Abstract
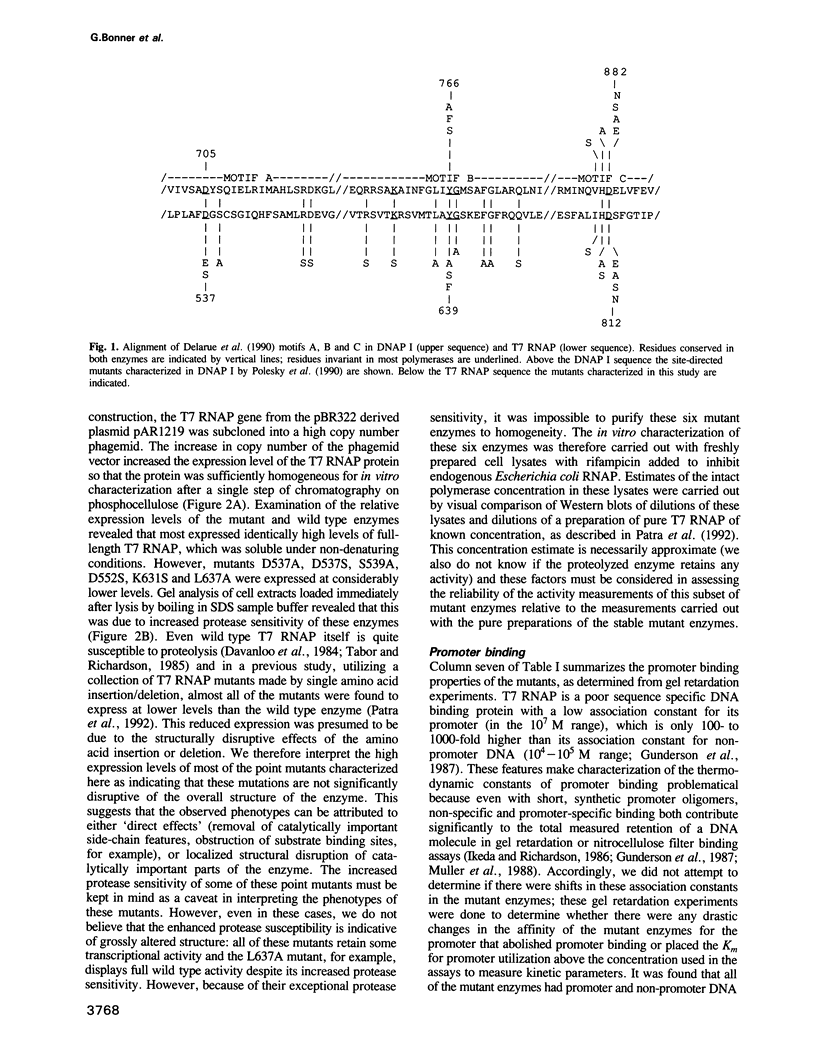
In order to test the proposal that most nucleotide polymerases share a common active site structure and folding topology, we have generated 22 mutations of residues within motifs A, B and C of T7 RNA polymerase (RNAP). Characterization of these T7 RNAP mutants showed the following: (i) most of the mutations resulted in moderate to drastic reductions in T7 RNAP transcriptional activity supporting the idea that motifs A, B and C identify part of the polymerase active site; (ii) the degree of conservation of an amino acid within these motifs correlated with the degree to which mutation of that amino acid reduced transcriptional activity, supporting the predictive ability of this alignment in identifying the most functionally critical residues; (iii) a comparison of DNAP I and T7 RNAP mutants revealed similarities (as well as differences) between corresponding mutant phenotypes; (iv) the Klenow fragment structure is shown to provide a reasonable basis for interpretation of the differential effects of mutating different amino acids within motifs A, B and C in T7 RNAP. These observations support the proposal that these polymerase active sites have similar three-dimensional structures.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Carroll S. S., Cowart M., Benkovic S. J. A mutant of DNA polymerase I (Klenow fragment) with reduced fidelity. Biochemistry. 1991 Jan 22;30(3):804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- Catalano C. E., Allen D. J., Benkovic S. J. Interaction of Escherichia coli DNA polymerase I with azidoDNA and fluorescent DNA probes: identification of protein-DNA contacts. Biochemistry. 1990 Apr 17;29(15):3612–3621. doi: 10.1021/bi00467a004. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Golomb M., Chamberlin M. Characterization of T7-specific ribonucleic acid polymerase. IV. Resolution of the major in vitro transcripts by gel electrophoresis. J Biol Chem. 1974 May 10;249(9):2858–2863. [PubMed] [Google Scholar]
- Gunderson S. I., Chapman K. A., Burgess R. R. Interactions of T7 RNA polymerase with T7 late promoters measured by footprinting with methidiumpropyl-EDTA-iron(II). Biochemistry. 1987 Mar 24;26(6):1539–1546. doi: 10.1021/bi00380a007. [DOI] [PubMed] [Google Scholar]
- Ikeda R. A., Richardson C. C. Enzymatic properties of a proteolytically nicked RNA polymerase of bacteriophage T7. J Biol Chem. 1987 Mar 15;262(8):3790–3799. [PubMed] [Google Scholar]
- Ikeda R. A., Richardson C. C. Interactions of the RNA polymerase of bacteriophage T7 with its promoter during binding and initiation of transcription. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3614–3618. doi: 10.1073/pnas.83.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C. T., Coleman J. E. Kinetic analysis of T7 RNA polymerase-promoter interactions with small synthetic promoters. Biochemistry. 1987 May 19;26(10):2690–2696. doi: 10.1021/bi00384a006. [DOI] [PubMed] [Google Scholar]
- Martin C. T., Muller D. K., Coleman J. E. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988 May 31;27(11):3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- McClure W. R., Chow Y. The kinetics and processivity of nucleic acid polymerases. Methods Enzymol. 1980;64:277–297. doi: 10.1016/s0076-6879(80)64013-0. [DOI] [PubMed] [Google Scholar]
- Mullen G. P., Shenbagamurthi P., Mildvan A. S. Substrate and DNA binding to a 50-residue peptide fragment of DNA polymerase I. Comparison with the enzyme. J Biol Chem. 1989 Nov 25;264(33):19637–19647. [PubMed] [Google Scholar]
- Muller D. K., Martin C. T., Coleman J. E. Processivity of proteolytically modified forms of T7 RNA polymerase. Biochemistry. 1988 Jul 26;27(15):5763–5771. doi: 10.1021/bi00415a055. [DOI] [PubMed] [Google Scholar]
- Polesky A. H., Steitz T. A., Grindley N. D., Joyce C. M. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem. 1990 Aug 25;265(24):14579–14591. [PubMed] [Google Scholar]
- Rush J., Konigsberg W. H. Photoaffinity labeling of the Klenow fragment with 8-azido-dATP. J Biol Chem. 1990 Mar 25;265(9):4821–4827. [PubMed] [Google Scholar]
- Shi Y. B., Gamper H., Hearst J. E. Interaction of T7 RNA polymerase with DNA in an elongation complex arrested at a specific psoralen adduct site. J Biol Chem. 1988 Jan 5;263(1):527–534. [PubMed] [Google Scholar]
- Sousa R., Rose J. P., Chung Y. J., Lafer E. M., Wang B. C. Single crystals of bacteriophage T7 RNA polymerase. Proteins. 1989;5(4):266–270. doi: 10.1002/prot.340050403. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]






