Abstract
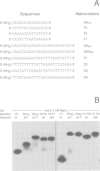
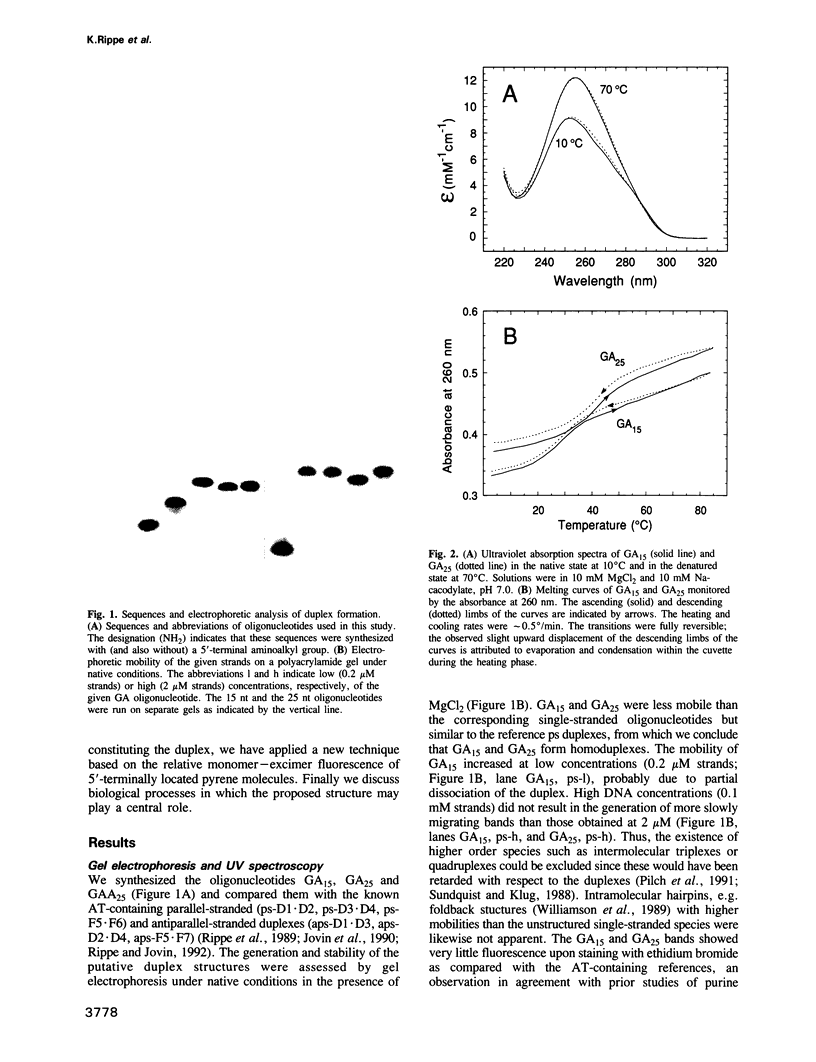
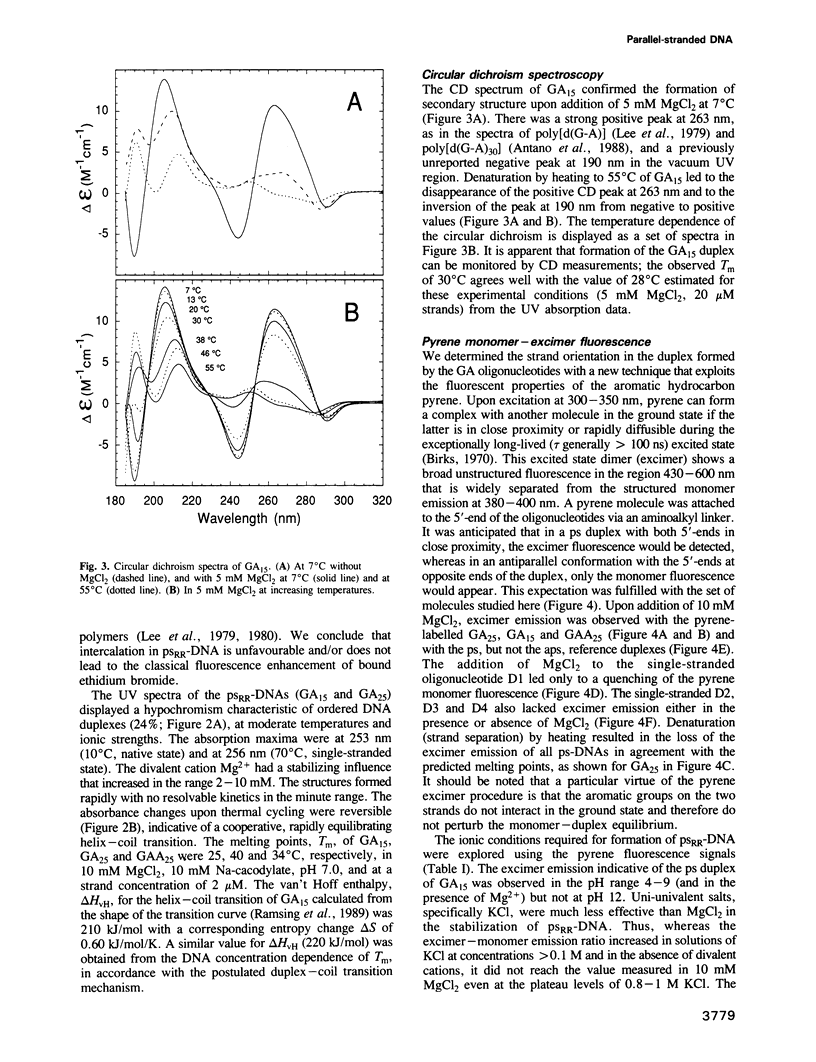
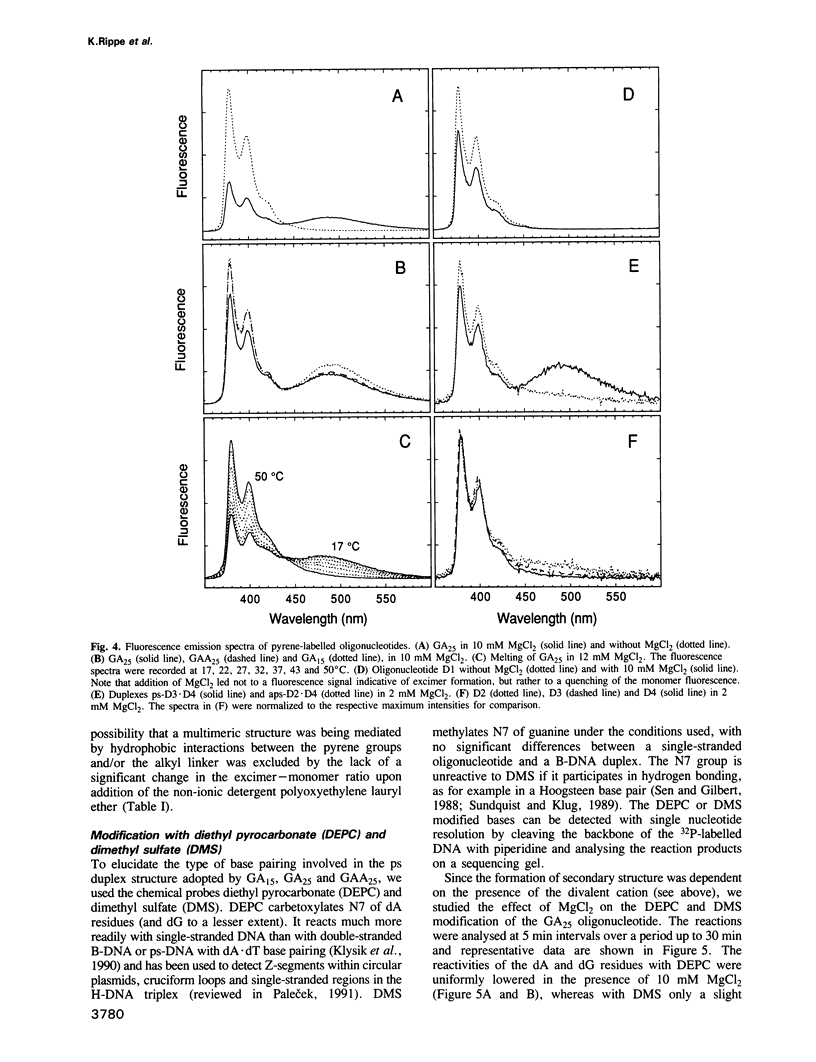
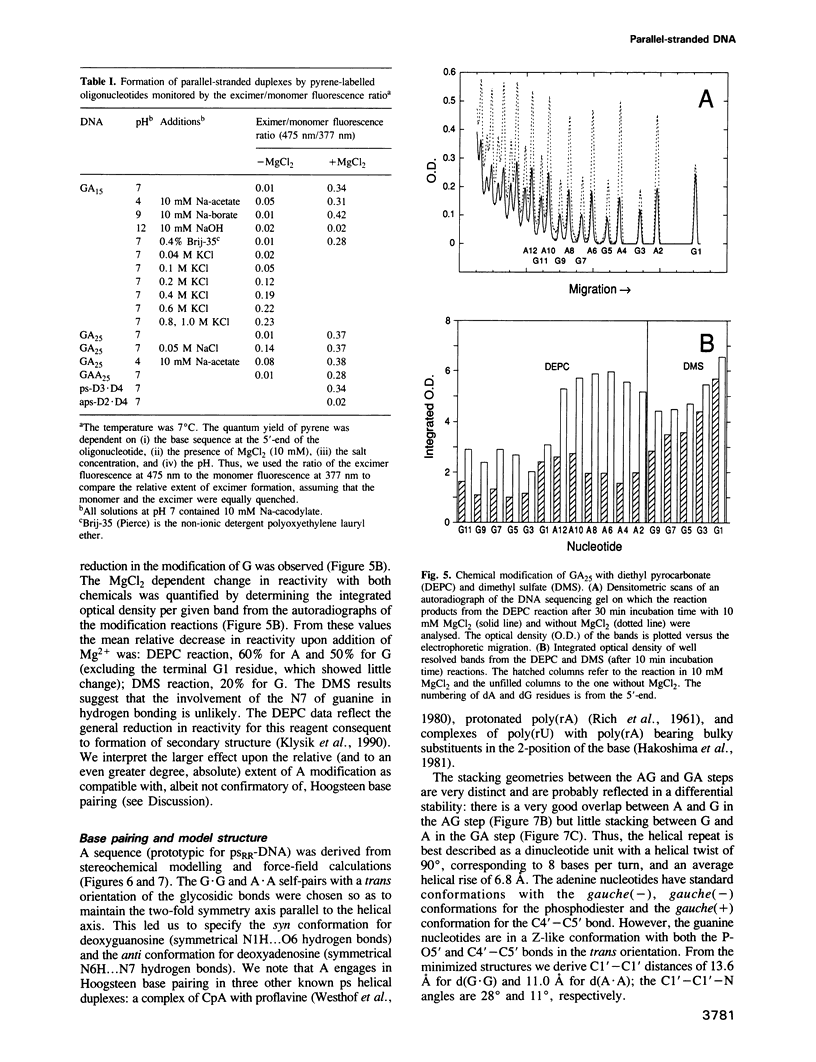
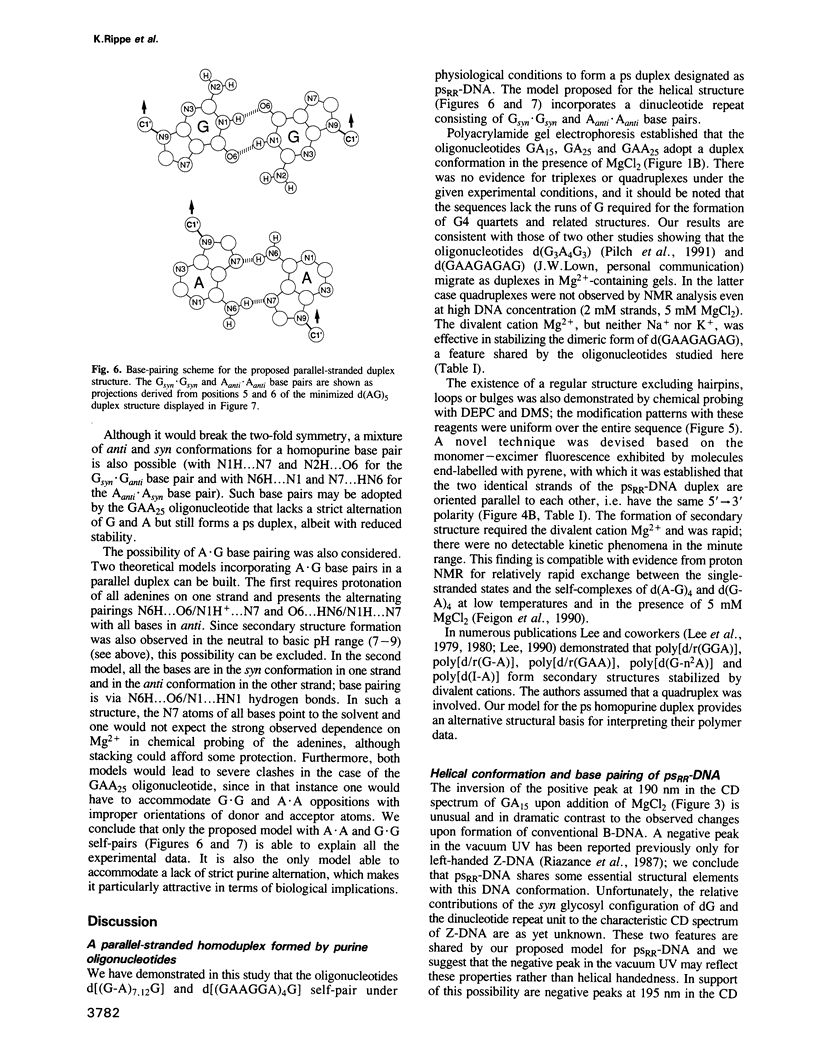
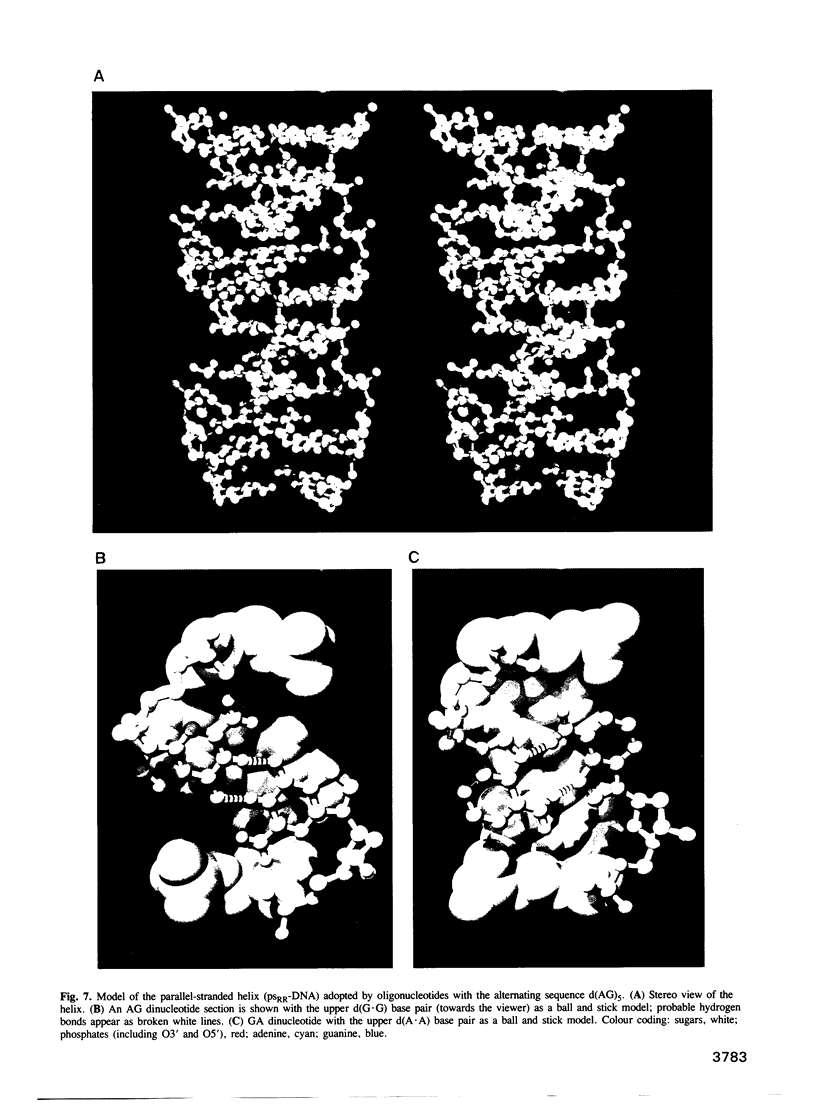
The oligonucleotides d[(G-A)7G] and d[(G-A)12G] self-associate under physiological conditions (10 mM MgCl2, neutral pH) into a stable double-helical structure (psRR-DNA) in which the two polypurine strands are in a parallel orientation in contrast to the antiparallel disposition of conventional B-DNA. We have characterized psRR-DNA by gel electrophoresis, UV absorption, vacuum UV circular dichroism, monomer-excimer fluorescence of oligonucleotides end-labelled with pyrene, and chemical probing with diethyl pyrocarbonate and dimethyl sulfate. The duplex is stable at pH 4-9, suggesting that the structure is compatible with, but does not require, protonation of the A residues. The data support a model derived from force-field analysis in which the parallel-stranded d(G-A)n helix is right-handed and constituted of alternating, symmetrical Gsyn.Gsyn and Aanti.Aanti base pairs with N1H...O6 and N6H...N7 hydrogen bonds, respectively. This dinucleotide structure may be the source of a negative peak observed at 190 nm in the vacuum UV CD spectrum, a feature previously reported only for left-handed Z-DNA. The related sequence d[(GAAGGA)4G] also forms a parallel-stranded duplex but one that is less stable and probably involves a slightly different secondary structure. We discuss the potential intervention of psRR-DNA in recombination, gene expression and the stabilization of genomic structure.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antao V. P., Gray D. M., Ratliff R. L. CD of six different conformational rearrangements of poly[d(A-G).d(C-T)] induced by low pH. Nucleic Acids Res. 1988 Jan 25;16(2):719–738. doi: 10.1093/nar/16.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOLLUM F. J., GROENIGER E., YONEDA M. POLYDEOXYADENYLIC ACID. Proc Natl Acad Sci U S A. 1964 May;51:853–859. doi: 10.1073/pnas.51.5.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernués J., Beltrán R., Casasnovas J. M., Azorín F. Structural polymorphism of homopurine--homopyrimidine sequences: the secondary DNA structure adopted by a d(GA.CT)22 sequence in the presence of zinc ions. EMBO J. 1989 Jul;8(7):2087–2094. doi: 10.1002/j.1460-2075.1989.tb03617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier D. A., Griffin J. A., Wells R. D. Non-B right-handed DNA conformations of homopurine.homopyrimidine sequences in the murine immunoglobulin C alpha switch region. J Biol Chem. 1988 May 25;263(15):7397–7405. [PubMed] [Google Scholar]
- Hakoshima T., Fukui T., Ikehara M., Tomita K. Molecular structure of a double helix that has non-Watson-Crick type base pairing formed by 2-substituted poly(A) and poly(U). Proc Natl Acad Sci U S A. 1981 Dec;78(12):7309–7313. doi: 10.1073/pnas.78.12.7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Htun H., Dahlberg J. E. Single strands, triple strands, and kinks in H-DNA. Science. 1988 Sep 30;241(4874):1791–1796. doi: 10.1126/science.3175620. [DOI] [PubMed] [Google Scholar]
- Kang C., Zhang X., Ratliff R., Moyzis R., Rich A. Crystal structure of four-stranded Oxytricha telomeric DNA. Nature. 1992 Mar 12;356(6365):126–131. doi: 10.1038/356126a0. [DOI] [PubMed] [Google Scholar]
- Klysik J., Rippe K., Jovin T. M. Reactivity of parallel-stranded DNA to chemical modification reagents. Biochemistry. 1990 Oct 23;29(42):9831–9839. doi: 10.1021/bi00494a012. [DOI] [PubMed] [Google Scholar]
- Kung H. J., Hu S., Bender W., Bailey J. M., Davidson N., Nicolson M. O., McAllister R. M. RD-114, baboon, and woolly monkey viral RNA's compared in size and structure. Cell. 1976 Apr;7(4):609–620. doi: 10.1016/0092-8674(76)90211-7. [DOI] [PubMed] [Google Scholar]
- Lee J. S., Evans D. H., Morgan A. R. Polypurine DNAs and RNAs form secondary structures which may be tetra-stranded. Nucleic Acids Res. 1980 Sep 25;8(18):4305–4320. doi: 10.1093/nar/8.18.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S., Johnson D. A., Morgan A. R. Complexes formed by (pyrimidine)n . (purine)n DNAs on lowering the pH are three-stranded. Nucleic Acids Res. 1979 Jul 11;6(9):3073–3091. doi: 10.1093/nar/6.9.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S. The stability of polypurine tetraplexes in the presence of mono- and divalent cations. Nucleic Acids Res. 1990 Oct 25;18(20):6057–6060. doi: 10.1093/nar/18.20.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyamichev V. I., Mirkin S. M., Frank-Kamenetskii M. D. Structures of homopurine-homopyrimidine tract in superhelical DNA. J Biomol Struct Dyn. 1986 Feb;3(4):667–669. doi: 10.1080/07391102.1986.10508454. [DOI] [PubMed] [Google Scholar]
- Lyamichev V. I., Voloshin O. N., Frank-Kamenetskii M. D., Soyfer V. N. Photofootprinting of DNA triplexes. Nucleic Acids Res. 1991 Apr 11;19(7):1633–1638. doi: 10.1093/nar/19.7.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor H., Rao B. S., Martin R. G. Abundance and degree of dispersion of genomic d(GA)n.d(TC)n sequences. J Mol Evol. 1988;27(2):96–101. doi: 10.1007/BF02138367. [DOI] [PubMed] [Google Scholar]
- Palecek E. Local supercoil-stabilized DNA structures. Crit Rev Biochem Mol Biol. 1991;26(2):151–226. doi: 10.3109/10409239109081126. [DOI] [PubMed] [Google Scholar]
- Parniewski P., Galazka G., Wilk A., Klysik J. Complex structural behavior of oligopurine-oligopyrimidine sequence cloned within the supercoiled plasmid. Nucleic Acids Res. 1989 Jan 25;17(2):617–629. doi: 10.1093/nar/17.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch D. S., Levenson C., Shafer R. H. Structure, stability, and thermodynamics of a short intermolecular purine-purine-pyrimidine triple helix. Biochemistry. 1991 Jun 25;30(25):6081–6088. doi: 10.1021/bi00239a001. [DOI] [PubMed] [Google Scholar]
- RICH A., DAVIES D. R., CRICK F. H., WATSON J. D. The molecular structure of polyadenylic acid. J Mol Biol. 1961 Feb;3:71–86. doi: 10.1016/s0022-2836(61)80009-0. [DOI] [PubMed] [Google Scholar]
- Ramsing N. B., Jovin T. M. Parallel stranded duplex DNA. Nucleic Acids Res. 1988 Jul 25;16(14A):6659–6676. doi: 10.1093/nar/16.14.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsing N. B., Rippe K., Jovin T. M. Helix-coil transition of parallel-stranded DNA. Thermodynamics of hairpin and linear duplex oligonucleotides. Biochemistry. 1989 Nov 28;28(24):9528–9535. doi: 10.1021/bi00450a042. [DOI] [PubMed] [Google Scholar]
- Reaban M. E., Griffin J. A. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990 Nov 22;348(6299):342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- Riazance J. H., Johnson W. C., Jr, McIntosh L. P., Jovin T. M. Vacuum UV circular dichroism is diagnostic for the left-handed Z form of poly [d(A-C).d(G-T)] and other polydeoxynucleotides. Nucleic Acids Res. 1987 Sep 25;15(18):7627–7636. doi: 10.1093/nar/15.18.7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe K., Jovin T. M. Parallel-stranded duplex DNA. Methods Enzymol. 1992;211:199–220. doi: 10.1016/0076-6879(92)11013-9. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Jovin T. M. Spectroscopic properties and helical stabilities of 25-nt parallel-stranded linear DNA duplexes. Biochemistry. 1989 Nov 28;28(24):9536–9541. doi: 10.1021/bi00450a043. [DOI] [PubMed] [Google Scholar]
- Rippe K., Ramsing N. B., Klement R., Jovin T. M. A parallel stranded linear DNA duplex incorporating dG.dC base pairs. J Biomol Struct Dyn. 1990 Jun;7(6):1199–1209. doi: 10.1080/07391102.1990.10508559. [DOI] [PubMed] [Google Scholar]
- Sarma M. H., Gupta G., Sarma R. H. A cytosine . cytosine base paired parallel DNA double helix with thymine . thymine bulges. FEBS Lett. 1986 Sep 15;205(2):223–229. doi: 10.1016/0014-5793(86)80902-4. [DOI] [PubMed] [Google Scholar]
- Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988 Jul 28;334(6180):364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- Shimizu M., Hanvey J. C., Wells R. D. Intramolecular DNA triplexes in supercoiled plasmids. I. Effect of loop size on formation and stability. J Biol Chem. 1989 Apr 5;264(10):5944–5949. [PubMed] [Google Scholar]
- Smith F. W., Feigon J. Quadruplex structure of Oxytricha telomeric DNA oligonucleotides. Nature. 1992 Mar 12;356(6365):164–168. doi: 10.1038/356164a0. [DOI] [PubMed] [Google Scholar]
- Son T. D., Guschlbauer W., Guéron M. Flexibility and conformations of guanosine monophosphates by the Overhauser effect. J Am Chem Soc. 1972 Nov 1;94(22):7903–7911. doi: 10.1021/ja00777a038. [DOI] [PubMed] [Google Scholar]
- Sprecher C. A., Johnson W. C., Jr Circular dichroism of the nucleic acid monomers. Biopolymers. 1977 Oct;16(10):2243–2264. doi: 10.1002/bip.1977.360161012. [DOI] [PubMed] [Google Scholar]
- Stavnezer J. Triple helix stabilization? Nature. 1991 Jun 6;351(6326):447–448. doi: 10.1038/351447b0. [DOI] [PubMed] [Google Scholar]
- Sundquist W. I., Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989 Dec 14;342(6251):825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- Tavale S. S., Sobell H. M. Crystal and molecular structure of 8-bromoguanosine and 8-bromoadenosine, two purine nucleosides in the syn conformation. J Mol Biol. 1970 Feb 28;48(1):109–123. doi: 10.1016/0022-2836(70)90222-6. [DOI] [PubMed] [Google Scholar]
- Tripathi J., Brahmachari S. K. Distribution of simple repetitive (TG/CA)n and (CT/AG)n sequences in human and rodent genomes. J Biomol Struct Dyn. 1991 Oct;9(2):387–397. doi: 10.1080/07391102.1991.10507919. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Wells R. D., Collier D. A., Hanvey J. C., Shimizu M., Wohlrab F. The chemistry and biology of unusual DNA structures adopted by oligopurine.oligopyrimidine sequences. FASEB J. 1988 Nov;2(14):2939–2949. [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Westhof E., Rao S. T., Sundaralingam M. Crystallographic studies of drug-nucleic acid interactions: proflavine intercalation between the non-complementary base-pairs of cytidilyl-3',5'-adenosine. J Mol Biol. 1980 Sep 25;142(3):331–361. doi: 10.1016/0022-2836(80)90276-4. [DOI] [PubMed] [Google Scholar]
- Williamson J. R., Raghuraman M. K., Cech T. R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989 Dec 1;59(5):871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- Yagil G. Paranemic structures of DNA and their role in DNA unwinding. Crit Rev Biochem Mol Biol. 1991;26(5-6):475–559. doi: 10.3109/10409239109086791. [DOI] [PubMed] [Google Scholar]
- Yee H. A., Wong A. K., van de Sande J. H., Rattner J. B. Identification of novel single-stranded d(TC)n binding proteins in several mammalian species. Nucleic Acids Res. 1991 Feb 25;19(4):949–953. doi: 10.1093/nar/19.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]