Abstract
Aims: Archived newborn bloodspots are valuable sample collections for genetic and epigenetic disease research. However, they have often been stored for long periods of time, under less than ideal circumstances, and nucleic acid yields can be low, particularly when samples become limiting. We wished to determine whether the quantity and quality of genomic DNA (gDNA) isolated from a single, surgical bloodspot punch (2 mm dia.) was adequate for accurate and reliable DNA methylome profiling on the Illumina HumanMethylation450 array.
Methods: A total of 25–750 ng of archived bloodspot or Jurkat cell gDNA were bisulfite converted and analyzed on the array without any additional DNA amplification steps. Methylation profiles were assessed for call rate, call confidence (detection p-value), and reproducibility.
Results: Using 25 ng gDNA from either Jurkat cells or dried bloodspots, array-wide call rates (∼99.9%) and detection p-values (99.9% with p < 5 × 10−6) were excellent. There was good agreement between methylation profiles generated from 25 ng gDNA and those generated from 750 ng (ρ > 0.98), although a fraction of CpG sites (2–8% depending on experiment) exhibited quantitative differences. Genome-wide methylation levels were strikingly reproducible from 25 ng DNA in both replicate and interindividual samples (ρ > 0.98).
Conclusions: Twenty-five nanograms of gDNA, isolated from a single, surgical punch (2 mm dia.) of an archived newborn bloodspot, generate a genome-wide methylation profile on the Illumina HumanMethylation450 array that is robust, reproducible, and suitable for differential methylation studies.
Keywords: : DNA methylation, epigenetics, bloodspot
Introduction
DNA methylation at CpG dinucleotides is an epigenetic modification that can affect gene expression and chromosome stability. The link between DNA methylation changes and disease in humans is well established (e.g., Ngollo et al., 2014). Archived newborn bloodspots are a valuable source of genomic DNA (gDNA) for epigenetic research because they reveal DNA methylation states established in utero. Such states may underlie adverse pregnancy outcomes (e.g., O'Neill et al., 2014) or contribute to clinical phenotypes observed in later life (Ramagopalan and Rakyan, 2013).
DNA methylation states are often assessed after bisulfite conversion of gDNA, which converts unmethylated cytosines into uracil. This process is harsh, resulting in DNA degradation, which necessitates starting with relatively large quantities of gDNA (hundreds of nanograms or more). It has previously been established that gDNA from archived bloodspots is suitable for genome-wide DNA methylation analysis, given such a sizeable starting quantity (Beyan et al., 2012; Hollegaard et al., 2013; Joo et al., 2013). However, because bloodspot material is often limited, researchers are sometimes forced to work with small portions (“punches”) of the original spot, thereby constraining gDNA yield. Adding to this, bloodspots may have been stored for long periods of time, possibly rendering these yields insufficient or inconsistent in quality for DNA methylation analysis.
Here we show that small quantities of gDNA (∼25 ng) isolated from a single surgical punch (2 mm dia.) from an archived bloodspot are suitable, with minimal drawbacks, for genome-wide methylation analysis on the Infinium HumanMethylation450K array (Illumina).
Materials and Methods
gDNA used for DNA methylation analysis came from two sources. Jurkat cell line gDNA (male) was purchased from BioChain, Inc. (Newark, CA). Bloodspot gDNA was extracted from dried, archived spots (2 mm dia.) using the QIAamp DNA Micro kit (QIAGEN) as described (Marini et al., 2011). Yields averaged 40 ng gDNA with most samples returning ≥25 ng. All gDNA preparations were bisulfite converted using the EpiTect Bisulfite kit (QIAGEN) using starting gDNA quantities indicated in the figures. Converted gDNA was eluted in a final volume of 16 μL. Four microliters of eluate was used for processing on the Illumina HumanMethylation450 bead array as per manufacturer's instructions.
Replicate samples (technical replicates) were from the same gDNA preparation, but were bisulfite converted and processed on the array independently. All comparative analyses were performed on non-normalized, raw data to minimize the impact of data transformation (exported from Genome Studio v.1.1; Illumina). Parameters analyzed were signal intensity (sum of methylated and unmethylated signals for a CpG site), detection p-value (confidence that signal is distinguishable from background), and β-value (fraction methylated).
Out of 485,577 CpG sites interrogated on the array, only loci with missing data were eliminated from analyses. For interindividual comparisons (Fig. 3), array probes known to overlap common polymorphisms were removed to minimize individual differences not related to DNA methylation (see Results section). Pair-wise comparisons were quantified by Pearson correlation coefficient (ρ).
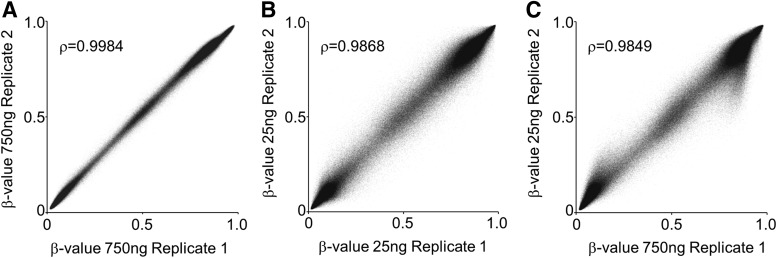
FIG. 3.
Reproducibility of DNA methylation profiles from 25 ng of gDNA isolated from small archived dried bloodspots. (A) Correlation plot of array β-values from 25 ng replicate samples. (B) Correlation plot of array β-values between two independent samples representing the median correlation coefficient (0.9873) from all pair-wise comparisons (n = 190) of 20 individuals. (C) Distribution of the standard deviations of mean β-values obtained from averaging array loci data from 20 individuals.
This study was approved by the California State Committee for the Protection of Human Subjects as well as Institutional Review Boards at Stanford University and the University of California, Berkeley. Bloodspots were collected from 1986 to 2003 and stored frozen.
Results
To determine the feasibility of using bloodspot surgical punches (2 mm dia.; ≥25 ng gDNA yields) for DNA methylation analysis on the Illumina 450K platform, we first assessed call rate (β-value), confidence (detection p-value), and reproducibility when starting with low quantities of a standard gDNA preparation (Jurkat cells; Figs. 1 and 2).
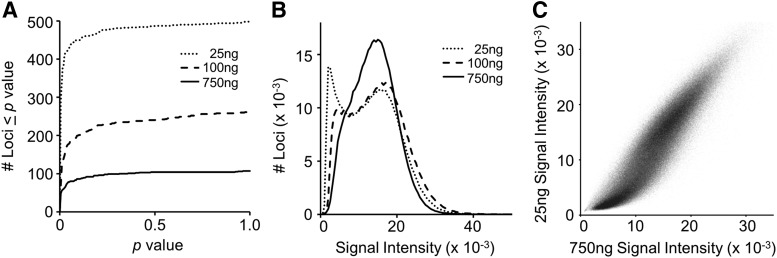
FIG. 1.
Signal detection attributes of array loci using different starting quantities (25, 100, and 750 ng) of Jurkat cell genomic DNA (gDNA). (A) The cumulative distributions of detection p-values for the subset of array loci with p > 5 × 10−6. (B) The distributions of raw signal intensities for 483,280 array positions present at all starting DNA concentrations. (C) Pair-wise comparison of the signal intensities in the 25 and 750 ng samples of the loci depicted in (B).
FIG. 2.
Correlation plots of DNA methylation profiles between replicates from high (750 ng) and low (25 ng) starting quantities of Jurkat cell gDNA. (A) Array β-values from 750 ng gDNA technical replicates (see Methods section for definition). (B) Array β-values from 25 ng replicates. (C) Array β-values from 750 ng replicate 1 compared with 25 ng replicate 2. The graphs depict 483,280 array positions present in all samples tested; ρ = correlation coefficient.
Surprisingly, the β-value call rates for CpG sites in samples starting with either 25, 100, or 750 ng gDNA were virtually unchanged at 99.9% (average number missing sites = 569, n = 6 samples; Fig. 1A). Furthermore, the signals from nearly all present CpGs were detected with the same, high degree of confidence (detection p < 5 × 10−6) irrespective of starting concentration. Although Figure 1A shows that the number of loci with detection p ≥ 5 × 10−6 increases with decreasing gDNA quantities, the number of these “lower confidence” CpG sites still represents only a small fraction of CpG sites queried (∼0.1%).
Figure 1B shows the distributions of signal intensities for array CpG sites as a function of gDNA input. At lower inputs, there was a shift of some probes toward the lower end of the intensity spectrum. A pair-wise comparison (Fig. 1C) shows that signal intensity was highly correlated between 25 and 750 ng (ρ = 0.9231) and that most loci had similar signal intensities (317,139 CpGs [66%] deviate by ≤20%).
Nevertheless, a subset of sites (39,729; 8%) showed a greater than twofold variation, most of which are on the low end of the intensity spectrum and account for the lower intensity shift seen in Figure 1B. Notably, these are almost exclusively CpG sites queried by Infinium II-type assays (single probe design versus the two-probe design of Infinium I assays, both of which are utilized in the array). Importantly, although these probes are more sensitive to intensity loss with lower starting DNA, these signals (and corresponding β-values) were called with high confidence and were reproducible in 25 ng gDNA replicates (Figs. 2 and 3).
A strong correlation was consistently observed between pair-wise comparisons of 25 ng gDNA replicates, although methylation levels are slightly more variable at some loci (reflected in marginally lower correlations; compare Fig. 2A, B). Figure 2C shows that the methylation pattern observed using 25 ng gDNA largely reflected that from the 750 ng input. However, this comparison revealed a subset of ∼11,000 CpG loci (<2.5% of array) with increased variability (most obvious in Fig. 2C as “clouds” emanating from the fully methylated or unmethylated clusters). Although this small fraction of sites (ranging from 2% to 6%, depending on experiment) is sensitive to input gDNA quantity, the β-values measured within replicates of the same gDNA input (even 25 ng) were quite consistent (Figs. 2B and 3). As already discussed, this group is enriched for Infinium type II assays with lower signal intensities (<3000). Thus, DNA methylation profiling with gDNA inputs as low as 25 ng is robust and reliable, although there are some minor differences and some increased variability.
To further examine the feasibility of DNA methylome analysis on small quantities of gDNA (∼25 ng) from bloodspots, we extracted and bisulfite converted gDNA from surgical punches (2 mm dia.) from 20 unrelated white newborns and processed these on the Illumina 450K array (see Supplementary Table S1 for individual bloodspot gDNA yields and quantities used; Supplementary Data are available online at www.liebertpub.com/gtmb). β-value call rates (mean = 99.9%), detection p-values (mean = 99.9% with p < 5 × 10−6), and technical reproducibility (Fig. 3A) were excellent and similar to those for the 25 ng Jurkat gDNA profiles.
We further compared interindividual DNA methylation patterns as a stringent test for sample-to-sample variation, with the expectation that profiles from these additional 20 newborns would be largely similar. To eliminate the contribution of genetic variation to these comparisons, we filtered 89,678 loci whose probes overlap known polymorphisms (annotated by Illumina) and, therefore, may display profile differences not related to methylation. We also eliminated 1160 sites not present in at least 18 of the 20 samples, leaving 394,739 CpG sites for analysis. Correlation coefficients were calculated for all pair-wise comparisons (n = 190) to demonstrate strong concordance in these samples (mean ρ = 0.9881; min = 0.9825, max = 0.9934). Figure 3B shows the interindividual comparison that represents the median correlation coefficient (ρ = 0.9873) within this set. Figure 3C shows the standard deviation of β-values at each CpG site and further demonstrates the tight correlation across samples: 90% of sites (n = 355,297) display a standard deviation <0.05.
Discussion
In this report, we show that (1) small quantities of gDNA (∼25 ng) provide a genome-wide methylation profile that is comparable with that achieved with larger gDNA quantities on the Infinium HumanMethylation450K array and (2) 25 ng gDNA isolated from a single, surgical punch (2 mm dia.) from an archived newborn bloodspot yields methylation profiles adequate for differential methylation studies on this platform.
Starting with only 25 ng gDNA, array-wide call rate and signal detection confidence (as reflected in a detection p-value) were excellent and virtually unchanged from higher concentrations (up to 750 ng; Fig. 1). This is true for commercially purchased gDNA (Jurkat cell) as well as gDNA isolated from small aliquots of archived bloodspots that, in some cases, have been stored up to 30 years.
Although signal intensities and consequent β-values were generally concordant between profiles derived from 25 ng or 750 ng gDNA for the majority of CpG sites on the array (Figs. 1 and 2), there was a small fraction of loci (2–8%) with more discordant values (see Results section). Importantly, these probe sets behaved quite consistently in all replicate profiles generated from the same input gDNA quantity, including 25 ng (compare Figs. 2B and 3A with Fig. 2C). Indeed, all methylation patterns derived from 25 ng gDNA were robust and highly correlated as determined by both replicate and interindividual analyses (Figs. 2B and 3).
An examination of variability in 20 Caucasian samples suggested that relatively small methylation changes can be measured in experimental groups (e.g., case–control studies). Measuring array-wide standard deviation (Fig. 3C), we estimated that sample sizes of 100 will give sufficient power (0.99) to detect 3% changes in β-value (α = 0.001) for >90% of CpGs on the array (standard deviation <0.05).
In conclusion, limiting quantities of gDNA from small archived bloodspots are not prohibitive for DNA methylome analysis through the Illumina Infinium array platform, including the recently introduced Infinium MethylationEPIC array, which expands coverage to >850,000 CpG sites. Thus, array-based methylome analysis can be an option for many researchers with limiting sample as a more straightforward and less informatically challenging alternative to whole-genome bisulfite sequencing in DNA methylation studies.
Supplementary Material
Acknowledgments
This research was partially supported by funds from the National Institutes of Health (R01HD074695 to N.J.M; R01GM072859 to J.R.) and the Centers for Disease Control and Prevention, Center of Excellence Award (U50/CCU913241) to G.M.S.
Author Disclosure Statement
No competing financial interests exist.
References
- Beyan H, Down TA, Ramagopalan SV, et al. (2012) Guthrie card methylomics identifies temporally stable epialleles that are present at birth in humans. Genome Res 22:2138–2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollegaard MV, Grauholm J, Norgaard- Pedersen B, et al. (2013) DNA methylome profiling using neonatal dried bloodspots: a proof-of-principle study. Mol Genet Metab 108:225–231 [DOI] [PubMed] [Google Scholar]
- Joo JE, Wong EM, Baglietto L, et al. (2013) The use of DNA from archival dried bloodspots with the Infinium HumanMethylation450 array. BMC Biotechnol 13:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini NJ, Hoffmann TJ, Lammer EJ, et al. (2011) A genetic signature of spina bifida risk from pathway-informed comprehensive gene-variant analysis. PLoS One 6:e28408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngollo M, Dagdemir A, Karsli-Ceppioglu S, et al. (2014) Epigenetic modifications in prostate cancer. Epigenomics 6:415–426 [DOI] [PubMed] [Google Scholar]
- O'Neill RJ, Vrana PB, Rosenfeld CS. (2014) Maternal methyl supplemented diets and effects on offspring health. Front Genet 5:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramagopalan SV, Rakyan VK. (2013) The promise and challenges of blood spot methylomics. Epigenetics 8:775–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.