Abstract
Background
The impacts of hybridization on the process of speciation are manifold, leading to distinct patterns across the genome. Genetic differentiation accumulates in certain genomic regions, while divergence is hampered in other regions by homogenizing gene flow, resulting in a heterogeneous genomic landscape. A consequence of this heterogeneity is that genomes are mosaics of different gene histories that can be compared to unravel complex speciation and hybridization events. However, incomplete lineage sorting (often the outcome of rapid speciation) can result in similar patterns. New statistical techniques, such as the D-statistic and hybridization networks, can be applied to disentangle the contributions of hybridization and incomplete lineage sorting. We unravel patterns of hybridization and incomplete lineage sorting during and after the diversification of the True Geese (family Anatidae, tribe Anserini, genera Anser and Branta) using an exon-based hybridization network approach and taking advantage of discordant gene tree histories by re-sequencing all taxa of this clade. In addition, we determine the timing of introgression and reconstruct historical effective population sizes for all goose species to infer which demographic or biogeographic factors might explain the observed patterns of introgression.
Results
We find indications for ancient interspecific gene flow during the diversification of the True Geese and were able to pinpoint several putative hybridization events. Specifically, in the genus Branta, both the ancestor of the White-cheeked Geese (Hawaiian Goose, Canada Goose, Cackling Goose and Barnacle Goose) and the ancestor of the Brent Goose hybridized with Red-breasted Goose. One hybridization network suggests a hybrid origin for the Red-breasted Goose, but this scenario seems unlikely and it not supported by the D-statistic analysis. The complex, highly reticulated evolutionary history of the genus Anser hampered the estimation of ancient hybridization events by means of hybridization networks. The reconstruction of historical effective population sizes shows that most species showed a steady increase during the Pliocene and Pleistocene. These large effective population sizes might have facilitated contact between diverging goose species, resulting in the establishment of hybrid zones and consequent gene flow.
Conclusions
Our analyses suggest that the evolutionary history of the True Geese is influenced by introgressive hybridization. The approach that we have used, based on genome-wide phylogenetic incongruence and network analyses, will be a useful procedure to reconstruct the complex evolutionary histories of many naturally hybridizing species groups.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-017-1048-2) contains supplementary material, which is available to authorized users.
Keywords: Hybridization, Phylogenetic Networks, D-statistic, PSMC, Phylogenomics
Background
The impacts of hybridization on the process of speciation are manifold [1]. Hybridization may slow down or even reverse species divergence. It may also accelerate speciation via adaptive introgression or contribute to species diversity through the formation of new hybrid taxa. These diverse effects occur at different spatial scales and during different stages across the speciation continuum [2]. The consequences of hybridization and its role in impeding or promoting speciation are thus expected to vary widely among hybridizing taxa and at different stages of divergence. In every case, the pattern of hybridization is only a single snapshot of a complex and continuously changing interaction.
Genomics has become a standard practise, also in ornithology [3, 4], opening avenues to answer longstanding questions in speciation and hybridization [2, 5]. Studies in speciation and hybridization genomics revealed that levels of genetic differentiation between species can be highly variable across the genome: genetic differentiation accumulates in certain genomic regions, while divergence is hampered in other regions by homogenizing gene flow, resulting in a heterogeneous genomic landscape [6–8]. A consequence of this heterogeneity is that genomes are mosaics of different gene histories [9–11] that can be compared to unravel complex speciation and hybridization events [12, 13].
Complex evolutionary histories with rapid speciation (leading to incomplete lineage sorting) and hybridization mostly result in high levels of phylogenetic incongruence (i.e. gene tree discordance), which can be difficult to capture in a traditional, bifurcating phylogenetic tree. Phylogenetic networks can be a powerful tool to display and analyse these evolutionary histories [14, 15]. For example, Suh et al. [16] quantified the amount of incomplete lineage sorting along the Neoaves phylogeny [17] using presence/absence data for 2118 retrotransposons and concluded that the “complex demographic history [of the Neoaves] is more accurately represented as local networks within a species tree.”
Here, we study patterns of hybridization and incomplete lineage sorting during and after the diversification of the True Geese (Table 1), a group of naturally hybridizing bird species [18, 19]. The True Geese are classified in the waterfowl tribe Anserini and have been traditionally divided over two genera: Anser and Branta [20]. Hybrids have been reported within each genus [21–27], but also intergeneric hybrids have been documented [28–31]. Previous studies suggested that the evolutionary history of the True Geese is heavily influenced by hybridization and rapid diversification [12, 32]. In this study, we explore this suggestion using a network approach and taking advantage of phylogenetic incongruence across the whole genome by fully re-sequencing all species of the True Geese clade. Moreover, we attempt to quantify the relative contributions of gene flow and incomplete lineage sorting during the evolution of this bird group.
Table 1.
Current taxonomy for the True Geese (tribe Anserini), previously published in Ottenburghs et al. (2016)
| English Name | Scientific Name | Subspecies |
|---|---|---|
| Genus ANSER | ||
| Swan Goose | Anser cygnoides | |
| Taiga Bean Goose | Anser fabalis |
A. f. fabalis
A. f. johanseni A. f. middendorffii |
| Tundra Bean Goose | Anser serrirostris |
A. s. rossicus
A. s. serrirostris |
| Pink-footed Goose | Anser brachyrhynchus | |
| Greater White-fronted Goose | Anser albifrons |
A. a. albifrons (Eurasian) A. a. flavirostris (Greenland) A. a. gambeli (Western) A. a. frontalis (Western) A. a. elgasi (Tule) |
| Lesser White-fronted Goose | Anser erythropus | |
| Greylag Goose | Anser anser |
A. a. anser (European) A. a. rubrirostris (Siberian) |
| Bar-headed Goose | Anser indicus | |
| Emperor Goose | Anser canagicus | |
| Snow Goose | Anser caerulescens |
A. c. caerulescens
A. c. atlantica |
| Ross’ Goose | Anser rossii | |
| Genus BRANTA | ||
| Brent Goose | Branta bernicla |
B. b. bernicla (Dark-bellied) B. b. hrota (Pale-bellied or Atlantic) B. b. nigricans (Black) B. b. orientalis |
| Barnacle Goose | Branta leucopsis | |
| Cackling Goose | Branta hutchinsii |
B. h. leucopareia (Aleutian) B. h. hutchinsii (Richardson’s) B. h. minima (Minima) B. h. taverneri (Taverner’s) |
| Canada Goose | Branta canadensis |
B. c. moffitti
B. c. maxima B. c. occidentalis B. c. fulva B. c. canadensis B. c. interior B. c. parvipes |
| Hawaiian Goose | Branta sandvicensis | |
| Red-breasted Goose | Branta ruficollis | |
The contrasting evolutionary histories of these closely related genera also provide an excellent opportunity to study the effects of hybridization on the speciation process. The Anser-clade can be regarded as an adaptive radiation and was probably affected more by hybridization compared to the more gradually diversifying Branta-clade [12]. Therefore, we hypothesize that the phylogenetic network of Anser will be more complex (i.e. contain more interconnections between the taxa) compared to the Branta-network. Moreover, statistics quantifying interspecific gene flow, such as the D-statistic [33], are expected to be higher for Anser compared to Branta.
Methods
Genomic dataset
We collected blood samples from 19 goose (sub)species (Additional file 1: Table S1). From these blood samples, genomic DNA was isolated using the Qiagen Gentra kit (Qiagen Inc.). DNA quantity and quality were assessed using Qbit (Invitrogen, Life Technologies). Sequence libraries were made following Illumina protocols and sequenced paired-end (100 bp) on the HiSeq2500 (Illumina Inc.).
Paired-end reads were mapped to the Mallard (Anas platyrhynchos) genome, version 73 [34] using SMALT (http://www.sanger.ac.uk/science/tools/smalt-0). Over 99% of the reads mapped successfully in all samples, but to decrease the incidence of off-site mapping only properly mapped reads were accepted, leading to mapping rates between 63% and 78% (Additional file 2: Table S2). Next, duplicate sequences were removed using SAMtools-dedup [35] and realigned with IndelRealigner in GATK 2.6 [36]. Variant sites calling was performed using UnifiedGenotyper in GATK 2.6 [36] with a heterozygosity value of 0.01 and a minimum base quality of 20. Heterozygous sites were coded following the IUPAC nucleotide codes (e.g., R for A and G). The genomic positions for exons that were one-to-one orthologous between Mallard and other bird species (chicken, turkey, flycatcher and zebra finch) were retrieved from the ENSEMBL database.
From whole genome sequence data, we thus filtered out high quality exonic sequences. The final dataset is comprised of 41,736 unique exons, representing 5887 genes. The total alignment (6,630,626 bp) was used in the neighbour-joining network and D-statistics analyses described below. In addition, we selected 3570 one-to-one orthologous genes with a minimum length of 500 bp. These genes were analysed separately under a GTR + Γ substitution model with 100 rapid bootstraps in RAxML 8.3 [37, 38]. The resulting gene trees were filtered on average bootstrap support (minimum >50). This final set of 3558 well-supported gene trees was used in the analysis to construct hybridization networks – which calculate evolutionary trees taking into account hybridization – and to determine the timing of gene flow. Ottenburghs et al. [19] provide the phylogenetic framework for the current study, which focuses on introgressive hybridization during the evolutionary history of the True Geese.
Gene flow analysis
The D-statistic is a statistical test that was first employed to quantify the amount of genetic exchange between Neanderthals and humans [39]. It exploits the asymmetry in frequencies of two nonconcordant gene trees in a three-population setting [33]. Consider three populations (P1, P2 and P3) and an outgroup (O), of which P1 and P2 are sister clades. In this ordered set op populations [P1, P2, P3, O], two allelic patterns are of interest: “ABBA” and “BABA”. The pattern ABBA refers to the situation in which P1 has the outgroup allele “A” and P2 and P3 share the derived allele “B”, while the pattern BABA refers to the situation in which P2 has the outgroup allele “A” and P1 and P3 share the derived allele “B”. Under the null hypothesis that P1 and P2 are more closely related to each other than to P3, and if the ancestral populations of P1, P2, P3 were panmictic, then it is expected that the derived alleles in P3 match the derived alleles in P1 and P2 equally often [40, 41]. In other words, the patterns ABBA and BABA should occur in equal frequencies and the D-statistic should equal zero:
A D-statistic equal to zero is expected under incomplete lineage sorting. Gene flow between P1 and P3 (indicated by an overrepresentation of BABA) or P2 and P3 (indicated by an overrepresentation of ABBA) result in a D-statistic that is significantly different from zero. For both genera, D-statistics were calculated for all possible combinations of three species in the program HybridCheck version 1.0.1 [42]. We combined all species of the other genus as the outgroup. To test for significance, we performed jackknife resampling using blocks of 50,000 bp. We did not quantify asymmetric gene flow between genera due to the lack of a proper outgroup.
To infer the timing of gene flow (during or after the diversification), we dated 3558 gene trees using the software PATHd8 version 1.0 [43], setting the divergence time between the genera at 9.5 million years ago (based on previous estimates, [44, 45]). For every species pair, histograms were constructed from the resulting divergence times [46]. The patterns expected under incomplete lineage sorting and when gene flow occurred during or after the diversification are presented in Fig. 1.
Fig. 1.
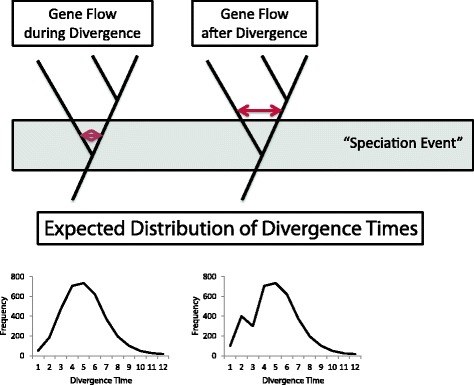
Expected distribution of divergence times. If gene flow occurred during the diversification process, it will be indistinguishable from genetic divergence at other loci, resulting in a single peak (left graph). A similar pattern is expected under incomplete lineage sorting, but to discriminate between gene flow and incomplete lineage sorting, other analyses are warranted. If, on the other hand, gene flow occurred after the diversification process, introgressed loci will show more recent divergence times, which becomes apparent as a recent, smaller peaks (right graph)
Network analyses
A previous phylogenomic analysis of the True Geese indicated high levels of gene tree discordance, which can be caused by hybridization and/or incomplete lineage sorting [12]. To visualize this phylogenetic incongruence, we constructed a phylogenetic neighbour-joining network using the ordinary least squares method (with default settings) in SplitsTree version 4.1.4.2 [15]. This network was based on genetic distances, which were calculated in RAxML 8.3 with a GTR + Γ substitution model [12, 37]. We calculated the degree distributions (i.e. the number of connections for each node in a network) for each genus to quantify the complexity of the networks using the R-package igraph [47]. The degree distributions for each genus were compared by means of a general linear model with Poisson distribution in R version 3.2.2.
Hybridization networks are networks that attempt to reconstruct a phylogenetic tree with the fewest amount of hybridization events [15, 48]. For each genus, we combined 3558 gene trees into hybridization networks using the Autumn algorithm [49] with default settings in Dendroscope version 3.4.4 [50].
Demographic analysis
We conducted a demographic analysis using a hidden Markov model approach as implemented in the software package PSMC [51]. A consensus sequence was generated from BAM files using the ‘pileup’ command in SAMtools [35]. For the PSMC analyses, we used the parameter settings suggested by Nadachowska-Brzyska et al. [52], namely “N30 –t5 –r5 –p 4 + 30*2 + 4 + 6 + 10.”
Results
Gene flow analysis
The D-statistic analysis supported gene flow between several goose species (Table 2 Additional file 5: Table S3). Although the D-statistics for Anser were slightly higher compared to Branta, there was no significant difference (Mann Whitney U, W = 4659, p = 0.088). To infer the timing of gene flow (during or after the diversification), we took advantage of gene tree discordance and constructed histograms based on divergence times of 3558 gene trees. All analyses supported a scenario of gene flow during divergence with low levels of recent gene flow because the histograms all displayed one peak corresponding to the initial species split. The divergence time of several gene trees was close to zero, suggesting low levels of recent gene flow between certain species. Figure 2 shows two examples, involving the Cackling Goose and the Lesser White-fronted Goose (for other species, see Additional file 3: Figure S1).
Table 2.
Significant D-statistics (Z-score > 3 and p < 0.0001) for different combination of three goose species
| P1 | P2 | P3 | D-statistics | Z-score |
|---|---|---|---|---|
| BRANTA | ||||
| Barnacle Goose | Cackling Goose | Canada Goose | 0.087 | 4.322 |
| Barnacle Goose | Cackling Goose | Red-breasted Goose | 0.146 | 6.022 |
| Barnacle Goose | Cackling Goose | Black Brent | 0.094 | 3.516 |
| Barnacle Goose | Canada Goose | Red-breasted Goose | 0.1 | 4.037 |
| Barnacle Goose | Hawaii Goose | Red-breasted Goose | 0.094 | 4.192 |
| Barnacle Goose | Black Brent | Red-breasted Goose | 0.073 | 3.022 |
| ANSER | ||||
| Lesser White-fronted | Greater White-fronted | Pink-footed Goose | 0.126 | 6.116 |
| Taiga Bean Goose | 0.09 | 4.413 | ||
| Pink-footed Goose | Greater White-fronted | Emperor Goose | 0.088 | 3.701 |
| Tundra Bean Goose | 0.079 | 3.383 | ||
| Taiga Bean Goose | 0.128 | 5.457 | ||
| Greylag Goose | 0.079 | 3.537 | ||
| Taiga Bean Goose | Greater White-fronted | Bar-headed Goose | 0.083 | 3.470 |
| Greater White-fronted | Lesser White-fronted | Swan Goose | 0.114 | 5.279 |
| Pink-footed Goose | 0.15 | 6.821 | ||
| Tundra Bean Goose | 0.16 | 8.15 | ||
| Taiga Bean Goose | 0.173 | 8.385 | ||
| Greater White-fronted | Lesser White-fronted | Ross’ Goose | 0.104 | 5.325 |
| Pink-footed Goose | 0.12 | 5.421 | ||
| Tundra Bean Goose | 0.103 | 4.682 | ||
| Taiga Bean Goose | 0.083 | 3.93 | ||
| Swan Goose | 0.103 | 3.984 | ||
| Emperor Goose | 0.102 | 4.51 | ||
| Greater White-fronted | Lesser White-fronted | Snow Goose | 0.165 | 7.336 |
| Pink-footed Goose | 0.174 | 7.032 | ||
| Tundra Bean Goose | 0.157 | 7.053 | ||
| Taiga Bean Goose | 0.137 | 6.224 | ||
| Swan Goose | 0.156 | 6.525 | ||
| Greylag Goose | 0.086 | 4.054 | ||
| Emperor Goose | 0.14 | 6.007 | ||
| Pink-footed Goose | Lesser White-fronted | Emperor Goose | 0.076 | 3.164 |
| Tundra Bean Goose | 0.07 | 3.0 | ||
| Taiga Bean Goose | 0.12 | 5.419 | ||
| Greylag Goose | 0.078 | 3.472 | ||
| Taiga Bean Goose | Pink-footed Goose | Bar-headed Goose | 0.082 | 3.436 |
| Greater White-fronted | Greylag Goose | Ross’ Goose | 0.087 | 3.719 |
| Pink-footed Goose | 0.107 | 4.496 | ||
| Tundra Bean Goose | 0.086 | 3.408 | ||
| Swan Goose | 0.084 | 3.049 | ||
| Emperor Goose | 0.163 | 6.67 | ||
| Greater White-fronted | Greylag Goose | Snow Goose | 0.072 | 3.055 |
| Pink-footed Goose | 0.091 | 3.416 | ||
| Emperor Goose | 0.139 | 5.59 | ||
| Emperor Goose | Tundra Bean Goose | Ross’ Goose | 0.078 | 3.692 |
| Emperor Goose | Tundra Bean Goose | Snow Goose | 0.074 | 3.317 |
| Emperor Goose | Taiga Bean Goose | Ross’ Goose | 0.139 | 6.346 |
| Emperor Goose | Taiga Bean Goose | Snow Goose | 0.139 | 6.655 |
| Taiga Bean Goose | Greylag Goose | Bar-headed Goose | 0.09 | 4.059 |
Asymmetric gene flow is between P2 and P3. The outgroup for Branta species was a consensus sequence based on all Anser species, while the outgroup for Anser species was a consensus sequence based on all Branta species
Fig. 2.
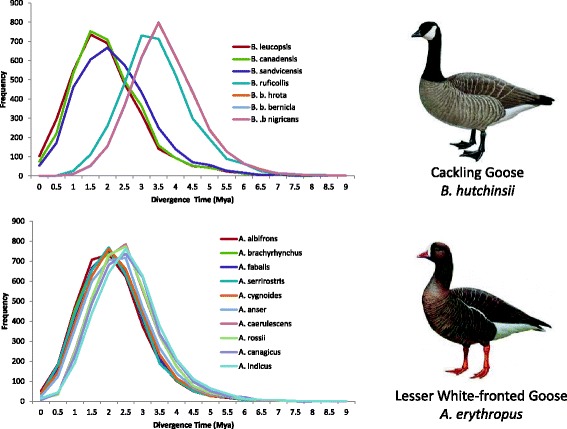
Distribution of divergence times for Lesser White-fronted Goose with all Anser species and for Cackling Goose with all Branta species. All distributions show a single peak, indicating gene flow during divergence. The divergence time of several gene trees was close to zero, suggesting low levels of recent gene flow between certain species
Network analyses
The phylogenetic neighbour-joining network (Fig. 3) based on genetic distances uncovered two main clades that corresponded to the genera Anser and Branta. Within these clades, the relationships correspond to previous phylogenetic analyses [12]. The comparison of degree distributions revealed that the Anser-network was more complex compared to the Branta-network (Poisson regression, SD = 0.1908, z-value = −5.08, p-value < 0.001), because the Anser-network contains more nodes with four or five edges compared to the Branta-network. The complexity of the networks was consistent with the suggestion that the evolutionary history of the Anser-clade is more heavily influenced by rapid diversification and hybridization compared to the Branta-clade.
Fig. 3.
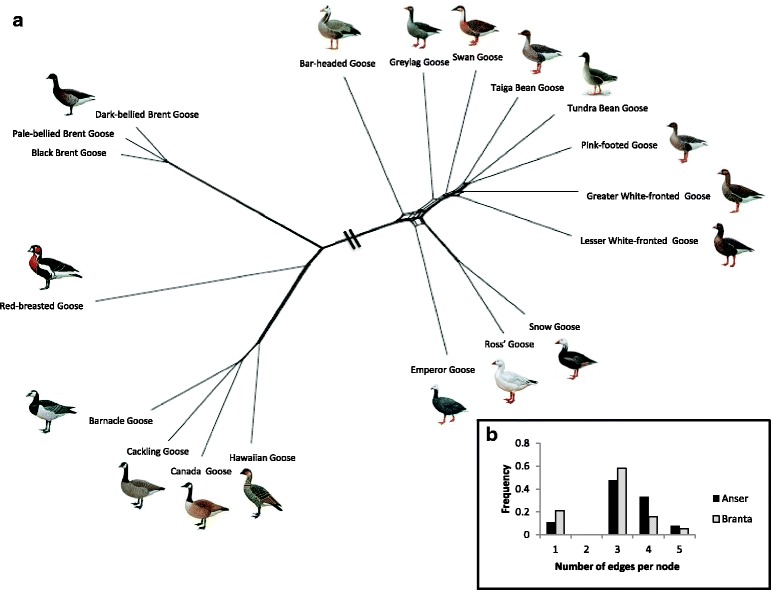
a Neighbour-joining Network of the True Geese using the ordinary least squares method (with default settings) in SplitsTree version 4.1.4.2 [15], based on genetic distances. b The comparison of degree distributions indicates that the Anser-network is more complex compared to the Branta-network as it contains relatively more nodes with four and five edges. Drawings used with permission of Handbook of Birds of the World [128]
We combined 3558 gene trees into hybridization networks for both genera. These networks attempt to reconstruct a phylogenetic tree with the fewest amount of hybridization events [15, 48]. Hybridization network analyses of the genus Anser did not result in most likely scenarios, underlining the complexity of introgression and incomplete lineage sorting among Anser species. In the genus Branta, the hybridization network analyses recovered three (not mutually exclusive) scenarios, indicating hybridization events between the Red-breasted Goose and the ancestor of the White-cheeked Geese (i.e. Hawaiian Goose, Canada Goose, Cackling Goose and Barnacle Goose) and between Red-breasted Goose and Brent Goose (Fig. 4a-b). In addition, one hybridization network (Fig. 4c) suggested a hybrid origin for the Red-breasted Goose. The network suggesting a hybrid origin for this species should not be regarded as definitive proof for hybrid speciation, but rather as a possible scenario that can serve as a starting point for further research.
Fig. 4.
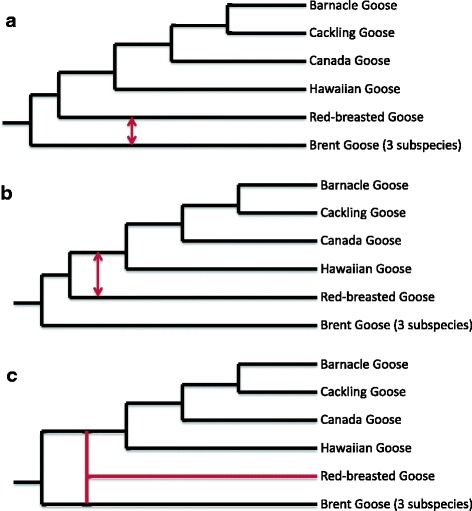
Hybridization networks for the genus Branta based on the Autumn algorithm [49] in Dendroscope version 3.4.4 [50]. Network a suggests hybridization between Red-breasted Goose and Brent Goose, network b between Red-breasted Goose and the ancestor of the White-cheeked Geese. Network c suggests a hybrid origin for the Red-breasted Goose
Demographic analysis
We reconstructed historical effective populations sizes (Ne) for all goose species using the pairwise sequentially Markovian coalescent (PSMC) approach over a range from 1 to 10 million years ago until about 10,000 years ago. Most Anser species (Greater White-fronted Goose, Lesser White-fronted Goose, Tundra Bean Goose, Taiga Bean Goose, Pink-footed Goose, Swan Goose, Greylag Goose, Bar-headed Goose, Snow Goose, and Ross’ Goose) and several Branta species (Canada Goose, Cackling Goose, Red-breasted Goose, Pale-bellied Brent Goose and Black Brent Goose) show a steady population increase followed by a dramatic expansion, which suggests population subdivision and occasional gene flow, leading to higher levels of heterozygosity and consequently higher estimates of Ne [51, 53]. Four species (Hawaiian Goose, Emperor Goose, Barnacle Goose and Dark-bellied Brent Goose) show clear signs of a bottleneck. Figure 5 shows these two patterns as illustrated by Greater White-fronted Goose and Hawaiian Goose (for other species, see Additional file 4: Figure S2).
Fig. 5.
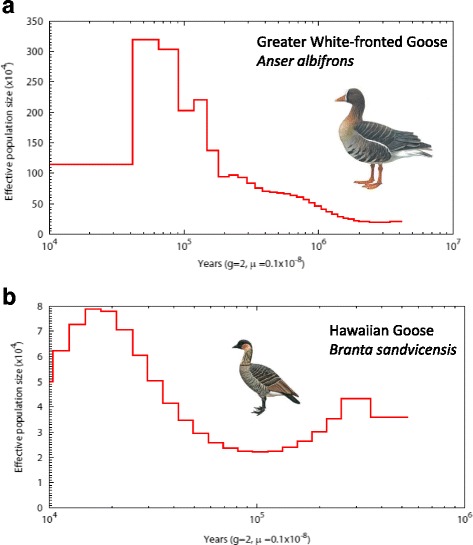
Examples of two general demographic patterns for the True Geese based on PSMC analyses. a Steady population increase followed by dramatic expansion which suggests population subdivision, as illustrated by Greater White-fronted Goose. b Population bottleneck after island colonization, as illustrated by Hawaiian Goose
Discussion
General patterns of introgression
Interspecific gene flow is an important aspect in avian speciation [54]. Based on hybridization networks and D-statistics, calculated from genome-wide data, we found indications for high levels of interspecific gene flow between several goose species. D-statistics allowed us to confidently discriminate between incomplete lineage sorting and interspecific gene flow. The significant D-statistics varied from 0.07 to 0.17, which is slightly higher compared to analyses on recent radiations, such as Darwin’s Finches (0.004–0.092; [55]) and butterflies of the genera Heliconius (0.04; [56]) and Papilio (0.04; [57]). These values do fall within the range of studies on other hybridizing species, such as pigs (0.11–0.23; [58]), bears (0.04–0.46; [59, 60]) and Xiphophorus fish (0.03–0.56; [61]).
A significant D-statistic does not necessarily indicate introgression between the species from which the genomes are being compared. There might have been gene flow with an extinct (not sampled) population or the signal might be a remnant from an older hybridization event [33, 62]. The latter possibility is probably the case for hybridization between Red-breasted Goose and three other species (Hawaiian Goose, Canada Goose and Cackling Goose). The hybridization network analysis supported the notion that significant D-statistics were caused by an ancient hybridization event between Red-breasted Goose and the ancestor of these three species. Many of the significant D-statistics in the Anser-clade can probably be explained in the same way, but the complexity of introgression patterns in this clade did not allow us to pinpoint putative hybridization events. In addition, the D-statistic only captures asymmetric gene flow [33]. Because we did not quantify symmetric gene flow, we are probably underestimating the amount of gene flow between the some goose species.
When did this gene flow occur? Further analyses, based on the divergence times of 3558 gene trees, indicated that this gene flow was largely due to ancient hybridization during the diversification of these species. Ancient gene flow has been reported for a variety of taxa [46, 63, 64], including several bird groups [55, 65–68]. For instance, Fuchs et al. [65] attributed a conflicting pattern between several loci to ancient hybridization between members of the woodpecker genus Campephilus and the melanerpine lineage (Melanerpes and Sphyrapicus). The increasing number of studies reporting ancient gene flow during species diversification [69] shows that the speciation process is often more complex than, for example, the classical allopatric speciation model [70, 71].
In the allopatric speciation model, populations become geographically isolated and diverge by genetic drift and/or differential selection pressures, resulting in intrinsic reproductive isolation due to the accumulation of Dobzhansky-Muller incompatibilities [72, 73]. This speciation model predicts that the distribution of interspecific divergence is largely determined by a single, shared species split [74]. But speciation is often more complex: in some cases, speciation may advance by divergent ecological or sexual selection in the face of ongoing gene flow [75], while, in other cases, allopatrically diverging populations may come into secondary contact and hybridize before reproductive isolation is complete [2]. These more complex speciation models predict that interspecific divergence varies considerably across the genome [6, 7], because some genomic regions reflect the initial species split time, whereas others indicate more recent genetic exchange [11, 13, 76].
With regard to the evolutionary history of geese, we found support for a complex speciation model with high levels of gene flow during species diversification. It is, however, not possible to determine whether this gene flow is the outcome of (repeated) secondary contact or divergence-with-gene-flow. ABC modelling based on multiple samples per species allows for the comparison of several scenarios that differ in the amount and timing of gene flow and can thus be used to confidently discriminate between divergence-with-gene-flow and secondary contact [77–79]. For example, Nadachowska-Brzyska et al. [80] compared 15 models (with different patterns and levels of gene flow) to assess the demographic history of Pied Flycatcher (Ficedula hypoleuca) and Collared Flycatcher (Ficedula albicollis). Whole genome re-sequencing data from 20 individuals supported a recent divergence with unidirectional gene flow from Pied Flycatcher into Collared Flycatcher after the Last Glacial Maximum, indicating that the hybrid zone between these species is a secondary contact zone. In this study, we were unable to perform an ABC modelling exercise because only one individual per species was sampled, while multiple samples per species are required.
Next to evidence for ancient gene flow, our results suggest low levels of recent gene flow, which can be explained in three ways. First, the D-statistic analysis may be unable to detect recent gene flow. Indeed, the D-statistic was developed to detect ancient gene flow and to estimate the extent of archaic ancestry in the genomes of extant populations [33]. The detection and quantification of recent gene flow warrants a population genomic approach whereby multiple individuals of one population are sequenced [3, 9, 81, 82]. Second, the relative rarity of goose hybrids diminishes the opportunity for backcrossing and introgression, leading to absence or low levels of recent gene flow [19]. Third, there may be little recent gene flow because of strong intrinsic and/or extrinsic selection against goose hybrids. Although most goose hybrids are viable and fertile [19], second generation hybrids or backcrosses may be impaired by genetic incompatibilities [83, 84], or hybrids might be ecologically maladapted (e.g., intermediate beak morphology) or unable to find a mate [73]. To answer these questions, field observations are needed, which is challenging given the relative rarity of hybrids [19] and the difficulty of identifying certain hybrids [85]. Strong selection against hybrids might also suggest that the diversification of the True Geese was partly driven by reinforcement [86].
Demographic patterns
The reconstruction of historical effective populations sizes (Ne) for all goose species using the pairwise sequentially Markovian coalescent (PSMC) approach indicated two main patterns. First, most species showed a steady increase during the Pliocene and Pleistocene followed by population subdivision (apparent as a dramatic increase in Ne) during the Last Glacial Maximum (LGM, about 110,000 to 12,000 years ago). The increase in population size during the Pliocene and Pleistocene can be explained by a global cooling trend which resulted in the formation of a circumpolar tundra and the emergence of temperate grasslands [87–89]. The tundra habitat acted as breeding ground [90], whereas the grasslands served as wintering grounds where mate choice occurred [91], enabling goose populations to proliferate. In addition, the climatic fluctuations during the Pliocene and Pleistocene might have instigated range expansions and shifts. This combination of large Ne and occasional range shifts might have facilitated contact between the diverging goose species, resulting in the establishment of numerous hybrids zones and consequent gene flow [92, 93].
During the LGM, many plant and animal populations were subdivided into separate refugia by the ice sheets that expanded from the north [94, 95]. This population subdivision has been described for several goose species [96] and the genetic signature of this subdivision has been uncovered for certain species, such as Pink-footed Goose [97], Bean Goose [98], Greater White-fronted Goose [99, 100], Canada Goose [101], and Snow Goose [26, 102].
Four species show a decrease in Ne and a consequent genetic bottleneck in the PSMC analyses, which suggests island colonization. Indeed, these four goose species have colonized island habitats: the Hawaiian Goose reached the Hawaiian archipelago [103], the Emperor Goose settled on the Aleutian Islands [104], and the Barnacle Goose and the Dark-bellied Brent Goose established populations on arctic islands in the North Atlantic, such as Spitsbergen and Novaya Zemlya [105]. It is well-established that island colonization leads to a reduction in heterozygosity and Ne [106], and that island populations have lower levels of genetic variation compared to mainland species [107]. Genetic bottlenecks following island colonization have been documented for numerous other bird species (e.g., [108, 109]). However, further analyses are warranted to confirm these scenarios of island colonization. For instance, comparing the genetic diversity of these four goose species with closely related mainland populations.
Comparing Anser and Branta
There is a striking contrast in the patterns of introgression between the two genera. As hypothesised, the general network analysis showed that the Anser-network is more complex than the Branta-network and D-statistics were slightly (although not significantly) higher in the Anser-clade. While high levels of gene flow hindered the precise reconstruction of hybridization events in the Anser-clade, it was possible to pinpoint several putative hybridization events within Branta-clade. The hybridization network analyses provided evidence for gene flow between the Red-breasted Goose and the ancestor of the White-cheeked Geese (i.e. Hawaiian Goose, Canada Goose, Cackling Goose and Barnacle Goose), between Red-breasted Goose and Brent Goose, and between Canada Goose and Cackling Goose. Past gene flow between the latter two species has been reported previously [23]. What factors can explain the differential introgression patterns between Anser and Branta? We will consider three possible factors: (1) macro-evolutionary dynamics, (2) morphological and behavioural differences, and (3) demographic dynamics.
First, these patterns of introgression were reconstructed by comparing the genomes of modern, extant species. The ancestors of these modern species may have interbred with unknown extinct species. It might thus be possible that the evolutionary history of the Branta-clade was as influenced by hybridization as much as the diversification of the Anser-clade, but that many Branta-species have become extinct. For example, the Hawaiian radiation of Branta geese consisted of at least three species, of which only the Hawaiian Goose remains today [103]. The different introgression patterns (as observed by comparing extant genomes) could then be attributed to differences in extinction rates between the genera. Unfortunately, the fossil record for geese is currently still too sparse to test this hypothesis [110, 111].
Second, differential introgression patterns may be explained by differences in behaviour [112, 113]. Although the behaviour of extant species does not necessarily correspond to the ancestral behaviour, we can speculate about possible differences between the genera. Pair formation, involving several pre-copulatory displays, and copulation vary little between the species and the genera [90, 114], which can explain the frequent occurrence of hybridization on the species level, but does not clarify the differences in introgression patterns between the genera. Are there differences in certain behaviours that lead to hybridization, such as interspecific nest parasitism or forced extra-pair copulations [115]? These behaviours have been observed in both genera, but the relative contribution of each behaviour to the occurrence of goose hybrids remains to be quantified [19].
Mate choice in waterfowl is largely determined by sexual imprinting [116]. Anser species are morphologically more similar compared to Branta species, which might increase the probability of heterospecific mate choice. Based on this reasoning, we expect more Anser hybrids compared to Branta. This expectation remains to be tested, but will be challenging because hybrids between morphologically similar species are difficult to identify [85] and many goose hybrids are probably of captive origin [19].
Third, differences in demographic dynamics, mediated by a particular biogeographical and climatic context, might determine the frequency of interspecific interactions, possibly leading to introgressive hybridization. The Anser-clade has a largely Eurasian distribution (with the exception of Snow Goose and Ross’ Goose). The open tundra landscape of Eurasia during the Pleistocene allowed for large effective population sizes and the climatic fluctuations during the Pliocene and Pleistocene might have instigated range expansions and shifts. In contrast to the Anser-clade, the Branta species are more widely distributed across the Northern Hemisphere: Canada Goose and Cackling Goose in North America, Hawaiian Goose on the Hawaiian islands, Barnacle Goose and Red-breasted Goose in Eurasia, and the circumpolar Brent Goose. This distribution limits the frequency of interspecific contact, although several species could achieve large effective population sizes.
The demographic differences between the genera might also lead to other speciation histories. The diversification of the Branta-clade was more gradual compared to the Anser-clade, which can be considered an adaptive radiation [12]. During an adaptive radiation the frequency of interspecific interactions increases, enhancing the probability of introgressive hybridization [117]. Moreover, as the radiation progresses, occasional hybridization could facilitate further ecological diversification [118]. Possibly, the diversification in beak morphology among Anser species was driven by hybridization, comparable to the radiation of Darwin’s Finches on the Galapagos Islands [55, 119].
A hybrid origin for the Red-breasted Goose?
The hybridization network analysis also suggested a possible alternative scenario in which the Red-breasted Goose is a hybrid species between the ancestors of the White-cheeked Geese and the Brent Goose. If so, the distinct morphology of this species, which is not intermediate between its putative parents, might be the outcome of transgressive segregation [120]. But indisputably demonstrating hybrid speciation is challenging and often the most likely scenario for the observed genomic pattern is introgressive hybridization [121]. To our knowledge, five bird species have been proposed to have hybrid origins: the Italian Sparrow (Passer italiae, [122]), the Audubon’s Warbler (Setophaga auduboni, [123]), the Genovesa Mockingbird (Mimus parvulus bauri, [124]), the Hawaiian Duck (Anas wylvilliana, [125]) and a recent lineage of Darwin’s finches on Daphne Major (referred to as ‘Big Bird’, [126]). However, the hybrid origin of these putative cases has not been unequivocally established [121]. Also, in the case of the Red-breasted Goose, the most parsimonious explanation seems to involve separate hybridization events between the Red-breasted Goose and the ancestor of the White-cheeked Geese and between Red-breasted Goose and Brent Goose. If the Red-breasted Goose is a hybrid species, one would expect significantly higher values for D-statistics. For example, a recent genomic study of the Italian Sparrow, a hybrid species between House Sparrow (Passer domesticus) and Spanish Sparrow (Passer hispaniolensis), uncovered D-statistics over 50% [127]. The highest value for Red-breasted Goose in our analysis was about 15% (even some Anser species displayed higher D-statistics). Hence, a hybrid origin for the Red-breasted Goose seems unlikely.
Conclusions
Using genomic datasets and modern analysis tools, such as the D-statistic and PSMC analysis, in combination with network analyses based on gene tree discordance, we were able to determine patterns of introgressive hybridization in the True Geese. High levels of ancient gene flow suggest a scenario of divergence-with-gene-flow. We found indications for low levels of recent gene flow, but the quantification of this recent gene flow warrants a population genomic approach whereby multiple individuals of one population are sequenced. The reconstruction of historical effective population sizes indicates that most species showed a steady increase during the Pliocene and Pleistocene followed by population subdivision during the Last Glacial Maximum about 110,000 to 12,000 years ago. The combination of large effective population sizes and occasional range shifts might have facilitated contact between diverging goose species, resulting in the establishment of numerous hybrid zones and consequent gene flow. Our approach, based on genome-wide phylogenetic incongruence and network analyses, will be a useful procedure to reconstruct the complex evolutionary histories of many naturally hybridizing species groups.
Additional files
Sampled goose species and sampling location. (DOCX 15 kb)
Mapping results of all goose samples to Mallard genome (version 73) using SMALT. (DOCX 20 kb)
D-statistics for all combinations of three species per genus. Significant D-statistics (Z-score > 4), suggesting gene flow between P2 and P3, are indicated in bold and colored in yellow. The outgroup for Branta was a consensus sequence based on all Anser species. Similarly, the outgroup for Anser was a consensus sequence based on all Branta species. (DOCX 19 kb)
Distribution of gene tree divergence times for all goose species. All distributions show a single peak, indicating gene flow during divergence. The divergence time of several gene trees was close to zero, suggesting low levels of recent gene flow between certain species. Final three figures represent the three subspecies of Brent Goose, which is depicted in the lower right panel. (ZIP 2715 kb)
Estimates of historical effective population sizes for all goose species, based on a PSMC analysis. Final three figures represent the three subspecies of Brent Goose, which is depicted in the lower right panel. (ZIP 766 kb)
Acknowledgments
We are indebted to Gerard Müskens, Jurre Brenders, Henk en Wim Meinen, Ouwehands Zoo (Gerard Meijer), Avifauna (Jan Harteman & Joost Lammers) and the Nederlands Instituut voor Ecologie (NIOO-KNAW) for their help during the collection of blood samples, and to Kimberley Laport for the DNA extraction.
Funding
This study was funded by Stichting de Eik.
Availability of data and materials
The dataset supporting the conclusions of this article are available at Bioproject PRJEB20373 (http://www.ebi.ac.uk/ena/data/view/PRJEB20373) at the European Nucleotide Archive (ENA).
Abbreviations
- ABC Modelling
Approximate Bayesian Computation Modelling
- Bp
Base pairs
- GATK
Genome Analysis Toolkit
- GTR
General Time-reversible
- IUPAC
International Union of Pure and Applied Chemistry
- LGM
Last Glacial Maximum
- Ne
Effective Population Size
- PSMC
Pairwise Sequentially Markovian Coalescent
- SD
Standard Deviation
Authors’ contributions
JO collected the blood samples, analysed the genomic data, and wrote the manuscript. RPMAC guided the DNA extraction and sequencing. H-JM, RHSK, PH, SEW, RCY, MAMG and HHTP. commented on earlier versions of the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The collection of blood samples (Submission 2,013,001.b) has been approved by the Ethical Committee for Animal Experiments (Dierexperimentencommissie [DEC]) at Wageningen University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-017-1048-2) contains supplementary material, which is available to authorized users.
References
- 1.Abbott R, et al. Hybridization and speciation. J Evol Biol. 2013;26(2):229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- 2.Seehausen O, et al. Genomics and the origin of species. Nat Rev Genet. 2014;15(3):176–192. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- 3.Toews DPL, et al. Genomic approaches to understanding population divergence and speciation in birds. Auk. 2016;133(1):13–30. doi: 10.1642/AUK-15-51.1. [DOI] [Google Scholar]
- 4.Kraus RH, Wink M. Avian genomics: fledging into the wild! J Ornithol. 2015;156(4):1–15. doi: 10.1007/s10336-015-1253-y. [DOI] [Google Scholar]
- 5.Jarvis ED. Perspectives from the Avian Phylogenomics Project: Questions that Can Be Answered with Sequencing All Genomes of a Vertebrate Class. Ann Rev Animal Biosci. 2016;4:45–59. doi: 10.1146/annurev-animal-021815-111216. [DOI] [PubMed] [Google Scholar]
- 6.Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18(3):375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- 7.Nosil P, Feder JL. Widespread yet heterogeneous genomic divergence. Mol Ecol. 2012;21(12):2829–2832. doi: 10.1111/j.1365-294X.2012.05580.x. [DOI] [PubMed] [Google Scholar]
- 8.Harrison RG, Larson EL. Heterogeneous genome divergence, differential introgression, and the origin and structure of hybrid zones. Mol Ecol. 2016;25:2454–2466. doi: 10.1111/mec.13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491(7426):756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- 10.Maddison WP. Gene trees in species trees. Syst Biol. 1997;46(3):523–536. doi: 10.1093/sysbio/46.3.523. [DOI] [Google Scholar]
- 11.Payseur BA. Using differential introgression in hybrid zones to identify genomic regions involved in speciation. Mol Ecol Resour. 2010;10(5):806–820. doi: 10.1111/j.1755-0998.2010.02883.x. [DOI] [PubMed] [Google Scholar]
- 12.Ottenburghs J, et al. A tree of geese: A phylogenomic perspective on the evolutionary history of True Geese. Mol Phylogenet Evol. 2016;101:303–313. doi: 10.1016/j.ympev.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 13.Harrison RG, Larson EL. Hybridization, Introgression, and the Nature of Species Boundaries. J Hered. 2014;105:795–809. doi: 10.1093/jhered/esu033. [DOI] [PubMed] [Google Scholar]
- 14.Ottenburghs J, et al. Birds in a bush: Toward an avian phylogenetic network. Auk. 2016;133:577–582. doi: 10.1642/AUK-16-53.1. [DOI] [Google Scholar]
- 15.Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 16.Suh A, Smeds L, Ellegren H. The Dynamics of Incomplete Lineage Sorting across the Ancient Adaptive Radiation of Neoavian Birds. PLoS Biol. 2015;13(8):e1002224. doi: 10.1371/journal.pbio.1002224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarvis ED, et al. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346(6215):1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy EM. Handbook of avian hybrids of the world. Oxford, New York: Oxford University Press. xiv; 2006. [Google Scholar]
- 19.Ottenburghs J, et al. Hybridization in geese: a review. Front Zool. 2016;13(1):1–9. doi: 10.1186/s12983-016-0153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delacour J, Mayr E. The family Anatidae. Wilson Bull. 1945;57(1):3–55. [Google Scholar]
- 21.Delnicki D. Ross Goose Snow Goose Hybrid in South Texas. Auk. 1974;91(1):174. [Google Scholar]
- 22.Hatch DRM, Shortt AH. Possible Intermediate Ross Goose and Snow Goose in Manitoba. Auk. 1976;93(2):391–392. [Google Scholar]
- 23.Leafloor JO, Moore JA, Scribner KT. A Hybrid Zone between Canada Geese (Branta canadensis) and Cackling Geese (B. hutchinsii) Auk. 2013;130(3):487–500. doi: 10.1525/auk.2013.12196. [DOI] [Google Scholar]
- 24.Nijman V, Aliabadian M, Roselaar CS. Wild hybrids of Lesser White-fronted Goose (Anser erythropus) x Greater White-fronted Goose (A. albifrons) (Aves: Anseriformes) from the European migratory flyway. Zool Anz. 2010;248(4):265–271. doi: 10.1016/j.jcz.2009.10.003. [DOI] [Google Scholar]
- 25.Trauger DL, Dzubin A, Ryder JP. White Geese Intermediate between Ross' Geese and Lesser Snow Geese. Auk. 1971;88(4):856. doi: 10.2307/4083843. [DOI] [Google Scholar]
- 26.Weckstein JD, et al. Hybridization and population subdivision within and between Ross's Geese and Lesser Snow Geese: A molecular perspective. Condor. 2002;104(2):432–436. doi: 10.1650/0010-5422(2002)104[0432:HAPSWA]2.0.CO;2. [DOI] [Google Scholar]
- 27.Lehmhus J, Gustavsson CG. Hybrids between Bar-headed Goose Anser indicus and Snow Goose Anser caerulescens. Ornis Scvecica. 2014;24:147–163. [Google Scholar]
- 28.Craven SR, Westemeier RL. Probable Canada Goose x White-fronted Goose Hybrids. Wilson Bulletin. 1979;91(4):628–629. [Google Scholar]
- 29.Nelson HK. Hybridization of Canada Geese with Blue Geese in the wild. Auk. 1952;69(4):425–428. doi: 10.2307/4081022. [DOI] [Google Scholar]
- 30.Prevett JP, Macinnes CD. Observations of Wild Hybrids between Canada and Blue Geese. Condor. 1973;75(1):124–125. doi: 10.2307/1366546. [DOI] [Google Scholar]
- 31.Gustavsson CG. Images of Barnacle Goose Branta leucopsis hybrids – a photo documentation of some crosses with different Anser species. Ornis Svecica. 2009;19(1):19–31. [Google Scholar]
- 32.Ruokonen M, Kvist L, Lumme J. Close relatedness between mitochondrial DNA from seven Anser goose species. J Evol Biol. 2000;13(3):532–540. doi: 10.1046/j.1420-9101.2000.00184.x. [DOI] [Google Scholar]
- 33.Durand EY, et al. Testing for Ancient Admixture between Closely Related Populations. Mol Biol Evol. 2011;28(8):2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang YH, et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat Genet. 2013;45(7):776–783. doi: 10.1038/ng.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 38.Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57(5):758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 39.Green RE, et al. A Draft Sequence of the Neandertal Genome. Science. 2010;328(5979):710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tajima F. Evolutionary Relationship of DNA-Sequences in Finite Populations. Genetics. 1983;105(2):437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hudson RR. Testing the Constant-Rate Neutral Allele Model with Protein-Sequence Data. Evolution. 1983;37(1):203–217. doi: 10.1111/j.1558-5646.1983.tb05528.x. [DOI] [PubMed] [Google Scholar]
- 42.Ward BJ, van Oosterhout C. hybridcheck: software for the rapid detection, visualization and dating of recombinant regions in genome sequence data. Mol Ecol Resour. 2016;16(2):534–539. doi: 10.1111/1755-0998.12469. [DOI] [PubMed] [Google Scholar]
- 43.Britton T, et al. Estimating divergence times in large phylogenetic trees. Syst Biol. 2007;56(5):741–752. doi: 10.1080/10635150701613783. [DOI] [PubMed] [Google Scholar]
- 44.Fulton TL, Letts B, Shapiro B. Multiple losses of flight and recent speciation in steamer ducks. Proc Royal Soc B-Biol Sci. 2012;279(1737):2339–2346. doi: 10.1098/rspb.2011.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jetz W, et al. The global diversity of birds in space and time. Nature. 2012;491(7424):444–448. doi: 10.1038/nature11631. [DOI] [PubMed] [Google Scholar]
- 46.Li G, et al. Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae) Genome Res. 2016;26(1):1–11. doi: 10.1101/gr.186668.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Csardi, G. and T. Nepusz, The igraph software package for complex network research. InterJournal, Complex Systems, 2006: p. 1695.
- 48.Chen ZZ, Wang L, Yamanaka S. A fast tool for minimum hybridization networks. BMC Bioinformatics. 2012;13:155. doi: 10.1186/1471-2105-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huson D, Linz S. Autumn Algorithm–Computation of Hybridization Networks for Realistic Phylogenetic Trees. 2016. [DOI] [PubMed] [Google Scholar]
- 50.Huson DH, Scornavacca C. Dendroscope 3: An Interactive Tool for Rooted Phylogenetic Trees and Networks. Syst Biol. 2012;61(6):1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475(7357):493–U84. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nadachowska-Brzyska K, et al. Temporal Dynamics of Avian Populations during Pleistocene Revealed by Whole-Genome Sequences. Curr Biol. 2015;25(10):1375–1380. doi: 10.1016/j.cub.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeong CW, et al. Admixture facilitates genetic adaptations to high altitude in Tibet. Nat Commun. 2014;5 [DOI] [PMC free article] [PubMed]
- 54.Rheindt FE, Edwards SV. Genetic Introgression: An Integral but Neglected Component of Speciation in Birds. Auk. 2011;128(4):620–632. doi: 10.1525/auk.2011.128.4.620. [DOI] [Google Scholar]
- 55.Lamichhaney S, et al. Evolution of Darwin's finches and their beaks revealed by genome sequencing. Nature. 2015;518(7539):371–375. doi: 10.1038/nature14181. [DOI] [PubMed] [Google Scholar]
- 56.Dasmahapatra KK, et al. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487(7405):94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang W, Kunte K, Kronforst MR. Genome-Wide Characterization of Adaptation and Speciation in Tiger Swallowtail Butterflies Using De Novo Transcriptome Assemblies. Genome Biol Evol. 2013;5(6):1233–1245. doi: 10.1093/gbe/evt090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frantz, L.A.F., et al., Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol, 2013. 14(9). [DOI] [PMC free article] [PubMed]
- 59.Liu SP, et al. Population Genomics Reveal Recent Speciation and Rapid Evolutionary Adaptation in Polar Bears. Cell. 2014;157(4):785–794. doi: 10.1016/j.cell.2014.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cahill JA, et al. Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears. Mol Ecol. 2015;24(6):1205–1217. doi: 10.1111/mec.13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cui RF, et al. Phylogenomics Reveals Extensive Reticulate Evolution in Xiphophorus Fishes. Evolution. 2013;67(8):2166–2179. doi: 10.1111/evo.12099. [DOI] [PubMed] [Google Scholar]
- 62.Schumer M, et al. Ancient hybridization and genomic stabilization in a swordtail fish. Mol Ecol. 2016;25(11):2661–2679. doi: 10.1111/mec.13602. [DOI] [PubMed] [Google Scholar]
- 63.Kutschera VE, et al. Bears in a forest of gene trees: phylogenetic inference is complicated by incomplete lineage sorting and gene flow. Mol Biol Evol. 2014;31(8):2004–2017. doi: 10.1093/molbev/msu186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brennan IG, Bauer AM, Jackman TR. Mitochondrial introgression via ancient hybridization, and systematics of the Australian endemic pygopodid gecko genus Delma. Mol Phylogenet Evol. 2016;94(Pt B):577–590. doi: 10.1016/j.ympev.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 65.Fuchs J, et al. A multi-locus phylogeny suggests an ancient hybridization event between Campephilus and melanerpine woodpeckers (Ayes: Picidae) Mol Phylogenet Evol. 2013;67(3):578–588. doi: 10.1016/j.ympev.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 66.McCormack JE, Venkatraman MX. A Distinctive Genetic Footprint of Ancient Hybridization. Auk. 2013;130(3):469–475. doi: 10.1525/auk.2013.12208. [DOI] [Google Scholar]
- 67.Rheindt FE, Christidis L, Norman JA. Genetic introgression, incomplete lineage sorting and faulty taxonomy create multiple cases of polyphyly in a montane clade of tyrant-flycatchers (Elaenia, Tyrannidae) Zool Scr. 2009;38(2):143–153. doi: 10.1111/j.1463-6409.2008.00369.x. [DOI] [Google Scholar]
- 68.Wang, W.J., et al., Past hybridization between two East Asian long-tailed tits (Aegithalos bonvaloti and A. fuliginosus). Front Zool, 2014. 11. [DOI] [PMC free article] [PubMed]
- 69.Pinho C, Hey J. Divergence with Gene Flow: Models and Data. Annu Rev Ecol Evol Syst. 2010;41:215–230. doi: 10.1146/annurev-ecolsys-102209-144644. [DOI] [Google Scholar]
- 70.Dobzhansky T. Genetics and the origin of species. New York: Columbia University Press; 1937. [Google Scholar]
- 71.Mayr, E., Systematics in the origin of species : from the viewpoint of a zoologist. Columbia biological series;no. 13. 1942, New York: Harvard University Press.
- 72.Muirhead CA, Presgraves DC. Hybrid Incompatibilities, Local Adaptation, and the Genomic Distribution of Natural Introgression between Species. Am Nat. 2016;187(2):249–261. doi: 10.1086/684583. [DOI] [PubMed] [Google Scholar]
- 73.Coyne, J.A. and H.A. Orr, Speciation. 2004, Sunderland, MA: Sinauer Associates, Inc.
- 74.Wilkinson-Herbots HM. The distribution of the coalescence time and the number of pairwise nucleotide differences in the "isolation with migration" model. Theor Popul Biol. 2008;73(2):277–288. doi: 10.1016/j.tpb.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 75.Nosil, P., Ecological speciation. Oxford series in ecology and evolution. 2012, Oxford: Oxford University Press.
- 76.Wu CI. The genic view of the process of speciation. J Evol Biol. 2001;14(6):851–865. doi: 10.1046/j.1420-9101.2001.00335.x. [DOI] [Google Scholar]
- 77.Smyth JF, Patten MA, Pruett CL. The evolutionary ecology of a species ring: a test of alternative models. Folia Zool. 2015;64(3):233–244. [Google Scholar]
- 78.Raposo do Amaral F, et al. Multilocus tests of Pleistocene refugia and ancient divergence in a pair of Atlantic Forest antbirds (Myrmeciza) Mol Ecol. 2013;22(15):3996–4013. doi: 10.1111/mec.12361. [DOI] [PubMed] [Google Scholar]
- 79.Yeung CKL, et al. Testing Founder Effect Speciation: Divergence Population Genetics of the Spoonbills Platalea regia and P. minor (Threskiornithidae, Aves) Mol Biol Evol. 2011;28(1):473–482. doi: 10.1093/molbev/msq210. [DOI] [PubMed] [Google Scholar]
- 80.Nadachowska-Brzyska, K., et al., Demographic Divergence History of Pied Flycatcher and Collared Flycatcher Inferred from Whole-Genome Re-sequencing Data. PLoS Genet, 2013. 9(11). [DOI] [PMC free article] [PubMed]
- 81.Lavretsky P, et al. Becoming pure: identifying generational classes of admixed individuals within lesser and greater scaup populations. Mol Ecol. 2016;25(3):661–674. doi: 10.1111/mec.13487. [DOI] [PubMed] [Google Scholar]
- 82.Poelstra JW, et al. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science. 2014;344(6190):1410–1414. doi: 10.1126/science.1253226. [DOI] [PubMed] [Google Scholar]
- 83.Arrieta RS, Lijtmaer DA, Tubaro PL. Evolution of postzygotic reproductive isolation in galliform birds: analysis of first and second hybrid generations and backcrosses. Biol J Linn Soc. 2013;110(3):528–542. doi: 10.1111/bij.12153. [DOI] [Google Scholar]
- 84.Lijtmaer DA, Mahler B, Tubaro PL. Hybridization and postzygotic isolation patterns in pigeons and doves. Evolution. 2003;57(6):1411–1418. doi: 10.1111/j.0014-3820.2003.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 85.Randler C. Frequency of bird hybrids: does detectability make all the difference? J Ornithol. 2004;145(2):123–128. doi: 10.1007/s10336-004-0022-0. [DOI] [Google Scholar]
- 86.Servedio MR, Noor MAF. The role of reinforcement in speciation: Theory and data. Annu Rev Ecol Evol Syst. 2003;34:339–364. doi: 10.1146/annurev.ecolsys.34.011802.132412. [DOI] [Google Scholar]
- 87.Zachos J, et al. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292(5517):686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 88.Prins HHT. The origins and development of grassland communities in northwestern Europe. In: Wallis de Vries MF, Bakker JP, van Wieren SE, editors. Grazing and Conservation Management. Boston: Kluwer Academic Publishers; 1998. pp. 55–105. [Google Scholar]
- 89.Kahlke RD. The origin of Eurasian Mammoth Faunas (Mammuthus coelodonta Faunal Complex) Quat Sci Rev. 2014;96:32–49. doi: 10.1016/j.quascirev.2013.01.012. [DOI] [Google Scholar]
- 90.Owen M. Wild geese of the world : their life history and ecology. London: Batsford; 1980. [Google Scholar]
- 91.Rodway MS. Timing of pairing in waterfowl I: Reviewing the data and extending the theory. Waterbirds. 2007;30(4):488–505. doi: 10.1675/1524-4695(2007)030[0506:TOPIWI]2.0.CO;2. [DOI] [Google Scholar]
- 92.Chunco AJ. Hybridization in a warmer world. Ecol Evol. 2014;4(10):2019–2031. doi: 10.1002/ece3.1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buggs RJA. Empirical study of hybrid zone movement. Heredity. 2007;99(3):301–312. doi: 10.1038/sj.hdy.6800997. [DOI] [PubMed] [Google Scholar]
- 94.Hewitt G. The genetic legacy of the Quaternary ice ages. Nature. 2000;405(6789):907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 95.Hewitt GM. Some genetic consequences of ice ages, and their role in divergence and speciation. Biol J Linn Soc. 1996;58(3):247–276. doi: 10.1111/j.1095-8312.1996.tb01434.x. [DOI] [Google Scholar]
- 96.Ploeger PL. Geographical Differentation in Artic Anatidae as a Result of Isolation During the Last Glacial. 1968. EJ Brill.
- 97.Ruokonen M, Aarvak T, Madsen J. Colonization history of the high-arctic pink-footed goose Anser brachyrhynchus. Mol Ecol. 2005;14(1):171–178. doi: 10.1111/j.1365-294X.2004.02380.x. [DOI] [PubMed] [Google Scholar]
- 98.Ruokonen M, Litvin K, Aarvak T. Taxonomy of the bean goose-pink-footed goose. Mol Phylogenet Evol. 2008;48(2):554–562. doi: 10.1016/j.ympev.2008.04.038. [DOI] [PubMed] [Google Scholar]
- 99.Eda M, et al. Phylogenetic relationship of the Greater White-fronted Goose Anser albifrons subspecies wintering in the Palaearctic region. Ornithol Sci. 2013;12(1):35–42. doi: 10.2326/osj.12.35. [DOI] [Google Scholar]
- 100.Ely CR, et al. Circumpolar variation in morphological characteristics of Greater White-Fronted Geese Anser albifrons. Bird Study. 2005;52:104–119. doi: 10.1080/00063650509461380. [DOI] [Google Scholar]
- 101.Scribner KT, et al. Phylogeography of Canada Geese (Branta canadensis) in western North America. Auk. 2003;120(3):889–907. doi: 10.1642/0004-8038(2003)120[0889:POCGBC]2.0.CO;2. [DOI] [Google Scholar]
- 102.Quinn T. The genetic legacy of Mother Goose–phylogeographic patterns of lesser snow goose Chen caerulescens caerulescens maternal lineages. Mol Ecol. 1992;1(2):105–117. doi: 10.1111/j.1365-294X.1992.tb00162.x. [DOI] [PubMed] [Google Scholar]
- 103.Paxinos EE, et al. mtDNA from fossils reveals a radiation of Hawaiian geese recently derived from the Canada goose (Branta canadensis) Proc Natl Acad Sci U S A. 2002;99(3):1399–1404. doi: 10.1073/pnas.032166399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eisenhauer DI, Kirkpatrick CM. Ecology of Emperor Goose in Alaska. Wildl Monogr. 1977;57:6–62. [Google Scholar]
- 105.Madsen, J., G. Cracknell, and T. Fox, Goose populations of the Western Palearctic : a review of status and distribution. Wetlands International publication. Vol. 48. 1999, Wageningen: Wetlands International.
- 106.Nei M, Maruyama T, Chakraborty R. Bottleneck Effect and Genetic-Variability in Populations. Evolution. 1975;29(1):1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- 107.Frankham R. Do island populations have less genetic variation than mainland populations? Heredity. 1997;78:311–327. doi: 10.1038/hdy.1997.46. [DOI] [PubMed] [Google Scholar]
- 108.Spurgin LG, et al. Genetic and phenotypic divergence in an island bird: isolation by distance, by colonization or by adaptation? Mol Ecol. 2014;23(5):1028–1039. doi: 10.1111/mec.12672. [DOI] [PubMed] [Google Scholar]
- 109.Clegg SM, et al. Genetic consequences of sequential founder events by an island-colonizing bird. Proc Natl Acad Sci U S A. 2002;99(12):8127–8132. doi: 10.1073/pnas.102583399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Mlíkovský J. Cenozoic birds of the world Part 1: Europe. Praha: Ninox Press; 2002. [Google Scholar]
- 111.Brodkorb, P., Catalogue of fossil birds: Part 2 (Anseriformes through Galliformes). 1964: University of Florida.
- 112.Wirtz P. Mother species-father species: unidirectional hybridization in animals with female choice. Anim Behav. 1999;58:1–12. doi: 10.1006/anbe.1999.1144. [DOI] [PubMed] [Google Scholar]
- 113.Randler C. Behavioural and ecological correlates of natural hybridization in birds. Ibis. 2006;148(3):459–467. doi: 10.1111/j.1474-919X.2006.00548.x. [DOI] [Google Scholar]
- 114.Johnsgard PA. Handbook of waterfowl behavior. Ithaca, N.Y: Comstock Pub. Associates. xiv; 1965. [Google Scholar]
- 115.Randler C. Do forced extrapair copulations and interspecific brood amalgamation facilitate natural hybridisation in wildfowl? Behaviour. 2005;142:477–488. doi: 10.1163/1568539054012029. [DOI] [Google Scholar]
- 116.Rohwer, F.C. and M.G. Anderson, Female-biased philopatry, monogamy, and the timing of pair formation in migratory waterfowl, in Current Ornithology. 1988, Springer. p. 187–221.
- 117.Seehausen O. Hybridization and adaptive radiation. Trends Ecol Evol. 2004;19(4):198–207. doi: 10.1016/j.tree.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 118.Gilbert, L., Adaptive novelty through introgression in Heliconius wing patterns: evidence for shared genetic “tool box” from synthetic hybrid zones and a theory of diversification. Ecology and Evolution Taking Flight: Butterflies as Model Systems, 2003: p. 281–318.
- 119.Almen MS, et al. Adaptive radiation of Darwin's finches revisited using whole genome sequencing. BioEssays. 2016;38(1):14–20. doi: 10.1002/bies.201500079. [DOI] [PubMed] [Google Scholar]
- 120.Rieseberg LH, Archer MA, Wayne RK. Transgressive segregation, adaptation and speciation. Heredity. 1999;83(4):363–372. doi: 10.1038/sj.hdy.6886170. [DOI] [PubMed] [Google Scholar]
- 121.Schumer M, Rosenthal GG, Andolfatto P. How common is homoploid hybrid speciation? Evolution. 2014;68(6):1553–1560. doi: 10.1111/evo.12399. [DOI] [PubMed] [Google Scholar]
- 122.Hermansen JS, et al. Hybrid speciation in sparrows I: phenotypic intermediacy, genetic admixture and barriers to gene flow. Mol Ecol. 2011;20(18):3812–3822. doi: 10.1111/j.1365-294X.2011.05183.x. [DOI] [PubMed] [Google Scholar]
- 123.Brelsford A, Mila B, Irwin DE. Hybrid origin of Audubon's warbler. Mol Ecol. 2011;20(11):2380–2389. doi: 10.1111/j.1365-294X.2011.05055.x. [DOI] [PubMed] [Google Scholar]
- 124.Nietlisbach P, et al. Hybrid ancestry of an island subspecies of Galapagos mockingbird explains discordant gene trees. Mol Phylogenet Evol. 2013;69(3):581–592. doi: 10.1016/j.ympev.2013.07.020. [DOI] [PubMed] [Google Scholar]
- 125.Lavretsky P, et al. Genetic admixture supports an ancient hybrid origin of the endangered Hawaiian duck. J Evol Biol. 2015;28(5):1005–1015. doi: 10.1111/jeb.12637. [DOI] [PubMed] [Google Scholar]
- 126.Grant PR, Grant BR. The secondary contact phase of allopatric speciation in Darwin's finches. Proc Natl Acad Sci U S A. 2009;106(48):20141–20148. doi: 10.1073/pnas.0911761106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Elgvin TO, et al. The genomic mosaicism of hybrid speciation. Sci Adv. 2017;3(6):e1602996. doi: 10.1126/sciadv.1602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.del Hoyo, J. and A. Elliott, Handbook of the birds of the world. 1992, Barcelona: Lynx.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sampled goose species and sampling location. (DOCX 15 kb)
Mapping results of all goose samples to Mallard genome (version 73) using SMALT. (DOCX 20 kb)
D-statistics for all combinations of three species per genus. Significant D-statistics (Z-score > 4), suggesting gene flow between P2 and P3, are indicated in bold and colored in yellow. The outgroup for Branta was a consensus sequence based on all Anser species. Similarly, the outgroup for Anser was a consensus sequence based on all Branta species. (DOCX 19 kb)
Distribution of gene tree divergence times for all goose species. All distributions show a single peak, indicating gene flow during divergence. The divergence time of several gene trees was close to zero, suggesting low levels of recent gene flow between certain species. Final three figures represent the three subspecies of Brent Goose, which is depicted in the lower right panel. (ZIP 2715 kb)
Estimates of historical effective population sizes for all goose species, based on a PSMC analysis. Final three figures represent the three subspecies of Brent Goose, which is depicted in the lower right panel. (ZIP 766 kb)
Data Availability Statement
The dataset supporting the conclusions of this article are available at Bioproject PRJEB20373 (http://www.ebi.ac.uk/ena/data/view/PRJEB20373) at the European Nucleotide Archive (ENA).


