SUMMARY
Tourette syndrome (TS) is a model neuropsychiatric disorder thought to arise from abnormal development and/or maintenance of cortico-striato-thalamo-cortical circuits. TS is highly heritable, but its underlying genetic causes are still elusive, and no genome-wide significant loci have been discovered to date. We analyzed a European ancestry sample of 2,434 TS cases and 4,093 ancestry-matched controls for rare (<1% frequency) copy-number variants (CNVs) using SNP microarray data. We observed an enrichment of global CNV burden that was prominent for large (>1 Mb), singleton events (OR=2.28, 95%CI [1.39–3.79], p=1.2×10−3) and known, pathogenic CNVs (OR=3.03 [1.85–5.07], p=1.5×10−5). We also identified two individual, genome-wide significant loci, each conferring a substantial increase in TS risk (NRXN1 deletions, OR=20.3, 95%CI [2.6–156.2]; CNTN6 duplications, OR=10.1, 95% CI [2.3–45.4]). Approximately 1% of TS cases carry one of these CNVs, indicating that rare structural variation contributes significantly to the genetic architecture of TS.
INTRODUCTION
Tourette syndrome (TS) is a complex neuropsychiatric disorder characterized by multiple chronic involuntary motor and vocal tics, with an estimated population prevalence of 0.3–0.9% (Scharf et al., 2015). Tics typically emerge during childhood and peak in adolescence, with a subsequent reduction in symptoms, supporting the notion that TS is neurodevelopmental in origin (Robertson et al., 2017). Most TS patients (>85%) present with additional neuropsychiatric comorbidities, typically attention deficit hyperactivity disorder (ADHD) and obsessive-compulsive disorder (OCD) (Hirschtritt et al., 2015), although the risk for mood, anxiety, major depressive, and autism spectrum disorders (ASD) is also elevated (Burd et al., 2009; Hirschtritt et al., 2015). Consequently, TS is often considered a model neuropsychiatric disorder in that identification of its underlying molecular, cellular, and neurophysiologic etiology may be broadly applicable to a wide range of psychiatric disorders.
Neuroimaging (Greene et al., 2016; Marsh et al., 2009) and neurophysiology (Draper et al., 2014; Gilbert et al., 2004) studies suggest that TS and its associated comorbidities (e.g., OCD and ADHD) arise from dysregulated development and/or maintenance of parallel cortico-striatal-thalamo-cortical (CSTC) motor, limbic, and cognitive circuits (Jahanshahi et al., 2015). Though non-genetic factors have been associated with increased TS risk (Browne et al., 2016; Leivonen et al., 2016), TS is primarily a genetic disorder. Family studies indicate that children of affected parents have a 60-fold higher risk of developing TS or chronic tics (CT), a closely related disorder, compared to the general population (Browne et al., 2015). TS heritability is estimated to be 0.77 (Mataix-Cols et al., 2015), making it one of the most heritable complex neuropsychiatric disorders. Despite this strong genetic component, the identification of bona-fide TS susceptibility genes has proven challenging. Although linkage analyses have identified several candidate regions, there is little consensus across studies, suggesting that, as with other neuropsychiatric disorders, TS is genetically complex and heterogeneous (Robertson et al., 2017). Similarly, analyses of TS genetic architecture using aggregated SNP data demonstrates that TS is highly polygenic, with the majority of inherited TS risk distributed throughout the genome (Davis et al., 2013), though an initial genome-wide association study (GWAS) did not yield any genome-wide significant loci, likely due to small sample size (Scharf et al., 2013).
Studies examining rare structural variation in individuals with TS have implicated several neurodevelopmental genes involved in neurite outgrowth and axonal migration. Rare chromosomal abnormalities affecting CNTNAP2 (Verkerk et al., 2003) and SLITRK1 (Abelson et al., 2005) have been found in isolated TS families, and exonic copy-number variants (CNVs) in NRXN1 are reported in small genome-wide studies (Nag et al., 2013; Sundaram et al., 2010), though no locus has yet survived genome-wide correction for multiple testing. Because of evidence suggesting that rare CNVs may have a role in TS etiology (Fernandez et al., 2012; McGrath et al., 2014), and since such variants contribute to susceptibility for other heritable neurodevelopmental disorders (NDDs) (Malhotra and Sebat, 2012), we assessed the impact of rare CNVs on TS disease risk in a large sample of 6,527 unrelated individuals of European ancestry. We demonstrate a global increase in the burden of large, rare CNVs in TS cases compared to controls driven primarily by large, singleton events, in particular large (>1Mb) deletions, consistent with marked genetic heterogeneity. We also report the first two TS susceptibility loci that meet genome-wide significance: deletions in NRXN1 and duplications in CNTN6. Each confers a substantial increase in disease risk and together are present in 1% of TS cases.
RESULTS
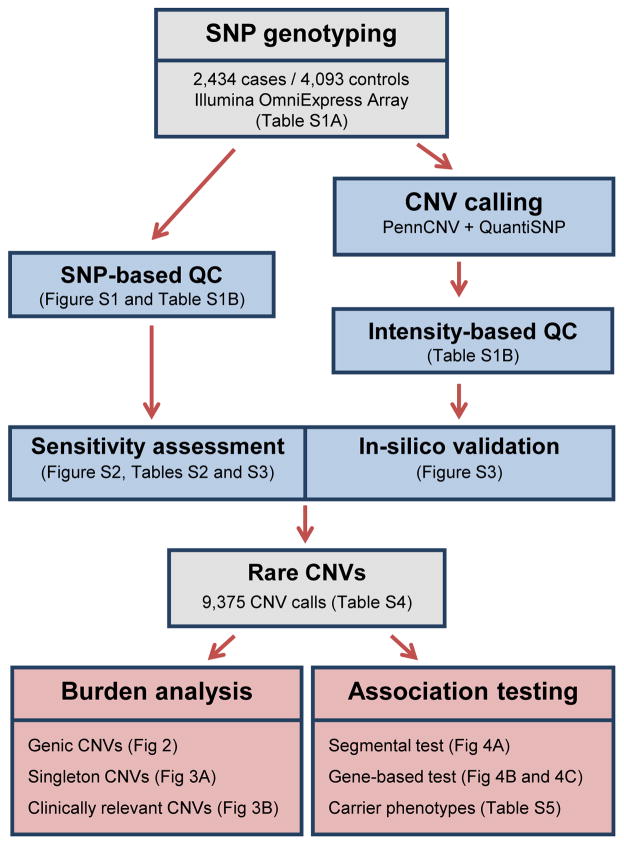
An overview of the sample selection, quality control, CNV detection, and data analysis performed in this study is presented in Figure 1 and described in detail in the STAR Methods. All TS cases and controls were recruited through the Tourette Syndrome Association International Consortium for Genetics (TSAICG) or through the Gilles de la Tourette Syndrome GWAS Replication Initiative (GGRI), with additional controls selected from external studies (STAR Methods). All DNA samples were genotyped on the Illumina OmniExpress SNP array platform (Table S1A). We restricted analysis to SNP assays common to all array versions. We conducted extensive quality control analyses including both SNP-based and CNV-based exclusion of outliers (Table S1B; STAR Methods) and genotype-based determination of ancestry (Figure S1). The final dataset consisted of 6,527 unrelated European ancestry samples: 2,434 individuals diagnosed with TS and 4,093 unselected controls.
Figure 1. Flow chart of experimental procedures and analyses.
CNVs were called from genome-wide SNP genotype data generated from 2,434 TS cases and 4,093 controls (grey). Data processing, CNV detection and quality control steps (blue) are described in the STAR Methods. An outline of the main analyses is presented in red. Figures or tables relevant to each outlined step are shown in parentheses.
Genome-wide detection of CNVs was performed using the consensus of two widely-used Hidden Markov Model (HMM)-based methods (STAR Methods). Additionally, we used a locus-specific, intensity-based clustering method to generate CNV genotypes in all samples across 11 common HapMap3 loci for sensitivity analysis (Figure S2; Table S2; STAR Methods). Using the proportion of concordant HMM-based calls at these loci as a sensitivity measure, we confirmed the absence of any bias in CNV detection between cases and controls across all loci (p=0.54, Fisher’s Exact test) and between individuals (p=0.15, Welch’s t-test; see Table S3). Post-call cleaning was performed and CNVs were annotated for gene content and frequency (STAR Methods). CNVs were considered “genic” if they overlapped the exon of a known protein-coding Refseq transcript. Frequencies were defined based on a 50% overlap with other CNVs as described elsewhere (CNV and SCZ Working Groups of the PGC, 2017); “singletons” denote CNVs with a frequency of one across the entire dataset. We filtered calls for rare (frequency <1% or <65 events) CNVs ≥30kb in length and spanning at least 10 probes. Finally, using a heuristically derived series of in silico validation metrics, we removed aberrant CNV calls due to mosaicism and misclassified rare events (Figure S3; STAR Methods). In total, we resolved 9,375 rare CNVs (Table S4).
Global burden analysis of rare CNVs in TS
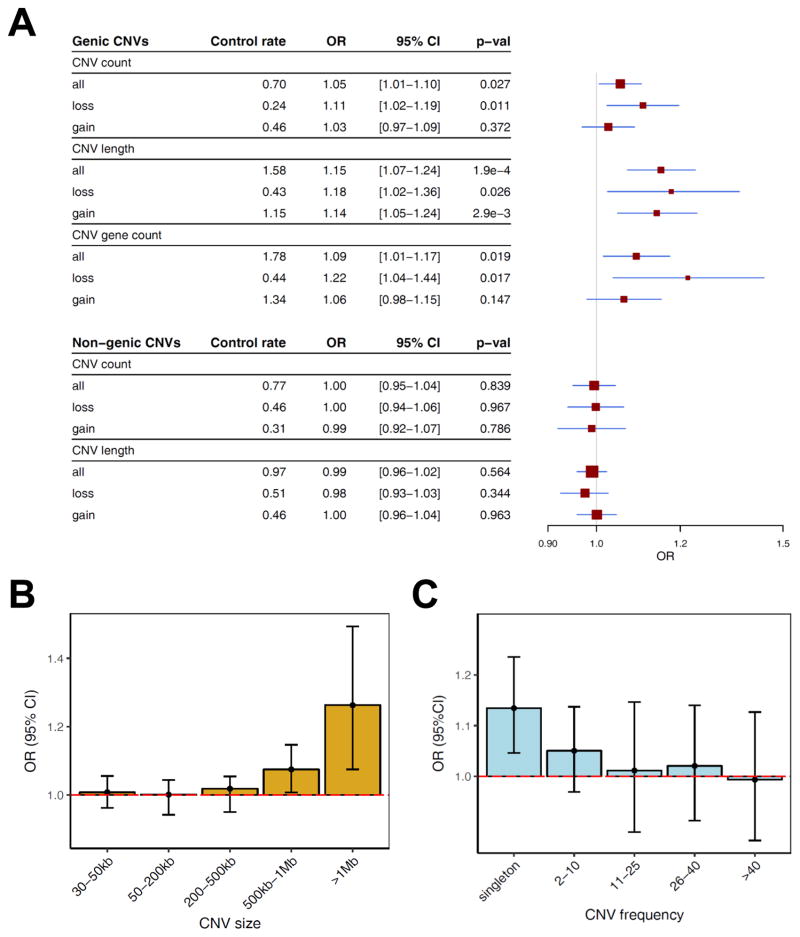
An increase in rare CNV burden has been consistently demonstrated in other NDDs (CNV and SCZ Working Groups of the PGC, 2017). Controlling for potential confounders, burden analysis was performed using logistic regression (STAR Methods) with three different burden metrics: 1) total number of CNVs (CNV count), 2) total genomic size of all CNVs (CNV length), and 3) number of genes affected (CNV gene count). For genic CNVs (n=4,604), we observed modest but significant increases in burden across all metrics (Figure 2A): CNV count (OR 1.05 [1.01–1.10], p=0.027), CNV gene count (OR 1.09 [1.01–1.17], p=0.019), and CNV length (OR 1.15 [1.07–1.24], p=1.9×10−4). By contrast, no enrichment was seen in a comparable number (n=4,771) of non-genic events. The increased burden in TS was most significant for CNV length and consistent across each control set individually (Figure S4). To explore the CNV length burden further, we partitioned the data across a range of CNV size and frequency bins and observed the enrichment was mainly attributable to large (>1Mb; OR 1.26 [1.08–1.49], p=5.3×10−3) (Figure 2B) and/or singleton CNVs (OR 1.13 [1.04–1.24], p=2.9×10−3) (Figure 2C).
Figure 2. Rare CNV burden in 2,434 TS cases and 4,093 controls.
(A) The global burden of all rare (<1% frequency) CNVs > 30kb is shown for genic (top) and non-genic (bottom) CNVs and stratified by CNV type (all, loss (deletions), gain (duplications)). Global CNV burden is compared using three different metrics: CNV count, total number of CNVs per subject; CNV length, aggregate length of all CNVs (in Mb); and CNV gene count, number of genes spanned by CNVs. Control rate, averaged baseline burden metric per control subject. Red boxes, odds ratios (box size is proportional to standard error); Blue lines indicate 95% confidence intervals. Genic CNVs are defined as those that overlap any exon of a known protein-coding gene (see STAR Methods).
(B) Analyses in (A) were assessed further by partitioning CNV length burden of all CNVs (deletions + duplications) into different CNV size categories. Whiskers represent 95% confidence intervals.
(C) The analysis in (B) was repeated for CNVs binned by frequency.
Odds ratios (OR) were calculated from logistic regression adjusted for covariates using standardized burden metrics (STAR Methods). ORs >1 indicate an increased TS risk.
Enrichment of large, singleton events and clinically relevant CNVs
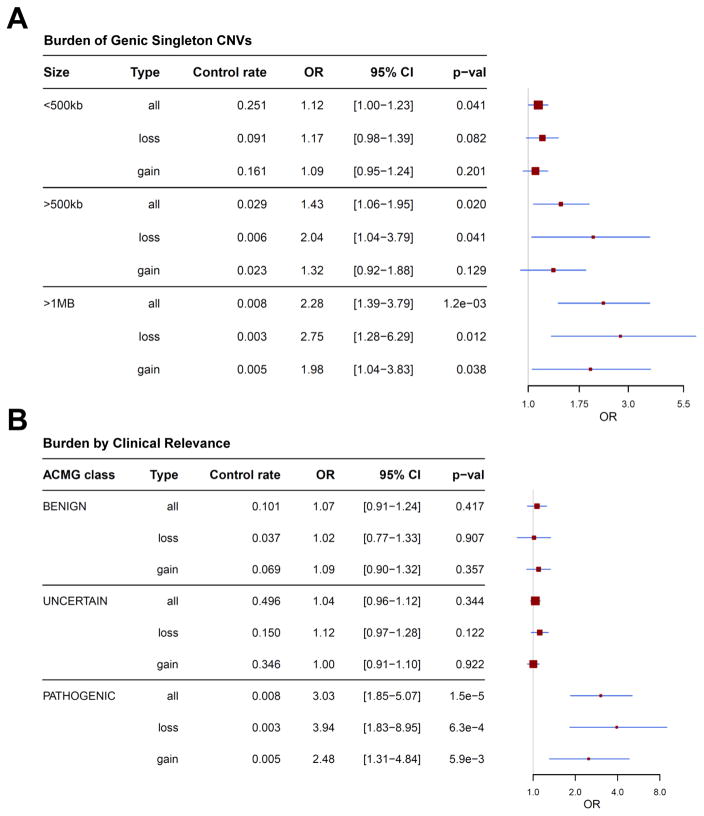
We next explored whether specific CNV classes were enriched in TS. Since the elevated TS CNV burden was confined to large and/or very rare events, we re-evaluated the CNV count burden restricted to genic singletons, stratified by CNV size. We observed a significant enrichment of singletons >500kb (OR 1.43 [1.06–1.95], p=0.020) that was further increased in the largest size category (>1Mb, OR 2.28 [1.39–3.79], p=1.2×10−3). The enrichment of >1Mb genic CNVs was greater for deletions (OR 2.75 [1.28–5.23], p=0.012) than duplications (OR 1.98 [1.04–3.83], p=0.038) (Figure 3A). Notably, the enrichment of singleton deletions >1Mb was driven by CNVs spanning genes under strong evolutionary constraint (probability of Loss-of-Function Intolerance (pLI) score>0.9; Lek et al., 2015) (Rate Ratio=2.65 [1.40–5.00], p=2.7×10−3; Poisson regression; STAR Methods).
Figure 3. Large, singleton CNVs and known pathogenic variants are overrepresented in TS.
(A) CNV count burden restricted to genic singleton events, stratified by CNV size and type (deletion/duplication).
(B) CNV burden of all rare CNVs, separated by clinical relevance (benign, uncertain, pathogenic) according to the American College of Medical Genetics guidelines (STAR Methods).
Red boxes, odds ratios; Blue lines, 95% CIs. ORs > 1 represent an increase in risk for TS per CNV.
It is well established that certain regions of the human genome are prone to rare, recurrent CNVs associated with a broad range of NDDs (Malhotra and Sebat, 2012). To evaluate if such pathogenic CNVs confer risk for TS, we classified all rare CNV calls by clinical relevance according to American College of Medical Genetics (ACMG) guidelines (Kearney et al., 2011) and assessed for enrichment between cases and controls. Known pathogenic CNVs were identified in 1.9% of TS cases vs. 0.8% of controls (OR 3.03 [1.85–5.07], p=1.5×10−5) (Figure 3B). Consistent with an increased pathogenicity of deletions compared to duplications, this enrichment was greater for deletions alone (OR per CNV 3.94 [1.83–8.95], p=6.3 ×10−4). By contrast, no increase in burden was observed among CNVs classified as either benign or of unknown clinical significance.
Deletions in NRXN1 and duplications in CNTN6 confer substantial risk for TS
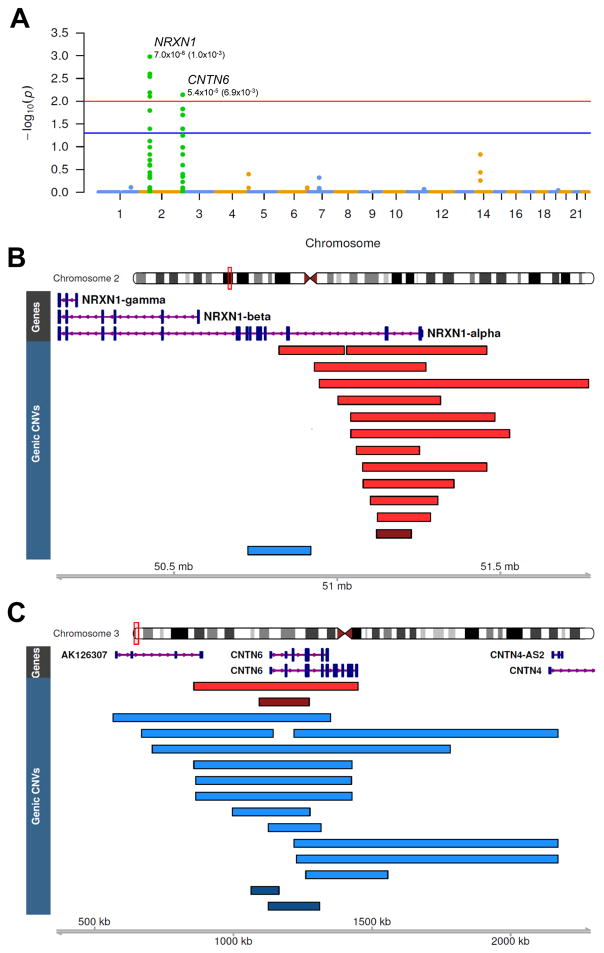
To test our sample for enrichment of rare CNVs at individual loci, we conducted an unbiased, point-wise (segmental) genome-wide association test, treating deletions and duplications independently (STAR Methods). As non-overlapping CNVs affecting the same gene would be unaccounted for by segmental assessments of enrichment, we also conducted a complementary gene-based test, conditioned on CNVs affecting exons. In contrast to SNP-based GWAS, there is no established p-value threshold to indicate genome-wide significance for CNVs, since the number of rare CNV breakpoints per genome varies across individuals and detection platforms. Therefore, for both tests, we established both locus-specific p-values (Pseg and Pgene for segmental and gene-based tests, respectively) and genome-wide corrected p-values (Pcorr) empirically through 1,000,000 label-swapping permutations, using the max(T) method (Westfall and Troendle, 2008; STAR Methods) to control for family-wise error rate (FWER). Both tests converged on the same two loci, one for deletions and another for duplications, which were enriched among TS cases and survived genome-wide correction for multiple testing.
For deletions, the peak segmental association signal was located on chromosome 2p16 (Pseg=7.0×10−6; Pseg-corr=1.0×10−3; Figure 4A), corresponding to heterozygous losses across the first two exons of NRXN1, and found exclusively among TS cases (N=10, Figure 4B). In the gene-based test of exonic CNVs, heterozygous NRXN1 deletions were also the most significant association genome-wide (Pgene=5.9×10−5; Pgene-corr=8.5×10−4), representing 12 cases (0.49%) and 1 control (0.02%), corresponding to a substantially increased TS risk (OR 20.3 [2.6–156.2]). Consistent with previously identified pathogenic NRXN1 deletions in other NDDs these exon-spanning CNVs clustered at the 5′ end and predominantly affected the NRXN1-α isoform (Dabell et al., 2013).
Figure 4. Segmental and gene-based tests converge on two distinct loci significantly enriched in TS cases.
(A) Manhattan plot of segmental association test results representing genome-wide corrected p-values calculated at each CNV breakpoint. The two genome-wide significant association peaks correspond to deletions at NRXN1 (Plocus=7.0×10−6, Pcorr=1.0×10−3) and duplications at CNTN6 (Plocus=5.4×10−5, Pcorr =6.9×10−3). Red and blue levels correspond to a genome-wide corrected α of 0.05 and 0.01, respectively.
(B) Heterozygous exonic deletions in NRXN1 found in 12 cases (0.49%) and 1 control (0.03%), corresponding to an OR=20.2, 95% CI (2.6–155.2). Exon-affecting CNVs cluster at the 5′ end with deletions across exons 1–3 found in 10 cases and no controls. Red, deletions in TS cases; Dark Red, deletion in controls; Blue, case duplication.
(C) Exon-spanning duplications over CNTN6 found in 12 cases and 2 controls (OR=10.2, [2.0–17.8]) CNTN6 duplications are considerably larger in cases compared to controls (640 vs. 143 kb, on average). Blue, case duplications; Dark blue, control duplications; Red, case deletion; Dark red, control duplication.
For duplications, the segmental association test identified one genome-wide significant locus on chromosome 3p26, within CNTN6 (Pseg=5.4×10−5, Pseg-corr=6.9×10−3), with a secondary peak located directly upstream (Pseg=5.9×10−5, Pseg-corr=6.9×10−3, Figures 4A and 4C). Closer inspection revealed an enrichment of large duplications spanning this gene. The gene-based test identified the same locus, exonic CNTN6 duplications, with heterozygous gains found in 12 cases (0.49%) and 2 controls (0.05%), corresponding to an OR=10.1 [2.3–45.4] (Pgene=2.5×10−4, Pgene-corr =8.3×10−3). Notably, the CNTN6 duplications in TS cases were considerably larger than those in controls (641 vs. 143 kb). 9 of 12 TS carriers harbored a duplication >500 kb in length, while duplications in controls were <200kb.
All genic CNV calls across NRXN1 and CNTN6 were verified by inspection of probe-level intensity plots (Figures S5 and S6). No additional loci were significant after controlling for FWER, under either segmental or gene-based tests of association, and we obtained similar results after pair-matching each case with its closest ancestrally matched control, suggesting that these results are not due to inter-European population stratification (Figure S7; STAR Methods).
DISCUSSION
In this study, we demonstrate a significant role for rare structural variation in the pathogenesis of TS, a still poorly understood neurodevelopmental disorder. We observe an increased global burden of rare CNVs and report two definitive TS risk loci that surpass empirical thresholds for genome-wide significance, deletions in NRXN1 and duplications in CNTN6.
NRXN1 is a highly-studied, pre-synaptic cell-adhesion molecule involved in synaptogenesis and synaptic transmission at both glutamatergic and GABAergic synapses (Pak et al., 2015). NRXN1 is primarily transcribed from two alternative promoters, resulting in a full-length NRXN1-α isoform and a short C-terminal NRXN1-β isoform (Ushkaryov et al., 1992). NRXN1-α contains six alternative splice sites which, in combination, generate hundreds of unique transcripts that segregate within specific brain regions and cell types (Fuccillo et al., 2015; Schreiner et al., 2014). NRXN1-α isoforms preferentially bind to various trans-synaptic partners, including neuroligins, cerebellins, neurexophilins and LRRTMs, each of which subserves different synaptic functions (de Wit and Ghosh, 2016). NRXN1-α trans-synaptic interactions play a critical role in thalamo-cortical synaptogenesis and plasticity (Singh et al., 2016), suggesting one possible mechanism in support of the prevailing theory that TS arises from abnormal sensorimotor CSTC circuit development (Jahanshahi et al., 2015).
Although previous studies have observed heterozygous exonic NRXN1 deletions in TS cases (Fernandez et al., 2012; Nag et al., 2013; Sundaram et al., 2010), small sample sizes precluded a definitive association of this deletion with TS. We demonstrate, in a large independent sample, that exonic NRXN1 deletions confer a substantial increase in TS risk. The association of heterozygous NRXN1 deletions with different NDDs is one of the most reliable findings in the neuropsychiatry CNV literature (Dabell et al., 2013). Consistent with this, 4 of the 12 TS cases with exonic NRXN1 deletions in our sample had another broadly-defined NDD (2 ASD, 1 DD, 1 Developmental Speech/Language Disorder unspecified) (Table S5), supporting the hypothesis that these deletions may interfere with a generalized neurodevelopmental process which, when combined with other disease-specific mutations and/or background polygenic risk, results in the observed phenotypic pleiotropy.
Like NRXN1, CNTN6 encodes a cell-adhesion molecule expressed primarily in the central nervous system (Ogawa et al., 1996). Contactins are members of the L1 immunoglobulin superfamily of proteins, and Cntn6 serves multiple functions in the developing mouse nervous system including orientation of apical dendrites in cortical pyramidal neurons, regulation of Purkinje cell development and synaptogenesis, and oligodendrocyte differentiation from neuroprogenitor cells (Oguro-Ando et al., 2017). Mice with homozygous inactivation of Cntn6 also exhibit reproducible motor impairment (Huang et al., 2012).
Duplications in CNTN6 represent a novel association for TS. CNVs affecting CNTN6 have been reported in isolated cases of intellectual disability/developmental delay (ID/DD) (Kashevarova et al., 2014), and deletions alone are enriched in ASD (Mercati et al., 2016). Notably, in a clinical series of 3,724 patients referred for cytogenetic testing, all 7 CNTN6 duplication carriers either presented with or had a first-degree relative with ADHD and/or OCD, while none of the 7 CNTN6 deletion carriers were diagnosed with these two common TS comorbidities (Hu et al., 2015). In our study, the rates of co-morbid OCD/ADHD were not increased in TS CNTN6 CNV carriers compared to non-carriers, and no TS CNTN6 carrier was noted to have ASD/ID/DD (Table S5).
Several limitations of the current study can inform future inquiry. First, although our sample represents the largest survey of CNVs in TS to date, it is still underpowered to detect extremely rare CNVs and/or those of moderate effect. While we show strong evidence for the involvement of deletions across NRXN1, our data also does not support other similarly implicated loci, including deletions in COL8A1 (Nag et al., 2013), which we observe only once in a single TS patient. This emphasizes the need for larger, independent samples for continued discovery and refinement of candidate TS loci. Second, while our TS cases were well characterized for OCD and ADHD, we did not formally assess ASD, ID, SCZ or epilepsy. Parents and/or adult subjects were queried about existing diagnoses of these NDDs as well as learning disorders/developmental delay, but cases with milder ASD/DD may not have been detected.
Additional efforts should focus on characterizing the full scope of phenotypes associated with NRXN1 and CNTN6 CNVs. A comprehensive molecular analysis of these CNVs is also needed to fully understand how they increase risk for such phenotype(s). Finally, the elevated burden observed here is largely confined to large singletons and known pathogenic CNVs, consistent with a global enrichment of CNVs under strong negative selection that possibly arose de novo or within the last few generations. This suggests that, in addition to substantial increases in sample size, alternative study designs that allow for discrimination of de novo CNVs will be fruitful for TS, as was recently shown for likely-gene-disrupting variants identified by exome sequencing in TS trios (Willsey et al., 2017).
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jeremiah M. Scharf (jscharf@partners.org)
Experimental Model and Subject Details
Sample Ascertainment
Tourette Syndrome (TS) cases (n=2,243) were ascertained primarily from TS specialty clinics through sites distributed throughout North America, Europe and Israel as part of an ongoing collaborative effort by the Tourette Syndrome Association International Consortium for Genetics (TSAICG; https://www.findtsgenes.org/) as described previously (Scharf et al., 2013). Subjects were assessed for a lifetime diagnosis of TS, Obsessive Compulsive Disorder (OCD) and Attention-Deficit/Hyperactivity Disorder (ADHD) using a standardized and validated semi-structured direct interview (TICS Inventory) (Darrow et al., 2015). TS case samples were also obtained through web-based recruitment of individuals with a prior clinical diagnosis of TS who subsequently completed an online questionnaire that has been validated against the gold-standard TS structured diagnostic interview with nearly 100% concordance for all inclusion/exclusion criteria as well as high correct classification rates for DSM-IV diagnoses of OCD and ADHD (Darrow et al., 2015). Individuals for web-based screening were solicited through the Tourette Association of America mailing list as well as from 4 TS specialty clinics in the United States. Individuals with a history of intellectual disability, seizure disorder, or a known tic disorder unrelated to TS were excluded.
Additional cases (n=628) and ancestry-matched controls (n=544) were collected by the Gilles de la Tourette Syndrome Genome-wide Association Study Replication Initiative (GGRI) through 9 TS specialty clinics in Austria, Canada, France, Germany, Greece, Hungary, Italy and the Netherlands by expert clinicians using Tourette Syndrome Study Group criteria for Definite TS (DSM-IV TS diagnosis plus tics observed by a trained clinician) as well as DSM-IV diagnostic criteria for OCD and ADHD as described previously (Paschou et al., 2014). We did not conduct formal standardized assessments for other neurodevelopmental disorders (NDDs) such as Intellectual Disability/Developmental Delay (ID/DD), Autism Spectrum Disorder (ASD), or schizophrenia/childhood psychosis; however, participants and their parents were asked about the presence of established or suspected diagnoses.
74.3% of TS cases were male, consistent with the 3- to 4-fold higher prevalence of the disorder in males compared to females (Robertson et al., 2017). The median age of cases was 17 (IQR, 12–31).
Method Details
External Control Sets
Additional control subjects were taken from four external large-scale genetic studies consisting of individuals sampled from similar geographic locations, specifically selected because intensity data was available and generated on the same Illumina OmniExpress platform as the TS cases and controls collected as part of this study:
Cardiff Controls (CC) (Green et al., 2010): UK Blood donors were recruited in Cardiff at the time of blood donation at centres in Wales and England. Although not explicitly screened for psychiatric disorders, these controls are likely to have low rates of severe neuropsychiatric illness, as blood donors in the UK are only eligible to donate if they are not taking any medications.
Consortium for Neuropsychiatric Phenomics (CNP) (Bilder et al., 2009): A collection of neuropsychiatric samples composed of patients with ADHD, bipolar disorder (BD), schizophrenia (SCZ), and psychologically normal controls, collected throughout North America as part of a large NIH Roadmap interdisciplinary research consortia centered at the University of California, Los Angeles (UCLA).
Genomic Psychiatry Cohort (GPC) (Pato et al., 2013): A large, longitudinal, population resource composed of clinically ascertained patients affected with BD, SCZ, their unaffected family members, and a large set of control samples with no family history of either disorder. Samples were collected at various sites throughout North America in a National Institute of Mental Health-sponsored study lead by the University of Southern California.
Wellcome Trust Case Control Consortium 2 (WTCCC2) (Power and Elliott, 2006): A subset of control samples from the National Blood Donors Cohort.
Genotyping
Samples selected for this study were all genotyped on the Illumina OmniExpress Exome v1.1 (Illumina, San Diego, CA, USA) in three separate batches (TS1–3), while samples from the CC, CNP, GPC, and WTCCC2 sets were genotyped on the Illumina OmniExpress 12v1.0 (Illumina, San Diego, CA, USA). A summary of the datasets, genotyping centers, and arrays used is provided in Table S1A. The OmniExpress Exome and OmniExpress arrays are identical except for the presence of exome-focused content on the former and additional intensity-only markers on the latter. We have observed that exome-specific assays in general exhibit a much higher variance overall in their Log-R Ratios (LRR) values. Therefore, in order to avoid detection biases due to this differential variance and to unequal probe density, only the SNP assays with common identifiers for all array versions across all datasets were used for quality control (QC) and CNV detection, a total of 689,077 markers.
To ensure the generation of the most reliable SNP calls, intensity measures, and B-allele frequencies (BAF), a custom cluster file was generated for each dataset separately and for each genotype batch when such information was available. Since the performance of Illumina’s proprietary normalization and cluster generation process improves with the number of samples, we processed all of the raw intensity data available, regardless of clinical phenotype. An initial round of QC was carried out using Beeline v1.0 (Illumina, San Diego, CA, USA) to determine baseline call rates and LRR standard deviation (LRR_SD) for each sample using the canonical cluster file (*.egt) provided by the manufacturer for each array version. Any sample with a call rate < 0.98 or an LRR_SD > 0.30 was deemed a failed assay and removed (Pre-cluster QC, Table S1B). SNP clustering and genotype calling was then performed with only the passing samples for each dataset individually with GenomeStudio v2.0 (Illumina, San Diego, CA, USA)
Genotype Sample QC
We performed an initial round of QC based on SNP genotype data. All samples at this stage had a minimal call rate >0.98. Samples with with discordant sex status were excluded. For samples run in duplicate, we retained the assay with the higher call rate. We filtered autosomal SNPs for missingness, minor-allele frequency, and deviation from Hardy-Weinberg equilibrium before pruning SNPs for LD using the following options in PLINK v1.9 (Chang et al., 2015):
A total of 96,350 SNPs remained for Identity-By-State relatedness testing. For all pairs of subjects (and duplicate/repeated samples) with PI_HAT > 0.185, we removed the sample with the lower call rate, with the exception that if a control individual was related to a subject with a neuropsychiatric phenotype, it was explicitly removed. Additionally, we excluded 44 subjects that were identical (PI_HAT > 0.99) between the CNP and GPC cohorts.
Ancestry Estimation
Following the exclusion of all clinical non-TS samples from external studies, genotype data for the remaining samples was combined with data from publicly available continental HapMap samples of CEU, YRI, and CHB/JPT ancestry genotyped on the OmniExpress array (Illumina, San Diego, CA, USA). Available European (EU) samples from the 1000 Genomes Project (http://www.internationalgenome.org/) genotyped on the Omni platform were also included to establish an appropriate calibration threshold for EU ancestry designation. We thinned our dataset randomly to 19,024 LD-independent markers for ancestry inference using fastStructure (Raj et al., 2014) with k=3. Samples were excluded if they contained > 0.0985 non-EU ancestry (Figure S1). A final round of ancestry exclusion was performed by removing all samples outside of the median +/− 3SD on the first three ancestry principal components (PCs).
CNV Calling
We created GC wave-adjusted LRR intensity files for all samples using PennCNV’s genomic_wave.pl script, and employed two widely-used HMM-based CNV calling algorithms, PennCNV v2011-05-03 (Wang et al., 2007), and QuantiSNP v2.0 (Colella et al., 2007) to initially detect structural variants in our dataset. For PennCNV, we generated a custom population B-allele frequency file for each dataset separately before calling CNVs using emission probabilities defined in the file hhall.hmm. QuantiSNP calls were generated from the GC-adjusted intensity files using the configuration file levels-hd.dat. A Perl script was used to merge concordant calls generated by both algorithms. CNVs were merged by taking the intersection of overlapping calls of the same CNV type (deletion or duplication). Additionally, adjacent CNV calls were merged if they were spanned by a CNV called by the other HMM algorithm. As HMMs have been shown to artificially break up large CNVs, we also merged CNV segments in the final concordant callset if they were of the same copy number and the number of intervening markers between them was less than 20% of the total of both segments combined using the PennCNV’s clean_cnv.pl script. We repeated this joining process iteratively until no more merging of segments occurred. Scripts and utility files used to generate CNV calls are available on bitbucket (http://bitbucket.org/ucla_coppolalab/tscnv).
Intensity Sample QC
The PennCNV calling algorithm generates a number of array intensity-based metrics with regard to CNV assay quality. Intensity-based QC was conducted based on the distribution of all available assays and subsequently combined with the results from the SNP-based QC. To remove samples with data unsuited for CNV detection, we used empirically defined thresholds across several different metrics:
Waviness factor (WF) - measures the waviness in intensity values, a known artifact caused by improper DNA concentration that can lead to spurious calls.
Log-R ratio standard deviation (LRR_SD) - a measure of the overall variance in intensity.
B-allele frequency drift (BAF_DRIFT) - summary of the deviation of BAF from expected values.
Thresholds for WF and LRR_SD were determined separately for each dataset by both manual examination of QC metrics and/or taking the mean +3x SD to determine outlying samples. Following intensity-based QC, all samples had an LRR-SD of <0.24, absolute value of WF < 0.04, and an BAF_DRIFT < 0.001, well within established limits required for reliable CNV detection.
CNV-load Sample QC
Although SNP and intensity-based QC removed most failed assays, we performed a final round of sample QC, removing eight additional samples with excessively high CNV load based on the total number of CNV calls (>45) or total CNV length (>10Mb). These thresholds were determined empirically by visual inspection of distributions across all datasets combined. Our final dataset after QC consisted 6,527 samples: 2,434 TS cases and 4,093 controls.
Data Handling and CNV Visualization
To facilitate further data processing and visualization of CNV events, we generated an HDF5 database consisting of sample metadata, CNV calls, probe information, LRR intensity and BAF values for all assays. Normalized intensity values were also generated by converting the GC-corrected, median-centered LRR measures into Z-scores within each sample and inserted into the database. Visualization of cluster plots (Figures S2A, S2B, S3C); median Z-score outlier detection (MeZOD) CNV calls (Vacic et al., 2011) (Figure 3B); and probe-level CNV plots (Figures S5 and S6) were generated from Z-scores of intensity data using in Matplotlib. Python code to create the HDF5 database and perform associated plotting functions is available on bitbucket (http://bitbucket.org/ucla_coppolalab/tscnv).
Sensitivity Assessment
We augmented a previously described method (Vacic et al., 2011) to investigate whether any difference in sensitivity to detect CNVs existed between cases and controls within the context of our study. Both HMM-based CNV callers we employed for genome-wide detection are univariate methods completely agnostic of intensity information across multiple samples and do not use known population frequency prior probabilities in their calling algorithms. Therefore, common CNVs act as an ideal proxy to evaluate the effectiveness to detect rare events accurately; they are detected in the same manner but are present at much higher frequencies, enabling an accurate estimation of the overall sensitivity of detection for rare events genome-wide.
We used the UCSC Genome Browser (https://genome.ucsc.edu) liftOver tool to translate a list of common HapMap3 CNVs to the hg19 reference. To match the thresholds used for our association tests in this study, we filtered the list of common CNVs to those that were >30 kbp in length. We reduced the number of markers required slightly to a minimum of 9 to ensure that an adequate number of events could be assessed. For each common CNV meeting these criteria, we examined the distribution of median-summarized normalized intensity measures within the CNV region across all study samples and retained only those loci that displayed no evidence of clustering intro different copy-number states. A total of 11 common CNV loci were retained for sensitivity analysis (Figure S2).
We generated locus-specific genotyping calls in the following manner. First, we extracted the LRR intensity Z-scores for all probes in the region across all samples. The Z-scores for all probes spanning the CNV locus were then subjected to second round of normalization across all samples. A Gaussian mixture model (GMM) was fit to this distribution of Z-scores using the SciKit-learn Python package. The optimum number of clusters was automatically determined by minimization of the Bayesian Information Criterion (BIC) and corrected, when necessary, by manual adjustment. Individuals were assigned to a cluster only if the posterior probability of assignment exceeded 0.95. Python code to perform GMM-based genotyping at specific loci is available on bitbucket (http://bitbucket.org/ucla_coppolalab/tscnv).
Copy number state was inferred by examining the original LRR intensity values for samples within each cluster. We inspected for allele frequency differences between controls and cases for all clusters and found no significant difference (Fisher Exact Test, Table S2). We collapsed the clusters at each locus into CNVs of the same type (deletion or duplication). As this locus-specific genotyping method is more sensitive than HMM segmentation methods, we used the proportion of concordant of HMM-based calls as a proxy for genome-wide detection sensitivity.
We found no significant difference in sensitivity to detect common CNVs between phenotypic groups at any of the 11 loci tested, either independently, or in concert (Fisher’s exact test, Table S3A and S3B). Furthermore, the mean sensitivity for each sample was calculated and collectively assessed for any systematic difference between phenotypic groups. Considering duplications, deletions, or both in concert, we observed no significant difference in the sensitivity of segmentation calls between case and control groups (Welch’s t-test, Table S3C).
Call Filtering and Delineation of Rare CNVs
Calls were removed from the dataset if they spanned less than 10 markers, were less than 30kb in length, or overlapped by more than 0.5 of their total length with regions known to generate artifacts in SNP-based detection of CNVs. This included immunoglobulin domain regions, segmental duplications, and regions that have previously demonstrated associations specific to Epstein-Barr virus immortalized cell lines (Shirley et al., 2012). In addition, we removed calls that spanned telomeric (defined as 100kb from the chromosome ends), centromeric regions, and gaps in the reference genome. As described elsewhere (CNV and SCZ Working Groups of the PGC, 2017), we assigned all CNV calls a specific frequency count using PLINK v.1.07 (Purcell et al., 2007), with the option --cnv-freq-method2 0.5. Here, the frequency count of an individual CNV is determined as 1 + the total number of CNVs overlap by at least 50% of its total length (in bp), irrespective of CNV type. We then filtered our callset for CNVs with MAF < 1% (a frequency of 65 or lower across 6,527 samples). Furthermore, we removed calls if they shared more than a 50% reciprocal overlap in length to common CNVs regions derived from several large, publically available SNP-array datasets, compiled by the Database of Genomic Variation and Phenotype in Humans using Ensembl Resources (DECIPHER; https://decipher.sanger.ac.uk/index):
845 population samples from the Deciphering Developmental Disorders study.
450 population samples from the 42M genotyped study.
5919 population samples from the Affy6 study.
Conservatively, CNV regions were only considered common if present at a frequency of >10% within any individual dataset above. Exclusion intervals and code to perform regional filtering of CNVs is available on bitbucket (http://bitbucket.org/ucla_coppolalab/tscnv).
In-silico Validation
For each putative rare CNV, we generated two different metrics based on LRR intensity and BAF banding. To qualifying CNVs based on intensity, we adopted a scoring methodology similar to the MeZOD method described elsewhere (Kirov et al., 2012), with modification. We observed that standardized intensity measures typically range from < −20 for homozygous deletions, [−6,−2.3] for heterozygous deletions, and > 1.3 for duplications. Because of the disproportionately large effect on intensity measures caused by homozygous deletions events in Illumina data, performing a second round of normalization across all samples within each putative CNV will skew the overall distribution when these events are present. Therefore, we only performed a single round of normalization of LRR intensity measures, within each sample. Each CNV was scored by calculating the median of LRR intensity Z-scores (LRR-Z) for all probes within the region. To qualify CNVs based on BAF banding, we calculated the proportion of probes within the CNV region that showed evidence of a duplication event (BAF of [0.25–04] or [0.6–0.8]), and denoted this measure “BAF-D.”
Based on thresholds established through manual inspection of large CNVs, we flagged deletions that had a LRR-Z > −2 or BAF-D > 0.02, and duplications with a LRR-Z < 1 or BAF-D < 0.1. To avoid differential missingness caused by subtle differences between datasets, we did not impose any hard cutoff for CNV exclusion based on these metrics. These thresholds were applied to flag CNV calls with marginal scores for manual inspection for the exclusion of obviously misclassified events. Through this in-silico validation process we discovered multiple instances of large CNV calls likely due to individual mosaicism (Figure S3A and S3B), and removed these events from subsequent analysis.
Furthermore, for each rare CNV call, we used distribution of summarized intensity information across all individuals For each rare CNV region, we quantified proportion of samples whose LRR-Z metric fell outside of [−2.3, 1.3] and further inspected these regions manually. Putative rare CNV loci that showed substantial evidence for extensive polymorphism were subsequently scored for frequency using the GMM genotyping method described above (Figure S3C) and conservatively, only removed if shown to be variant in more than 10% of the samples across the entire dataset. Python code to generate in-silico metrics for CNV calls from intensity data stored in an HDF5 database is available on bitbucket (http://bitbucket.org/ucla_coppolalab/tscnv).
Annotation of Rare CNVs
Rare CNVs were annotated for gene content according to RefSeq provided by the hg19 assembly of the UCSC Genome Browser. We only considered a CNV as “genic” if it overlapped any exon of a known protein-coding transcript (as designated by the RefSeq transcript accession prefix “NM”). The “gene count” of a CNV represents the total number of non-redundant genes whose respective transcripts it overlaps. “Non-genic CNVs” represent all variants that are not genic according to the definition above.
In addition, all rare CNVs were assessed for clinical relevance in accordance with guidelines set forth by the American College of Medical Genetics (ACMG) (Kearney et al., 2011). This was accomplished through the use of the Scripps Genome Advisor (https://genomics.scripps.edu/ADVISER/); inspection of curated resources including ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/), DECIPHER (https://decipher.sanger.ac.uk/index), and Online Mendelian Inheritance in Man (https://www.omim.org/); and followed by confirmation through review of primary literature. Conservatively, we only considered a CNV as “pathogenic” if directly supported by more than one primary publication. To screen for additional relevant pathogenic variants, we included variants that overlapped compiled lists defined in the literature (Malhotra and Sebat, 2012; McGrath et al., 2014). All non-pathogenic variants were automatically annotated by SGA: those classed as Category 2–4 were assigned as a variant of “unknown clinical significance”, and those assigned to Category 5 were considered “benign”. Note that, as only rare CNVs were considered, the number of “benign” variants is small.
Rare genic CNVs were further annotated using pLI (probability of LoF Intolerance) scores (http://exac.broadinstitute.org/). A CNV was marked as “constrained” if it overlapped any exon of a gene with a pLI score of > 0.9, as per the approach by Ruderfer and colleagues (Ruderfer et al., 2016).
Global CNV Burden Analysis
We measured global CNV burden using three separate metrics: the number of rare CNVs (CNV count), the total length of all CNVs (CNV length) and the total number of genes intersected by CNVs (CNV gene count). To examine the effect of different covariates on different metrics of global CNV burden, we first fit a linear regression model for each burden metric, using the glm function in R:
In the above model, ancestry_PCs included the first ten PCs derived from SNP data and, as various assay intensity quality metrics are highly correlated, LRR_SD was used as a single measure of assay quality. Across the entire dataset, only PC2 and LRR_SD were associated (P<0.05) with global CNV burden as measured by CNV count. To assess for a global burden difference between TS cases and controls, we fit the following general logistic regression model:
Independent variables included burden metric, subject sex, and the first 4 ancestry PCs. Even though assay quality (LRR_SD) was associated exclusively with small CNV burden (<100kbp), it was included in all burden analysis, regardless of size. For all burden analyses, ORs, 95% CIs, and significance were calculated using the R function glm (family=”binomial”). ORs were calculated by taking the exponential of the logistic regression coefficient.
To examine whether evolutionarily constrained CNVs are enriched in TS patients, we assumed a Poisson distribution of such rare events, and conducted a Poisson regression using the R function glm (family=”poisson”) with the same covariates as in the logistic regression above, including adjustment of subject sex, the first four ancestry PCs, and LRR_SD:
We tested large (>1Mb), singleton CNVs that affect conserved genes (pLI>0.9), stratified by CNV type, and compared the relative risk of such events to all large, singleton genic CNVs.
In Figure 2, global burden was analyzed separately for genic CNVs and non-genic CNVs. Since we were interested in comparing the relative contribution to TS risk by different measures of burden and across various categories, sizes, and frequency classes, the ORs presented in Figure 2 and Figure S4 are calculated from standardized CNV burden metrics. For Figure 3 and elsewhere, we calculated ORs using the unstandardized value of actual CNV counts, as this is directly interpretable. R code to perform burden analysis is available on bitbucket (http://bitbucket.org/ucla_coppolalab/tscnv).
Locus-specific Tests of Association
The segmental test of association was performed by quantifying the frequencies of case and control CNV carriers at all unique CNV breakpoint locations; the unique set of CNV breakpoints defines all locations genome-wide where the frequency of CNVs can differ between cases and controls. For gene-based association tests, we restricted our analysis to genic CNVs (CNVs that intersect an exon of any protein-coding transcript, as defined above) and quantified the frequencies of cases and control CNVs across each gene. Locus-specific p-values for both tests of association were determined by 1,000,000 permutations of phenotype labels, and genome-wide corrected p-values were obtained using the max(T) permutation method (Westfall and Troendle, 2008) as implemented in PLINK v1.07, which controls for family-wise error rate by comparing the locus-specific test statistic to all test statistics genome-wide within each permutation. Association tests were conducted separately for deletions and duplications.
Sensitivity Analysis of Association Results
The segmental association test was repeated after carefully pair-matching each case with a control such that the global difference between each pair was minimized using GemTools (Lee et al., 2010) (Figure S7). For the matched segmental association analysis, because of the drastic reduction in sample size, a genome-wide corrected alpha < 0.05 was used as a cutoff to indicate significance.
Quantification and Statistical Analysis
All statistical analyses were performed using R v3.4.0 or PLINK v1.07. R code and example commands to perform all statistical analyses are available on bitbucket (http://bitbucket.org/ucla_coppolalab/tscnv). For sensitivity assessment (see “Sensitivity Assessment”), as also indicated in the main text and/or Tables S2 and S3, we used a Fisher Exact test to test for a difference between cases and controls in: the frequency of non-reference genotypes at common CNV loci (Table S2), the sensitivity of HMM-based CNV calls at these loci individually (Table S3A), and the sensitivity of HMM-based CNV across all loci (Table S3B). For Table S3C, we used Welch’s t-test to test if the average sensitivity per individual differed between cases and controls, assuming that the variance of this measure was not necessarily equal between groups. For common CNVs, we did not expect allele frequency or detection sensitivity to differ between cases and controls and therefore, significance was assessed using a two-sided test.
To allow for the inclusion of covariates, we used a logistic regression framework to evaluate the contribution of global CNV burden on TS risk. ORs above 1 indicate an increased risk for TS per unit of CNV burden, 95% CIs are provided for all estimates (see “Global Analysis of CNV Burden”). For Figure 2 and S4, global burden was measured by CNV count, CNV length, or CNV gene count as indicated, and by CNV count for Figure 3 and elsewhere. Burden measures were aggregated per individual, genome-wide, and restricted to specific CNV categories as described. To test whether evolutionarily constrained CNVs are enriched in TS patients, we assumed a Poisson distribution of such rare events, and compared the rates of constrained CNVs between TS cases and controls using Poisson regression.
For association testing, we performed 1×106 label-swapping permutations to determine both locus-specific and genome-wide p-values empirically using the max(T) method (Westfall and Troendle, 2008) as implemented in PLINK v1.07. This method is appropriate given all samples were processed through the same CNV calling pipeline on identical assays. For the segmental test, case and control frequencies were calculated at each unique CNV breakpoint. For the gene-based test, frequencies were based on the number of genic CNVs at each gene locus. Deletions and duplications were tested independently, using a 1-sided test as we expect an increased frequency of CNVs in cases compared to controls.
Data and Software Availability
Data
CNV calls for all samples are provided in Table S1. Intensity files for the TSAICG datasets are deposited in dbGaP at [PENDING].
Software
Utility files and custom code written in BASH, Perl, Python, and R used to conduct this analysis and generate figures from this manuscript are available on bitbucket (http://bitbucket.org/ucla_coppolalab/tscnv).
Supplementary Material
Document S1. Figures S1–S7, Tables S1–S3 and S5, Consortia Author Lists and Disclosures
Table S4, Related to Figures 1–4. Rare CNV calls from 2,434 TS cases and 4,093 controls
This table contains a list of all rare CNVs detected in this study. For each CNV, the subject’s sex, phenotype (TS), and presence of comorbid disorders (ADHD, OCD) is indicated. Genomic locations are provided in hg19 coordinates. Frequencies were assigned to CNVs as described in the STAR Methods. The number of genes spanned by a CNV (GENE_COUNT) and the associated genes (GENE_SYMBOLS) are determined by overlap with exons of protein-coding genes (see STAR Methods for details).
Acknowledgments
The authors thank the patients with Tourette Syndrome and their families, and all the volunteers who participated in this study. This study was supported by the US NIH U01 NS040024 to Drs. Pauls, Mathews, and Scharf and the TSAICG, ARRA Grant NS040024-09S1, K23 MH085057, and K02 NS085048 to Dr. Scharf, ARRA Grants NS040024-07S1 and NS016648 to Dr. Pauls, MH096767 to Dr. Mathews and NINDS Informatics Center for Neurogenetics and Neurogenomics grant P30 NS062691 to Drs. Coppola and Freimer, by grants from the Tourette Association of America to Drs. Paschou, Pauls, Mathews, and Scharf and from the German Research Society to Dr. Hebebrand.
Footnotes
AUTHOR CONTRIBUTIONS
All authors were involved in the conception and design of this study. A.H., P.P., C.A.M., J.M.S., and G.C. designed and oversaw the analyses. A.H., D.Y., L.K.D., J.H.S., F.T., V.R., I.Z., E.M.R., L.O., J.A.C., L.M.M., B.M.N., N.B.F., P.P., C.A.M., J.M.S., and G.C. conducted the analyses. Major contributions to writing and editing were made by A.H., C.A.M., J.M.S, and G.C. All authors assisted with critically revising the manuscript.
Additional Resources
TSAICG website: https://www.findtsgene.org
Bitbucket repository: http://bitbucket.org/ucla_coppolalab/tscnv
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abelson JF, Kwan KY, O’Roak BJ, Baek DY, Stillman AA, Morgan TM, Mathews CA, Pauls DL, Rasin MR, Gunel M, et al. Sequence variants in SLITRK1 are associated with Tourette’s syndrome. Science. 2005;310:317–320. doi: 10.1126/science.1116502. [DOI] [PubMed] [Google Scholar]
- Bilder RM, Sabb FW, Cannon TD, London ED, Jentsch JD, Parker DS, Poldrack RA, Evans C, Freimer NB. Phenomics: the systematic study of phenotypes on a genome-wide scale. Neuroscience. 2009;164:30–42. doi: 10.1016/j.neuroscience.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne HA, Hansen SN, Buxbaum JD, Gair SL, Nissen JB, Nikolajsen KH, Schendel DE, Reichenberg A, Parner ET, Grice DE. Familial clustering of tic disorders and obsessive-compulsive disorder. JAMA Psychiatry. 2015;72:359–366. doi: 10.1001/jamapsychiatry.2014.2656. [DOI] [PubMed] [Google Scholar]
- Browne HA, Modabbernia A, Buxbaum JD, Hansen SN, Schendel DE, Parner ET, Reichenberg A, Grice DE. Prenatal Maternal Smoking and Increased Risk for Tourette Syndrome and Chronic Tic Disorders. J Am Acad Child Adolesc Psychiatry. 2016;55:784–791. doi: 10.1016/j.jaac.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Burd L, Li Q, Kerbeshian J, Klug MG, Freeman RD. Tourette syndrome and comorbid pervasive developmental disorders. J Child Neurol. 2009;24:170–175. doi: 10.1177/0883073808322666. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CNV and Schizophrenia Working Groups of the Psychiatric Genomics Consortium, and Psychosis Endophenotypes International Consortium. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49:27–35. doi: 10.1038/ng.3725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colella S, Yau C, Taylor JM, Mirza G, Butler H, Clouston P, Bassett AS, Seller A, Holmes CC, Ragoussis J. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabell MP, Rosenfeld JA, Bader P, Escobar LF, El-Khechen D, Vallee SE, Dinulos MBP, Curry C, Fisher J, Tervo R, et al. Investigation of NRXN1 deletions: clinical and molecular characterization. Am J Med Genet A. 2013;161A:717–731. doi: 10.1002/ajmg.a.35780. [DOI] [PubMed] [Google Scholar]
- Darrow SM, Illmann C, Gauvin C, Osiecki L, Egan CA, Greenberg E, Eckfield M, Hirschtritt ME, Pauls DL, Batterson JR, et al. Web-based phenotyping for Tourette Syndrome: Reliability of common co-morbid diagnoses. Psychiatry Res. 2015;228:816–825. doi: 10.1016/j.psychres.2015.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LK, Yu D, Keenan CL, Gamazon ER, Konkashbaev AI, Derks EM, Neale BM, Yang J, Lee SH, Evans P, et al. Partitioning the heritability of Tourette syndrome and obsessive compulsive disorder reveals differences in genetic architecture. PLoS Genet. 2013;9:e1003864. doi: 10.1371/journal.pgen.1003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper A, Stephenson MC, Jackson GM, Pépés S, Morgan PS, Morris PG, Jackson SR. Increased GABA contributes to enhanced control over motor excitability in Tourette syndrome. Curr Biol. 2014;24:2343–2347. doi: 10.1016/j.cub.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez TV, Sanders SJ, Yurkiewicz IR, Ercan-Sencicek AG, Kim YS, Fishman DO, Raubeson MJ, Song Y, Yasuno K, Ho WSC, et al. Rare copy number variants in tourette syndrome disrupt genes in histaminergic pathways and overlap with autism. Biol Psychiatry. 2012;71:392–402. doi: 10.1016/j.biopsych.2011.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccillo MV, Földy C, Gökce Ö, Rothwell PE, Sun GL, Malenka RC, Südhof TC. Single-Cell mRNA Profiling Reveals Cell-Type-Specific Expression of Neurexin Isoforms. Neuron. 2015;87:326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert DL, Bansal AS, Sethuraman G, Sallee FR, Zhang J, Lipps T, Wassermann EM. Association of cortical disinhibition with tic, ADHD, and OCD severity in Tourette syndrome. Mov Disord. 2004;19:416–425. doi: 10.1002/mds.20044. [DOI] [PubMed] [Google Scholar]
- Green EK, Grozeva D, Jones I, Jones L, Kirov G, Caesar S, Gordon-Smith K, Fraser C, Forty L, Russell E, et al. The bipolar disorder risk allele at CACNA1C also confers risk of recurrent major depression and of schizophrenia. Mol Psychiatry. 2010;15:1016–1022. doi: 10.1038/mp.2009.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene DJ, Williams AC, Iii, Koller JM, Schlaggar BL, Black KJ. Brain structure in pediatric Tourette syndrome. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschtritt ME, Lee PC, Pauls DL, Dion Y, Grados MA, Illmann C, King RA, Sandor P, McMahon WM, Lyon GJ, et al. Lifetime prevalence, age of risk, and genetic relationships of comorbid psychiatric disorders in Tourette syndrome. JAMA Psychiatry. 2015;72:325–333. doi: 10.1001/jamapsychiatry.2014.2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Liao J, Sathanoori M, Kochmar S, Sebastian J, Yatsenko SA, Surti U. CNTN6 copy number variations in 14 patients: a possible candidate gene for neurodevelopmental and neuropsychiatric disorders. J Neurodev Disord. 2015;7:26. doi: 10.1186/s11689-015-9122-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Yu Y, Shimoda Y, Watanabe K, Liu Y. Loss of neural recognition molecule NB-3 delays the normal projection and terminal branching of developing corticospinal tract axons in the mouse. J Comp Neurol. 2012;520:1227–1245. doi: 10.1002/cne.22772. [DOI] [PubMed] [Google Scholar]
- Jahanshahi M, Obeso I, Rothwell JC, Obeso JA. A fronto-striato-subthalamic-pallidal network for goal-directed and habitual inhibition. Nat Rev Neurosci. 2015;16:719–732. doi: 10.1038/nrn4038. [DOI] [PubMed] [Google Scholar]
- Kashevarova AA, Nazarenko LP, Schultz-Pedersen S, Skryabin NA, Salyukova OA, Chechetkina NN, Tolmacheva EN, Rudko AA, Magini P, Graziano C, et al. Single gene microdeletions and microduplication of 3p26.3 in three unrelated families: CNTN6 as a new candidate gene for intellectual disability. Mol Cytogenet. 2014;7:97. doi: 10.1186/s13039-014-0097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, Moran J, Chambert K, Toncheva D, Georgieva L, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AB, Luca D, Klei L, Devlin B, Roeder K. Discovering genetic ancestry using spectral graph theory. Genet Epidemiol. 2010;34:51–59. doi: 10.1002/gepi.20434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivonen S, Voutilainen A, Chudal R, Suominen A, Gissler M, Sourander A. Obstetric and Neonatal Adversities, Parity, and Tourette Syndrome: A Nationwide Registry. J Pediatr. 2016;171:213–219. doi: 10.1016/j.jpeds.2015.10.063. [DOI] [PubMed] [Google Scholar]
- Malhotra D, Sebat J. CNVs: harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148:1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh R, Maia TV, Peterson BS. Functional disturbances within frontostriatal circuits across multiple childhood psychopathologies. Am J Psychiatry. 2009;166:664–674. doi: 10.1176/appi.ajp.2009.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mataix-Cols D, Isomura K, Pérez-Vigil A, Chang Z, Rück C, Larsson KJ, Leckman JF, Serlachius E, Larsson H, Lichtenstein P. Familial Risks of Tourette Syndrome and Chronic Tic Disorders. A Population-Based Cohort Study. JAMA Psychiatry. 2015;72:787–793. doi: 10.1001/jamapsychiatry.2015.0627. [DOI] [PubMed] [Google Scholar]
- McGrath LM, Yu D, Marshall C, Davis LK, Thiruvahindrapuram B, Li B, Cappi C, Gerber G, Wolf A, Schroeder FA, et al. Copy number variation in obsessive-compulsive disorder and tourette syndrome: a cross-disorder study. J Am Acad Child Adolesc Psychiatry. 2014;53:910–919. doi: 10.1016/j.jaac.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercati O, Huguet G, Danckaert A, André-Leroux G, Maruani A, Bellinzoni M, Rolland T, Gouder L, Mathieu A, Buratti J, et al. CNTN6 mutations are risk factors for abnormal auditory sensory perception in autism spectrum disorders. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Bochukova EG, Kremeyer B, Campbell DD, Muller H, Valencia-Duarte AV, Cardona J, Rivas IC, Mesa SC, Cuartas M, et al. CNV analysis in Tourette syndrome implicates large genomic rearrangements in COL8A1 and NRXN1. PLoS One. 2013;8:e59061. doi: 10.1371/journal.pone.0059061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa J, Kaneko H, Masuda T, Nagata S, Hosoya H, Watanabe K. Novel neural adhesion molecules in the Contactin/F3 subgroup of the immunoglobulin superfamily: isolation and characterization of cDNAs from rat brain. Neurosci Lett. 1996;218:173–176. doi: 10.1016/s0304-3940(96)13156-6. [DOI] [PubMed] [Google Scholar]
- Oguro-Ando A, Zuko A, Kleijer KTE, Burbach JPH. A current view on contactin-4, -5, and -6: Implications in neurodevelopmental disorders. Mol Cell Neurosci. 2017 doi: 10.1016/j.mcn.2016.12.004. [DOI] [PubMed] [Google Scholar]
- Pak C, Danko T, Zhang Y, Aoto J, Anderson G, Maxeiner S, Yi F, Wernig M, Südhof TC. Human Neuropsychiatric Disease Modeling using Conditional Deletion Reveals Synaptic Transmission Defects Caused by Heterozygous Mutations in NRXN1. Cell Stem Cell. 2015;17:316–328. doi: 10.1016/j.stem.2015.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschou P, Yu D, Gerber G, Evans P, Tsetsos F, Davis LK, Karagiannidis I, Chaponis J, Gamazon E, Mueller-Vahl K, et al. Genetic association signal near NTN4 in Tourette syndrome. Ann Neurol. 2014;76:310–315. doi: 10.1002/ana.24215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pato MT, Sobell JL, Medeiros H, Abbott C, Sklar BM, Buckley PF, Bromet EJ, Escamilla MA, Fanous AH, Lehrer DS, et al. The genomic psychiatry cohort: partners in discovery. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:306–312. doi: 10.1002/ajmg.b.32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power C, Elliott J. Cohort profile: 1958 British birth cohort (National Child Development Study) Int J Epidemiol. 2006;35:34–41. doi: 10.1093/ije/dyi183. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Stephens M, Pritchard JK. fastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics. 2014;197:573–589. doi: 10.1534/genetics.114.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson MM, Eapen V, Singer HS, Martino D, Scharf JM, Paschou P, Roessner V, Woods DW, Hariz M, Mathews CA, et al. Gilles de la Tourette syndrome. Nat Rev Dis Primers. 2017;3:16097. doi: 10.1038/nrdp.2016.97. [DOI] [PubMed] [Google Scholar]
- Ruderfer DM, Hamamsy T, Lek M, Karczewski KJ, Kavanagh D, Samocha KE, MacArthur DG, Fromer M, et al. Exome Aggregation Consortium Daly MJ. Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nat Genet. 2016 doi: 10.1038/ng.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf JM, Yu D, Mathews CA, Neale BM, Stewart SE, Fagerness JA, Evans P, Gamazon E, Edlund CK, Service SK, et al. Genome-wide association study of Tourette’s syndrome. Mol Psychiatry. 2013;18:721–728. doi: 10.1038/mp.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf JM, Miller LL, Gauvin CA, Alabiso J, Mathews CA, Ben-Shlomo Y. Population prevalence of Tourette syndrome: a systematic review and meta-analysis. Mov Disord. 2015;30:221–228. doi: 10.1002/mds.26089. [DOI] [PubMed] [Google Scholar]
- Schreiner D, Nguyen TM, Russo G, Heber S, Patrignani A, Ahrné E, Scheiffele P. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron. 2014;84:386–398. doi: 10.1016/j.neuron.2014.09.011. [DOI] [PubMed] [Google Scholar]
- Shirley MD, Baugher JD, Stevens EL, Tang Z, Gerry N, Beiswanger CM, Berlin DS, Pevsner J. Chromosomal variation in lymphoblastoid cell lines. Hum Mutat. 2012;33:1075–1086. doi: 10.1002/humu.22062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Stogsdill JA, Pulimood NS, Dingsdale H, Kim YH, Pilaz LJ, Kim IH, Manhaes AC, Rodrigues WS, Jr, Pamukcu A, et al. Astrocytes Assemble Thalamocortical Synapses by Bridging NRX1α and NL1 via Hevin. Cell. 2016;164:183–196. doi: 10.1016/j.cell.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram SK, Huq AM, Wilson BJ, Chugani HT. Tourette syndrome is associated with recurrent exonic copy number variants. Neurology. 2010;74:1583–1590. doi: 10.1212/WNL.0b013e3181e0f147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Südhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- Vacic V, McCarthy S, Malhotra D, Murray F, Chou HH, Peoples A, Makarov V, Yoon S, Bhandari A, Corominas R, et al. Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature. 2011;471:499–503. doi: 10.1038/nature09884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJMH, Mathews CA, Joosse M, Eussen BHJ, Heutink P, Oostra BA Tourette Syndrome Association International Consortium for Genetics. CNTNAP2 is disrupted in a family with Gilles de la Tourette syndrome and obsessive compulsive disorder. Genomics. 2003;82:1–9. doi: 10.1016/s0888-7543(03)00097-1. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SFA, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall PH, Troendle JF. Multiple testing with minimal assumptions. Biom J. 2008;50:745–755. doi: 10.1002/bimj.200710456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Fernandez TV, Yu D, King RA, Dietrich A, Xing J, Sanders SJ, Mandell JD, Huang AY, Richer P, et al. De Novo Coding Variants Are Strongly Associated with Tourette Disorder. Neuron. 2017;94:486–499. e9. doi: 10.1016/j.neuron.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit J, Ghosh A. Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci. 2016;17:22–35. doi: 10.1038/nrn.2015.3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Figures S1–S7, Tables S1–S3 and S5, Consortia Author Lists and Disclosures
Table S4, Related to Figures 1–4. Rare CNV calls from 2,434 TS cases and 4,093 controls
This table contains a list of all rare CNVs detected in this study. For each CNV, the subject’s sex, phenotype (TS), and presence of comorbid disorders (ADHD, OCD) is indicated. Genomic locations are provided in hg19 coordinates. Frequencies were assigned to CNVs as described in the STAR Methods. The number of genes spanned by a CNV (GENE_COUNT) and the associated genes (GENE_SYMBOLS) are determined by overlap with exons of protein-coding genes (see STAR Methods for details).