Abstract
Autophagy is a highly conserved self-digestion process, which is essential for maintaining homeostasis and viability in response to nutrient starvation1–4. Although the components of autophagy in the cytoplasm have been well studied5,6, the molecular basis for the transcriptional and epigenetic regulation of autophagy is poorly understood. Here we identify co-activator-associated arginine methyltransferase 1 (CARM1) as a crucial component of autophagy in mammals. Notably, CARM1 stability is regulated by the SKP2-containing SCF (SKP1-cullin1-F-box protein) E3 ubiquitin ligase in the nucleus, but not in the cytoplasm, under nutrient-rich conditions. Furthermore, we show that nutrient starvation results in AMP-activated protein kinase (AMPK)-dependent phosphorylation of FOXO3a in the nucleus, which in turn transcriptionally represses SKP2. This repression leads to increased levels of CARM1 protein and subsequent increases in histone H3 Arg17 dimethylation. Genome-wide analyses reveal that CARM1 exerts transcriptional co-activator function on autophagy-related and lysosomal genes through transcription factor EB (TFEB). Our findings demonstrate that CARM1-dependent histone arginine methylation is a crucial nuclear event in autophagy, and identify a new signalling axis of AMPK–SKP2–CARM1 in the regulation of autophagy induction after nutrient starvation.
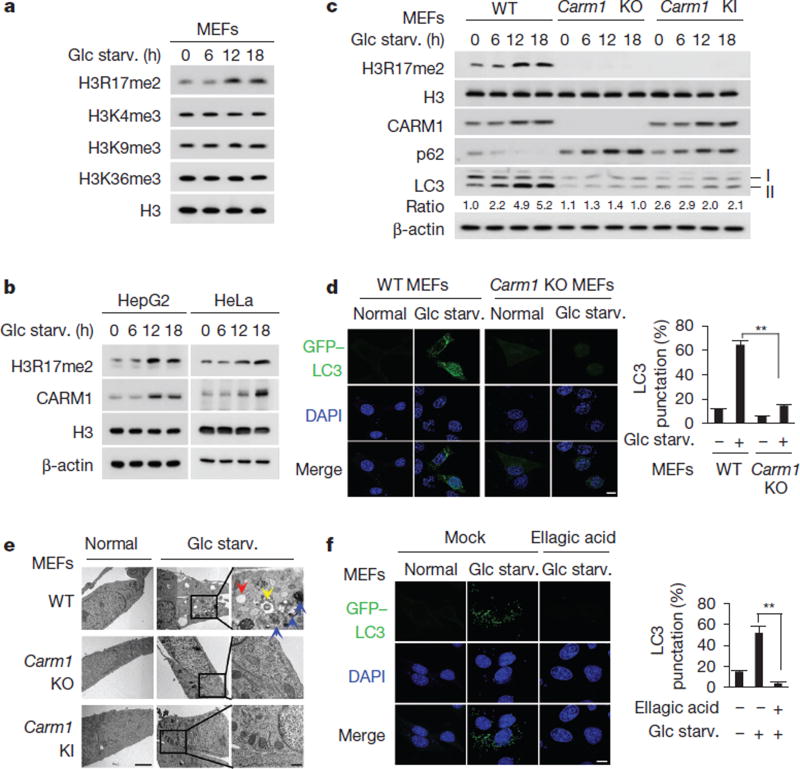
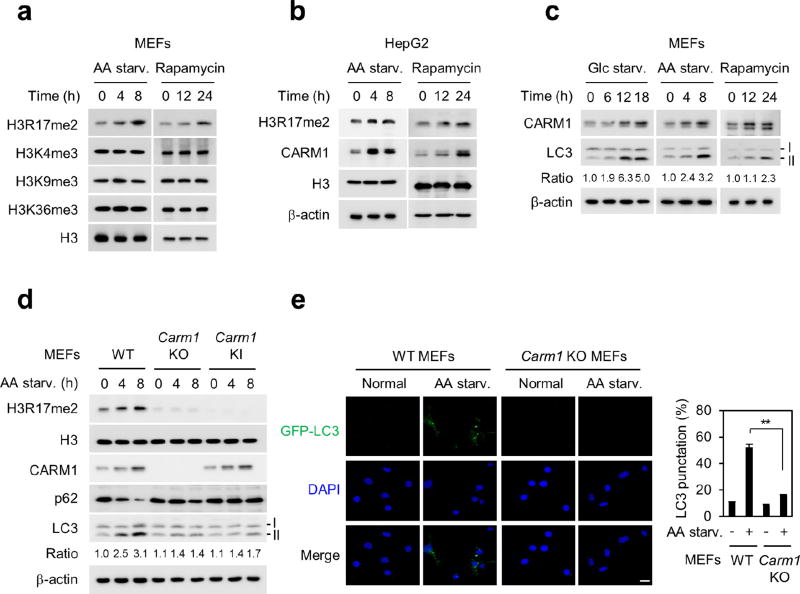
To explore the importance of nuclear events in autophagy, we proposed that specific histone marks are involved in the epigenetic and transcriptional regulation of autophagy in the nucleus leading to the fine-tuning of the autophagy process. We induced autophagy in mouse embryonic fibroblasts (MEFs) by glucose starvation, and sought to identify altered specific histone marks. We observed an increase in histone H3 Arg17 dimethylation (H3R17me2) levels in response to glucose starvation (Fig. 1a), which also occurred when autophagy was triggered by amino acid starvation or rapamycin (Extended Data Fig. 1a). Notably, nutrient starvation resulted in increased levels of CARM1 protein (Fig. 1b and Extended Data Fig. 1b).
Figure 1. Increased H3R17 dimethylation by CARM1 is critical for proper autophagy.
a, b, Immunoblot analysis of various histone marks and CARM1 in response to glucose starvation (Glc starv.). c, Wild-type (WT), Carm1 knockout (KO) or knock-in (KI) MEFs were subject to immunoblot analysis. The LC3-II/LC3-I ratio is indicated. d, Representative confocal images of GFP–LC3 puncta formation. Graph shows quantification of LC3-positive punctate cells (right). Nuclei counterstained with DAPI. Scale bar, 10 µm. e, Representative TEM images. Scale bar, 2 µm. High magnification of boxed areas is shown on the right. Scale bar, 0.5 µm. Autophagosomes (blue arrows), autolysosomes (red arrows) and multilamellar body (yellow arrow). f, Representative confocal images of GFP–LC3 puncta formation. Ellagic acid (100 µM). Scale bar, 10 µm. Bars, mean ± s.e.m.; n = 5, with over 100 cells; **P < 0.01 (one-tailed t-test) (d, f).
To examine whether CARM1 induction and subsequent increases in H3R17me2 are related to autophagy occurrence, we analysed the conversion of non-lipidated LC3-I to lipidated LC3-II, as a common marker of autophagic activity7. The increase in CARM1 was associated with an increase in LC3-II (Fig. 1c and Extended Data Fig. 1c, d). To confirm that the decrease in LC3-II reflects decreases in functional autophagic degradation, autophagic flux was also analysed using the levels of p62 (also known as SQSTM1)8,9. Glucose starvation induced p62 degradation and LC3-II accumulation in wild-type MEFs but not in Carm1 knockout and knock-in MEFs expressing the enzymatic activity-deficient mutant (Fig. 1c).
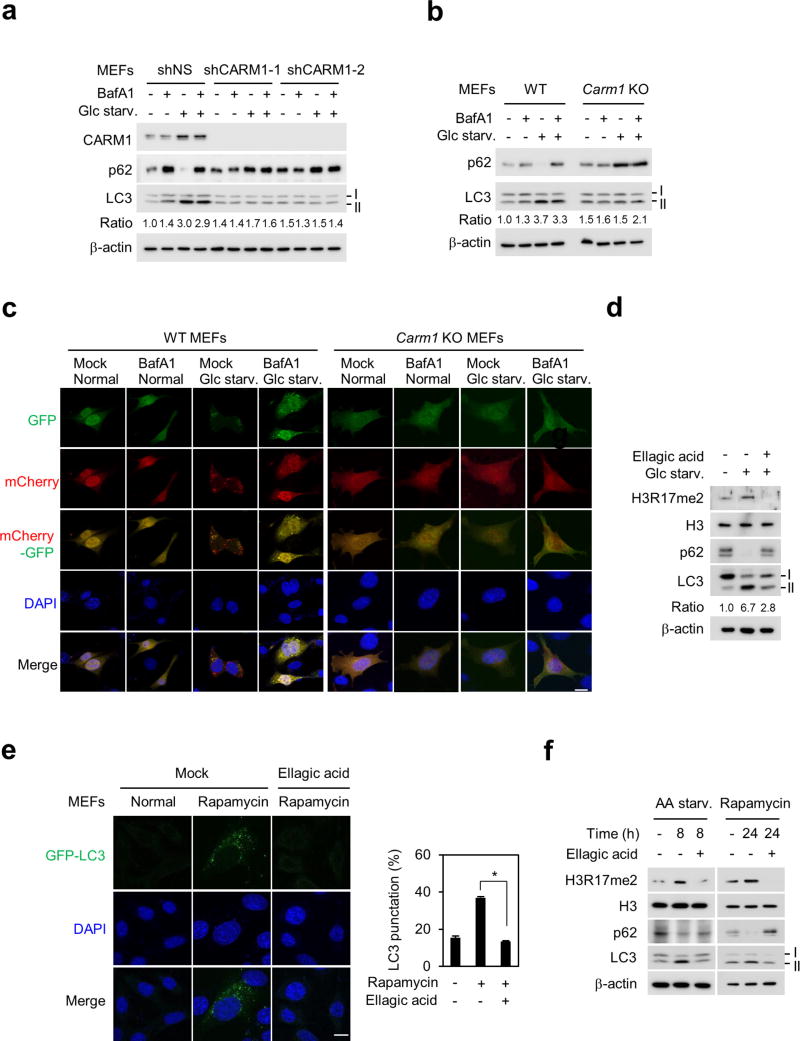
To evaluate the role of CARM1 in the autophagic process, the formation of green fluorescent protein (GFP)-tagged LC3-positive autophagosome was examined. The increase in GFP–LC3 punctate cells was notably attenuated in Carm1 knockout compared to wild-type MEFs (Fig. 1d and Extended Data Fig. 1e). Transmission electron microscopy (TEM) further showed an increase in the number of autophagic vesicles in wild-type MEFs, but not in Carm1 knockout and knock-in MEFs (Fig. 1e). We performed LC3 flux analysis using bafilomycin A1, an inhibitor of the late phase of autophagy. Defects in autophagic flux caused by the loss of CARM1 were confirmed by immunoblot analysis (Extended Data Fig. 2a, b) and imaging experiments using mCherry-GFP–LC3, which provides a simultaneous readout of autophagosome formation and maturation (Extended Data Fig. 2c). In addition, ellagic acid, a naturally occurring polyphenol reported to selectively inhibit H3R17me2 (ref. 10), greatly compromised the autophagic process (Fig. 1f and Extended Data Fig. 2d–f).
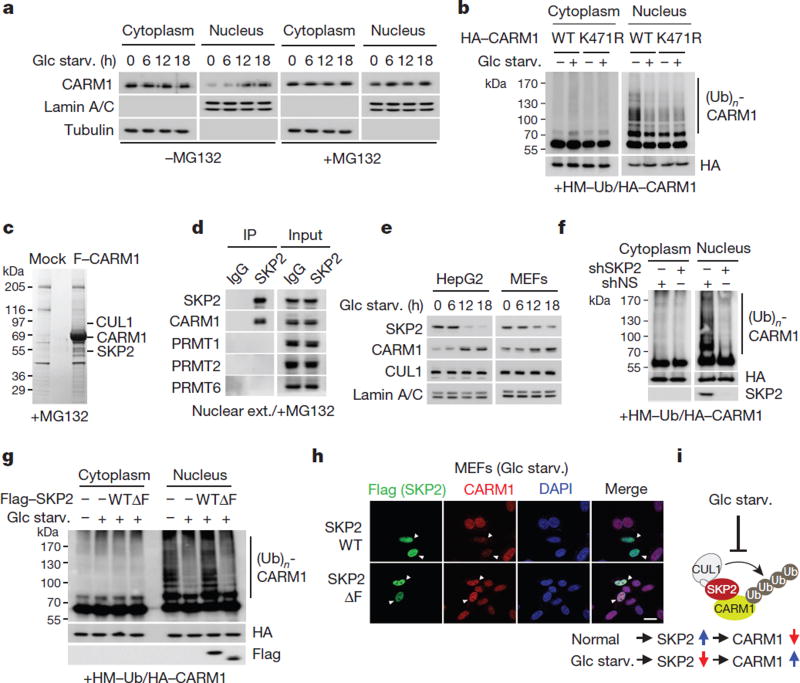
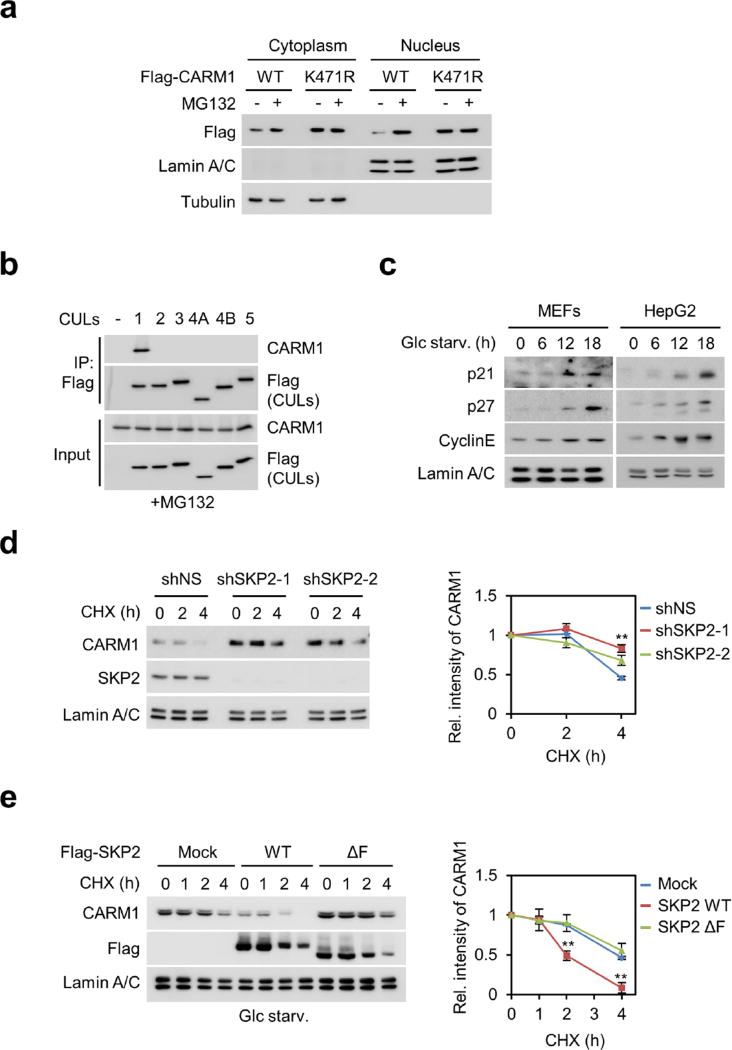
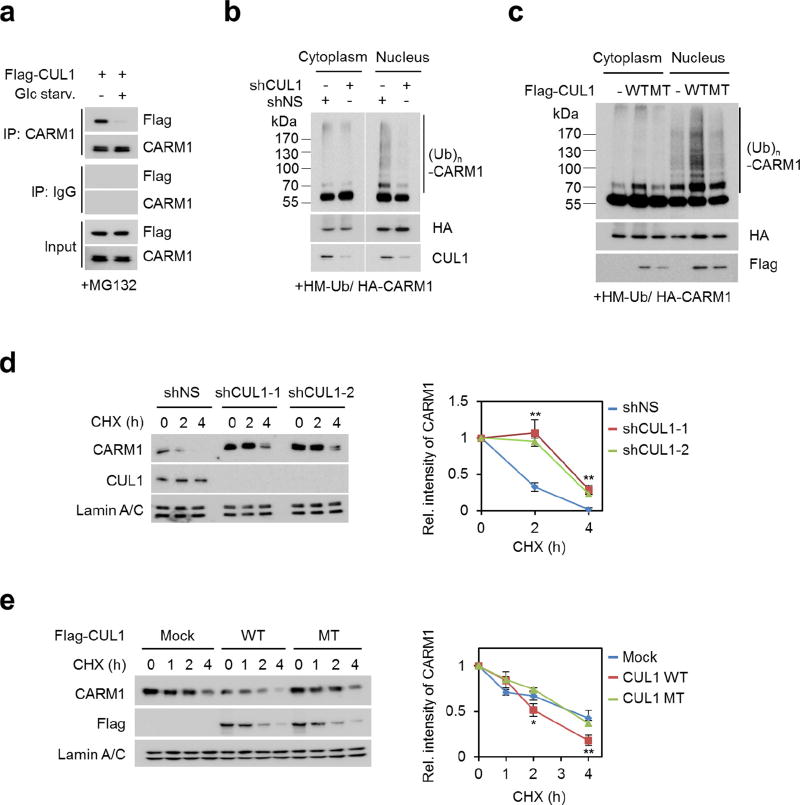
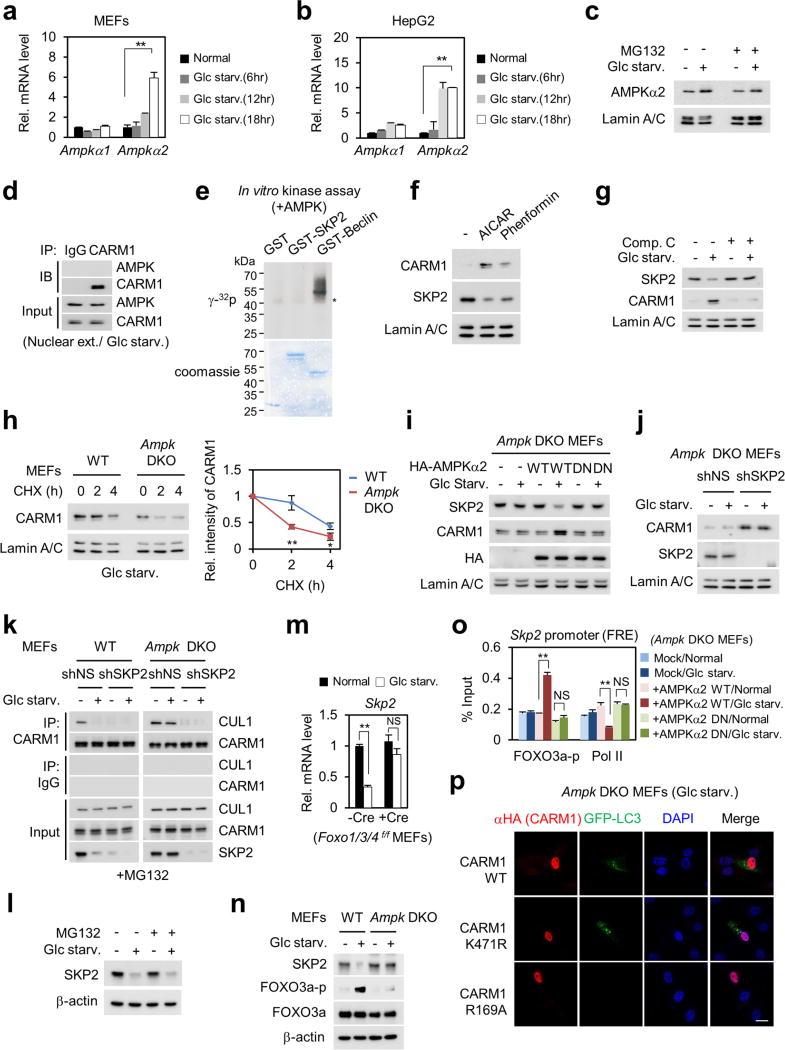
Next, we examined how CARM1 induction is regulated after glucose starvation. We found that CARM1 protein levels were increased only in the nucleus after glucose starvation (Fig. 2a, left). Treatment of MG132, a 26S proteasome inhibitor, inhibited nuclear CARM1 degradation (Fig. 2a, right). Glucose starvation markedly reduced the ubiquitination of CARM1 in the nucleus, whereas CARM1(K471R) failed to be ubiquitinated, indicating that K471 is the ubiquitination-targeting site (Fig. 2b and Extended Data Fig. 3a). We then sought to identify the E3 ubiquitin ligase responsible for CARM1 ubiquitination. Notably, SKP2, an F-box protein of the SCF E3 ubiquitin ligase complex, was identified as a CARM1-binding protein along with cullin 1 (CUL1) (Fig. 2c and Supplementary Table 1). CARM1 exhibited specific binding to SKP2 (Fig. 2d) and CUL1 (Extended Data Fig. 3b).
Figure 2. CARM1 is degraded by the SKP2-containing SCF E3 ligase in the nucleus under nutrient-rich conditions.
a, MEFs were deprived of glucose in the absence (left) or presence (right) of MG132 (5 µg ml−1) and subject to immunoblotting. b, In vivo ubiquitination assay of wild-type CARM1 or ubiquitination-defective K471R mutant CARM1. HA, haemagglutinin; HM, HisMax tag. c, Identification of CARM1-interacting proteins. F–CARM1 denotes Flag-tagged CARM1 construct. d, Interactions between SKP2 and protein arginine methyltransferases (PRMTs) were analysed. e, Glucose-starved cells were subjected to immunoblotting. f, g, In vivo ubiquitination assay of CARM1. h, Representative confocal images. Scale bar, 20 µm. i, Schematic of SKP2-containing SCF E3 ligase-dependent degradation of CARM1.
Since CARM1 is stabilized after glucose starvation and possibly ubiquitinated by the SKP2-containing E3 ligase complex under nutrient- rich condition, we checked for changes in SKP2 protein levels. A reduction in SKP2 and an increase in CARM1 protein levels were observed in glucose-starved cells (Fig. 2e). Decreased levels of SKP2 resulted in the stabilization of other known SKP2-SCF substrates (Extended Data Fig. 3c). Furthermore, SKP2 knockdown attenuated CARM1 ubiquitination in the nucleus (Fig. 2f) and markedly increased the half-life of CARM1 (Extended Data Fig. 3d). By contrast, overexpression of wild-type SKP2, but not the SKP2ΔF mutant that is not able to form a SKP2–SCF complex11, decreased the half-life and protein levels of CARM1 in cells deprived of glucose (Fig. 2g, h and Extended Data Fig. 3e). We speculate that exclusive nuclear localization of SKP2 results in selective CARM1 ubiquitination in the nucleus. As a result of SKP2 downregulation, the interaction between CUL1 and CARM1 significantly decreased after glucose starvation (Extended Data Fig. 4a). Also, as a component of the SCF complex, CUL1 regulated CARM1 protein levels (Extended Data Fig. 4b–e). Collectively, these data indicate that the SKP2-containing SCF E3 ligase complex is responsible for CARM1 degradation in the nucleus under nutrient-rich conditions (Fig. 2i).
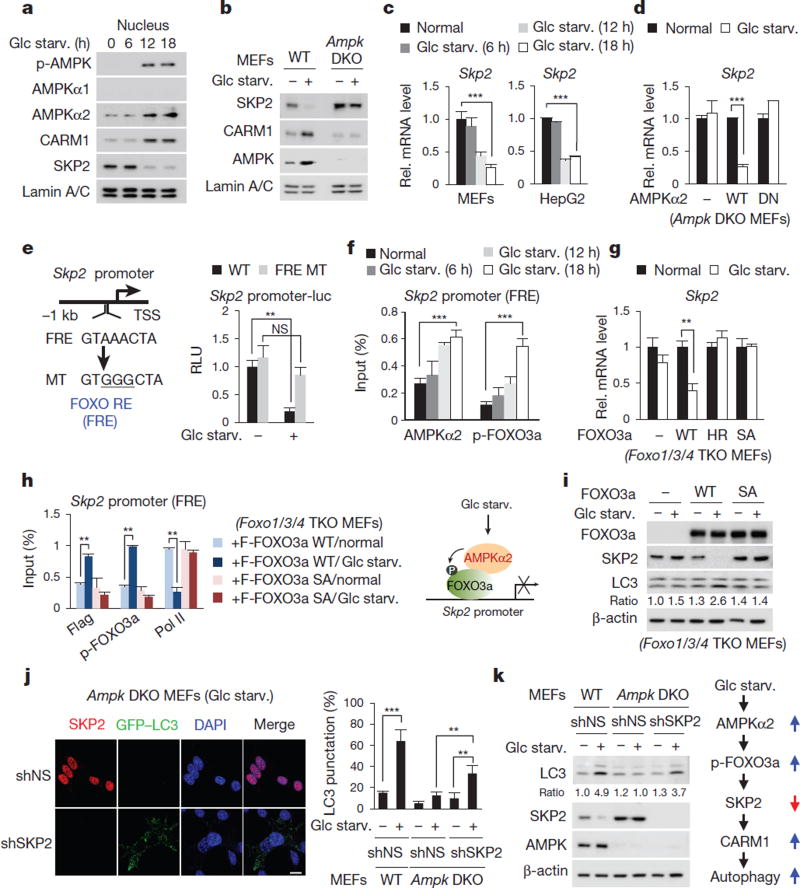
It has been shown that AMPK is activated during glucose starvation and leads to starvation-induced autophagy12–14. As the role of nuclear AMPK in autophagy outcome has not been defined thus far, we aimed to examine whether AMPK is involved in the transcriptional regulation of autophagy. We found that AMPKα2 and phosphorylated AMPK, the activated form of AMPK, increased in the nucleus after glucose starvation (Fig. 3a). Increased AMPKα2 resulted from transcription induction rather than post-translational regulation (Extended Data Fig. 5a–c). AMPKα2 has been shown to be preferentially expressed in the nucleus15, suggesting that it might perform distinct roles in the nucleus. AMPK failed to directly bind or phosphorylate CARM1 and SKP2 (Extended Data Fig. 5d, e). However, AMPK activation by aminoimidazole carboxamide ribonucleotide (AICAR) and phenformin resulted in the increase of CARM1 and reduction of SKP2 (Extended data Fig. 5f), and this was compromised when AMPK activity was blocked by compound C (Extended data Fig. 5g).
Figure 3. Decrease in SKP2 after glucose starvation is AMPK dependent.
a, MEFs deprived of glucose were analysed with the indicated antibodies. b, Nuclear fractions from wild-type and Ampk double knockout (DKO) MEFs were subjected to immunoblotting. c, d, qRT–PCR of Skp2. DN, dominant negative. e, Left, schematic of Skp2 promoter. Right, luciferase activities of wild-type Skp2 or FOXO response element (FRE) mutant promoter were measured. MT, mutant; RE, response element; RLU, relative light units; TSS, transcription start site. f, ChIP assays of the Skp2 promoter. g, Skp2 mRNA levels were analysed in Foxo1/3/4 triple knockout (TKO) MEFs. HR, H212R mutant; SA, sextuple T179A/ S399A/ S413A/S555A/S588A/S626A mutant. h, Left, ChIP assays of the Skp2 promoter. Right, schematic of SKP2 regulation by the AMPK–FOXO axis (right). Bars, mean ± s.e.m.; n = 3. NS, not significant. **P < 0.01, ***P < 0.001 (one-tailed t-test) (c–h). i, Immunoblot analysis in Foxo1/3/4 TKO MEFs. j, Representative confocal images of GFP–LC3 puncta formation. shSKP2, short-hairpin RNA (shRNA) against SKP2; shNS, nonspecific shRNA. Scale bar, 20 µm. Bars, mean ± s.e.m.; n = 5, with over 80 cells; **P < 0.01, ***P < 0.001 (one-tailed t-test). k, Left, immunoblot analysis from whole-cell extracts. Right, schematic of the AMPK–SKP2–CARM1 signalling cascade in autophagy.
We then used wild-type and Ampka1 and Ampkα2 (also known as Prkaa1 and Prkaa2) double knockout (DKO) MEFs to check for the expression of CARM1 and SKP2. In the nucleus, CARM1 induction and SKP2 reduction after glucose starvation were abrogated in Ampk DKO MEFs (Fig. 3b).The half-life of CARM1 in the nucleus was decreased in Ampk DKO MEFs (Extended Data Fig. 5h). Introduction of wild-type AMPKα2, but not the dominant-negative form, in Ampk DKO MEFs resulted in a recovered expression pattern of SKP2 and CARM1, similar to wild-type MEFs (Extended Data Fig. 5i). SKP2 depletion in Ampk DKO MEFs led to increased CARM1 protein levels, indicating that the reduction of CARM1 in Ampk DKO MEFs is mediated by SKP2 (Extended Data Fig. 5j). Furthermore, since binding of CARM1 to CUL1 is mediated by SKP2, the CARM1–CUL1 interaction was maintained upon glucose starvation in Ampk DKO MEFs (Extended Data Fig. 5k).
Reduction of SKP2 expression after glucose starvation is not mediated by proteasomal degradation (Extended Data Fig. 5l), but instead regulated at the transcription level (Fig. 3c). Glucose starvation failed to decrease Skp2 mRNA levels in Ampk DKO MEFs, but reconstitution of wild-type AMPKα2 restored the reduction in Skp2 mRNA (Fig. 3d). Therefore, we were prompted to search for a possible regulatory mechanism of SKP2 downregulation by AMPKα2. Recent studies have emphasized the AMPK–FOXO axis as a highly conserved nutrient-sensing pathway crucial for cellular and organismal homeostasis16. AMPK directly phosphorylates FOXO3a and regulates FOXO3a transcriptional activity17. Although mainly known as a transcriptional activator, FOXO also functions as a transcriptional repressor18–21. Skp2 promoter analysis revealed a highly conserved FOXO response element (FRE) (Fig. 3e). We proposed that FOXO might function as a transcriptional repressor of SKP2 and performed luciferase reporter assay driven by the Skp2 promoter. Glucose starvation attenuated Skp2 promoter luciferase activity, but not the Skp2 promoter containing an FRE mutation (Fig. 3e). Skp2 mRNA levels failed to decrease in Foxo1/3/4 triple knockout (TKO) MEFs (Extended Data Fig. 5m), indicating that FOXO is a crucial transcription factor in the repression of SKP2.
Glucose starvation resulted in AMPK-dependent FOXO3a phosphorylation (Extended Data Fig. 5n). In addition, AMPKα2 and phosphorylated FOXO3a were co-recruited to the Skp2 promoter upon glucose starvation (Fig. 3f). The recruitment of phosphorylated FOXO3a accompanied by a decrease in RNA polymerase II was also observed in Ampk DKO MEFs reconstituted with wild-type AMPKα2 (Extended Data Fig. 5o). Notably, reconstitution of wild-type FOXO3a in Foxo1/3/4 TKO MEFs significantly reduced the Skp2 mRNA level, but neither the FOXO3a(H212R) DNA-binding mutant22 nor the FOXO3a sextuple SA mutant, which is not phosphorylated by AMPK17, reduced Skp2 mRNA levels (Fig. 3g). Furthermore, after glucose starvation, phosphorylated FOXO3a, but not the FOXO3a SA mutant, was recruited to the Skp2 promoter (Fig. 3h), indicating that AMPK-dependent FOXO3a phosphorylation is crucial for the recruitment of FOXO3a at the Skp2 promoter. SKP2 expression failed to decrease and autophagy occurrence was impaired in FOXO3a SA mutant-reconstituted Foxo1/3/4 TKO MEFs (Fig. 3i).
We observed a marked increase in autophagy occurrence in Ampk DKO MEFs after SKP2 knockdown (Fig. 3j, k). We also tested whether CARM1 overexpression could restore autophagy in Ampk DKO MEFs. Introduction of wild-type or K471R mutant CARM1 restored the number of GFP–LC3 punctate cells, whereas enzymatic-dead mutant CARM1(R169A) failed to do so (Extended Data Fig. 5p). Collectively, we found a signalling axis in autophagy induction in which glucose starvation activates AMPKα2 in the nucleus, leading to transcriptional repression of Skp2 via FOXO3a phosphorylation. Reduction of SKP2 expression in turn leads to increased levels of CARM1.
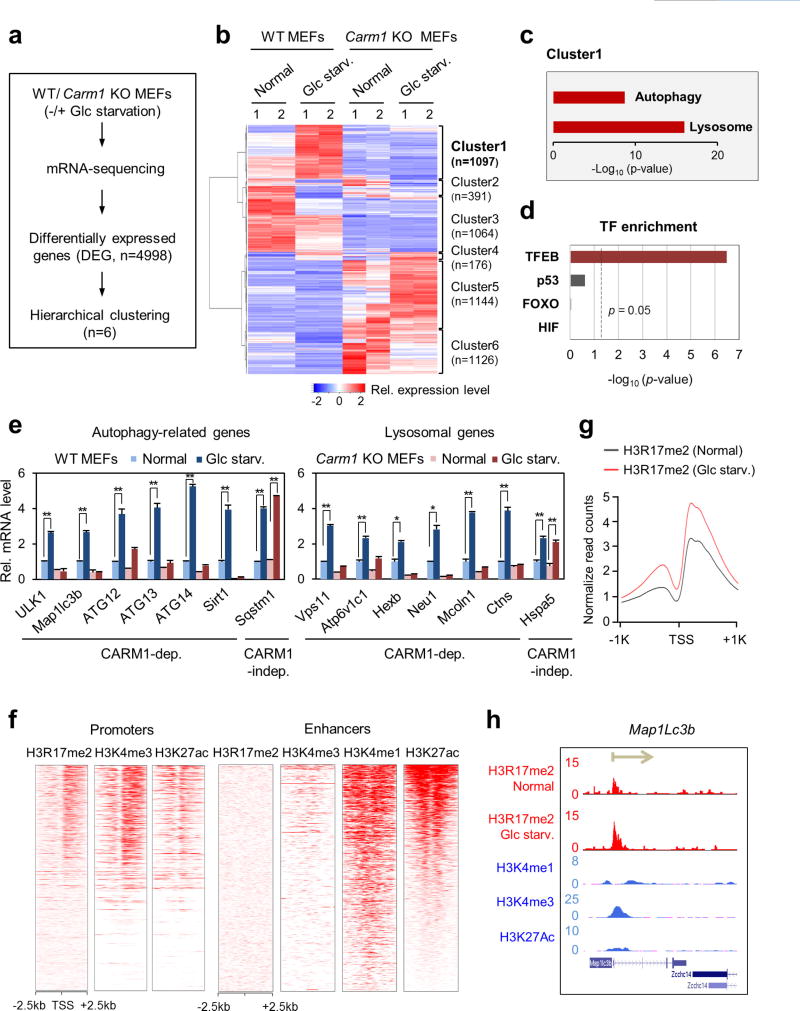
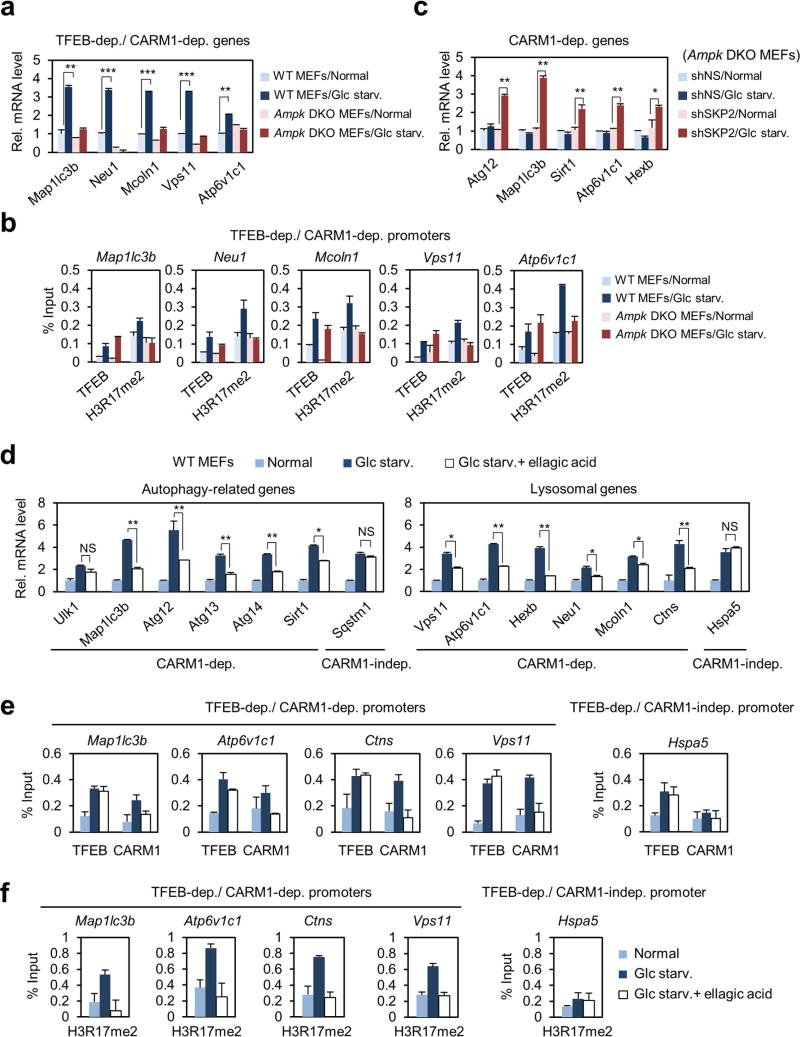
To gain insight into the role of CARM1 in transcriptional regulation of autophagy, we performed RNA-sequencing (RNA-seq) in wild-type and Carm1 knockout MEFs after glucose starvation (Extended Data Fig. 6a, b). Using a comprehensive list of known autophagy-related and lysosomal genes (Supplementary Table 2), we found that potential CARM1 target genes (cluster 1) are significantly enriched for autophagy-related and lysosomal genes (Extended Data Fig. 6c). Transcription factor motif analysis indicated TFEB as a putative major transcription factor for CARM1 (Extended Data Fig. 6d). We validated CARM1 dependency of the autophagy-related and lysosomal genes by quantitative reverse transcription PCR (qRT–PCR) (Extended Data Fig. 6e). Furthermore, we performed chromatin immunoprecipitation with high-throughput sequencing (ChIP–seq) of H3R17me2 in wild-type MEFs after glucose starvation and observed enriched H3R17me2 as well as activating H3K4me3 signals at active promoters (Extended Data Fig. 6f–h and Supplementary Table 3).
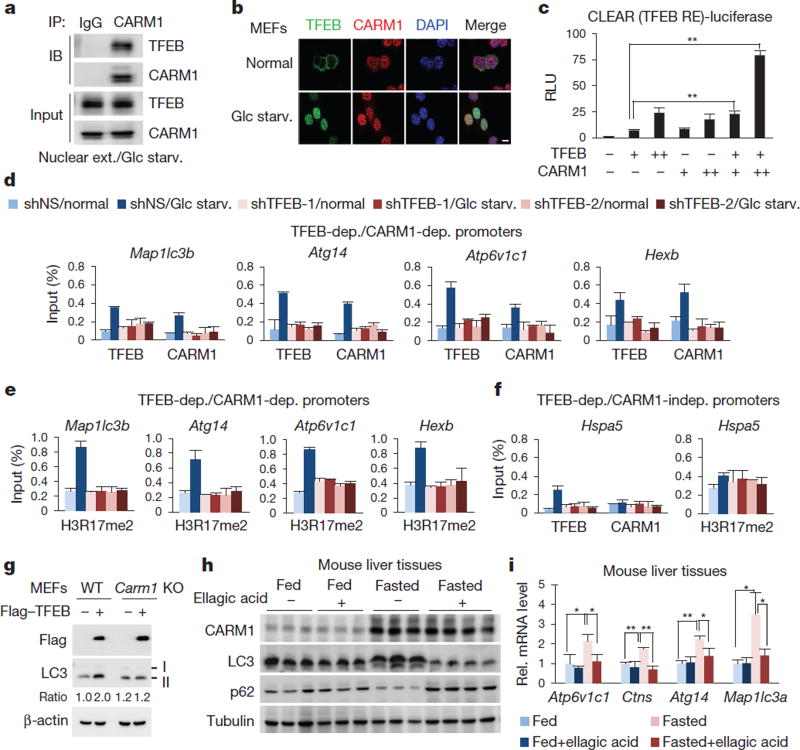
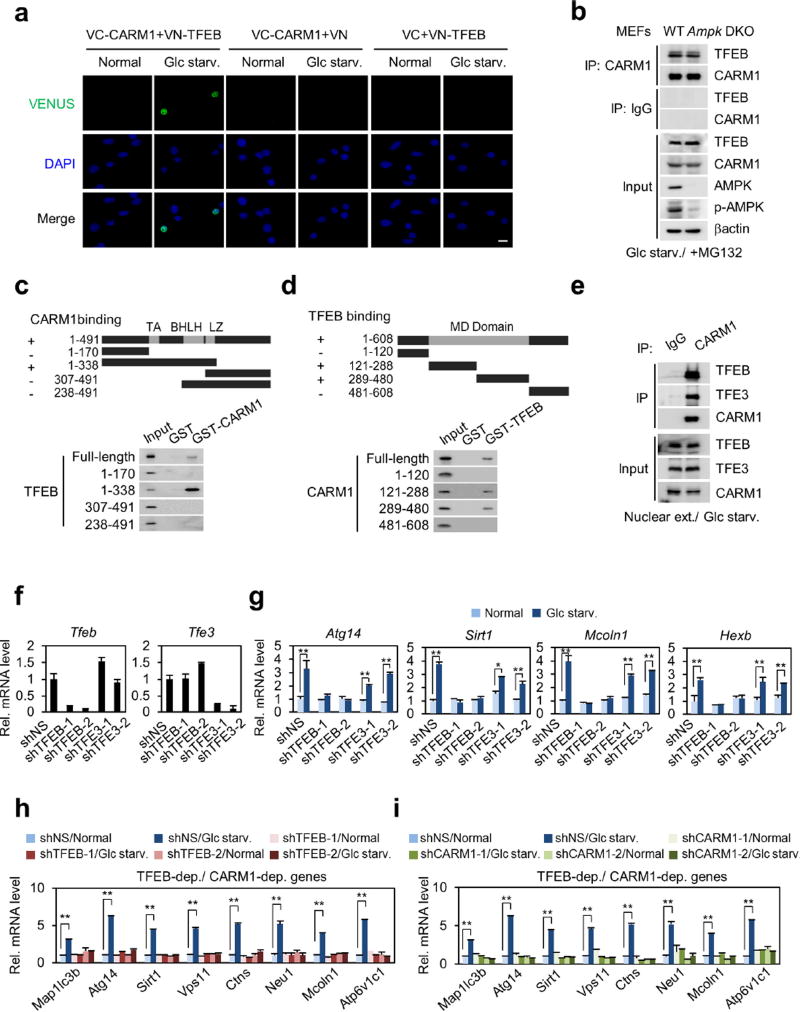
TFEB functions as a master regulator of lysosomal biogenesis and autophagy23–25. After glucose starvation, CARM1 and TFEB exhibited mutual binding in the nucleus (Fig. 4a, b and Extended Data Fig. 7a). The binding of CARM1 to TFEB was not affected by AMPK (Extended Data Fig. 7b). CARM1 binds to the transcriptional activation domain of TFEB, whereas TFEB binds to the methyltransferase domain of CARM1 (Extended Data Fig. 7c, d). Although CARM1 also interacts with TFE3, TFEB knockdown, but not TFE3 knockdown, markedly altered the transcription induction of various target genes (Extended Data Fig. 7e–h).
Figure 4. CARM1 exerts a transcriptional co-activator function on autophagy-related and lysosomal genes through TFEB.
a, Binding between CARM1 and TFEB. b, Representative confocal images. Scale bar, 10 µm. c, 2× CLEAR (TFEB RE)-luciferase reporter assays. Bars, mean ± s.e.m.; n = 3. **P < 0.01 (one-tailed t-test). d–f, ChIP assays on TFEB-dependent, CARM1-dependent (d, e) or CARM1-independent (f) promoters after knockdown of TFEB. Bars, mean ± s.e.m.; n = 3. g, Wild-type and Carm1 knockout MEFs transfected with Flag–TFEB were subject to immunoblot analysis. h, Liver tissues from fed or fasted mice treated with vehicle or ellagic acid were subjected to immunoblot analysis (n = 3 per group). i, Expression of autophagy-related genes and lysosomal genes in wild-type mouse livers. Bars, mean ± s.e.m.; n = 3 per group. *P < 0.05, **P < 0.01 (two-tailed t-test).
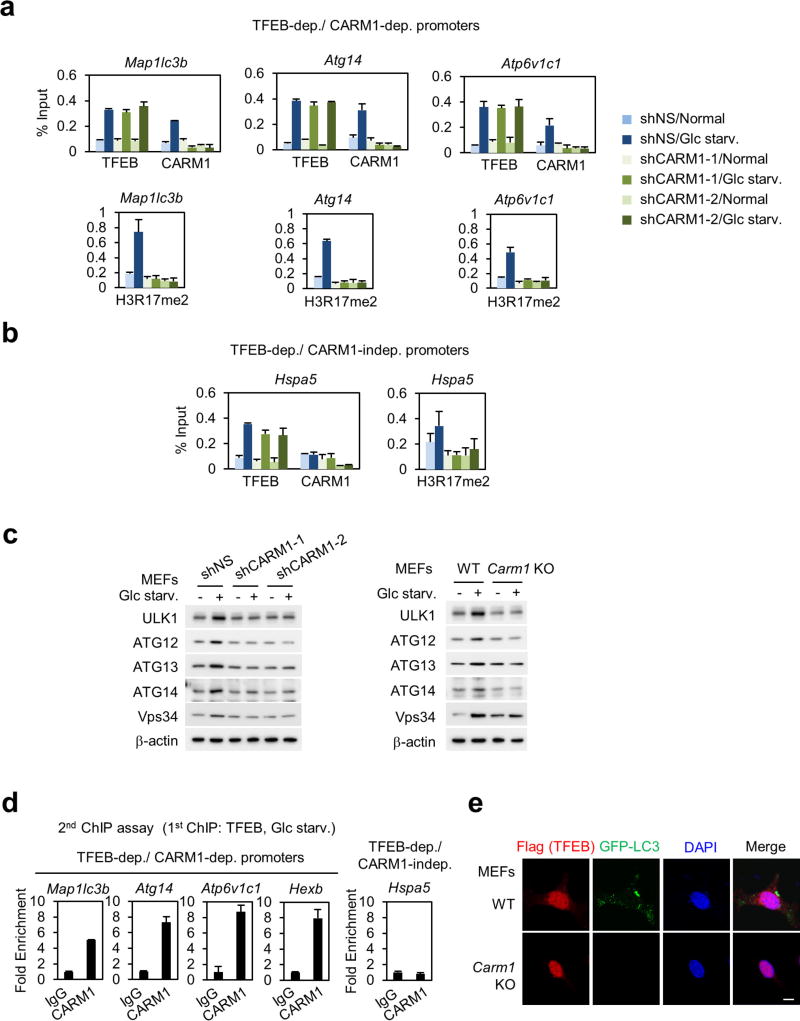
Introduction of TFEB increased CLEAR-element-containing luciferase reporter activity and overexpression of CARM1 further enhanced its activity (Fig. 4c). To examine whether CARM1-dependent target genes are regulated by TFEB, we searched for putative CLEAR motif (Supplementary Table 2) and performed ChIP assays. Knockdown of TFEB abolished the recruitment of CARM1 to its target promoters, subsequently leading to the failure of H3R17me2 induction (Fig. 4d, e). CARM1 recruitment was not observed on CARM1-independent promoters (Fig. 4f). Conversely, a subset of TFEB target genes failed to increase upon glucose starvation after CARM1 knockdown (Extended Data Fig. 7i). CARM1 depletion was accompanied by a reduction in H3R17me2 on TFEB-dependent, CARM1-dependent target promoters, with little or no effect on TFEB recruitment (Extended Data Fig. 8a, b). Immunoblot analysis confirmed several key autophagy regulators that are transcriptionally regulated by CARM1 were induced by glucose starvation in wild-type MEFs, but not in Carm1 knockdown or knock-out MEFs (Extended Data Fig. 8c). Furthermore, a two-step ChIP assay confirmed the recruitment of CARM1 at TFEB-bound genes (Extended Data Fig. 8d).
Previous studies reported that overexpression of TFEB induces autophagy24. However, introduction of TFEB in Carm1 knockout MEFs failed to increase the formation of autophagosomes and levels of LC3-II (Fig. 4g and Extended Data Fig. 8e). As CARM1 fails to increase upon glucose starvation in Ampk DKO MEFs, TFEB-dependent, CARM1-dependent target gene expression and induction of H3R17me2 were dampened in Ampk DKO MEFs (Extended Data Fig. 9a, b). However, SKP2 knockdown significantly increased the mRNA levels of CARM1 target genes (Extended Data Fig. 9c), indicating that partial recovery of autophagy in Ampk DKO MEFs by SKP2 knockdown is due to transcriptional activation of autophagy-related and lysosomal genes. Collectively, these data indicate CARM1 as a crucial co-activator of TFEB.
To examine whether CARM1 and subsequent H3R17me2 are important for autophagy occurrence in vivo, we analysed hepatic autophagy in wild-type mice. Livers of fasted mice showed a marked increase in CARM1 levels, as well as an increase in LC3 conversion. However, LC3 conversion was greatly attenuated in fasted livers of mice treated with ellagic acid (Fig. 4h). Furthermore, the mRNA expression of various CARM1-dependent autophagy-related and lysosomal genes failed to increase (Fig. 4i). Ellagic acid treatment inhibited the induction of a subset of autophagy-related and lysosomal genes regulated by CARM1, and blocked the recruitment of CARM1, but not TFEB, along with reduced H3R17me2 levels at CARM1-dependent promoters (Extended Data Fig. 9d–f). Given that the inhibition of H3R17me2 by ellagic acid almost completely blocks CARM1-induced autophagy occurrence, ellagic acid might have the potential to be developed as a therapeutic agent in autophagy-related diseases.
Here, we provide a link between energy sensing, chromatin modifications and transcriptional and epigenetic regulation of autophagy (Extended Data Fig. 10). Although our current work is focused on CARM1 stabilization, we speculate that this type of regulation in the nucleus might be an efficient way to regulate target gene expression, and could be a prototype of protein stabilization for histone modifiers. In addition, our data indicate that when glucose starvation persists and transcription of various autophagy-related genes is needed to sustain autophagy, AMPK accumulates in the nucleus and actively controls transcription. Our findings shed light on the potential therapeutic targeting of a new signalling axis of AMPK–SKP2–CARM1 in autophagy-related diseases.
Methods
Antibodies and reagents
The following commercially available antibodies were used: anti-AMPKα1 (ab110036), anti-AMPKα2 (ab3760), anti-ATG14 (ab173943), anti-FOXO3a (ab12162), anti-histone H3 (ab1791), anti-H3R17me2 (ab8284), anti-H3K4me3 (ab8580), anti-H3K9me3 (ab8898), anti-H3K36me3 (ab9050), anti-PI3K class 3 (ab124905), and anti-TFEB (ab2636) antibodies were purchased from Abcam. Anti-AMPK (2532), anti-ATG12 (4180), anti-CARM1 (3379 for immunblotting, 12495 for immunoprecipitation and ChIP), anti-LC3 (2775), anti-phospho-AMPKαT172 (2535), anti-phospho-FOXO3a S413 (8174), anti-SQSTM1/p62 (5114), and anti-TFE3 (14779) antibodies were from Cell Signaling Technology. Anti-SKP2 (sc-7164), anti-CUL1 (sc-17775), anti-tubulin (sc-8035), and anti-Lamin A/C (sc-6215) were from Santa Cruz Biotechnology. Anti-Flag (F3165), anti-ULK1 (A7481) and anti-β-actin (A1978) antibodies were from Sigma, anti-HA antibody (MMS-101R) from Covance, and anti-tubulin antibody (LF-PA0146A) from Abfrontier. The following chemicals were used in this study: rapamycin (R-5000) was purchased from LC laboratories, cycloheximide (C4859), AICAR (A9978) and phenformin (P7045) from Sigma, bafilomycin A1 (11038) and ellagic acid from Cayman (10569), compound C from Calbiochem (171260), and MG132 (M-1157) from A.G. Scientific.
Cell culture and generation of shRNA knockdown cells
HEK293T, HeLa and HepG2 cells, and wild-type, Carm1 knockout, Carm1 knock-in, Ampk DKO and Foxo1/3/4f/f MEFs were cultured at 37 °C in DMEM containing 10% fetal bovine serum (FBS) and antibiotics in a humidified incubator with 5% CO2. All cell lines used in the study were regularly tested for mycoplasma contamination. For glucose starvation, cells were washed with PBS, then incubated with glucose-free DMEM supplemented with 10% dialysed FBS. Transfection was performed with Turbofect (Fermentas) or Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. To generate knockdown cells, lentiviral shRNA constructs were first transfected along with viral packaging plasmids (psPAX2 and pMD2.G) into HEK293T cells. Three days after transfection, viral supernatant was filtered through 0.45-µm filter and infected into targeting cells. Infected cells were then selected with 5 µg ml−1 puromycin. The targeting sequences of shRNAs are as follows. mCARM1-1; 5′-TCAGGGACATGTCTGCTTATT-3′, mCARM1-2; 5′-GCCTGAGCAAGTGGACATTAT-3′, mTFE3-1; 5′-GTGGATTACATCCGCAAATTA-3′, mTFE3-2; 5′-TGTGGATTACATCCGCAAATT-3′, mTFEB-1; 5′-GCAGGCTGTCATGCATTATAT-3′, mTFEB-2; 5′-CCAAGAAGGATCTGGACTTAA-3′, mSKP2; 5′-GCAAGACTTCTGAACTGCTAT-3′, hCUL1-1; 5′-GATTTGATGGATGAGAGTGTA-3′, hCUL1-2; 5′- CCCGCAGCAAATAGTTCATGT-3′, hSKP2-1; 5′-TTCCGCTGCCCACGATCATTT-3′, hSKP2-2; 5′-AGTCGGTGCTATGATATAATA-3′.
Animal studies
All animal studies and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of Seoul National University. Eight-to-ten-week-old male wild-type C57BL/6J mice were injected with vehicle (PEG400) or ellagic acid (10 mg kg−1 day−1) intraperitoneally for four consecutive days. Mice were then fed ad libidum or fasted for 24 h. Liver tissues were collected after mice were euthanized. Sample sizes were at least n = 3 to allow for statistical analysis.
Whole-cell lysate preparation and subcellular fractionation
All cells were briefly rinsed with ice-cold PBS before collection. For whole-cell lysates, the cells were resuspended in RIPA buffer (150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl (pH 7.5), and 2 mM EDTA (pH 8.0)) supplemented with protease inhibitors and sonicated using a Branson Sonifier 450 at output 3 and a duty cycle of 30 for five pulses. For cytosolic and nuclear fractions, cells were lysed in harvest buffer (10 mM HEPES (pH 7.9), 50 mM NaCl, 0.5 M sucrose, 0.1 mM EDTA, 0.5% Triton X-100 and freshly added DTT, PMSF and protease inhibitors), incubated on ice for 5 min and spun at 120g for 10 min at 4 °C. The supernatant (cytosolic fraction) was removed to a separate tube. The nuclear pellet was rinsed twice with 500 µl of buffer A (10 mM HEPES (pH 7.9), 10 mM KCl, 0.1 mM EDTA, and 0.1 mM EGTA) and spun down at 120g for 10 min at 4 °C. The supernatant was discarded and the pellet (nuclear fraction) were resuspended in RIPA buffer and sonicated as for the whole-cell lysates. All lysates were quantified by the Bradford method and analysed by SDS–PAGE.
Electron microscopy
Cells were fixed in 0.1 M sodium cacodylate containing 4% glutaraldehyde, 1% paraformaldehyde for 1 h at room temperature. After washing three times with 0.1 M sodium cacodylate, cells were dehydrated through a gradient series of ethanol, 20 min each step, starting from 50% ethanol and ending with 100% ethanol. Afterwards, cells were incubated with progressively concentrated propylene oxide dissolved in ethanol then infiltrated with increasing concentration of Eponate 812 resin. Samples were baked in a 65 °C oven overnight then sectioned using an Ultra microtome. Sections were viewed with an energy filtering TEM unit (LEO- 192AB OMEGA, Carl Zeiss) at the Korean Basic Science Institute, South Korea.
Immunofluorescence
Immunocytochemistry was performed as previously described26. Cells grown on coverslips at a density of 7 × 104 cells were washed three times with PBS and then fixed with 2% paraformaldehyde in PBS for 10 min at room temperature. Fixed cells were permeabilized with 0.1% Triton X-100 in PBS (PBS-T) for 10 min at room temperature. Blocking was performed with 3% bovine serum in PBS-T for 30 min. For staining, cells were incubated with antibodies for 2 h at room temperature, followed by incubation with fluorescent labelled secondary antibodies for 1 h (Invitrogen). Cells were mounted and visualized under a confocal microscope (Zeiss, LSM700). For autophagy studies, MEFs were transfected with GFP–LC3 and sub-cultured onto coverslips. The following day, cells were incubated with either complete media or glucose starvation media for 18 h. Cells were treated with rapamycin or ellagic acid for 18 h. For BiFC experiments, pHA-CARM1-VC155 and pFlag-TFEB-VN173 constructs were used.
Ubiquitination assay
Ubiquitination assay was performed as previously described27. Cells were transfected with combinations of plasmids including HisMax-tagged ubiquitin. After incubation for 48 h, cells were treated with 5 µg ml−1 of MG132 for 4 h, lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-HCl (pH 8.0), 5 mM imidazole, and 10 mM β-mercaptoethanol), and incubated with Ni2+-NTA beads (QIAGEN) for 4 h at room temperature. The beads were sequentially washed with buffer A, buffer B (8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris-HCl (pH 8.0), and 10 mM β- mercaptoethanol), and buffer C (8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris-HCl (pH 6.3), and 10 mM β-mercaptoethanol). Bound proteins were eluted with buffer D (200 mM imidazole, 0.15 M Tris-HCl (pH 6.7), 30% glycerol, 0.72 M β- mercaptoethanol, and 5% SDS), and subject to immunoblot analysis. Ubiquitination site prediction software was used for CARM1 ubiquitination site prediction28.
Bacterial expression and GST pull-down assay
Glutathione S-transferase (GST)- tagged constructs were transformed in Rosetta Escherichia coli and purified with glutathione beads (GE Healthcare). 35S-methionine-labelled TFEB deletions or CARM1 deletions were generated using TNT Quick Coupled Transcription/ Translation system (Promega) according to the manufacturer’s guidance. Purified proteins and in vitro translated proteins were diluted in binding buffer (125 mM NaCl, 20 mM Tris (pH 7.5), 10% glycerol, 0.1% NP-40, 0.5 mM DTT supplemented with protease inhibitors) for GST pull-down experiment. Samples were then washed four times with dilution buffer and boiled with SDS sample buffer for immunoblotting analysis.
In vitro kinase assay
GST–SKP2 and beclin (1–148 amino acids) were purified using glutathione bead and eluted in elution buffer (50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM l-glutathione reduced (Sigma)). HA–AMPKα1 constitutively active (CA) was co-transfected in HEK293T cells with Flag–AMPKβ and HA–AMPKγ , and the complex was immunoprecipitated using Flag-M2 beads (Sigma) and eluted through 3×-Flag peptide in elution buffer (0.1 mg ml−1 in TBS). Then 1 µg of each substrate was reacted with AMPK complexes in kinase reaction buffer containing 20 mM HEPES (pH 7.4), 5 mM MgCl2, 1 mM EGTA, 0.4 mM EDTA and 0.05 mM DTT, as previously described29. Reactions were incubated with 150 µM AMP and 2 µCi of radiolabelled [γ-32P]ATP at 30 °C for 15 min. The reactions were terminated by adding SDS sampling buffer, and phosphorylation was detected by SDS–PAGE and autoradiography.
Construction of reporter plasmids and luciferase assays
The Skp2 promoter region (from 1 kb upstream of transcription start site to 200 bp downstream) and 2× CLEAR (GTCACGTGACCCCAGGGTCACGTGAC) sequence (underlined bases denote the known sequence of the CLEAR element) were cloned into pGL2-luciferase reporter vector (Promega). FOXO response element (FRE) mutant at the Skp2 promoter was constructed by site-directed mutagenesis. MEFs were transiently transfected with luciferase reporter plasmids and luciferase activity was measured 36 h after transfection and normalized by β-galactosidase expression.
qRT–PCR
Total RNAs were extracted using Trizol (Invitrogen) and reverse transcription was performed from 2.5 µg total RNAs using the M-MLV cDNA Synthesis kit (Enzynomics). The abundance of mRNA was detected by an ABI prism 7500 system or BioRad CFX384 with SYBR TOPreal qPCR 2× PreMix (Enzynomics). The quantity of mRNA was calculated using the ΔΔCt method and Hprt, Gapdh and Actb were used as controls. mRNA levels from mouse liver tissues were normalized by the 36B4 (also known as Rplp0) gene. All reactions were performed as triplicates.
The following mouse primers were used in this study. Actb; forward (fwd) 5′-TAGCCATCCAGGCTGTGCTG-3′, reverse (rev) 5′-CAGGATCTTCATGAGGTAGTC-3′; Gapdh; fwd 5′-CATGGCCTTCCGTGTTCCTA-3′, rev 5′-CCTGCTTCACCACCTTCTTG A-3′; Hprt; fwd 5′-GCTGGTGAAAAGGACCTCTCG-3′, rev 5′-CCACAGGACTAGAACACCTGC-3′; 36B4; fwd 5′-CAACCCAGCTCTGGAGAAAC-3′, rev 5′-CCAACAGCATATCCCGAATC-3′; Ulk1; fwd 5′-GCTCCGGTGACTTACAAAGCTG-3′, rev 5′-GCTGACTCCAAGCCAAAGCA-3′; Map1lc3b; fwd 5′-CACTGCTCTGTCTTGTGTAGGTTG-3′, rev 5′-TCGTTGTGCCTTTATTAGTGCATC-3′; Atg12; fwd 5′-TCCGTGCCATCACATACACA-3′ , rev 5′-TAAGACTGCTGTGGGGCTGA-3′; Atg13; fwd 5′-CCAGGCTCGACTTGGAGAAAA-3, rev 5′-AGATTTCCAC ACACATAGATCGC-3′; Atg14; fwd 5′-AGCGGTGATTTCGTCTATTTCG-3′, rev 5′-GCTGTTCAATCCTCATCTTGCAT-3′; Sirt1; fwd 5′-GATACCTTGGAGCAGGTTGC-3′ , rev 5′-CTCCACGAACAGCTTCACAA-3′; Sqstm1; fwd 5′- ATGTGGAACATGGAGGGAAGA-3′ , rev 5′-GGAGTTCACCTGTAGATGGGT-3′; Vps11; fwd 5′-AAAAGAGAGACGGTGGCAATC-3′ , rev 5′-AGCCCAGTAACGGGATAGTTG-3′; Atp6v1c1; fwd 5′-ACTGAGTTCTGGCTCATATCTGC-3′ , rev 5′-TGGAAGAGACGGCAAGATTATTG-3′; Hexb; fwd 5′-CTGGTGTCGCTAGTGTCGC-3′ , rev 5′-CAGGGCCATGATGTCTCTTGT-3′; Neu1; fwd 5′-GGACCGCTGAGCTATTGGG-3′, rev 5′-CGGGATGCGGAAAGTGTCTA-3′; Mcoln1; fwd 5′-CTGACCCCCAATCCTGGGTAT-3′ , rev 5′-GGCCCGGAACTTGTCACAT-3′; Ctns; fwd 5′-ATGAGGAGGAATTGGCTGCTT-3′ , rev 5′-ACGTTGGTTGAACTGCCATTTT-3′; Hspa5; fwd 5′-ACTTGGGGACCACCTATTCCT-3′ , rev 5′-ATCGCCAATCAGACGCTCC-3′; Skp2; fwd 5′-CCTCCAAGGAAACGAGTCAAG-3′ , rev 5′-CAGGAGACACCTGGAAAGTTC-3′ , Tfeb; fwd 5′-AAGGTTCGGGAGTATCTGTCTG-3′ , rev 5′-GGGTTGGAGCTGATATGTAGCA-3′; Tfe3; fwd 5′-TGCGTCAGCAGCTTATGAGG-3′ , rev 5′-AG ACACGCCAATCACAGAGAT-3′ , Ampka1; fwd 5′-GTCAAAGCCGACCCAATGATA-3′ , rev 5′-CGTACACGCAAATAATAGGGGTT-3′; Ampka2; fwd 5′-CAGGCCATAAAGTGGCAGTTA-3′ , rev 5′-AAAAGTCTGTCGGAGTGCTGA-3′.
The following human primers were used in this study. ACTB; fwd 5′-ATTGCCGACAGGATGCAGAA-3′, rev 5′-ACATCTGCTGGAAGGTGGACAG-3′; GAPDH; fwd 5′-CGACCACTTTGTCAAGCTCA-3′, rev 5′-AGGGGAGATTCAGTGTGGTG-3′; HPRT; fwd 5′-TGACACTGGCAAAACAATGCA-3′, rev 5′-GGTCCTTTTCACCAGCAAGCT-3′; SKP2; fwd 5′-ATGCCCCAATCTTGTCCATCT-3’, rev 5′-CACCGACTGAGTGATAGGTGT-3′; AMPKA1; fwd 5′-TTTGCGTGTACGAAGGAAGAAT-3′, rev 5′-CTCTGTGGAGTAGCAGTCCCT-3′; AMPKA2; fwd 5′-CTGTAAGCATGGACGGGTTGA-3′, rev 5′-AAATCGGCTATCTTGGCAT TCA-3′.
RNA-seq and ChIP–seq analyses
The TruSeq method was used to generate RNA-seq libraries. ChIP–seq libraries were prepared using the NEXTflex ChIP–seq kit (Bioo Scientific), according to the manufacturer’s instructions. RNA-seq libraries were pair-end sequenced and ChIP–seq libraries were single-end sequenced on an Illumina Hi-seq 2500 (NICEM, Seoul National University). All the RNA-seq data were mapped using Tophat package30 against the mouse genome (mm9). Differential analysis has been done via EdgeR package31. Differentially regulated genes were identified using a false discovery rate (FDR) cut-off of 1 × 10−5 for knockout against knockout-glucose starvation (KO-GS), wild type against wild-type-glucose starvation (WT-GS), wild type against knockout, and WT-GS against KO-GS. We did hierarchical clustering analysis using the gene expression values from all conditions and replicates for previously selected differential genes. Specifically, we used Ward’s criterion for genes with 1 − (correlation coefficient) as a distance measure. Clustering heatmap was drawn using z-score that is scaled across samples for each gene. ChIP–seq data were mapped to the mouse genome using Bowtie. The tracks were generated using uniquely aligned reads. At promoters, genes were sorted based on the expression levels, indicating that H3R17me2 as well as H3K4me3 were enriched at active promoters. We used 8,398 distal (<2.5 kb from annotated TSSs) CBP and MED12 binding sites for enhancers, which were sorted based on H3K27ac levels. H3R17me2 was not detected at enhancers. The data on H3R17me2, H3K4me1, H3K4me3 and H3K27ac were obtained from MEFs under normal conditions.
ChIP, two-step ChIP assays, and qRT–PCR analyses
The ChIP and sequential two-step ChIP assays were conducted as previously described32. In brief, cells were crosslinked with 1% formaldehyde for 10 min at room temperature. After glycine quenching, the cell pellets were lysed in buffer containing 50 mM Tris-HCl (pH 8.1), 10 mM EDTA, 1% SDS, supplemented with complete protease inhibitor cocktail (Roche), and sonicated. Chromatin extracts containing DNA fragments with an average of 250 bp were then diluted ten times with dilution buffer containing 1% Triton X-100, 2 mM EDTA, 150 mM NaCl and 20 mM Tris-HCl (pH 8.1) with complete protease inhibitor cocktail, pre-cleared with protein A/G sepharose and subjected to immunoprecipitations overnight at 4 °C. Immunocomplexes were captured by incubating 45 µl of protein A/G sepharose for 2 h at 4 °C. Beads were washed with low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 150 mM NaCl), highsalt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.1), 500 mM NaCl), buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 10 mM Tris-HCl (pH 8.1), 1 mM EDTA), TE buffer (10 mM Tris-HCl (pH 8.0), 0.5M EDTA) and eluted in elution buffer (1% SDS, 0.1 M NaHCO3). The supernatant was incubated overnight at 65 °C to reverse-crosslink, and then digested with RNase A for 2 h at 37 °C and proteinase K for 2 h at 55 °C. ChIP and input DNA were then purified and analysed for qRT–PCR analysis or used for constructing sequencing libraries. For the two-step ChIP assays, components were eluted from the first immunoprecipitation reaction by incubation with 10 mM DTT at 37 °C for 30 min and diluted 1:50 in ChIP dilution buffer followed by re-immunoprecipitation with the second antibodies. Two-step ChIP assay was performed in essentially the same way as the first immunoprecipitation. qPCR was used to measure enrichment of bound DNA, and the value of enrichment was calculated relative to input and the ratio to IgG. All reactions were performed in triplicates. The following primers were used in ChIP assays. Skp2 (FRE); fwd 5′-CCTTAGGACTGGGTCTGTGG-3′, rev 5′-GCACGCTGATTTGATCTTCA-3′; Map1lc3b; fwd 5′-AGCCAGTGGGATATTGGTCT-3′, rev 5′-AGAGCCTGCGGTACCCTAC-3′; Atg14; fwd 5′-GAGACGCCATGATGATCTGA-3′, rev 5′-GCCAAGGAGTGTGGGAAGTA-3′; Atp6v1c1; fwd 5′-ACTCAGTGGCAGAAGGGAGA-3′, rev 5′-AAACACCCAGTGGAGACTGC-3′; Hexb; fwd 5′-GAATTGGGACTGTGGTCGAT-3′, rev 5′-CTAGTGTCGCTGGCCCTAGT-3′; Hspa5; fwd 5′-ATTGGTGGCCGTTAAGAATG-3′, rev 5′-TGAAGTCGCTACTCGTTGGA-3′; Ctns; fwd 5′-CCTCTGGTAGCGTAGGT-3′, rev 5′-GCTTTTGGTGAGGTCTGTCC-3′; Vps11; fwd 5′-GGGCCGATCTTAACCTTTGT-3′, rev 5′-AGCCCAGATGTCTTTTGTGG-3′; Neu1; fwd 5′-AGGATGACTTCAGCCTGGTG-3′, rev 5′-AGGATAGTATGGGCCGAACC-3′; Mcoln1; fwd 5′-TACTGGAAGATGGGCTTTCG-3′, rev 5′-TGCCCAGATTCTAGGAGGAA-3′.
Statistical analysis
All experiments were performed independently at least three times. For GFP–LC3 puncta counting, five random confocal images were chosen and the number of cells with GFP-positive dots was counted. An average of 80 cells was examined for each group and P values were calculated using one-tailed t-tests. For animal studies, sample size for experiments were determined empirically based on previous studies to ensure appropriate statistical power. Mice in the study were randomly chosen for ellagic acid treatment and fasting. No animals were excluded from statistical analysis, and the investigators were not blinded in the studies. Values are expressed as mean ± s.e.m. Significance was analysed using two-tailed, unpaired t-test. P < 0.05 was considered statistically significant.
Extended Data
Extended Data Figure 1. Increased H3R17me2 by CARM1 in amino acid starvation-induced autophagy.
a, b, Immunoblot analysis of various histone marks in response to amino acid (AA) starvation or rapamycin (100 nM). c, Immunoblot analysis of CARM1 and LC3 conversion (LC3-II). d, Amino acid-starved wild-type, Carm1 knockout or knock-in MEFs were analysed by immunoblot. e, Representative confocal images of GFP–LC3 puncta formation. GFP–LC3 (green); DAPI (blue). Scale bar, 20 µm. The graph shows quantification of LC3-positive punctate cells (right). Bars, mean ± s.e.m.; n = 5, with over 100 cells. **P < 0.01 (one-tailed t-test).
Extended Data Figure 2. Loss of CARM1 and inhibition of H3R17me2 impair autophagy.
a, LC3 flux was analysed in MEFs infected with nonspecific shRNA (shNS) or CARM1 shRNAs (shCARM1-1 and-2). Bafilomycin A1 (BafA1; 200 nM, 2 h). The LC3-II/LC3-I ratio is indicated. b, LC3 flux was analysed in wild-type and Carm1 knockout MEFs in the absence or presence of Bafilomycin A1. The LC3-II/LC3-I ratio is indicated. c, mCherry-GFP–LC3 was transfected in wild-type and Carm1 knockout MEFs and the formation of autophagosome (mCherry-positive; GFP-positive) and autolysosome (mCherry-positive; GFP-negative) was examined. Scale bar, 20 µm. d, Immunoblot analysis in MEFs. e, Representative confocal images of GFP–LC3 puncta formation. Scale bar, 10 µm. Bars, mean ± s.e.m.; n = 5, over 150 cells. *P < 0.05 (one-tailed t-test). f, Immunoblot analysis in MEFs.
Extended Data Figure 3. CARM1 is degraded by SKP2-containing SCF E3 ligase in the nucleus.
a, Wild-type CARM1 and ubiquitination-defective mutant K471R were analysed for their expression in MEFs after MG132 treatment. b, Interaction between CARM1 and CUL proteins was analysed. c, Lysates were analysed by immunoblot. d, Left, HepG2 cells infected with two different SKP2 shRNAs were subject to cycloheximide (CHX) experiment. Right, protein half-life of CARM1 was quantitatively defined (right). e, Left, CHX experiment in HepG2 expressing wild-type SKP2 or ΔF mutant. Right, protein half-life of CARM1 was quantitatively defined. Data are mean ± s.e.m.; n = 3. **P < 0.01 (one-tailed t-test) (d, e).
Extended Data Figure 4. CARM1 is degraded by CUL1-containing SCF E3 ligase in the nucleus under nutrient-rich condition.
a, HepG2 cells transfected with Flag–CUL1 were deprived of glucose for 18 h and treated with MG132 before collecting. Interaction between CARM1 and CUL1 was analysed. b, c, In vivo ubiquitination assay of CARM1 after knockdown of CUL1 (b) or overexpression of wild-type or K720R mutant (MT) CUL1 (c). d, e, Left, HepG2 cells infected with two different CUL1 shRNAs (d) or overexpressing wild-type or mutant CUL1 (e) were subject to cycloheximide treatment. Right, protein half-life of CARM1 was quantitatively defined. Data are mean ± s.e.m.; n = 3. *P < 0.05, **P < 0.01 (one-tailed t-test) (d, e).
Extended Data Figure 5. AMPKα2 accumulates in the nucleus leading to repression of SKP2 and stabilization of CARM1 under nutrient-starved conditions.
a, b, qRT–PCR of Ampka1 and Ampka2 in MEFs (a) and HepG2 cells (b) upon glucose starvation. c, The nuclear AMPKα2 expression level was analysed in the absence or presence of MG132. d, Binding between CARM1 and AMPK was assessed. e, In vitro kinase assay with AMPK. f, MEFs were treated with AICAR (1 mM) or phenformin (2 mM) for 4 h. The nuclear fraction was analysed by immunoblot. g, MEFs were deprived of glucose in the absence or presence of 10 µM compound C and the nuclear fraction was analysed by immunoblot. h, Left, cycloheximide treatment in wild-type and Ampk DKO MEFs. Right, protein half-life of CARM1 was quantitatively defined. i, j, Ampk DKO MEF lysates were analysed by immunoblot. k, CARM1– CUL1 interaction was analysed after SKP2 knockdown in wild-type and Ampk DKO MEFs. l, SKP2 expression levels were analysed in the absence or presence of MG132. m, Foxo1/3/4f/f MEFs infected with Cre virus were analysed for Skp2 mRNA. n, SKP2 and phosphorylated FOXO3a were analysed by immunoblot. o, ChIP assay of the Skp2 promoter. Data are mean ± s.e.m.; n = 3. *P < 0.05, **P < 0.01 (one-tailed t-test) (a, b, h, m, o). p, Representative confocal images. Scale bar, 20 µm.
Extended Data Figure 6. Identification of CARM1 target genes by RNA-seq and ChIP–seq analyses.
a, Flow chart showing the strategy of RNA-seq analysis. b, Hierarchical clustering results applied to 4,998 differentially expressed genes (DEGs). c, Autophagy-related and lysosomal genes significantly observed in cluster 1. Hyper-geometric P values were calculated. d, Genes from cluster 1 were analysed for transcription factor (TF) motif enrichment at their promoter region (− 500–100). Hypergeometric P values were calculated. e, qRT–PCR analysis of CARM1-dependent autophagy-related and lysosomal genes. Data are mean ± s.e.m.; n = 3. *P < 0.05, **P < 0.01 (one-tailed t-test). f, Enrichment of H3R17me2 at promoters (left) and enhancers (right). The data on H3R17me2, H3K4me1, H3K4me3 and H3K27ac were obtained from MEFs under normal condition. g, Increase in H3R17me2 at promoters of genes from cluster 1 after glucose starvation. h, Increased H3R17me2 levels in response to 18 h of glucose starvation at the autophagy-related gene Map1lc3b. The direction of transcription is indicated by the arrow and the beginning of the arrow indicates the TSS.
Extended Data Figure 7. Binding mapping of CARM1 and TFEB and their target gene regulation in glucose starvation.
a, Bimolecular fluorescence complementation (BiFC) analysis of the CARM1–TFEB interaction. Scale bar, 20 µm. b, Interaction between CARM1 and TFEB was analysed in wild-type and Ampk DKO MEFs after glucose starvation. c, d, In vitro GST pull-down assays for domain mapping of CARM1–TFEB interaction. BHLH, basic helix–loop–helix; LZ: leucine zipper. MD, methyltransferase domain; TA, transcription activation domain. e, Endogenous co-immunoprecipitation from nuclear fraction of wild-type MEFs. f, g, qRT–PCR analysis in MEFs after knockdown of TFEB or TFE3. h, i, qRT–PCR analysis showing mRNA levels of TFEB-dependent and CARM1-dependent genes after knockdown of TFEB (h) or CARM1 (i). Bars, mean ± s.e.m.; n = 3. *P < 0.05, **P < 0.01 (one-tailed t-test) (f–i).
Extended Data Figure 8. CARM1 functions as a co-activator of TFEB.
a, ChIP assays on TFEB-dependent, CARM1-dependent promoters after knockdown of CARM1. b, ChIP assays of the Hspa5 promoter, a TFEB-dependent, CARM1-independent target promoter. c, MEFs were analysed with indicated antibodies. d, Two-step ChIP assays were performed on promoters of TFEB-dependent, CARM1-dependent target genes or TFEB-dependent, CARM1-independent target genes in MEFs after 18 h of glucose starvation. The chromatin fractions were first subject to pull-down with anti-TFEB antibody, eluted from immunocomplexes and applied for the second pull-down with control IgG or anti-CARM1 antibody. Bars, mean ± s.e.m.; n = 3 (a, b, d). e, Representative confocal images. Scale bar, 10 µm.
Extended Data Figure 9. A subset of autophagy-related and lysosomal genes regulated by TFEB requires CARM1.
a, qRT–PCR analysis showing mRNA levels of TFEB-dependent and CARM1-dependent autophagy-related and lysosomal genes in wild-type and Ampk DKO MEFs in response to glucose starvation. b, ChIP assays on TFEB-dependent, CARM1-dependent target genes in wild-type and Ampk DKO MEFs. c, qRT–PCR analysis of CARM1-dependent genes after knockdown of SKP2 in Ampk DKO MEFs. d, qRT–PCR analysis was performed in MEFs deprived of glucose in the absence or presence of H3R17me2- specific inhibitor, ellagic acid. e, f, ChIP assays on TFEB-dependent, CARM1-dependent promoters. Hspa5 promoter was also analysed as a CARM1-independent promoter. Bars, mean ± s.e.m.; n = 3. *P < 0.05, **P < 0.01, ***P < 0.001 (one-tailed t-test) (a–f).
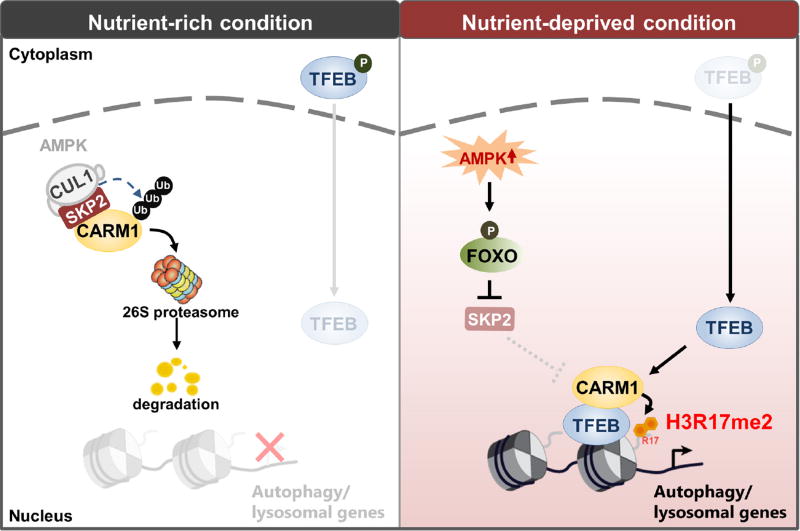
Extended Data Figure 10. Graphical summary of the AMPK–SKP2–CARM1 signalling cascade.
Proposed model depicting the AMPK–SKP2– CARM1 signalling axis in the transcriptional and epigenetic regulation of autophagy. The SKP2-containing SCF E3 ubiquitin ligase complex degrades CARM1 under nutrient-rich conditions, but in nutrient-deprived conditions, AMPK-dependent phosphorylation of FOXO3a downregulates SKP2 and stabilizes CARM1, which in turn functions as a co-activator of TFEB in regulation of autophagy.
Supplementary Material
Acknowledgments
We thank members of the Chromatin Dynamics Research Center for technical assistance and discussions and J. Kim and J. Chung for valuable reagents and discussions. We thank Y. S. Yu for illustrations. The TEM data were analysed in the Korean Basic Science Institute. Carm1 knockout and knock-in MEFs were provided by M. T. Bedford. Ampk DKO MEFs was a gift from B. Viollet, and Foxo1.3.4 f/f MEFs were a gift from R. DePinho and J.-H. Paik.
This work was supported by Creative Research Initiatives Program (Research Center for Chromatin Dynamics, 2009-0081563) to S.H.B.; the National Junior Research Fellowship (NRF-2011-A01496-0001806) to H.-J.R.S.; the Basic Science Research Program (NRF-2014R1A6A3A0405 7910) to H.K. from the National Research Foundation (NRF) grant funded by the South Korean government (MSIP); NIH grant (R01DK106027) to K.-J.W.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Supplementary Information is available in the online version of the paper.
Author Contributions H.-J.R.S., H.K., S.O., J.-G.L. and M.K. performed the cell biology and biochemistry experiments; H.-J.K. and M.-N.K. provided TEM analysis and critical comments; H.-J.R.S. and K.J.W. performed RNA and ChIP–seq preparation and systemic analysis; H.-J.R.S., H.K., K.J.W. and S.H.B. organized and analysed the data; H.-J.R.S., K.J.W. and S.H.B. wrote the manuscript.
Author Information The RNA-seq and H3R17me2 ChIP–seq data sets have been deposited in the NCBI Gene Expression Omnibus (GEO) database underthe accession number GSE72901.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N. Engl. J. Med. 2013;368:651–662. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 5.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 6.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjørkøy G, et al. Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol. 2009;452:181–197. doi: 10.1016/S0076-6879(08)03612-4. [DOI] [PubMed] [Google Scholar]
- 10.Selvi BR, et al. Identification of a novel inhibitor of coactivator-associated arginine methyltransferase 1 (CARM1)-mediated methylation of histone H3 Arg-17. J. Biol. Chem. 2010;285:7143–7152. doi: 10.1074/jbc.M109.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- 12.Hardie DG. AMPK and autophagy get connected. EMBO J. 2011;30:634–635. doi: 10.1038/emboj.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat. Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoki K, Kim J, Guan KL. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 2012;52:381–400. doi: 10.1146/annurev-pharmtox-010611-134537. [DOI] [PubMed] [Google Scholar]
- 15.Salt I, et al. AMP-activated protein kinase: greater AMP dependence, and preferential nuclear localization, of complexes containing the α2 isoform. Biochem. J. 1998;334:177–187. doi: 10.1042/bj3340177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat. Rev. Mol. Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 17.Greer EL, et al. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J. Biol. Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- 18.Potente M, et al. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J. Clin. Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Li P-F. Foxo3a regulates apoptosis by negatively targeting miR-21. J. Biol. Chem. 2010;285:16958–16966. doi: 10.1074/jbc.M109.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y-C, et al. DNMT3B overexpression by deregulation of FOXO3a-mediated transcription repression and MDM2 overexpression in lung cancer. J. Thorac. Oncol. 2014;9:1305–1315. doi: 10.1097/JTO.0000000000000240. [DOI] [PubMed] [Google Scholar]
- 21.Lam EW-F, Brosens JJ, Gomes AR, Koo C-Y. Forkhead box proteins: tuning forks for transcriptional harmony. Nat. Rev. Cancer. 2013;13:482–495. doi: 10.1038/nrc3539. [DOI] [PubMed] [Google Scholar]
- 22.Tsai K-L, et al. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. 2007;35:6984–6994. doi: 10.1093/nar/gkm703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sardiello M, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 24.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Settembre C, Medina DL. TFEB and the CLEAR network. Methods Cell Biol. 2015;126:45–62. doi: 10.1016/bs.mcb.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Kim IS, et al. Roles of Mis18α in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol. Cell. 2012;46:260–273. doi: 10.1016/j.molcel.2012.03.021. [DOI] [PubMed] [Google Scholar]
- 27.Kim H, et al. DNA damage-induced RORα is crucial for p53 stabilization and increased apoptosis. Mol. Cell. 2011;44:797–810. doi: 10.1016/j.molcel.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Zhou Y, Song J, Zhang Z. hCKSAAP_UbSite: improved prediction of human ubiquitination sites by exploiting amino acid pattern and properties. Biochim. Biophys. Acta. 2013;1834:1461–1467. doi: 10.1016/j.bbapap.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boo K, et al. Pontin functions as an essential coactivator for Oct4-dependent lincRNA expression in mouse embryonic stem cells. Nat. Commun. 2015;6:6810. doi: 10.1038/ncomms7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.