E, G, H, I, J, K produced viral neutralizing antibodies, whereas lines B, F and L did not (Figures 1A and 1B). Consistent with a lack of an Ab response, lines B, F and L continued to transmit infectious virus to their progeny (Figure 1A). Additionally, BALB/cJ congenic mouse lines E, G, H and I, but not F, generated an Ab response to MuLV that resulted in the elimination of the virus (Figures 1A and 1C). Thus, following the anti-retroviral immune response in the infected congenic mice allowed for narrowing the vic1 locus to a ~0.71 Mb region located between 33.60 and 34.31 Mbs on the Chromosome 17 (Figure 1A).
vic1 is not a classical MHC-I or class MHC-II gene
The 0.71 Mb vic1 region contained 39 genes, including the classical MHC-I and MHC-II genes. To determine whether vic1R, were encoded by classical MHC-I or MHC-II, we used B6 mice with targeted deletion of MHC-I or MHC-II genes. If either the I/LnJ MHC-I or MHC-II genes encoded vic1R, then the F1 MHCi/− hemizygous mice must exhibit the resistant phenotype and produce virus-neutralizing antibodies. However, if vic1R were encoded by a different gene, mice should remain sensitive due to recessive nature of vic1R.
MMTV-infected F1 progeny of I/LnJ mice crossed to B6.H2-Kb−/−H2-Db−/− mice deficient for classical MHC-I genes (Vugmeyster et al., 1998) (MHC-Ii/− hemizygous mice) were screened for anti-virus Abs. Whereas control (B6vic1i/i × I/LnJ) F1 mice produced anti-virus Abs, MHC-Ii/−(B6.H2-Kb−/−H2-Db−/− × I/LnJ) F1 mice did not, indicating that I/LnJ MHC-I was not encoding vic1 (Figure 2).
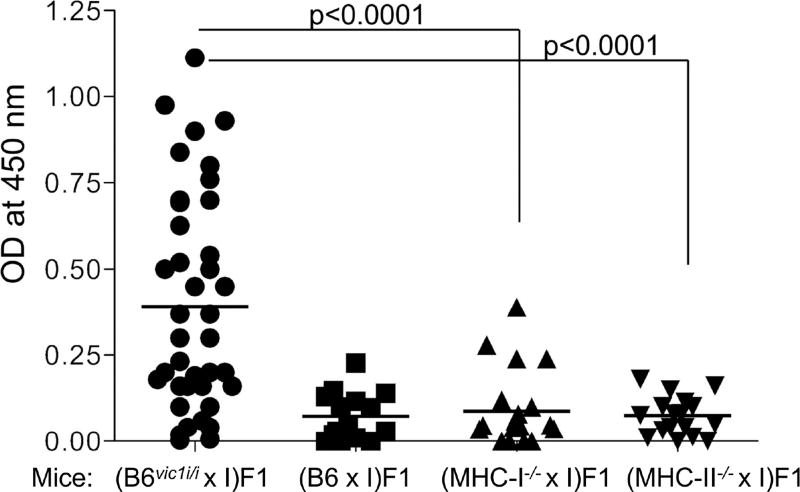
Figure 2. vic1 is not a classical MHC-I or MHC-II gene.
Mice were infected with MMTV via fostering by infected females and screened for anti-virus Abs three months later. (B6 × I) F1 and (B6vic1i/ix I) F1 mice were used as negative and positive controls for Ab production, respectively. Means are derived from combined data from 4 independent experiments. (MHC-II−/− × I) F1 mice include 20 (H2-Ab−/− × I) F1 and 10 (MHC-II del × I) F1 mice. I, I/LnJ mice. Significance was calculated using an unpaired t test.
A similar approach was used to rule out the possibility that classical MHC-II encodes vic1. B6 mice express only I-A MHC-II molecules due to a deletion in the promoter of H2-Ea gene (Dembic et al., 1985). Thus, we first crossed B6 mice carrying a ‘floxed’ allele of the H2-Ab gene (Hashimoto et al., 2002) with ZP3-Cre B6 transgenic mice (de Vries et al., 2000) to generate mice with a permanently deleted H2-Ab gene and crossed them to I/LnJ mice to produce F1 mice that carry only MHC-II genes of the I/LnJ origin (MHC-IIi/− hemizygous mice). In addition, we crossed I/LnJ mice to B6.MHC-IIdel/del mice (Madsen et al., 1999), which lack all classical MHC-II genes. MMTV infected mice from both groups were screened for anti-virus Abs. Unlike the positive control MHC-IIi/i (B6vic1i/i × I/LnJ) F1 mice, negative control MHC-IIb6/i (B6 × I/LnJ) F1 mice and the experimental MHC-IIi/− (B6MHC-II null × I/LnJ) F1 mice did not produce Abs against MMTV (Figure 2). Together, these data definitively eliminated classical MHC-I and MHC-II genes as encoding vic1.
Identification of a non-classical MHC-II gene as the candidate gene encoding vic1
To further reduce the size of the vic1 region, a bacterial artificial chromosome (BAC) phenotype rescue experiment was performed. The expression of the dominant susceptible B6 vic1 allele by a transgenic BAC should oppose the ability of virus-resistant B6vic1i/i congenic mice to produce anti-virus Abs. Overlapping BACs spanning the entire B6 vic1 locus were used to make four independent transgenic lines on the B6vic1i/i congenic background (Figure 3A). Transgenic and non-transgenic littermates from all lines were infected with MMTV and screened for anti-virus Abs (Figure 3B). Only two BACs (J18 and L18) reversed the resistant phenotype of B6vic1i/i mice (Figures 3A and 3B). The overlap between the two BACs positioned the vic1 gene within a region of 32.9 Kb (between bp 34,211,575 and 34,244,528) (Figure 3A) that contained a partial sequence of Tap2, a complete sequence of H2-Ob and a complete sequence of a predicted gene, Gm15821.
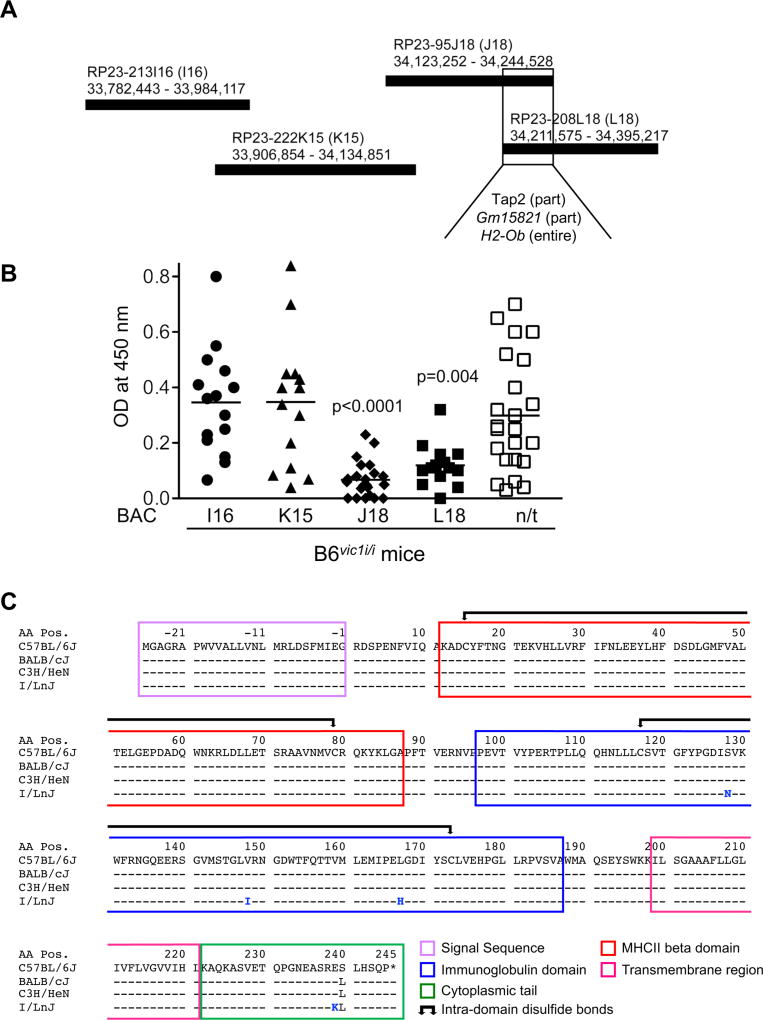
Figure 3. Putative identification of vic1 as a non-classical MHC gene.
(A) Production of B6vic1i/i transgenic mice expressing overlapping regions of B6 BACs. BACs are shown with their corresponding chromosomal coordinates (Kb). The region of overlap between BACs J18 and L18 shows common genes.
(B) Phenotypes of BAC transgenic mice. Transgenic and non-transgenic (n/t, data combined from all strains) littermates were infected with MMTV at 6–8 weeks of age and screened for antiviral IgG Abs three months later. Means are combined from 5 independent experiments. p values were calculated using unpaired t test and are shown relative to non-transgenic mice. ≥13 mice per strain were used.
(C) Amino acid alignment of B6, BALB/cJ, C3H/HeN and I/LnJ Ob. Amino acid number 1 corresponds to the first amino acid after signal sequence cleavage. Negative numbers correspond to the amino acid residues of the signal sequence. Protein domains are marked by colored frames.
We reasoned that only mutations found uniquely in the I/LnJ genome but not in the genomes of susceptible mice would be meaningful. Thus, whole genome sequencing of DNA from I/LnJ, BALBc/J and C3H/HeN mice was performed followed by alignment of the sequences to the reference B6 genome to screen for polymorphic mutations within the new shortened 32.9Kb vic1 critical region of chromosome 17. Several unique non-synonymous variants were identified in the coding regions of I/LnJ H2-Ob compared to similar regions in mice from all three retrovirus-susceptible mouse strains. Four mutations leading to amino acid substitutions (S128N, V148I, L167H, and E239K) were identified in the H2-Ob gene (Figure 3C) and eight and four nucleotide substitutions were found in 5’ and 3’ non-coding regions of the H2-Ob gene, respectively (Tables S1–S3). In addition, two substitutions (N424S and D585A) were found in the Tap2 gene (Tables S1–S3). However, Tap2 was not further pursued because BAC L18 encoded only part of Tap2 (Table S1c) and yet suppressed the production of anti-virus Abs (Figures 3A and 3B). Gm15821 was also eliminated as a candidate for vic1 since no mutations were found within the coding or non-coding regions of the I/LnJ Gm15821 allele.
H2-O-deficient mice produce virus-neutralizing antibodies
The recessive mechanism of the Ab response in I/LnJ mice suggested that I/LnJ vic1 might be a loss of function allele. If so, MMTV-infected H2-Ob-deficient mice are expected to emulate the I/LnJ anti-virus immune response. The H2-Ob gene encodes the Oβ protein, which together with Oα (encoded by H2-Oa) forms the relatively non-polymorphic MHC-II-like molecule, H2-O (Karlsson et al., 1991). H2-O was a promising candidate for vic1, as it functions as a negative regulator of MHC-II presentation (Denzin et al., 1997). As H2-O is an obligate heterodimer, the loss of either chain leads to degradation of the other chain and to H2-O-deficiency (Liljedahl et al., 1996). Thus, we evaluated the ability of both B6.H2-Oa-deficient mice (Liljedahl et al., 1998) and B6.H2-Ob-deficient mice that were generated using CRISPR/Cas9 technology (Figure S1) to produce anti-virus Abs following infection. H2-O-deficient mice and their littermate controls were infected and screened for anti-MMTV Abs by ELISA. Both H2-Oa-and H2-Ob-deficient infected mice produced anti-virus Abs (Figure 4). Similar to sera from infected control B6vic1i/i mice, sera from infected H2-Oa- and H2-Ob-deficient mice neutralized virus infectivity (Figures 4, S2A and S2B). Furthermore, like infected B6vic1i/i congenic mice, infected H2-Ob-deficient mice produced uninfected offspring (Figure S2C). Thus, Abs produced by infected H2-O-deficient mice were virus-neutralizing. These data definitively identify vic1 as H2-Ob. Furthermore, B6.H2-Oa−/−, B6.H2-Ob−/− and I/LnJ mice carry different MHC haplotypes (H2b in B6 and H2j in I/LnJ mice) yet all generated virus-neutralizing Abs, demonstrating that Ab production was independent of the specific classical MHC haplotype.
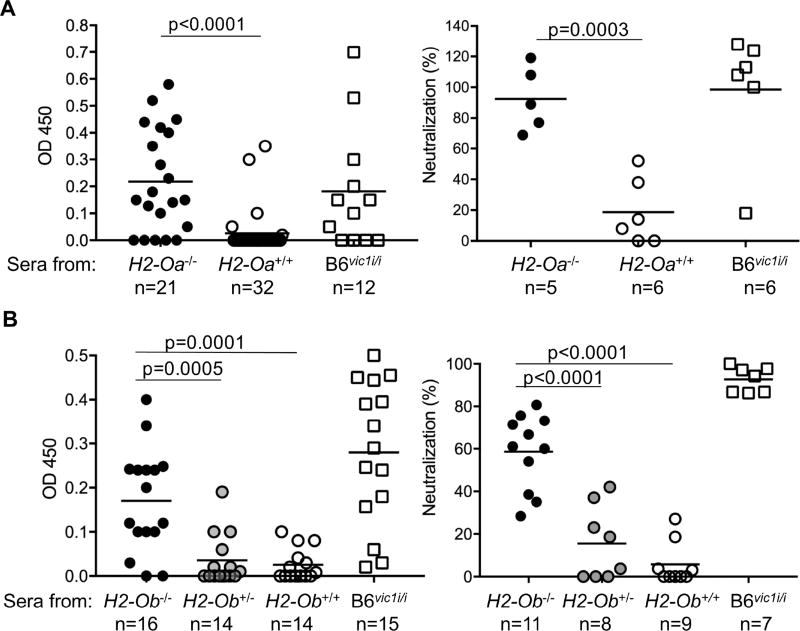
Figure 4. vic1 is H2-Ob.
Production of anti-virus antibodies by infected B6.H2-Oa−/− (A) and B6.H2-Ob−/− mice (B). Mice were infected with MMTV at 6–8 weeks of age, and at three months (H2-Oa−/− mice) and two months (H2-Ob−/−) post infection their sera were screened for the presence of anti-virus IgG2c Abs by ELISA (left graphs) and for neutralization of MMTV infection in BALB/cJ mice (right graphs). B6 (H2-Oa+/+ and H2-Ob+/+) and B6vic1i/i mice were used as negative and positive controls, respectively. Means are derived from combined data from more than 3 independent experiments. Sera that showed reactivity above the background in ELISA were used for pre-incubation with MMTV virions before injection into susceptible BALB/cJ mice. Neutralization (%) was calculated as described in Methods. n, number of mice (left graphs) or number of sera used (right graphs). Significance was calculated using an unpaired t test. See also Figures S1 and S2.
I/LnJ Oβ is functionally null
The process of loading of MHC-II molecules with peptides is mediated by the interaction of MHC-II with another non-polymorphic MHC-II-like molecule, H2-M [HLA-DM (DM) in humans] [reviewed in (Blum et al., 2013; Denzin, 2013; Mellins and Stern, 2014)]. H2-M replaces MHC-II-associated invariant chain peptides (CLIP) that protect the MHC-II peptide-binding groove until MHC-II arrival in endosomal compartments with high affinity, pathogen-derived peptides. H2-M function is opposed by H2-O, which acts as an MHC-II mimic, blocking the ability of H2-M to catalyze MHC-II peptide loading (Guce et al., 2013).
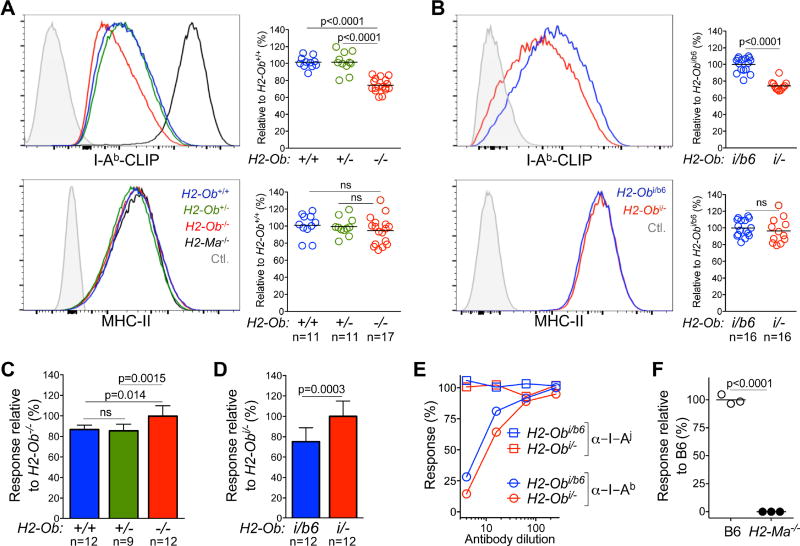
Since H2-O-deficient mice generated virus-neutralizing Ab responses similar to I/LnJ mice we hypothesized that I/LnJ H2-Ob was a non-functional allele. Thus, we directly measured the impact of I/LnJ H2-Ob on MHC-II peptide presentation. Cells that lack H2-O have lower levels of MHC-II-CLIP complexes as H2-M function is enhanced in the absence of inhibition by H2-O (Gu et al., 2013) and MHC-II-CLIP levels on the cell surface can be used as a read-out to measure H2-M activity (Chen et al., 2002; Denzin et al., 1997; Glazier et al., 2002). Therefore, we used an Ab that specifically recognizes I-Ab-CLIP complexes (Liljedahl et al., 1998) to measure H2-M activity in B cells from B6, B6.H2-Ob−/−, B6.H2-Ob+/− and F1 mice generated by crossing B6 or B6.H2-Ob+/− mice with I/LnJ mice. As expected, I-Ab-CLIP levels, but not total MHC-II levels, were significantly reduced on B cells from H2-Ob−/− mice compared to B cells from H2-Ob+/+ and H2-Ob+/− control mice (Figure 5A). Most importantly, a similar reduction in I-Ab-CLIP levels was observed on B cells from (B6. H2-Ob−/− × I/LnJ) F1 mice (expressing only the I/LnJ H2-Ob allele), but not on B cells from (B6 × I/LnJ) F1 mice (expressing both I/LnJ and B6 alleles of H2-Ob) (Figure 5B).
Figure 5. I/LnJ Oβ is functionally null.
(A) and (B), Comparison of MHC-II expression in mice with various H2-Ob genotypes. Representative FACS analysis showing the expression level of I-Ab-CLIP (top) and MHC-II (bottom) on the cell surface of CD19 positive B cells from B6 mice with indicated H2-Ob genotypes (A) or from (I/LnJ × B6) F1 (H2-Obi/b6) and (I/LnJ × B6.H2-Ob−/−) F1 (H2-Obi/−) (B) mice. In (A) I-Ab-CLIP levels for B6.H2-Ma−/− B cells and MHC-II-negative CD8+ T cells were used as positive and negative staining controls (Ctl), respectively; unstained B cells were used as a negative staining control in (B). Graphs show quantification of the geometrical mean fluorescence intensity of I-Ab-CLIP and MHC-II for the indicated strains of mice relative to that obtained for H2-Ob+/+ (A) or H2-Obi/b6 (B) B cells. n, number of mice used in 7 independent experiments.
(C) and (D) Enhanced antigen presentation by B cells expressing I/LnJ Oβ. Antigen presentation by purified B cells from the indicated strains of mice was measured by co-culture with the I-Ab-restricted 8-1-6 T cell line. IL-2 release was measured after overnight incubation by color conversion of Alamar Blue by an IL-2–dependent cell line. Data is presented as response relative to the level of IL-2 produced in response to H2-Ob−/− (C) or Obi/− (D) B cells. n, number of mice used in 5 independent experiments.
(E) The 8-1-6 T cell line is restricted to I-Ab. Activation of I-Ab-restricted 8-1-6 T cell line was measured in the presence of titrated doses of a monoclonal Ab specific for I-Ab (Y3JP) or of a control monoclonal Ab specific for I-Aj (10.2.6).
(F) The 8-1-6 T cell line is activated by a H2-M dependent peptide. IL-2-secretion by 8-1-6 T cell line was measured in response to B6 and B6.H2-Ma−/− splenocytes. Data are representative of 2 similar experiments. Significance was calculated using an unpaired t test. ns, non-significant; n, number of mice.
Next, we measured the presentation of I-Ab-peptide complexes by B cells carrying different alleles of H2-Ob to T cells. Several I-Ab-reactive T cell lines (Logunova et al., 2005) were compared for IL-2 production in response to H2-Ob-sufficient and H2-Ob-deficient B cells. One T cell lymphoma line, 8-1-6, detected increased antigen presentation by H2-Ob-deficient B cells (Figure 5C). B cells from (I/LnJ × B6.H2-Ob−/−) F1 mice stimulated the 8-1-6 cell line significantly better than B cells from mice carrying either one B6 H2-Ob allele (H2-Ob+/− mice), or both B6 and I/LnJ H2-Ob alleles [(I/LnJ × B6) F1 mice] (Figure 5D). Activation of 8-1-6 was dependent on both MHC-II (I-Ab), and the presence of H2-M (Figures 5E and 5F). Thus, compared to B cells expressing B6 H2-Ob, B cells that expressed I/LnJ H2-Ob had increased H2-M activity. In fact, B cells expressing only I/LnJ H2-Ob had the same phenotype as B cells that expressed no H2-Ob (Figure 5A–D). These functional data showed that I/LnJ H2-Ob is a loss of function allele.
Human HLA-DOB alleles with altered function
Next, to determine whether humans also carry HLA-DOB and HLA-DOA alleles with altered function, over 60,000 exomes from the ExAC database were analyzed (Tables S4 and S5). Ninety-four HLA-DOB and 107 HLA-DOA allelic variants were identified that had altered protein-coding sequences with the frequencies ranging from 8.24 × 10−6 to 0.114 (Figure 6A and Tables S4–S5). Seven of 94 HLA-DOB alleles had frameshift/stop codon mutations and were predicted to be null. The frequencies of these alleles were very low and none were found to be homozygous. At the same time, thirty-two HLA-DOB alleles with varying frequencies carried missense mutations that could potentially produce proteins with altered function. Twenty missense HLA-DOB mutation-carrying alleles, as well as five previously identified common alleles and the seven frameshift/stop codon mutants were chosen for further functional studies (Table S4).
Figure 6. HLA-DOB variants have altered function.
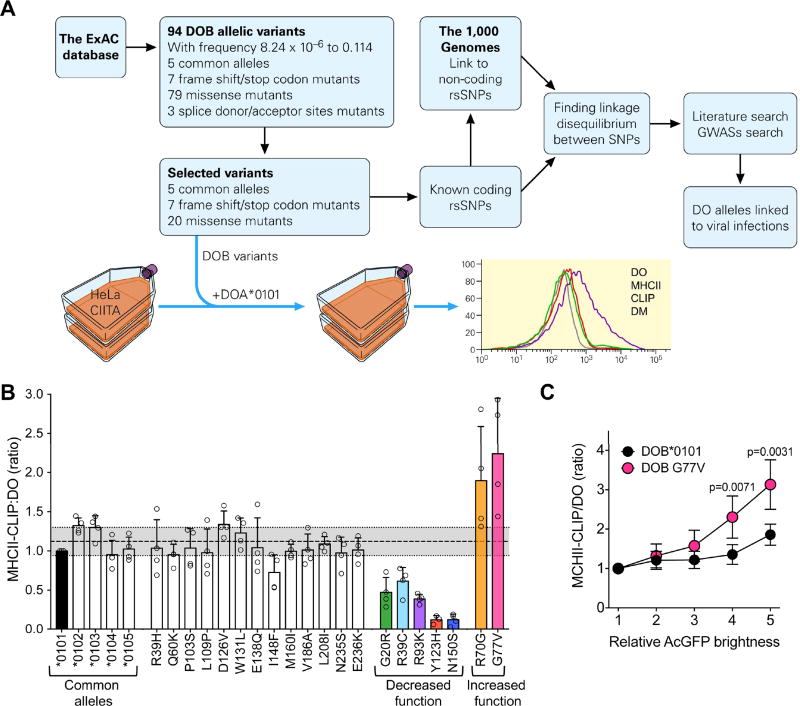
(A) Scheme for identification of HLA-DOB alleles, characterization of their function and search for association between alleles and viral infection outcome. See text for details.
(B) Identification of HLA-DOB variants with altered function. HeLa cells stably expressing CIITA (MHC-II+, DM+ and DOαlow) were transiently co-transfected with a plasmid encoding HLA-DOA*0101 (most common allele) together with plasmids expressing each of the 32 identified HLA-DOB variants. Cells were analyzed 72h later by flow cytometry to determine MHC-II-CLIP, DO, DM and MHC-II levels. Geometric mean fluorescence intensity (gMFI) obtained for MHC-II-CLIP divided by gMFI obtained for DO was used as a measurement of the ability of each resulting DO to inhibit DM mediated peptide loading (removal of CLIP). For comparison across individual experiments, results were normalized to that obtained for the most common HLA-DOB allele (HLA-DOB*0101; black bar). The seven stop and frameshift DOβ variants did not produce any detectable DO protein (Figure S4A). The shaded grey area represents the average MHC-II-CLIP:DO ratio +/− one standard deviation for the five most common known DOβ alleles. DOβ variants with altered function (increased or decreased; colored bars) were defined as possessing a MHC-II-CLIP:DO ratio that fell outside this range. Circles represent results from independent experiments and bars represent the mean +/− the standard deviation. Data were combined from three to four individual experiments. Experimental controls and individual levels obtained for DO, MHC-II-CLIP, DM and MHC-II are shown in Figure S3.
(C) Comparative inhibition of DM function by the DOβ 0101 and DOβ G77V alleles. HeLa.CIITA cells transfected with plasmids encoding HLA-DOA*0101 and either HLA-DOB*0101 or HLA-DOB G77V were stained as in B. For analysis, mRuby (DOα)+ and MHC-II+ (MHC-II-CLIP analysis) or DM+ (DO analysis) cells were parsed into bins (1–5) based on AcGFP levels which reports DOβ expression as detailed in Figure S4B. The ratio of the MHC-II-CLIP gMFI to the DO gMFI was determined for each AcGFP bin as a measure of relative DM-mediated peptide loading (CLIP removal from MHC-II) and plotted relative to the AcGFP brightness (bin). Data were normalized relative to the value obtained for bin 1 for each of the DOβ variants. Data were combined from 5 individual experiments. Symbols and error bars represent the mean +/−SD. Significance was calculated using an unpaired, two-tailed student’s t-test.
As in mouse cells, the ability of DO to inhibit DM in human MHC-II expressing cells can be measured by monitoring MHC-II-CLIP levels. Cells lacking DO have low levels of MHC-II-CLIP whereas cells expressing DO have increased levels of MHC-II-CLIP (Denzin et al., 1997). Therefore, to determine if the mutations encoded by the HLA-DOB alleles resulted in DO proteins with altered function, an in vitro system was used. HeLa cells stably transfected with the Class II transcriptional activator (HeLa.CIITA) that drives the expression of MHC-II, DM and invariant chain (Khalil et al., 2002), were transiently transfected with plasmids expressing HLA-DOA*0101 (most common allele of HLA-DOA) and the variant HLA-DOB alleles (Figure 6A). Three days later MHC-II-CLIP, DO, DM and MHC-II levels were measured by flow cytometry. MHC-II-CLIP levels were low on cells expressing empty vectors or only HLA-DOA*0101 or HLA-DOB*0101 individually, but increased on cells transfected with HLA-DOA*0101 and HLA-DOB*0101 together (Figure S3A), validating this approach for measurement of the impact of HLA-DOB mutations on DO activity.
As predicted, the seven frameshift/stop codon HLA-DOB mutants produced no detectable protein after expression with HLA-DOA*0101 in HeLa.CIITA cells and MHC-II-CLIP levels remained low (Figure S3B and S4B). Expression of the five known common HLA-DOB alleles (HLA-DOB*0101-*0105) resulted in similar levels of DO and MHC-II-CLIP (Figure S3B) demonstrating that they were functionally similar. The other alleles could be grouped into four different cohorts when compared to DOβ*0101: 1) nine alleles (R39H, P103S, E138Q, I148F, M160I, V186A, L208I, N235S, and E236K) that expressed both DO and MHC-II-CLIP at similar levels; 2) six alleles that resulted in reduced levels of both DO and MHC-II-CLIP levels (G20R, R70G, Y123H, D126V, W131L and N150S); 3) four alleles that produced similar amounts of DO but reduced MHC-II-CLIP (R39C, Q60K, R93K, and L109P); and 4) two alleles that resulted in reduced DO but showed no difference in MHC-II-CLIP levels (R70G and G77V) (Figures 6B and S3B). MHC-II and DM levels were unchanged after expression of the various DOβ alleles (Figure S3B).
To identify DOβ alleles with altered function, the ratio of MHC-II-CLIP to DO was determined. Since the five known DOβ alleles all expressed similar levels of DO and MHC-II-CLIP, we considered alleles that had a MHC-II-CLIP:DO ratio more than one standard deviation away from the average obtained for the five known alleles (1.12 ± 0.177) to be alleles that possessed altered function. While G20R, R39C, R93K, Y123H and N150S exhibited decreased DO function, R70G and G77V showed enhanced DO activity (Figure 6B). Enhanced DO activity of the G77V variant was maintained even at lower levels of DO expression (Figures 6C and S4B). Interestingly, alleles with both reduced (G20R and R39C) and enhanced (G77V and R70G) DO activity had mutations that were mapped to the MHC-II beta-like domain of DOβ (Figure S5). The other DOβ variants with reduced function (Y123H and N150S) were mapped to the immunoglobulin domain, while R93K was mapped in between the two domains (Figure S5).
Some human HLA-DOB alleles can be linked to specific outcomes of persistent viral infections
To determine whether the HLA-DOB alleles with altered function have relevance to viral clearance or persistence, bioinformatics and literature searches were performed. First, we found that the coding regions of some HLA-DOB alleles used in our studies possessed previously documented single nucleotide polymorphism (SNP) IDs (Figure 6A, Table S4). We then tested for linkage of these SNPs to the known non-coding nearby SNPs using the 1,000 Genomes database. This approach allowed us to establish that two SNPs, rs144814623 and rs2071469, mapped to the coding region and the 5’ UTR of DOB were in linkage disequilibrium (LD) with three SNPs, rs9276370, rs7756516, and rs7453920 encompassing the HLA-DQA2-DQB2 locus (Chang et al., 2014) (Figure S6), which was previously linked to HCV (Duggal et al., 2013) and HBV (Chang et al., 2014) persistence. The ‘C’ allele of rs2071469 (in HLA-DOB) was found associated with increased susceptibility to HCV infection (Huang et al., 2015). These results indicate that HLA-DOB is co-inherited with DQA2-B2 genes, and thus may contribute to HCV and HBV persistence. Indeed, we found that ‘A’ in rs144814623 (resulting in G77V DOβ allele) was exclusively associated with ‘C’ in rs4273729 (D’ = 1; P < .0016), which was previously linked to HCV persistence (Duggal et al., 2013), whereas ‘G’ in rs4273729 was more often associated with virus clearance. Since the DOβ G77V variant has gain of function properties, it is possible that it impairs viral clearance by negatively influencing MHC-II presentation. The 1000 genomes database reports DOβ G77V as uniquely present in people of African ancestry. Since HCV-infected patients of African descent are less likely to spontaneously clear the virus than patients of European or Asian genetic backgrounds (Thomas et al., 2000) our data provide potential insights into why that takes place.
I/LnJ H2-O and DO variants with altered function are still capable of interacting with DM
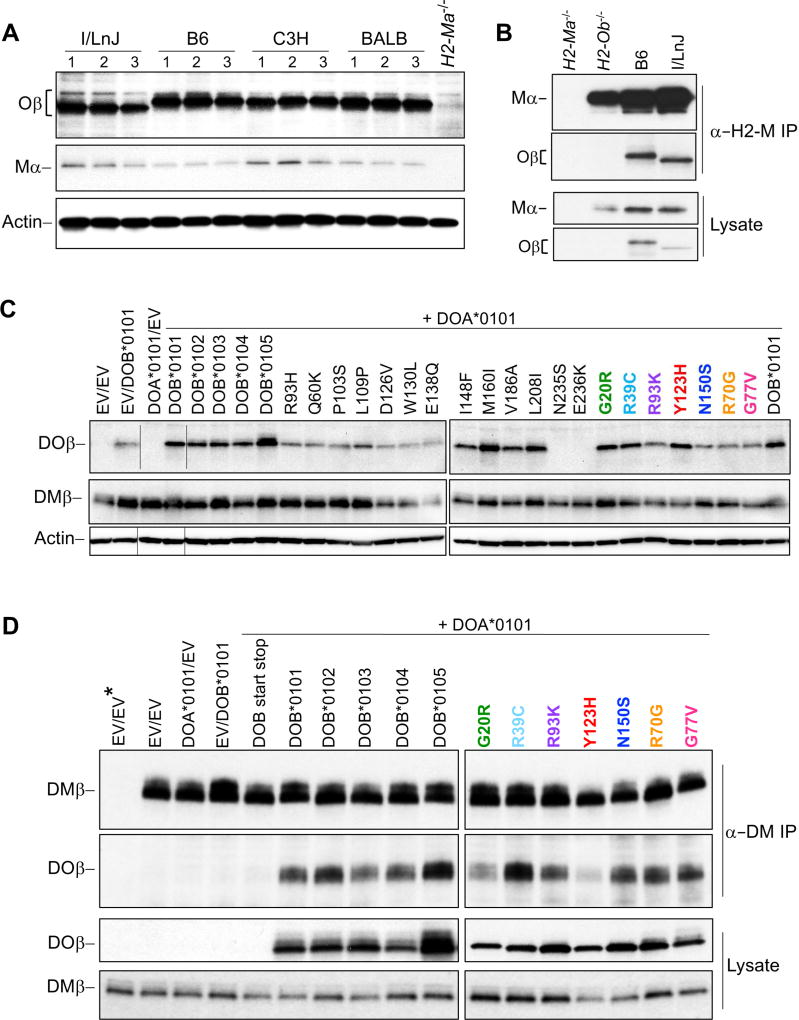
We have described a range of rare mutant alleles of the H2-Ob and HLA-DOB genes present in mice and humans and have used the ability of H2-M or DM to remove CLIP from MHC-II complexes as a proxy to measure the function of the resulting variant H2-O or DO proteins (Figures 5 and 6). Mutations found in mouse I/LnJ H2-Ob and human HLA-DOB genes resulted in enhanced or reduced inhibition of H2-M or DM. H2-M/DM and H2-O/DO are heterodimers formed by the association of Mα/DMα and Mβ/DMβ or Oα/DOα and Oβ/DOβ, respectively (Alfonso and Karlsson, 2000). H2-M/H2-O and DM/DO complexes assemble in the endoplasmic reticulum (ER) and remain associated during and after transport through the Golgi and into endosomal compartments (Liljedahl et al., 1996). Western blot analyses (Figure 7A, 7C, and S4A) and flow cytometry analyses (for DO; Figure S3B) showed that, with the exception of the frameshift and stop codon DOβ variants, I/LnJ Oβ and human DOβ variants produced detectable protein. Moreover, many of the variants, including I/LnJ Oβ, were expressed at levels similar to the common alleles (B6, BALB/cJ and C3H/HeN Oβ for mice, Figures 7A and S7A and the five DOβ known alleles for human, Figures 7D and S3B).
Figure 7. Human and I/LnJ Ob/DOB alleles interact with H2-M/DM.
(A) Oβ and Mα protein levels. B cell lysates from the indicated mouse strains were probed with Oβ luminal domain (top), Mα luminal domain (middle), or β-actin (loading control; bottom) specific antibodies. Purified H2-Ma−/− B cells were used as a negative control. Quantitation of protein levels for multiple mice are shown in Figure S7A. Note that the faster migration of I/LnJ Oβ on SDS-PAGE was restored after electrophoresis in the presence of 8M urea indicating that the observed aberrant migration was likely due to amino acid substitutions in I/LnJ that resulted in more SDS binding and not due to protein truncation (Figures S7B and S7C).
(B) H2-O and H2-M are associated in I/LnJ B cells. H2-M captured from lysates of purified B6, I/LnJ, H2-Ob−/− or H2-Ma−/− splenic B cells was analyzed by western blotting with Abs specific for Mα or for the Oβ luminal domain (top). Western blotting of lysates used for the immunoprecipitations are included as controls (bottom). Data are representative of 4 similar experiments.
(C) Conformation of DOβ variant protein expression by western blot analysis. Whole cell lysates from HeLa.CIITA cells transiently transfected with plasmids expressing HLA-DOA*0101 and the individual HLA-DOB variants (as indicated) were probed with polyclonal serum to the cytoplasmic tails of DOβ (top) or DMα (middle) or to α-actin as a control. Cells transfected with empty vectors (EV) or with HLA-DOA*0101 plus EV or HLA-DOB*0101 plus EV were included as controls. Data are representative of 4–5 similar experiments. The DOβ N235S and E236K variants have cytoplasmic tail missense mutations that abrogate recognition by the DOβ tail specific sera. Lanes (indicated by horizontal lines) in the DOβ and actin blots were spliced to present data in a logical order.
(D) All but one DOβ variant with altered function produced DO proteins that interacted with DM. DM captured from lysates of HeLa.CIITA cells transfected with plasmids encoding HLA-DOA*0101 and the seven HLA-DOB variants with altered function, with the five common alleles (positive controls) or with negative control plasmids was probed by western blotting with Abs specific for the cytoplasmic tails of DMβ or DOβ (top blots). Western blot analyses of lysates used for the immunoprecipitations are included (bottom). Data are representative of 3 similar experiments. DOB alleles with altered activity are color coded as in Figure 6. Empty vector; EV, *; indicates immunoprecipitation performed with mouse IgG as a negative control.
The most likely mechanism for reduced or enhanced H2-O/DO inhibition of H2-M/DM by the H2-O/DO variants would be altered H2-O/DO interactions with H2-M/DM. To test this idea, H2-M/DM molecules were captured from B cell lysates of I/LnJ and control B6 mice and from HLA-DOA/HLA-DOB transfected HeLa.CIITA cell lysates followed by western blotting for co-associated Oβ and DOβ proteins (Figures 7B and 7D). Results showed that I/LnJ H2-O and all but one (DOβ Y128H) of the human DOβ variants with altered function were efficiently co-precipitated with H2-M/DM (Figures 7B and 7D). Functionally, I/LnJ Oβ was most similar to the human R93K and R39C DOβ alleles. All three molecules inefficiently inhibited DM (Figures 5B, 5D and 6B) but were expressed at near normal levels and interacted with H2-M/DM (Figures 7, S3B, S7A). These data indicated that H2-M/H2-O and DO/DM binding is necessary but not sufficient for H2-O/DO inhibition of H2-M/DM and suggest that H2-O/DO function is more complex than currently appreciated.
DISCUSSION
The highly polymorphic MHC locus defines adaptive immune responses to viral infections. A candidate gene approach, based on SNPs located within the human MHC region, implicated specific alleles of the classical MHC-I genes (HLA-B and HLA-C), in HIV control for a group of ‘elite controllers’ (Kosmrlj et al., 2010; Kulkarni et al., 2011; Miura et al., 2009; Pereyra et al., 2010), and MHC-II (HLA-DQ) genes in HCV clearance (Duggal et al., 2013; Osburn et al., 2014; Pestka et al., 2007) and HBV infection outcomes (Chang et al., 2014). However, all GWASs that link a given phenotype to the HLA locus typically identify a region spread over a relatively wide area. The overall low level of recombination within the HLA locus (Kauppi et al., 2004) underlies the reason for this and makes it nearly impossible to accurately map the genes responsible for specific phenotypes. Furthermore, the definitive identification of causal genetic variants within the HLA locus is extremely difficult since almost all genes (~140) within this region are polymorphic, and approximately 40% of them participate in immune responses. Whereas the roles of classical MHC genes in anti-viral defenses are well defined, possible contributions of other immune-related genes within the locus are highly likely and should not be ignored. Uncovering the roles of these other genes requires genome manipulation, which is not experimentally possible in humans.
Although laboratory animals are not susceptible to many human pathogens, they are infected with their own pathogens that use similar strategies. Importantly, mouse genomes, unlike human genomes are easily manipulated. Early on in our studies, we found that I/LnJ mice were capable of controlling two different mouse retroviruses via the production of virus-neutralizing Abs (Case et al., 2008; Purdy et al., 2003). This finding led us to probe for the role of classical MHC molecules responsible for the phenotype as the monogenic trait responsible for the response mapped to the MHC locus (Kane et al., 2011). However, subsequent traditional positional cloning that involved virus-resistant I/LnJ and virus susceptible mice from three different backgrounds ruled out the classical MHC and allowed for discovery of a null allele of the H2-Ob gene as responsible for retrovirus control. Like I/LnJ mice, both B6 H2-Oa- and H2-Ob-deficient mice produced virus-neutralizing antibodies capable of clearing viral infection. Since I/LnJ and B6 mice have different MHC haplotypes, H2j and H2b, respectively, the virus-specific Ab response we discovered was independent of the specific MHC haplotype.
Together with Oα, Oβ forms an obligate heterodimer, H2-O, which was first discovered as an H2-M interacting protein by the Karlsson laboratory (Liljedahl et al., 1996). The tight association of H2-O/DO with H2-M/DM during assembly in the ER and during transport to and residence in endosomal compartments suggested that H2-O/DO might regulate H2-M/DM function. Such regulation was established by in vitro biochemical analyses and in experiments in which expression of DO resulted in changes in MHC-II-CLIP and MHC-II-peptide levels on the cell surface (Chen et al., 2002; Denzin et al., 1997; Liljedahl et al., 1998; van Ham et al., 1997). In vivo studies have demonstrated that H2-O impedes the ability of B cells to gain efficient T-cell help and participate in the germinal center reaction (Draghi and Denzin, 2010). Thus, it was predicted that H2-O has evolved to control humoral immunity to self. However, although some elements of autoimmunity were previously reported in aged mice from one colony of H2-O-deficient (H2-Oa−/−) mice (Gu et al., 2013) it was not supported by other studies (Liljedahl et al., 1998). These contradictory results may indicate that the Ab response observed by Gu et al. (Gu et al., 2013) might be driven by a colony-specific pathogen. The Jackson Laboratory, which has maintained the I/LnJ colony for many years, has not reported any signs of autoimmunity in these mice in any of their databases. We also have maintained a colony of I/LnJ mice for over 18 years and have not found any sign of systemic or organ-specific autoimmunity. A low degree of nuclear staining was found when sera from I/LnJ mice that were over 6 months of age were used for staining of Hep2 cells, but similar staining was also observed in age-matched BALB/cJ mice. Whereas control lpr/lpr sera produced a brightness of 10 [according to a standard scale, (Stranges et al., 2007)] at 1:100, I/LnJ (n=6) and BALB/cJ (n=5) sera showed brightness of 2.16±0.5 and 1.4±0.5, respectively at 1:10 dilution. The weak staining and a lack of difference between I/LnJ and BALB/cJ mice (p=0.3) along with the lack of abnormalities during histological examination of various tissues, let us to conclude that the absence of functional Oβ does not predispose for autoimmunity.
The highly-conserved MHC-linked H2-O-dependent mechanism of immune regulation we discovered in I/LnJ mice was unlikely to be relevant to just one type of pathogen or one species. Thus, we searched for potential contributions of HLA-DOB on the outcomes of viral infection in humans. Approximately 20% of HCV- and 90% of HBV-infected adults who received viruses via horizontal transmission spontaneously clear virus while the rest are persistently infected (Huang et al., 2006; Villano et al., 1999). The early presence of a broadly neutralizing Ab response correlates significantly with viral clearance in both HBV and HCV infected individuals (Huang et al., 2006; Osburn et al., 2014; Pestka et al., 2007). The outcome of HBV and HCV infections was linked to the HLA-DQ locus (Chang et al., 2014; Duggal et al., 2013). HLA-DOB is mapped to the LD block, which contains the DQA2-B2 locus (Figure S6) suggesting that HLA-DOB could contribute to the control of both viruses. Furthermore, the HLA-DP locus (SNP rs3077 on HLA-DPA1) which was previously linked to chronic hepatitis in HBV-infected people (Kamatani et al., 2009), was found in the LD block with HLA-DOA (SNP rs34987694 within the coding region of the gene, Table S5) (D’=1, p<0.0001). Thus, both HLA-DOB and HLA-DOA could contribute to (or even be mediating) the control of human viruses as much as the classical MHC-II genes do.
We used bioinformatics to identify HLA-DOB and HLA-DOA alleles present within the ExAc 60,000 exomes to show that these genes are more polymorphic than previously appreciated. HLA-DOB and HLA-DOA alleles included mutations in untranslated and translated regions of both genes, some leading to amino acid changes, frameshifts and stop codon insertions. More frequently represented HLA-DOB alleles with missense mutations as well as alleles predicted to produce no protein or predicted to produce DO proteins with functional implications were further analyzed for their ability to inhibit DM by monitoring MHC-II-CLIP levels. This led to the discovery of multiple HLA-DOB alleles with altered function. Coding SNPs characteristic of some of these alleles were mapped to genomic sequences in the 1000 Genomes database. The small size of this database was a limiting factor in finding linkages between coding and non-coding SNPs that are commonly used for the GWASs. Nevertheless, the DOβ G77V allele that resulted in a DO protein with enhanced DO activity was linked to HCV persistence. It is possible that the enhanced DM inhibitory activity exerted by DOβ G77V variant results in negative regulation of the anti-viral Ab response thereby allowing HCV to persist.
Since our exploration of DO function in virus control began with investigating its role in retroviral infection in a mouse, it would be highly logical to analyze the potential role of DO in HIV infections. However, in contrast to HCV, HIV infected individuals do not spontaneously clear the virus and both HIV disease-progressors and non-progressors produce neutralizing Abs. Therefore, it is not possible to genetically link general presence of HIV virus-neutralizing antibodies and viral clearance. This obstacle, of course, does not exclude the existence of DO-mediated HIV control in humans. However, proper human cohorts must be established to perform GWAS that may reveal potential contributions of HLA-DO to virus control. For example, it is possible that ‘high risk’ HIV-free people (excluding virus co-receptor-deficient individuals) with multiple documented exposures to the virus have a DO-mediated mechanism of virus control. However, this possibility remains to be explored.
How does H2-O/DO control pathogen-specific Aby responses? Loading of MHC-II molecules with peptides is mediated by the interaction of MHC-II with H2-M/DM [reviewed in (Denzin, 2013; Mellins and Stern, 2014)]. These MHC-II-peptide complexes are recognized by CD4+ T cells, which serve to help antigen-specific B cells. H2-O/DO interferes with H2-M/DM-MHC-II binding as it acts as an MHC-II mimic (Guce et al., 2013). However, our studies indicate that the precise mechanism by which H2-O/ functions is not as simple as competition between DO and MHC-II for DM binding. DOβ alleles with both enhanced and reduced function interact with H2-M/DM as does the functionally null I/LnJ Oβ allele. Thus, the mechanism by which DO and H2-O regulate MHC-II presentation is independent of their ability to bind to DM and H2-M, respectively. We have also found that I/LnJ H2-O matured normally indicating that it associated with H2-M in post-Golgi compartments. Importantly, mutations in different domains of DOβ led to similar functional consequences suggesting that the current knowledge of H2-O/DO function may be insufficient for understanding how exactly DO negatively controls virus-specific immunity.
Perhaps one of the most intriguing questions raised by these studies is why do the H2-Ob alleles expressed by most mouse strains prevent the generation of a functional adaptive immune response that neutralizes retroviral infection? The same is likely true for the human DO common variants that efficiently downregulated DM activity. The negative regulatory role of these HLA-DOB alleles suggests that their evolutionary conservation could be driven by pathogens that take advantage of exaggerated Ab responses. For example, Abs can facilitate the uptake of intracellular pathogens, such as Mycobacteria tuberculosis (Armstrong and Hart, 1975). Interestingly, the high sensitivity of I/St mice to Mycobacteria tuberculosis, which have common ancestry and identical MHC and H2-O loci with I/LnJ mice (Eruslanov et al., 2004; Nikonenko et al., 2000; Radaeva et al., 2005) is associated with a strong anti-bacterial Ab response (Radaeva et al., 2005). Thus, it is possible that the anti-mycobacterial Abs may facilitate pathogen uptake by Fc-receptor expressing macrophages and worsen disease outcome in mice (and potentially humans) who have non-functional H2-Ob or HLA-DOB.
In sum, our studies have defined an essential role for H2-O in the negative regulation of the immune response to mouse retroviruses. Results of our studies also implicate alleles of HLA-DOB as potentially influencing the control of HCV and HBV infections. Importantly, H2-O, which is mapped in the vicinity of classical MHC-II, influences anti-retrovirus Ab responses independently of the MHC-II haplotype. These results do not diminish the importance of classical MHC genes in virus clearance, but certainly implicate non-classical MHC genes in the process. Determining the precise mechanism by which the loss of DO function in controlling DM-mediated peptide loading leads to sustained neutralizing Ab responses will be essential to manipulate DO/DM interactions for therapeutic control of viral infections in humans.
STAR Methods
Contact for Reagent and Method Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Tatyana Golovkina (tgolovki@bsd.uchicago.edu).
Experimental Model and Subject Details
Mice
H2-Oa−/− (Liljedahl et al., 1998) B6 mice (Fallas et al., 2007), H2-Ma−/− (Miyazaki et al., 1996) (The Jackson Laboratory), B6.CD45.1 (The Jackson Laboratory) and B6.CD45.1.H2-Oa−/− mice were bred and maintained at the animal facility of Rutgers University. I/LnJ, C57BL/6J and BALB/cJ (The Jackson Laboratory) were bred and maintained at The University of Chicago. C3H/HeN MMTV-free mice were originally purchased from the National Cancer Institute Frederick Cancer Research Facility, Frederick, MD, and maintained in colony at The University of Chicago.
To generate the C3H/HeNvic1i/i and BALB/cJvic1i/i congenic lines, we first crossed C3H/HeN or BALB/cJ mice to I/LnJ mice to produce F1 mice. F1 mice were then backcrossed to parental C3H/HeN or BALB/cJ mice and resulting N2 females were genotyped with the markers D17Mit143 at 8.3 Mb, D17Mit89 at 63 Mb (for C3H/HeN mice) and D17Mit185 at 68.4 (for BALB/cJ mice). Mice inheriting the I/LnJ region between these 2 markers were backcrossed to C3H/HeN (or BALB/cJ mice) to produce N3 offspring. At each generation, only those offspring that had received the I/LnJ region were selected for the next round of backcrossing. Two 10th-generation carriers of the I/LnJ region were intercrossed, and offspring homozygous for both I/LnJ chromosomes were selected to continue the line through brother-sister mating (Line A, Figure 1). B6 mice congenic for the region between Mbs 25 and 63 have been published (Kane et al., 2011). All recombinant lines shown in Figure 1a were produced from the original lines carrying the region between Mbs 8.3 and 63–68.4 (for BALB/cJ and C3H/HeN mice) and between Mbs 25 and 63 (for B6 mice).
To generate MHC-I hemizygous mice, I/LnJ mice were crossed to B6. H2-Kb−/−H2-Db−/− mice, which are deficient for all classical MHC-I genes (Vugmeyster et al., 1998). The resultant F1 mice have only I/LnJ alleles of MHC-I genes (expression was confirmed by staining of F1 mouse splenic cells with anti-Kb, BD Bioscience, San Jose, CA).
To generate MHC-II hemizygous mice, we crossed I/LnJ mice to B6.MHC-II−/− mice (Madsen et al., 1999), which lack all classical MHC-II genes (MHC-IIdel/ del mice). The resultant F1 mice have only I/LnJ alleles of all classical MHC-II genes. In addition, I-E-negative B6 mice (Dembic et al., 1985) carrying a ‘floxed’ allele of the H2-Ab gene (Hashimoto et al., 2002) were first crossed to ZP3-Cre B6 transgenic females, which express Cre recombinase in oocytes (de Vries et al., 2000) and intercrossed to generate mice with H2-Ab permanently deleted (confirmed by lack of staining of splenic cells with anti-I-Ab Ab, BioLegend). These MHC-II-negative B6 mice were then crossed to I/LnJ mice to produce F1 mice that carry only MHC-II genes of the I/LnJ origin. Splenic cells of F1 mice generated in both crosses were stained with 10.2.16 (Oi et al., 1978), an anti-I-Ak Ab (ATCC), cross reactive with I-AJ to confirm expression of the I/LnJ MHC-II allele.
Overlapping B6 BACs (Children’s Hospital Oakland Resources) spanning the entire vic1 locus (from Kbs 33.7 – 34.4) were used to make 4 distinct transgenic lines with 2 founders per line (Fig. 1b). B6 BAC transgenic mice were crossed to MMTV-resistant congenic B6vic1i/i mice from Line J (Fig. 1a) and then intercrossed to generate B6vic1i/i BAC transgenic and non-transgenic mice. RP23–222K15 (K15) and RP23–208L18 (L18) contain the MHC-I and MHC-II genes, respectively. Expression of K15 and L18 transgenes was detected in peripheral blood lymphocytes and splenocytes stained with anti-I-Kb (α-MHC-I) and anti-I-Ab (α-MHC-II) Abs, respectively (not shown). To verify expression of R23–213I16 (I16) and RP23–95J18 (J18), we used our genomic sequence data to design primers specific for B6 alleles of rgl2 (I16) and tap2 (J18) and performed RT-PCR to establish that both BACs were expressed in B6vic1i/i BAC transgenic mice. Mice of both genders were used at 1:1 ratios in all experiments.
C57BL6/J (B6) H2-Ob−/− mice were produced at The Jackson Laboratory using CRISPR/Cas9 technology. Two independent lines were produced. Line 134 had a 43 bp deletion, whereas line 4905 had a 19 bp deletion in exon 3. Founder mice were crossed to B6 mice for 2 generations and then intercrossed to establish homozygous lines. Western blot analysis confirmed that H2-Ob−/− did not express any detectable Oβ protein (Figure S1) and FACS analysis showed that Oβ-expressing antigen presenting cells developed normally in H2-Ob-deficient mice (Figure S2).
The studies described here have been reviewed and approved by the Animal Care and Use Committees at The University of Chicago and Rutgers University which are both accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Viral strains and infection
Mice were infected with MMTV (LA) a naturally occurring exogenous virus consisting of three different exogenous MMTVs, BALB2, BALBLA, and BALB14 (Piazzon et al., 1994) either via fostering by viremic females (the natural way of infecting mice as the virus is transmitted via the milk) or via intraperitoneal (i.p.) injection of 6–8-week-old mice with milk-borne MMTV as previously described (Kane et al., 2011; Purdy et al., 2003). MMTV(LA), a naturally occurring exogenous virus (Piazzon et al., 1994), was used for infection. The virus was propagated in C3H/HeN mice. MMTV(LA) consists of three different exogenous MMTVs, BALB2, BALBLA, and BALB14, with Vβ2-, Vβ6-, and Vβ14-specific superantigens (SAgs), respectively (Golovkina et al., 1997; Piazzon et al., 1994). The Vβ6-specific SAg encoded by BALBLA can be presented by both the I-E and I-A molecules of MHC-II and thus is capable of efficiently infecting I-E-negative mice, like B6 mice (Buggiano et al., 1999). Deletion of CD4+/Vβ6 SAg-cognate T cells was used to confirm MMTV infection. FACS analysis of peripheral blood lymphocytes was used to measure deletion rates.
Rauscher-like MuLV (RL-MuLV), a mixture consisting of NB-tropic, ecotropic and mink lung cell focus-forming viruses, was described previously (Hook et al., 2002). The virus was propagated in BALB/cJ mice and was isolated from spleens of chronically infected animals. Ecotropic (Eco) viral titers were determined by an infectious center assay (Rowe et al., 1970). Experimental mice were injected i.p. with 2.3 × 103 Eco PFUs at 6–8 weeks of age and screened for anti-virus antibodies and PFUs 3 months later.
Cell lines
HeLa (Khalil et al., 2002) and L (Chang et al., 1994) cells transfected with the class II transactivator (CIITA; HeLa.CIITA and L-CIITA, respectively) were grown in DMEM supplemented with 5% FBS at 5% CO2 at 37°C. CIITA expression allowed for the expression of DM/H2-M and DOα/Oα but not DOβ/Oβ. CTLL-2 (ATTC) and 8-1-6 expressing T cells (Logunova et al., 2005) were grown in Click’s containing 5% FBS cultured at 5% CO2 at 37°C.
Method details
ELISA
To detect MMTV and MuLV Abs in mouse sera, an enzyme-linked immunosorbent assay (ELISA) was performed as previously described (Case et al., 2008; Chirgwin et al., 1979; Purdy et al., 2003). All sera were used at 1 × 10−2 and 5 × 10−3 dilution. I/LnJ mice produced anti-virus Abs of predominantly the IgG2a isotype, as the response is interferon gamma dependent (Case et al., 2008; Purdy et al., 2003). Therefore, mouse IgG2a- (for screening BALB/cJ, C3H/HeN congenic mice) or IgG2c–specific (for screening B6 mice), as well as total IgG-specific (for screening of all mice) secondary antibodies coupled to horseradish peroxidase (HRP) (Jackson ImmunoResearch) were used to detect anti-virus antibodies. Backgrounds obtained from incubation with secondary antibodies alone were subtracted from the values obtained from sera of infected mice.
Genome sequence of the vic1 critical region
To identify mutations unique to the virus-resistant I/LnJ strain, genomic DNA from BALB/cJ, C3H/HeN and I/LnJ mice was subjected to high-throughput whole genome sequencing using the Illumina HiSeq platform with >20X coverage. SNPs and insertions/deletions (indels) across all three strains of mice were identified using a multistage pipeline. First, reads were aligned to the reference B6 genome (GRCm38/mm10) using Bowtie2 (Langmead and Salzberg, 2012). SNPs and indels called in each strain were relative to the same reference genome. Second, alignments were base quality recalibrated and realigned to the Chr17: 33,700,000 – 34,400,000 critical region using Novoalign (http://www.novocraft.com). In addition, PCR duplicates were removed with samtools (http://samtools.sourceforge.net). Third, SNP and indels were called using the Unified Genotype and Haplotype Caller functions in GATK (http://www.broadinstitute.org/gatk/). Finally, ANNOVAR (Wang et al., 2010) was used to identify non-synonymous SNPs using the mm10 genome annotation (http://genome.ucsc.edu). The resulting SNP and indel calls were qualitatively assessed and compared to SNPs previously identified in BALB/cJ mice (Yalcin et al., 2012) to estimate sensitivity. Polymorphic changes unique to virus-resistant I/LnJ mice were identified by excluding common changes in virus-susceptible B6, C3H/HeN, and BALB/cJ mice. Gm15821 is a Gene Model that has only been computationally predicted by one gene annotation group (Ensembl) and is not included in the standard gene annotations of UCSC and NCBI. Therefore, we performed a separate analysis focused on the predicted Gm15821.
Purification of B cells
B cells were purified from the spleens of the indicated strains of mice by staining cells with biotin-conjugated Ab specific for CD19 (eBiosciences) followed by MACS (Miltenyi Biotec) using streptavidin-conjugated microbeads according to the manufacturer’s protocol. Purified B cells were >95% pure as determined by FACS analysis.
Luminal domain specific anti-Oβ sera
Rabbit polyclonal antisera (R.Obeta1) were produced by injecting rabbits with the luminal domain of Oβ that had been purified from E. coli as a His-tagged fusion protein.
Functional analyses of HLA-DOB variants
HLA-DOB gene variants and HLA-DOA*0101 (most common HLA-DOA gene variant) were synthesized and cloned into pEF1α-MCS-IRES-AcGFP1 (Clonetech Laboratories) and pCDH-EF1-MCS-IRES-RFP (System Biosciences, Inc.), respectively by GenScript. HeLa.CIITA cells seeded into 6 well plates 12–24 hours earlier were transiently transfected with 1 ug of pCDH-EF1-DOA*0101-IRES-RFP (mRuby) plasmid and 1 ug of the individual 32 pEF1α-DOB varient-IRES-AcGFP1 plasmid mixed in Lipofectamine 2000 (ThermoFischer). Transfection of HeLa.CIITA with both empty vectors (pCDH-EF1-MCS-IRES-RFP and pEF1α-MCS-IRES-AcGFP1), with pCDH-EF1-DOA*0101-IRES-RFP and pEF1α-MCS-IRES-AcGFP1 with no HLA-DOB gene inserted (empty vector) or with pEF1α-DOB*0101-IRES-AcGFP1 with pCDH-EF1-MCS-IRES-RFP with no HLA-DOA gene inserted (empty vector) were used as controls. After 72 hr, transfected cells were harvested and analyzed by flow cytometry, western blotting or immunoprecipitation. The percentage of HeLa.CIITA cells expressing both mRuby and AcGFP was measured by flow cytometry and used to determine transfection efficiency. In general, transfection efficiencies (mRuby+ AcGFP+) consistently ranged from 25–50%.
Western blot analyses and quantification
Splenocytes, purified B cells or transiently transfected L-CIITA or HeLa.CIITA cells were extracted for 30 min on ice in 20 mM Tris-HCl, 130 mM NaCl pH 8.0 containing 1% Triton X-100 and protease inhibitors (Roche Life Science). After the removal of nuclear material by centrifugation, supernatants were mixed with Laemmli sample buffer containing 20 mM DTT and incubated at 95°C for 5 min prior to separation by 10–20% gradient SDS-PAGE gels (Criterion or Criterion TGX; Bio-Rad) and transferred to polyvinylidene fluoride membrane (Millipore). Blocked membranes were incubated with a rabbit serum to the luminal domain of Oβ (R.Obeta1, see above), a monoclonal Ab specific for Mα [YoDMA.1(Fallas et al., 2004)], a hamster monoclonal Ab to the cytoplasmic tail of Oβ [Mag.Ob1(Fallas et al., 2007)], an anti-GFP (F56-GA1.2.3; cross-reacts with YFP; Frank W. Fitch Monoclonal Ab Facility, The University of Chicago ), β-actin (clone AC-74; Sigma) or rabbit serums specific for the cytoplasmic tails of DOβ [R.DOB/c (Denzin et al., 1997)] or DMβ [R.DMB/c (Denzin et al., 1994)] followed by detection with HRP-conjugated donkey or goat anti-rabbit, -mouse, or -hamster antibodies (Jackson ImmunoResearch). Blots were developed with SuperSignal West Pico chemiluminescent peroxidase substrate (Pierce Biotechnology) followed by exposure to film or by image capture using a ChemiDoc Touch Imaging System (Bio-Rad). Where indicated, protein levels were quantified by measuring relative band intensities with Image Lab software (Bio-Rad).
For the quantification of B cell Oβ and Mα levels, the relative densities obtained for Oβ and Mα were normalized to those obtained for β-actin to control for unequal sample loading. The normalized values were then used to calculate Oβ and Mα levels relative to those obtained for B6 (or H2-Ob+/+). This was accomplished by dividing the normalized Oβ and Mα levels from B cells isolated the various strains by normalized Oβ and Mα levels from B cells from B6 mice. The number of mice analyzed for each experiment are denoted in the individual Figures.
FACS analysis
The following monoclonal Abs for FACs analysis were purchased from eBiosciences, BD Biosciences or Miltenyi Biotec: B220-v450 (clone RA3–6B2), CD8-APC-CY7 (clone 53–6.7), CD11c–PE (clone HL3), CD19-Alexa 488 (clone 1D3), MHC-II-APC (clone M1/114), CD205-PE-Cy7 (clone NLDC-145) and HLA-DR, -DP, DQ-PE-Vio770 (clone REA332). The I-Ab-CLIP-specific monoclonal Ab 15G4 (Liljedahl et al., 1998) (provided by A. Rudensky, Memorial Sloan Kettering Cancer Center), the human CLIP specific monoclonal Ab CerCLIP.1 (Denzin et al., 1994) and the human DM and DO heterodimer specific monoclonal Abs MaP.DM1 and MagsDO5, respectively (Glazier et al., 2002; Hammond et al., 1998) were purified from bioreactor supernatants using standard Protein G chromatography and conjugated with Alexa 647 (15G4 and Mags.DO5) or biotin (CerCLIP.1 and MaP.DM1) according to the manufacturer’s protocol (Invitrogen).
For FACS analysis of splenic B cells and dendritic cells spleens were digested in 400 U/ml collagenase D and 100 µg/ml DNase I for 30 min at 37°C. The resulting spl enocytes were blocked with Mouse BD Fc Block (BD Biosciences), incubated on ice with antibodies specific for surface proteins for 30 min followed by washing and analysis. Data was acquired using a BD LSRII cytometer and analyzed using FlowJo software (Tree Star). Dead cells (DAPI or propidium iodide) and doublets were excluded from analyses. APC populations were identified on the basis of surface markers as follows: CD8α+ DCs: CD11c+, MHC-II+, CD8α+, CD205+; CD8α− DCs: CD11c+, MHC-II+, CD8α−, CD205−; B cells: CD19+ or CD19+, B220+.
HeLa.CIITA cells transiently transfected with plasmids encoding HLA-DOA*0101 and the various HLA-DOB variants were harvested and split for surface staining (for MHC-II-CLIP and MHC-II) and for intracellular staining (for DM and DO). For surface staining, HeLa.CIITA cells were blocked with normal mouse serum, incubated on ice with Abs specific for CLIP (CerCLIP.1-Biotin) and MHC-II for 30 min on ice and washed extensively. After the addition of streptavidin-Alexa 647 (Invitrogen), cells were incubated for 20 min on ice, washed and analyzed by flow cytometry. DAPI was added prior to analysis to allow for dead cell exclusion. For the intracellular measurement of DM and DO levels, the transiently transfected HeLa.CIITA cells were fixed and permeabilized with Cytofix/Cytoperm (BD Biosciences) for 20 min on ice. After washing cells were blocked with normal mouse serum and cells were stained for DM (MaP.DM1-biotin) and DO (Mags.DO5-Alexa 647) for 1 hour at room temperature. After washing, cells were incubated with streptavidin-Alexa 700 for 20 min at room temperature, washed and analyzed by flow cytometry. Transfection of HeLa.CIITA cells with empty vectors expressing only mRuby or AcGFP (pEF1α-MCS-IRES-AcGFP1 or pCDH-EF1-MCS-IRES-RFP) were performed to provide compensation controls for the fluorescence of mRuby and AcGFP. Data was acquired on a BD LSRII cytometer and analyzed using FlowJo software (Tree Star).
Immunoprecipitation and glycan digestions
Purified B cells (10×106; B6, I/LnJ, H2-Ma−/−, H2-Ob−/− or H2-Oa−/−) were extracted in 20 mM Tris-HCl, 130 mM NaCl pH 7.4 containing 1% Triton X-100 and protease inhibitors (Roche Life Science) for 30 min on ice. Following the removal of nuclei and cellular debris by centrifugation, lysates were precleared for 30 min at 4°C by incuba tion with rat or hamster IgG (α-H2-M or Ob1 immunoprecipitations, respectively) and 40 µl of Protein G-Sepharose (GE Healthcare Life Sciences). After centrifugation, H2-M and any co-associated H2-O was immunoprecipated from the supernatants by the addition of the α-H2-M heterodimer specific monoclonal Ab 2C3A (Fallas et al., 2007) and 40 µl of Protein G-Sepharose. Oβ was collected from H2-Oa−/− B cell lysates by immunoprecipitation with Mags.Ob1 (Fallas et al., 2007) and 40 µl of Protein G-Sepharose. Precipitates were incubated for 2 hr at 4°C and Protein-G-Sepharose pellets were washed three times with lysis buffer. Precipitated proteins were released from the immunoprecipitation pellets by heating the samples for 5 min at 95°C in 0.5% SDS and 40 mM DTT. Samples were split into three equal aliquots and digested with Endo H (NEB BioLabs), Peptide-N-Glycosidase F (NEB BioLabs) or mock digested according to the supplied protocols. Proteins were separated on 10–20% gradient SDS-PAGE gels (Criterion; Bio-Rad), transferred to polyvinylidene difluoride membranes, and analyzed by immunoblotting with antibodies specific for Mα (YoDMA.1) or for Oβ luminal domain (R.Obeta1). H2-Ma−/− and H2-Ob−/− B cells were used for negative immunoprecipitation controls. Oβ immunoprecipitates from H2-Oa−/− B cells were used as a positive control to show that the Endo H digestion worked as Oβ remains sensitive to Endo H digestion in the absence of Oα (Supplemental Figure S4).
Co- immunoprecipitation of DM co-associated DO variants from transiently transfected HeLa.CIITA cells was performed as described above for mouse H2-M and H2-O with the following modifications. Preclearing was performed with mouse IgG and the DM-DO complex was precipitated using a mixture of monoclonal antibodies specific for the cytoplasmic tail of DMβ [Map.DMB/c, (Robbins et al., 1996)] and to the DM heterodimer (Map.DM1). After washing, DM and any co-associated DO was released from the Protein G beads by the addition of Laemmli sample buffer lacking any reducing agent and heating for 5 min at 95°C. The Protein G beads were pelleted and the supernatant containing the proteins was transferred to a new tube and dithiothreitol (Sigma) was added to a final concentration of 5mM prior to separation by SDS-PAGE, transfer to membranes and analysis by western blotting.
In vitro Transcription and Translation
I/LnJ and B6 H2-Ob cDNAs cloned in pTnT (Promega) were transcribed and translated in vitro according to the manufactures protocol using the TnT SP6 coupled Wheat Germ Extract System (Promega).
Tris-Tricine SDS-PAGE with urea
B6 and I/LnJ Oβ proteins were separated in standard 10% polyacrylamide gel or gels comprised of 10% polyacrylamide, 1M Tris-HCl, pH 8.45, 0.1% SDS and 8 M Urea separating gels and a stacking gels composed of 6% polyacrylamide, 1.15 M Tris-HCl, pH 8.45 and 8 M Urea. Samples were solubilized for 5 min at 95°C in loadi ng buffer containing 8% SDS, 24% glycerol, 100 mM Tris-HCl, pH 6.8, 10 mM DTT, 0.02% Coomassie Brilliant Blue G250 and 8 M Urea prior to running gels using a cathode buffer containing 0.1M Tricine, 0.1 M Tris, 01% SDS and an anode buffer of 0.2 M Tris-HCl, pH 8.9. Electrophoresis was performed at 30–40 V until the samples completely entered the stacking gel and then the voltage was increased to 150 V and run until the Coomassie Brilliant Blue ran off the gel. Gels were transferred to polyvinylidene difluoride membranes, and analyzed by immunoblotting with appropriate antibodies as described above.
Oβ-YFP fusion proteins and analysis
YFP was fused to the C-terminus of B6 and I/LnJ H2-Ob utilizing standard PCR and cloning procedures. The resulting construct contained a 15 amino acid linker (Leu-Glu-Gly-Gly-Ser-Gly-Gly-Ser-Gly-Gly Ser-Gly-Gly-Ser-Gly) between Oβ and YFP. The final constructs were confirmed by DNA sequencing and subcloned into the pUB-EGFP-Thy1.1 vector from which EGFP had been excised (Zhou et al., 2009). Analysis of the I/LnJ and B6 Oβ-YFP fusion proteins was performed by Lipofectamine (Invitrogen) mediated transfection of the plasmids into L-CIITA cells. Forty-eight hr later cells were harvested, lysed and analyzed by western blotting as described above.
T cell activation assay
Purified B cells (3×105) and 5×104 8-1-6 expressing T cells [lymphoma cell line transfected with T cell receptor chains isolated from a previously described hybridoma (Logunova et al., 2005)] were plated in flat bottom 96-well-plates. Eighteen hours later, culture supernatants were frozen and screened for IL-2 by color conversion of Alamar Blue (Thermo Fisher) by an IL-2– dependent cell line (CTLL-2, ATCC) as described (Logunova et al., 2005). For Ab blocking studies, titrated doses (1:4 to 1:250) of the I-Ab specific monoclonal Ab Y3JP (Janeway et al., 1984) or a control Ab reactive with I-Aj (10.2.16) (Oi et al., 1978) were added to the assay and analyzed for IL-2 secretion as above.
Virus neutralization
Sera from MMTV-infected H2-Oa−/− and H2-Ob−/− and control mice were tested for their ability to neutralize virus in two types of assays. First, and as performed for H2-Ob−/− and their corresponding control mice, each serum diluted at 1/10 with PBS was incubated with purified MMTV(LA) for 2 h at room temperature and injected into 3 footpads of BALB/cJ mice. Four days after injection, cells isolated from the draining popliteal lymph node were analyzed by FACS for the percentage of CD4+/Vβ6+ T cells among CD4+ T cells. MMTV-encoded SAg stimulates cognate T cells to proliferate during initial stages of infection (Acha-Orbea and MacDonald, 1995). Proliferating T cells subsequently undergo clonal deletion (Acha-Orbea and MacDonald, 1995). The proliferation of SAg cognate T cells was used as indicator of virus infectivity (Finke et al., 2003). The amount of MMTV(LA) used was titrated to give an increase from 10% to 25% of SAg-reactive T cells four days after virus injection. Neutralization (%) was calculated as follows: (a–c)-(b–c):(a–c)X100 where a= mean of CD4+/Vβ6+ (%) in mice injected with virus plus sera from uninfected mice, b= mean of CD4+/Vβ6+ (%) in experimental mice and c=mean of CD4+/Vβ6+(%) in uninfected mice (Figure 4b, right panel).
Alternatively, and as performed for H2-Oa−/− and their corresponding control mice, serum diluted at 1/10 with PBS was incubated with purified MMTV(LA) virions for 2 h at room temperature and then injected i.p. into 3 BALB/cJ mice as described (Purdy et al., 2003). Five weeks after injection, mice were bled and peripheral T cells were analyzed by FACS for the percentage of CD4+/Vβ6+ T cells among CD4+ T cells. The deletion of SAg-cognate T cells was used as indicator of virus infectivity. Neutralization (%) was calculated as follows: 1- [(a–c)-(b–c):(a–c)]X100, where a = mean of CD4+/Vβ6+ (%) in uninfected mice, b = mean of CD4+/Vβ6+ (%) in experimental mice and c = mean of CD4+/Vβ6+ (%) in mice injected with virus plus sera from uninfected mice (Figure 4a, right panel).
Quantification and Statistical Analysis
Significance was calculated using unpaired t tests as indicated in relevant figure legends.
LD analysis
The original LD block in the MHC-II region identified by three SNPs, rs9276370, rs7756516 and rs7453920 (Chang et al., 2014) did not include HLA-DOB specific SNPs in the analysis. We used LDlink (Machiela and Chanock, 2015) to calculate a new heatmap of pairwise LD statistics in the MHC-II region. Data from all populations in 1000 Genomes Project was used to calculate D’ scores. We calculated the D’ score or measure of LD (normalized for allele frequency) for each pair of variants. The analysis reproducibly detected the original LD block identified in the MHC-II region. We also found an association between rs144814623 and rs4273729 with D’ = 1 and P < 0.0016; chi square test, suggesting both SNPs were associated. Variation in rs144814623 was observed only in individuals with African ancestry in 1000 Genomes, including GWD (Gambian in Western Divisions in the Gambia), LWK (Luhya in Webuye, Kenya), MSL (Mende in Sierra Leone) and YRI (Yoruba in Ibadan, Nigeria).
Supplementary Material
I/LnJ mice are resistant to retroviruses by producing virus-neutralizing antibodies
Resistance is conferred by mutant non-classical MHC class II molecule H2-O
Mutations in several alleles of human HLA-DO affected antigen presentation
Wild-type H2-O and HLA-DO serve as negative regulators of anti-viral immunity
Genetics affects sensitivity of animals and humans to viral infections. Denzin et al. show mice from the I/LnJ strain produce neutralizing antibodies against retroviruses due to inactivation of a beta subunit of non-classical Major Histocompatibility Complex (MHC) protein H2-O. Mutations found in human orthologous HLA-DO genes may explain resistance or sensitivity to Hepatitis B and C viruses.
Acknowledgments
We are thankful to Derek Sant’Angelo, Alexander Mankin and members of the Golovkina, Chervonsky and Denzin laboratories for discussions and Louis Osorio, Emmily Shanks and Kelly O’Grady for technical assistance. This work was supported by PHS grant CA134667 to T.V.G., AI117535 to T.V.G. and L.K.D., AI061484 to L.K.D., by T32 AI 007090 to J.W., by P30 CA014599 to the University of Chicago, by the National Center for Advancing Translational Sciences of the National Institutes of Health through Grant Number UL1 TR000430 to the University of Chicago and by the Robert Wood Johnson Foundation (grant number 67038) to the Child Health Institute of New Jersey. Whole genome sequencing of mouse strains was performed by the University of Chicago Genomics Facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
L.K.D. designed and performed the human DOβ functional screen and biochemical experiments, assisted with other experiments and contributed to the discussion of results and the writing of the manuscript. J.W., M.K., H.A.B., S.D. and L.K.C. performed genetic mapping studies. A.A.K. performed the mouse and human genome data analyses. D.R. assisted in the generation of H2-Ob−/− mice. F.V. contributed to biochemical experiments and performed the DOb functional screen. M.W. contributed to biochemical experiments. A.V.C contributed to T cell activation experiments, discussion of the results and writing of the manuscript. T.V.G. conceived the project, guided the genetic mapping, contributed to all experiments and to writing of the manuscript.
References
- Acha-Orbea H, MacDonald HR. Superantigens of mouse mammary tumor virus. Annu Rev Immunol. 1995;13:459–486. doi: 10.1146/annurev.iy.13.040195.002331. [DOI] [PubMed] [Google Scholar]
- Alfonso C, Karlsson L. Nonclassical MHC class II molecules. Annu Rev Immunol. 2000;18:113–142. doi: 10.1146/annurev.immunol.18.1.113. [DOI] [PubMed] [Google Scholar]
- Armstrong JA, Hart PD. Phagosome-lysosome interactions in cultured macrophages infected with virulent tubercle bacilli. Reversal of the usual nonfusion pattern and observations on bacterial survival. J Exp Med. 1975;142:1–16. doi: 10.1084/jem.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggiano V, Goldman A, Nepomnaschy I, Bekinschtein P, Berguer P, Lombardi G, Deroche A, Francisco MV, Piazzon I. Characterization of two infectious mouse mammary tumour viruses: superantigenicity and tumorigenicity. Scand J Immunol. 1999;49:269–277. doi: 10.1046/j.1365-3083.1999.00502.x. [DOI] [PubMed] [Google Scholar]
- Case LK, Petell L, Yurkovetskiy L, Purdy A, Savage KJ, Golovkina TV. Replication of beta- and gammaretroviruses is restricted in I/LnJ mice via the same genetic mechanism. Journal of virology. 2008;82:1438–1447. doi: 10.1128/JVI.01991-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LK, Purdy A, Golovkina TV. Molecular and cellular basis of the retrovirus resistance in I/LnJ mice. J Immunol. 2005;175:7543–7549. doi: 10.4049/jimmunol.175.11.7543. [DOI] [PubMed] [Google Scholar]
- Chang CH, Fontes JD, Peterlin M, Flavell RA. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SW, Fann CS, Su WH, Wang YC, Weng CC, Yu CJ, Hsu CL, Hsieh AR, Chien RN, Chu CM, et al. A genome-wide association study on chronic HBV infection and its clinical progression in male Han-Taiwanese. PLoS One. 2014;9:e99724. doi: 10.1371/journal.pone.0099724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Laur O, Kambayashi T, Li S, Bray RA, Weber DA, Karlsson L, Jensen PE. Regulated expression of human histocompatibility leukocyte antigen (HLA)-DO during antigen-dependent and antigen-independent phases of B cell development. J Exp Med. 2002;195:1053–1062. doi: 10.1084/jem.20012066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirgwin JM, Prxybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;18:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Ciupe SM, Ribeiro RM, Perelson AS. Antibody responses during hepatitis B viral infection. PLoS Comput Biol. 2014;10:e1003730. doi: 10.1371/journal.pcbi.1003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis. 2000;26:110–112. [PubMed] [Google Scholar]
- Dembic Z, Ayane M, Klein J, Steinmetz M, Benoist CO, Mathis DJ. Inbred and wild mice carry identical deletions in their E alpha MHC genes. EMBO J. 1985;4:127. doi: 10.1002/j.1460-2075.1985.tb02326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzin LK. Inhibition of HLA-DM Mediated MHC Class II Peptide Loading by HLA-DO Promotes Self Tolerance. Frontiers in immunology. 2013;4:465. doi: 10.3389/fimmu.2013.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzin LK, Robbins NF, Carboy-Newcomb C, Cresswell P. Assembly and intracellular transport of HLA-DM and correction of the class II antigen-processing defect in T2 cells. Immunity. 1994;1:595–606. doi: 10.1016/1074-7613(94)90049-3. [DOI] [PubMed] [Google Scholar]
- Denzin LK, Sant’Angelo DB, Hammond C, Surman MJ, Cresswell P. Negative regulation by HLA-DO of MHC class II-restricted antigen processing. Science. 1997;278:106–109. doi: 10.1126/science.278.5335.106. [DOI] [PubMed] [Google Scholar]
- Dudley JP, Golovkina TV, Ross SR. Lessons Learned from Mouse Mammary Tumor Virus in Animal Models. ILAR J. 2016;57:12–23. doi: 10.1093/ilar/ilv044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal P, Thio CL, Wojcik GL, Goedert JJ, Mangia A, Latanich R, Kim AY, Lauer GM, Chung RT, Peters MG, et al. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann Intern Med. 2013;158:235–245. doi: 10.7326/0003-4819-158-4-201302190-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eruslanov EB, Majorov KB, Orlova MO, Mischenko VV, Kondratieva TK, Apt AS, Lyadova IV. Lung cell responses to M. tuberculosis in genetically susceptible and resistant mice following intratracheal challenge. Clinical and experimental immunology. 2004;135:19–28. doi: 10.1111/j.1365-2249.2004.02328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallas JL, Tobin HM, Lou O, Guo D, Sant’Angelo DB, Denzin LK. Ectopic expression of HLA-DO in mouse dendritic cells diminishes MHC class II antigen presentation. J Immunol. 2004;173:1549–1560. doi: 10.4049/jimmunol.173.3.1549. [DOI] [PubMed] [Google Scholar]
- Fallas JL, Yi W, Draghi NA, O’Rourke HM, Denzin LK. Expression patterns of H2-O in mouse B cells and dendritic cells correlate with cell function. J Immunol. 2007;178:1488–1497. doi: 10.4049/jimmunol.178.3.1488. [DOI] [PubMed] [Google Scholar]
- Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke D, Luther SA, Acha-Orbea H. The role of neutralizing antibodies for mouse mammary tumor virus transmission and mammary cancer development. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:199–204. doi: 10.1073/pnas.0134988100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazier KS, Hake SB, Tobin HM, Chadburn A, Schattner EJ, Denzin LK. Germinal center B cells regulate their capability to present antigen by modulation of HLA-DO. J Exp Med. 2002;195:1063–1069. doi: 10.1084/jem.20012059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkina TV. A novel mechanism of resistance to mouse mammary tumor virus infection. Journal of virology. 2000;74:2752–2759. doi: 10.1128/jvi.74.6.2752-2759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkina TV, Piazzon I, Nepomnaschy I, Buggiano V, de Olano Vela M, Ross SR. Generation of a tumorigenic milk-borne mouse mammary tumor virus by recombination between endogenous and exogenous viruses. J Virol. 1997;71:3895–3903. doi: 10.1128/jvi.71.5.3895-3903.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Jensen PE, Chen X. Immunodeficiency and autoimmunity in H2-O-deficient mice. J Immunol. 2013;190:126–137. doi: 10.4049/jimmunol.1200993. [DOI] [PubMed] [Google Scholar]
- Guce AI, Mortimer SE, Yoon T, Painter CA, Jiang W, Mellins ED, Stern LJ. HLA-DO acts as a substrate mimic to inhibit HLA-DM by a competitive mechanism. Nature structural & molecular biology. 2013;20:90–98. doi: 10.1038/nsmb.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C, Denzin LK, Pan M, Griffith JM, Geuze HJ, Cresswell P. The tetraspan protein CD82 is a resident of MHC class II compartments where it associates with HLA-DR, -DM, and -DO molecules. J Immunol. 1998;161:3282–3291. [PubMed] [Google Scholar]
- Hasenkrug KJ, Valenzuela A, Letts VA, Nishio J, Chesebro B, Frankel WN. Chromosome mapping of Rfv3, a host resistance gene to Friend murine retrovirus. J Virol. 1995;69:2617–2620. doi: 10.1128/jvi.69.4.2617-2620.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Joshi SK, Koni PA. A conditional null allele of the major histocompatibility IA-beta chain gene. Genesis. 2002;32:152–153. doi: 10.1002/gene.10056. [DOI] [PubMed] [Google Scholar]
- Hook LM, Jude BA, Ter-Grigorov VS, Hartley JW, Morse HC, 3rd, Trainin Z, Toder V, Chervonsky AV, Golovkina TV. Characterization of a novel murine retrovirus mixture that facilitates hematopoiesis. Journal of virology. 2002;76:12112–12122. doi: 10.1128/JVI.76.23.12112-12122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, Lin SS, Ho YC, Chen FL, Yang CC. The immune response induced by hepatitis B virus principal antigens. Cell Mol Immunol. 2006;3:97–106. [PubMed] [Google Scholar]
- Huang P, Zhang Y, Lu X, Xu Y, Wang J, Zhang Y, Yu R, Su J. Association of polymorphisms in HLA antigen presentation-related genes with the outcomes of HCV infection. PLoS One. 2015;10:e0123513. doi: 10.1371/journal.pone.0123513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA, Jr, Conrad PJ, Lerner EA, Babich J, Wettstein P, Murphy DB. Monoclonal antibodies specific for Ia glycoproteins raised by immunization with activated T cells: possible role of T cellbound Ia antigens as targets of immunoregulatory T cells. J Immunol. 1984;132:662–667. [PubMed] [Google Scholar]
- Kamatani Y, Wattanapokayakit S, Ochi H, Kawaguchi T, Takahashi A, Hosono N, Kubo M, Tsunoda T, Kamatani N, Kumada H, et al. A genome-wide association study identifies variants in the HLA-DP locus associated with chronic hepatitis B in Asians. Nat Genet. 2009;41:591–595. doi: 10.1038/ng.348. [DOI] [PubMed] [Google Scholar]
- Kane M, Case LK, Wang C, Yurkovetskiy L, Dikiy S, Golovkina TV. Innate Immune Sensing of Retroviral Infection via Toll-like Receptor 7 Occurs upon Viral Entry. Immunity. 2011;35:135–145. doi: 10.1016/j.immuni.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson L, Surh CD, Sprent J, Peterson PA. A novel class II MHC molecule with unusual tissue distribution. Nature. 1991;351:485–488. doi: 10.1038/351485a0. [DOI] [PubMed] [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- Khalil H, Deshaies F, Bellemare-Pelletier A, Brunet A, Faubert A, Azar GA, Thibodeau J. Class II transactivator-induced expression of HLA-DO(beta) in HeLa cells. Tissue Antigens. 2002;60:372–382. doi: 10.1034/j.1399-0039.2002.600504.x. [DOI] [PubMed] [Google Scholar]
- Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Savan R, Qi Y, Gao X, Yuki Y, Bass SE, Martin MP, Hunt P, Deeks SG, Telenti A, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Si L, Zhai Y, Hu Y, Hu Z, Bei JX, Xie B, Ren Q, Cao P, Yang F, et al. Genome-wide association study identifies 8p21.3 associated with persistent hepatitis B virus infection among Chinese. Nat Commun. 2016;7:11664. doi: 10.1038/ncomms11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljedahl M, Kuwana T, Fung-Leung WP, Jackson MR, Peterson PA, Karlsson L. HLA-DO is a lysosomal resident which requires association with HLA-DM for efficient intracellular transport. The EMBO journal. 1996;15:4817–4824. [PMC free article] [PubMed] [Google Scholar]
- Liljedahl M, Winqvist O, Surh CD, Wong P, Ngo K, Teyton L, Peterson PA, Brunmark A, Rudensky AY, Fung-Leung WP, et al. Altered antigen presentation in mice lacking H2-O. Immunity. 1998;8:233–243. doi: 10.1016/s1074-7613(00)80475-6. [DOI] [PubMed] [Google Scholar]
- Logunova NN, Viret C, Pobezinsky LA, Miller SA, Kazansky DB, Sundberg JP, Chervonsky AV. Restricted MHC-peptide repertoire predisposes to autoimmunity. J Exp Med. 2005;202:73–84. doi: 10.1084/jem.20050198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen L, Labrecque N, Engberg J, Dierich A, Svejgaard A, Benoist C, Mathis D, Fugger L. Mice lacking all conventional MHC class II genes. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:10338–10343. doi: 10.1073/pnas.96.18.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellins ED, Stern LJ. HLA-DM and HLA-DO, key regulators of MHC-II processing and presentation. Current opinion in immunology. 2014;26:115–122. doi: 10.1016/j.coi.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura T, Brockman MA, Brumme ZL, Brumme CJ, Pereyra F, Trocha A, Block BL, Schneidewind A, Allen TM, Heckerman D, et al. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. Journal of virology. 2009;83:140–149. doi: 10.1128/JVI.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Mice lacking H2-M complexes, enigmatic elements of the MHC class II peptide-loading pathway. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- Miyazawa M, Tsuji-Kawahara S, Kanari Y. Host genetic factors that control immune responses to retrovirus infections. Vaccine. 2008;26:2981–2996. doi: 10.1016/j.vaccine.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Nikonenko BV, Averbakh MM, Jr, Lavebratt C, Schurr E, Apt AS. Comparative analysis of mycobacterial infections in susceptible I/St and resistant A/Sn inbred mice. Tubercle and lung disease : the official journal of the International Union against Tuberculosis and Lung Disease. 2000;80:15–25. doi: 10.1054/tuld.1999.0225. [DOI] [PubMed] [Google Scholar]
- Oi VT, Jones PP, Goding JW, Herzenberg LA, Herzenberg LA. Properties of monoclonal antibodies to mouse Ig allotypes, H-2, and Ia antigens. Curr Top Microbiol Immunol. 1978;81:115–120. doi: 10.1007/978-3-642-67448-8_18. [DOI] [PubMed] [Google Scholar]
- Osburn WO, Snider AE, Wells BL, Latanich R, Bailey JR, Thomas DL, Cox AL, Ray SC. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology. 2014;59:2140–2151. doi: 10.1002/hep.27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka JM, Zeisel MB, Blaser E, Schurmann P, Bartosch B, Cosset FL, Patel AH, Meisel H, Baumert J, Viazov S, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:6025–6030. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazzon I, Goldman A, Torello S, Nepomnaschy I, Deroche A, Dran G. Transmission of an Mls-1a–like superantigen to BALB/c mice by foster-nursing on F1 Mls-1bxa mothers. Sex-influenced onset of clonal deletion. J Immunol. 1994;153:1553–1562. [PubMed] [Google Scholar]
- Purdy A, Case L, Duvall M, Overstrom-Coleman M, Monnier N, Chervonsky A, Golovkina T. Unique resistance of I/LnJ mice to a retrovirus is due to sustained interferon gamma-dependent production of virus-neutralizing antibodies. J Exp Med. 2003;197:233–243. doi: 10.1084/jem.20021499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radaeva TV, Nikonenko BV, Mischenko VV, Averbakh MM, Jr, Apt AS. Direct comparison of low-dose and Cornell-like models of chronic and reactivation tuberculosis in genetically susceptible I/St and resistant B6 mice. Tuberculosis (Edinb) 2005;85:65–72. doi: 10.1016/j.tube.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Robbins NF, Hammond C, Denzin LK, Pan M, Cresswell P. Trafficking of major histocompatibility complex class II molecules through intracellular compartments containing HLA-DM. Hum Immunol. 1996;45:13–23. doi: 10.1016/0198-8859(95)00152-2. [DOI] [PubMed] [Google Scholar]
- Rowe WP, Pugh WE, Hartley JW. Plaque assay techniques for murine leukemia viruses. Virology. 1970;42:1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- Stranges PB, Watson J, Cooper CJ, Choisy-Rossi CM, Stonebraker AC, Beighton RA, Hartig H, Sundberg JP, Servick S, Kaufmann G, et al. Elimination of antigen-presenting cells and autoreactive T cells by Fas contributes to prevention of autoimmunity. Immunity. 2007;26:629–641. doi: 10.1016/j.immuni.2007.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DL, Astemborski J, Rai RM, Anania FA, Schaeffer M, Galai N, Nolt K, Nelson KE, Strathdee SA, Johnson L, et al. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA. 2000;284:450–456. doi: 10.1001/jama.284.4.450. [DOI] [PubMed] [Google Scholar]
- van Ham SM, Tjin EP, Lillemeier BF, Gruneberg U, van Meijgaarden KE, Pastoors L, Verwoerd D, Tulp A, Canas B, Rahman D, et al. HLA-DO is a negative modulator of HLA-DM-mediated MHC class II peptide loading. Curr Biol. 1997;7:950–957. doi: 10.1016/s0960-9822(06)00414-3. [DOI] [PubMed] [Google Scholar]
- Villano SA, Vlahov D, Nelson KE, Cohn S, Thomas DL. Persistence of viremia and the importance of long-term follow-up after acute hepatitis C infection. Hepatology. 1999;29:908–914. doi: 10.1002/hep.510290311. [DOI] [PubMed] [Google Scholar]
- Vugmeyster Y, Glas R, Perarnau B, Lemonnier FA, Eisen H, Ploegh H. Major histocompatibility complex (MHC) class I KbDb −/− deficient mice possess functional CD8+ T cells and natural killer cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12492–12497. doi: 10.1073/pnas.95.21.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nature Reviews Immunology. 2013;13:487–498. doi: 10.1038/nri3478. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin B, Wong K, Bhomra A, Goodson M, Keane TM, Adams DJ, Flint J. The fine-scale architecture of structural variants in 17 mouse genomes. Genome biology. 2012;13:R18. doi: 10.1186/gb-2012-13-3-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Kurt-Jones EA, Cerny AM, Chan M, Bronson RT, Finberg RW. MyD88 intrinsically regulates CD4 T-cell responses. Journal of virology. 2009;83:1625–1634. doi: 10.1128/JVI.01770-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.