Abstract
The balance between myeloid and lymphoid populations must be well controlled. Here, we report that a protein, osteopontin (OPN), skews the balance between myeloid and lymphoid populations during pathogenic conditions, such as infection and autoimmunity. Importantly, two OPN isoforms exert distinct effects in shifting the balance through cell type-specific regulation of apoptosis. Intracellular OPN (iOPN) reduces population sizes of myeloid progenitors and myeloid cells, and secreted OPN (sOPN) increases population sizes of lymphoid cells. The total impact of OPN in skewing leukocyte population balance was observed as host sensitivity to early systemic Candida infection and T cell-mediated colitis. This study suggests novel detrimental roles of two OPN isoforms causing the imbalance of leukocyte populations.
Skewing a population balance between myeloid and lymphoid cells impacts immune responses. Thus, the hematopoietic process is tightly controlled. In contrast to steady-state hematopoiesis, physiological insults that require an acute supply of leukocytes temporarily alter patterns of hematopoiesis. Such demand-adapted hematopoiesis is observed during severe infections, inflammation, and irradiation, and myelopoiesis becomes highly active to compensate the loss of myeloid cells1, 2, 3, 4. This response is called ‘emergency myelopoiesis’ (or ‘emergency granulopoiesis’ especially for the acute generation of neutrophils). Emergency granulopoiesis is triggered by stimulating pattern-recognition receptors (PRRs), reactive oxygen species, and cytokines, such as IL-6, GM-CSF, G-CSF, and others1, 2, 3, 4, 5, 6, 7, 8, 9, 10. Decreased cell density by depleting neutrophils can also promote granulopoiesis in the bone marrow (BM)10. Lymphocytes have distinct mechanisms from myeloid cells to regulate their population sizes, and a normal immune system keeps an optimal balance between myeloid and T cells.
OPN is a phosphoglycoprotein expressed in various tissues and cell types. OPN controls various immune responses and is involved in the pathogenesis of a wide variety of diseases11, 12, 13, 14, 15, 16, 17. OPN is expressed by BM stroma cells18 and negatively regulates stem cell pool size and function of Lin−Sca-1+c-kit+ (LSK) cells, including hematopoietic stem cells (HSCs)19, 20, 21. However, the impact of OPN on myeloid or lymphoid progenitors has not been explored.
OPN exists as two translational isoforms, secreted OPN (sOPN) and intracellular OPN (iOPN). They have distinct functions due to their localization22. The majority of OPN studies have focused on sOPN, which interacts with receptors such as integrins and CD44. In contrast, iOPN was later found as a product of alternative translation23 and resides in the cytoplasm and occasionally in the nucleus. iOPN functions as an adaptor or scaffold protein in signal transduction pathways, as well as stabilizing other intracellular proteins11, 13, 14, 24, 25. Although sOPN in the hematopoietic stem cell niche in the BM is a negative regulator of HSC proliferation19, 20, the role of iOPN in hematopoiesis is entirely unknown.
In this study, we report that OPN skews the balance of cell populations towards a decrease of myeloid and an increase of lymphoid populations. However, this happens only during demand-adapted myelopoiesis (elicited by such as irradiation and systemic fungal infection) and lymphoid cell expansion in lymphopenic recipients. We found that iOPN is responsible for the negative regulation of myelopoiesis. In contrast, sOPN enhances lymphoid cell expansion. Thus, two different OPN isoforms play distinct roles but, as a total, work together to decrease myeloid progenitors and increase lymphoid cells during demand-adapted myelopoiesis and lymphoid cell expansion in lymphopenic hosts.
RESULTS
Cell population balance in irradiation BM chimeric mice
In naïve mice, OPN-deficiency does not affect numbers of total splenocytes, total BM cells, lineage negative (Lin−) progenitors, differentiated leukocytes in the BM19, 26, as well as compositions of BM progenitor and differentiated leukocyte populations (Supplementary Fig. 1a–e). No impact of OPN was also identified in proportions of embryonic leukocyte and their progenitor populations in fetal livers among littermate embryos (E13–15) from Spp1 (gene encoding OPN) heterozygous breeders (Supplementary Fig. 1f, g).
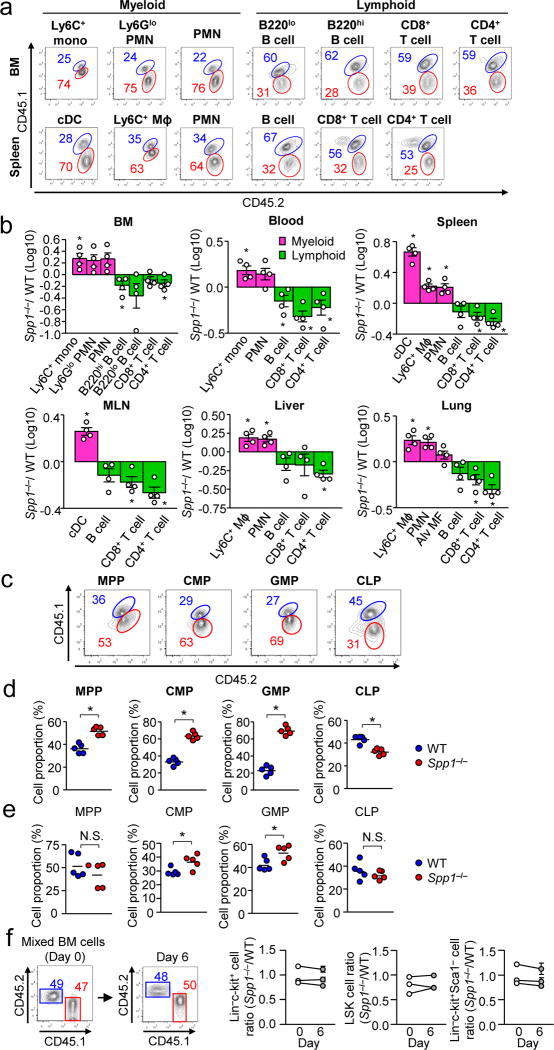
Next, we examined whether OPN affects the cell population balance in mixed BM radiation chimeras transferred with WT and Spp1−/− BM cells (Supplementary Fig. 2a, b). Serum OPN (i.e., secreted OPN; sOPN) in the chimera were comparable to those in naive WT mice (Supplementary Fig. 2c). Seven weeks after BM cell transplant, Spp1−/− donor cells showed increased myeloid cell populations and decreased lymphoid cell populations in multiple organs including BM, spleen, blood, mesenteric lymph nodes (MLNs), liver, and lungs (Fig. 1a, b). Spp1−/− donor cells had larger populations in multipotent progenitors (MPPs), common myeloid progenitors (CMPs), and granulocyte-macrophage progenitors (GMPs), but slightly a smaller common lymphoid progenitor (CLPs) cell populations, compared to WT donor cells (Fig. 1c, d). To confirm the BM cell transfer results, we also used mixed LSK (Lin−Sca-1+c-kit+) cells for transfer (Supplementary Fig. 2d, e), and again Spp1−/−CMP and GMP populations were larger than those of WT (Fig. 1e). Furthermore, we ruled out differential homing of transferred Spp1−/− cells to BM, as shown by the unaltered donor cell ratio (1:1 of WT and Spp1−/−) in HSC and progenitor populations (Lin−c-kit+), the LSK population (Lin−c-kit+Sca-1+, including HSCs and MPPs), and the Lin−c-kit+Sca-1− population (including megakaryocyte-erythroid progenitors, GMPs, and CMPs) on Day 6 in BM (Fig. 1f). Taken together, in irradiated recipients, cell-intrinsic OPN expression in hematopoietic progenitors is sufficient to reduce myeloid progenitor populations.
Figure 1. Analysis of cell population in irradiation mix BM chimera mice generated with WT vs. Spp1−/− donor cells.
WT (CD45.1/CD45.2) and Spp1−/− mice (CD45.2) to irradiated WT recipients (CD45.1). (a) Flow cytometry to detect indicated cell populations of WT (labeled with blue) and Spp1−/− (labeled with red) donor origins in the BM and spleens 7-wk after BM cell transfer. (b) Ratios (Spp1−/−/WT) of donor cell numbers in the indicated organs. Mɸ: macrophages. Datasets are representative of two independent experiments. (c, d) Analyzing hematopoietic progenitors in the BM of the mixed BM-chimera mice at 3 weeks after BM transplantation. Representative flow plots (c) and frequencies of WT and Spp1−/− cells in indicated progenitors (d). (e) Instead of total BM cells, LSK cells from WT (CD45.1) and Spp1−/− (CD45.2) were transferred to WT recipients (CD45.1/CD45.2). Ratios of WT and Spp1−/− progenitor cells were assessed by flow cytometry at 3 weeks after transfer. Each dataset is a representative of two independent repeats (5 mice per group)(c–e). (f) Equal numbers of BM cells from naive WT (CD45.2) and Spp1−/− (CD45.1) mice were transferred to irradiated WT recipients (CD45.1/CD45.2). Six days after BM cell transfer, the ratio of WT and Spp1−/− in Lin−c-kit+, LSK (Lin−Sca-1+c-kit+), and Lin−Sca-1+c-kit− cells in the BM was examined. n=3 per each circle on day 6. Data were from three independent experiments. Error bars indicate ± SEM. * p<0.05 (Student’s t-Test).
OPN limits emergency granulopoiesis
Peritoneal thioglycollate injection decreased the neutrophil population in the BM within 12 hr with recovery by 70 hrs (Supplementary Fig. 3a). At 24 hrs after injection, the CMP and GMP populations were significantly increased and proliferated in BM, while such behaviors were not observed in CLPs (Supplementary Fig. 3b, c). These observations reflect emergency granulopoiesis after thioglycollate injection.
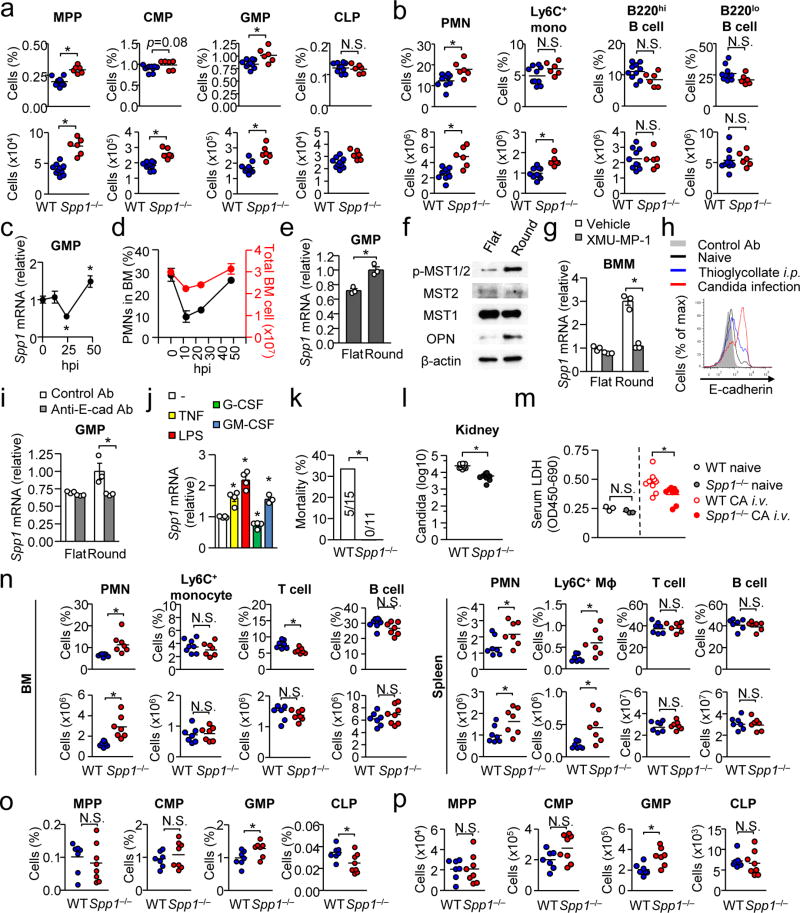
In the setting of thioglycollate injection, Spp1−/− mice again showed increased GMPs and neutrophils, compared to WT mice, in BM 24 hrs after injection (Fig. 2a, b). Here, WT and Spp1−/− mice showed similar kinetics in the decrease of BM neutrophils and numbers of neutrophils recruited in the peritoneal cavity in the first 12 hrs (Supplementary Fig. 3d, e), suggesting that OPN may not affect the egress and migration of BM neutrophils. However, enhanced GMP cellularity in the Spp1−/− BM and apparently reduced neutrophils in the BM suggested OPN downregulates granulopoiesis, while the impact of OPN on CLPs and B cells was minimal in the BM (Fig. 2a, b). Thus, OPN selectively regulates granulopoiesis by reducing the GMP population.
Figure 2. OPN inhibits emergency myelopoiesis.
(a, b) Proportions and numbers of indicated progenitors (a) and differentiated cell subsets (b) in the BM 24 hrs after thioglycollate i.p. injection. Data were pooled from two independent experiments with 3–5 mice per experiment. (c) Spp1 (OPN) mRNA levels in GMPs from BM of WT mice at indicated time points. hpi: hrs post injection. n=4 per group. (d) Total cell numbers and frequency of neutrophils in the BM after thioglycollate i.p. injection. n=4 per group. (e) Spp1 mRNA levels in GMPs at 16 hrs after starting cell culture in flat- vs. round-bottom plates. (f) Western blotting of lysates of BMMs cultured in flat- or round-bottom wells for 12 hr. (g) Reduction of Spp1 expression by inhibiting MST1/2. BMMs were cultured on flat- or round-bottom plates (105 per well) with or without an Mst1/2 inhibitor (XMU-MP-1; 5 µg/ml). (h) E-cadherin expression levels on GMPs, which were harvested at 0 or 24 hr from WT mice after thioglycollate i.p. injection and Candida infection. (i) Reduction of Spp1 mRNA by blockade of E-cadherin. Similar setting to (g) but GMPs and anti-E-cadherin antibody (10 µg/ml) were used. (j) Spp1 mRNA levels in GMPs stimulated with TNF-α, LPS, G-CSF and GM-CSF (10 ng/ml for all) for 6 hrs in flat- or round-bottom wells. Datasets are representative of two (d–j) or three (c) independent experiments. (k) Mortality of mice i.v.-injected with 2×106 Candida albicans spores at 24 hrs after infection. Data were pooled from three independent experiments. (l) Candida CFU in the kidney 24 hrs after i.v.-injecting Candida spores (106 spores/mouse). (m, n) Comparison of serum levels of lactate dehydrogenase (LDH), indicating tissue damage (m), and cell numbers and frequencies in the BM and spleen 24 hrs after systemic Candida infection (n). (o, p) Proportions (o) and numbers (p) of BM progenitors during at 15 hrs after Candida infection. Error bars indicate ± SEM (c–e, g, i, j). * p<0.05. N.S.; not significant. Data were analyzed by the Student’s t-test (a, b, c, e, g, i, j, l–p) or by the Chi-square test (k).
Although serum OPN amounts were increased during the first 24 hrs after thioglycollate injection (Supplementary Fig. 3f), Spp1 mRNA in GMPs was temporarily reduced (Fig. 2c). Based on the enhanced outcome of myelopoiesis in Spp1−/− mice (Fig. 1), we hypothesized that the temporary decrease of OPN expression in GMPs positively impacted enhanced myelopoiesis and set out to determine what downregulated the OPN expression in the myeloid progenitor cells during emergency granulopoiesis. Here, we first evaluated the cell density because we reasoned that massive cell egress by acute inflammation reduced the cell density in the stem cell niche (Fig. 2d). To recapitulate a high cell density condition ex vivo, we plated GMPs in a round-bottom plate, in which cells are crowded at the bottom of wells. Indeed, GMPs in a round-bottom plate expressed higher levels of Spp1 mRNA than GMPs in a flat-bottom plate (Fig. 2e). For further evaluation, we used BM-derived macrophages (BMMs) because of the paucity of GMPs. With BMMs, we confirmed that Spp1 expression depends on cell density in a dosage-dependent manner (Supplementary Fig. 3g). Spp1 expression was positively correlated to cell density, while gene expression of Tnf, Il6, Il10, and Cxcl1 had a negative correlation to cell density (Supplementary Fig. 3h).
Hippo signaling induces OPN through Mst1/2 activation
To examine how cells sense cell density to regulate Spp1 expression, we first tested the involvement of soluble factors by comparing BMM culture supernatants from round-bottom or flat-bottom plates. However, no difference was found in Spp1 expression in cells cultured in the supernatants (Supplementary Fig. 3i). Then, we evaluated a signaling pathway activated by cell-cell contacts, which activates the Hippo pathway27, 28, 29. We found that BMMs cultured in round-bottom wells increased OPN protein accompanied with the activation of Mst1/2, a critical signaling molecule in the Hippo pathway30 (Fig. 2f). Inhibition of Mst1/2 abolished cell density-mediated upregulation of Spp1 expression (Fig. 2g), strongly suggesting the Hippo signaling induces OPN expression. Blockade of E-cadherin, known to trigger the Hippo pathway29, decreased Spp1 expression at least partially in BMMs plated in round-bottom wells (Supplementary Fig. 3j). Thioglycollate treatment and systemic Candida infection induced E-cadherin expression, and blockade of E-cadherin also prevented cell density-mediated Spp1 upregulation in GMPs (Fig. 2h, i). Thus, high cell density increased Spp1 expression through the Hippo pathway.
We next evaluated whether inflammatory stimuli downregulate Spp1 expression by stimulating GMPs with TNF, G-CSF, GM-CSF, or lipopolysaccharide (LPS), which are known to promote granulopoiesis, but none of them greatly decreased Spp1 expression in GMPs (Fig. 2j). Then, we evaluated whether cell egress from BM without inflammation downregulated Spp1 expression. Mice treated with a CXCR4 antagonist decreased total BM cell counts and Spp1 expression in GMPs (Supplementary Fig. 3k, l). Although factors other than cellularity in the BM may also be involved in downregulating Spp1 mRNA, these results suggest that the decrease of Spp1 mRNA in vivo requires low cell densities in the BM.
Emergency myelopoiesis during systemic Candida infection
Emergency granulopoiesis plays an important role in protecting hosts from systemic fungal infections such as Candida albicans31, 32 by quickly generating neutrophils, which are critical for fungal clearance at an early stage of the infection. Candida infection enhanced proliferation of CMPs and GMPs, and increased their population sizes, while CLPs did not respond as CMPs and GMPs did (Supplementary Fig. 3c, m). Here, we hypothesized that OPN is detrimental at an early stage of systemic Candida infection because OPN restricts emergency granulopoiesis. Indeed, WT host survival was significantly worse than Spp1−/− host survival at 24 hrs after Candida infection (Fig. 2k) with increased fungal loads in the kidney and spleen (Fig. 2l and Supplementary Fig. 3n) in addition to a sign of elevated tissue damage (Fig. 2m). Infected WT mice showed less abundant neutrophils and Ly6C+ monocytes/macrophages compared to Spp1−/− mice, while lymphoid cell populations were not altered between WT and Spp1−/− mice (Fig. 2n). WT mice also showed decreased sizes of the GMP population compared to Spp1−/− mice without affecting MPP, CMP, and CLP population sizes (Fig. 2o, p). Here, no difference was found between WT and Spp1−/− mice in major anti-fungal responses, including serum TNF, neutrophil chemoattractants (CXCL1 and CXCL2), cytokine and chemokine expression in renal neutrophils and macrophages, as well as cell-intrinsic functions such as phagocytosis and fungicidal activities in neutrophils and BMMs (data not shown). Taken together, these results suggest that OPN is detrimental in an early stage of acute candidemia due to restricted emergency granulopoiesis.
Impact of OPN on MPPs at the single-cell level
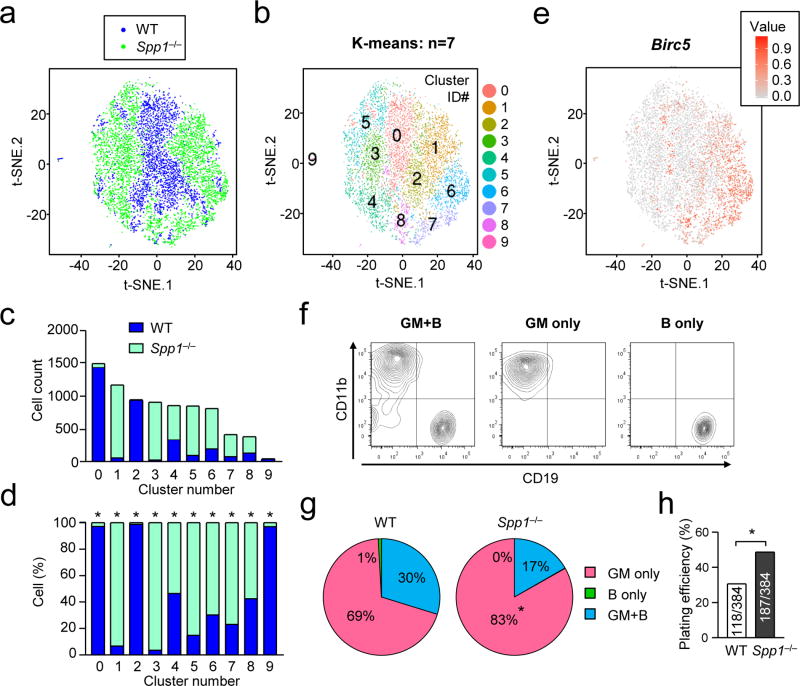
Since MPPs consists of a small and highly defined population, we analyzed gene expression patterns of MPPs at the single-cell level. Twenty hours after thioglycollate injection, MPPs were flow-sorted from the BM of WT and Spp1−/− mice and submitted for single-cell sequencing (Supplementary Table 1). Dimensionality reduction indicated Spp1−/− MPPs to have distinct t-SNE plot distribution from WT MPPs (Supplementary Fig. 4a, b). Next, the two sets of data (WT and Spp1−/−) were normalized by sequencing depth and overlaid on a t-SNE plot (Fig. 3a; Supplementary Fig. 4c), and generated ten distinct clusters (Fig. 3b; Supplementary Fig. 4d). Each cluster was dominated by either WT or Spp1−/− cells (Fig. 3c, d). Expression of Birc5, encoding survivin, was high in certain clusters, particularly ones consisting with Spp1−/− MPPs, such as clusters 1, 6, 7, and 8 (Fig. 3b, e).
Figure 3. Single cell sequencing and Ex vivo MPP differentiation assay.
(a–f) Single-cell RNA sequencing of MPPs from BMs of WT and Spp1−/− mice 20 hrs after intraperitoneal injection of thioglycollate. Numbers of sequenced MPPs of WT and Spp1−/− are 3,294 and 4,574, respectively. Two sets of data from WT (blue) and Spp1−/− (green) MPPs were normalized by sequencing depth and analysis of batch effects visualized by reduction to 2D by PCA and t-distributed stochastic neighbor embedding (t-SNE) plot (a). The cells in the t-SNE plot in (a) were clustered as K-means of 10 (b). Numbers in (b) denote cluster ID. Numbers of WT and Spp1−/− MPPs in each cluster in (b) are shown in (c). Ratios of WT and Spp1−/− cells in each cluster are shown in (d). Birc5 mRNA expression levels in single cells are indicated (e). (f–h) Ex vivo single-cell MPP differentiation assay. Representative flow plots showing the MPP development to granulocyte-myeloid (GM) and B cells both (“GM+B”, left panel), GM cells (middle), and B cells (right)(f). Percentages of MPPs developed into the indicated cell types (g). Plating efficiency of WT and Spp1−/− MPPs denotes percentages of wells that included proliferated and survived cells at the time of cell harvest (h). Data are representative of two independent experiments. * p<0.05. Data were analyzed by the Chi-squared test (c, d, h).
Next, we sought to determine whether OPN negatively regulates myelopoiesis and increases lymphopoiesis by an ex vivo cell differentiation assay33. This assay is also single cell-based by plating one cell per well to observe the cell development into granulo-macrophage (GM) cells and/or B cells (Fig. 3f). Significantly more Spp1−/− MPPs included unipotent myeloid progenitors (‘GM only’ in Fig. 3g) than WT MPPs did. In addition, ‘plating efficiency’ (i.e., how many cells survived in the tissue culture) was high in Spp1−/− MPPs (Fig. 3h), reflecting high Birc5 expression in Spp1−/− dominant clusters (Fig. 3e). The results also implicate better survival of MPPs without OPN.
OPN and apoptosis in myeloid vs. lymphoid progenitors
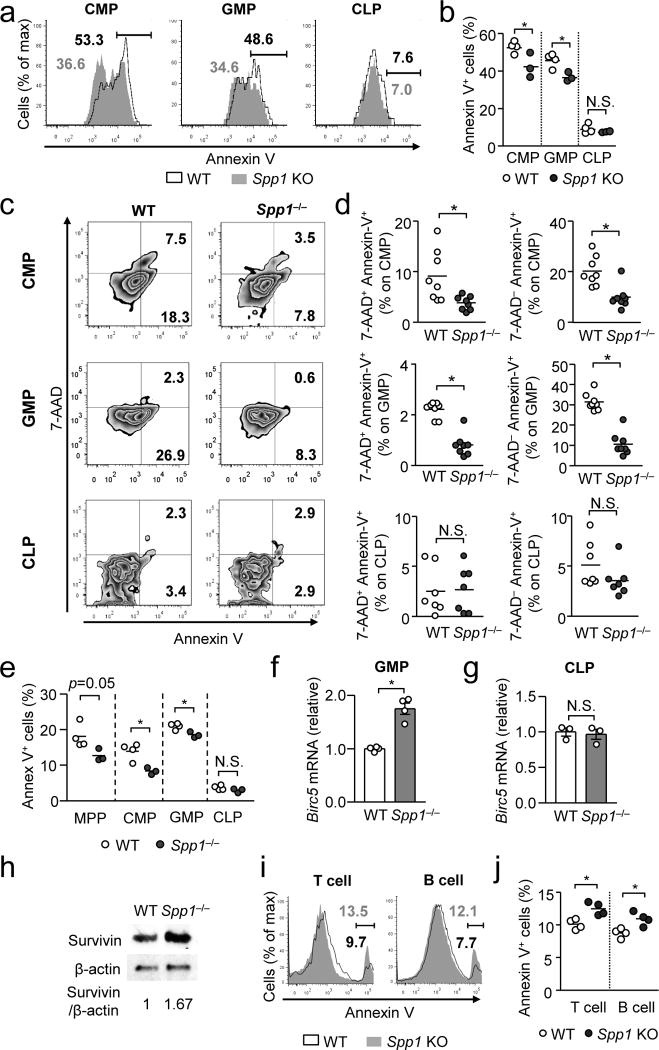
We asked how OPN inhibits emergency myelopoiesis. Gene expression levels of transcription factors (Ikaros, Spi1, Notch1, Notch2, Gata2, Flt3, Pbx1, Scl, Hoxb3, Hoxb4) critical for the developmental dichotomy from MPPs to either lymphoid and myeloid progenitors were comparable between WT and Spp1−/− MPPs after thioglycollate injection (Supplementary Fig. 4e). OPN also did not impact the proliferation of CMPs, GMPs and CLPs determined during emergency myelopoiesis induced by thioglycollate (Supplementary Fig. 5a, b). Therefore, we next examined if OPN affected apoptosis in the hematopoietic progenitors. At the steady-state, OPN had no impact on apoptosis of CMPs, GMPs, and CLPs (Supplementary Fig. 5c, d). However, in irradiation BM chimeras (Supplementary Fig. 5e), OPN significantly enhanced apoptosis in donor CMPs and GMPs, but not in CLPs (Fig. 4a, b). The similar pattern – OPN promotes apoptosis in CMPs and GMPs, but not CLPs – was observed in mice that had intraperitoneal thioglycollate injection (Fig. 4c, d) and systemic Candida infection (Fig. 4e). In contrast, apoptosis in differentiated myeloid cells, such as neutrophils and monocytes, was not affected by OPN (Supplementary Fig. 5f, g), suggesting that the pro-apoptotic role of OPN is restricted to myeloid progenitors. As the Birc5 transcript encoding survivin was identified in a single cell sequence analysis (Fig. 3e), enhanced expression of Birc5 was also identified in GMPs from Spp1−/− mice 15 hr after Candida infection (Fig. 4f). Importantly, the expression of Birc5 was not altered in CLPs, Gr-1+ cells and B cells in the BM (Fig. 4g; Supplementary Fig. 5h). Upregulation of survivin in Spp1−/− GMPs was also confirmed at the protein level (Fig. 4h).
Figure 4. OPN plays opposing roles in apoptosis of myeloid progenitors and lymphoid cells.
(a, b) Annexin-V staining of indicated progenitors at 3 weeks after BM cell transfer (WT or Spp1−/−) to WT recipients, as shown in Supplementary Fig. 5e. Representative flow panels (a) and data from a single experiment from two independent experiments. One circle denotes data from one recipient mouse. (c, d) Annexin-V and 7-AAD staining in indicated progenitors in the BM at 24 hrs after i.p. thioglycollate injection (not BM chimera). Representative flow plots (c) and data pooled from two independent experiments (d) are shown. (e) Apoptotic progenitors 15-hrs after systemic Candida infection (2×106 spores/mouse) in BM of WT and Spp1−/− mice (not BM chimera). (f, g) Birc5 mRNA levels of in GMPs (f) and CLPs (g) 15 hr after systemic Candida infection (2×106 spores/mouse). (h) A representative Western image of survivin expression in GMPs 15 hr after Candida infection (left). Survivin expression levels normalized β-actin expression (right). (i, j) Annexin-V staining of lymphoid cells in the spleen of recipients described in (a, b). Representative histograms (i) and frequencies of Annexin-V+ T and B cells (j) are shown. Data are representatives of independent two (a, b, e–j) or three (c, d) experiments. Error bars denote ± SEM. * p<0.05 (Student’s t-Test). N.S.; not significant.
Since OPN does not affect apoptosis in lymphoid progenitors (Fig. 4a–e), we examined whether differentiated lymphoid cells are affected by OPN. In the presence of OPN, apoptosis of T and B cells was inhibited in irradiation BM chimera (Fig. 4i, j). Even at steady state, apoptosis of B cells was slightly inhibited by OPN (Supplementary Fig. 5i). In contrast, OPN did not alter lymphocyte proliferation in irradiation BM chimera (Supplementary Fig. 5j, k). Thus, OPN plays an anti-apoptotic role in differentiated lymphoid cells in peripheral lymphoid organs. This result indicates a clear contrast in OPN’s roles: OPN is pro-apoptotic in myeloid progenitors but anti-apoptotic in differentiated lymphoid cells.
Apoptosis of donor lymphocytes in lymphopenic recipients
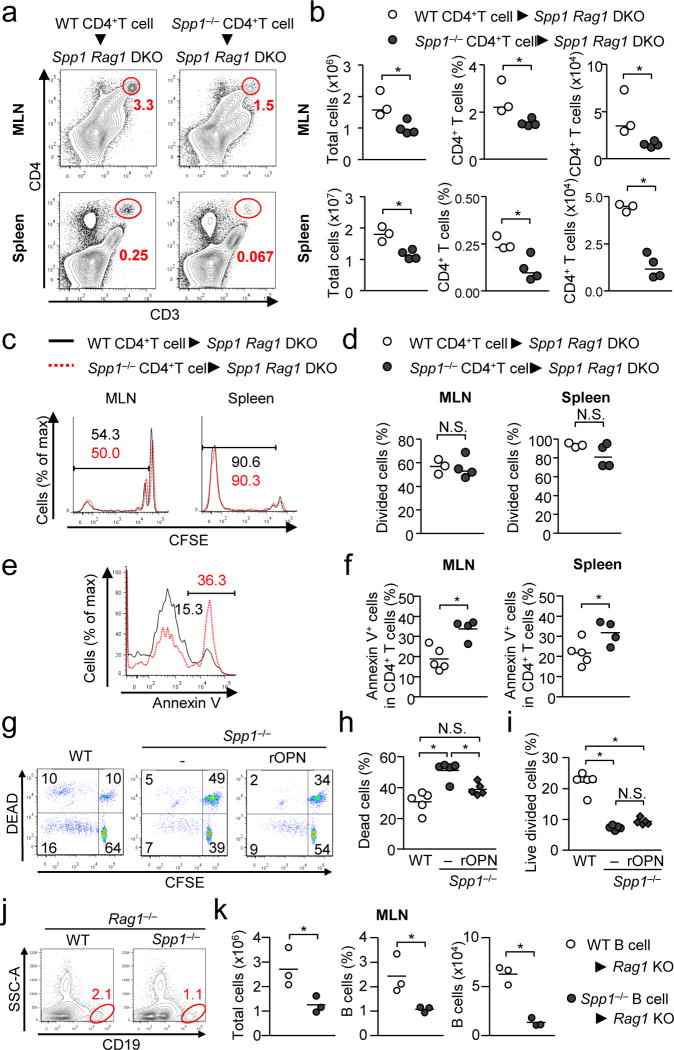
Numbers and proportions of WT CD4+ T cells were significantly higher than those of Spp1−/− CD4+ T cells during chronic lymphopenia-induced proliferation (cLIP)34, in which naive T cells were transferred to Spp1−/−Rag1−/− (Fig. 5a, b). Despite comparable proliferation between WT and Spp1−/− donor CD4+ T cells (Fig. 5c, d), WT cells were less prone to apoptosis than Spp1−/− cells (Fig. 5e, f). We obtained similar results in transferred T cells in OPN-sufficient Rag1−/− recipients (Supplementary Fig. 6a–d), suggesting donor T cell-derived OPN protects T cells from apoptosis. Systemic OPN is determined by recipient-derived OPN (Supplementary Fig. 6e); thus, either T cell autocrine OPN or iOPN within T cells could in principle inhibit T cell survival.
Figure 5. OPN-mediated enhancement of donor lymphocyte survival in lymphopenic recipients.
(a–f) CD62LhiCD4+ T cells (WT or Spp1−/−) (106 per mouse) were transferred to Spp1−/−Rag1−/− mice. Proportion and numbers of CD4+ T cells in the MLN and spleen (a, b), proliferation (c, d) and apoptosis of CD4+ T cells (e, f) were examined at day 6 (a–d) or day 3 (e, f). Representative flow plots (a, c, e) and analyzed data (b, d, and f) are shown. (g–i) Analyzing T cell expansion and death in ex vivo. CFSE-labeled CD62LhiCD4+ T cells obtained from WT and Spp1−/− mice were co-cultured with total MLN cells from Spp1−/−Rag1−/− mice for 7 days in the absence or presence of rOPN (10 ng/ml). Representative flow images (g), and frequencies of the dead (h) and proliferating CD4+ T cells (i) are shown. (j, k) B cells obtained from WT and Spp1−/− mice were transferred to Rag1−/− mice, and B cells in the MLNs were detected at 4 weeks after transfer. CD19+B220+CD93− mature B cells (2×106 per mouse) were used to exclude the contribution of immature B cells, which may develop into mature B cells during experiments Representative flow images (j), and proportions and numbers of B cells in the MLN (k) are shown. All the data is a representative of successfully repeated two independent experiments. * p<0.05. Data were analyzed by Student’s t-Test (b, d, f, k) and one-way ANOVA (h, i). N.S.; not significant.
Because T cell expansion in cLIP is mediated by gut commensal microbes35, we first tested if CD4+ T cells from WT mice (i.e., polyclonal) were proliferated ex vivo by total MLN cells without supplying exogenous antigens (Supplementary Fig. 6f). T cells were proliferated and survived better with MLN cells than those without MLN cells or with splenocytes (Supplementary Fig. 6g). In this ex vivo system, WT CD4+ T cells also proliferated better than Spp1−/− CD4+ T cells and were significantly protected from apoptosis, resulting in increased live and divided WT T cells (Fig. 5g–i). Importantly, the survival of Spp1−/− CD4+ T cells was recovered, at least partially, by supplying recombinant OPN (rOPN) in the cell culture (Fig. 5g, h), suggesting that anti-apoptotic function of OPN is mediated by sOPN. In contrast, OPN does not affect apoptosis and proliferation in T cells upon strong T cell receptor (TCR) stimulation with anti-CD3 plus CD28 antibodies (Supplementary Fig. 6h, i). These results indicated that sOPN supports T cell survival in activated T cells but not with strong TCR stimulation.
We also found that B cell-derived OPN significantly increased frequencies and numbers of B cells after being transferred into Rag1−/− lymphopenic recipients (Fig. 5j, k). Since B cells generally do not proliferate without T cells, this system without T cells evaluates B cell survival rather than proliferation. Taken together, OPN expressed in T or B cells promote survival of the cells, and sOPN appears to be sufficient for the effect.
OPN exacerbates T cell-mediated colitis
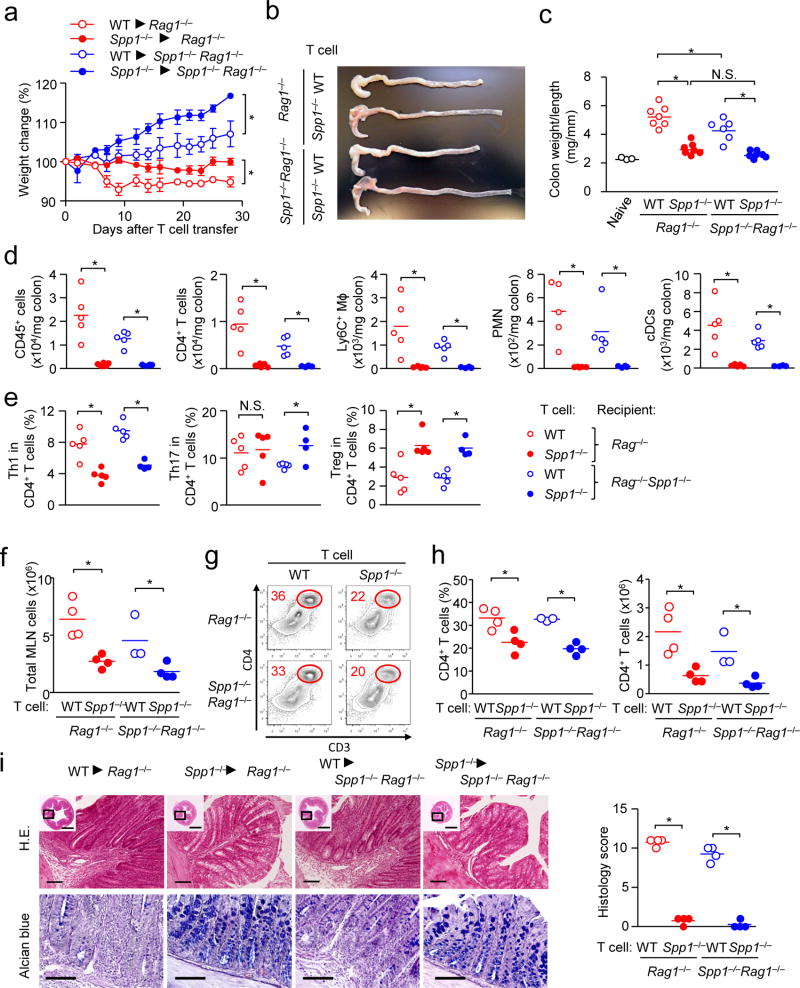
Colitis was induced in Rag1−/− and Spp1−/−Rag1−/− recipients by transferring CD62LhiCD4+ T cells from naive WT and Spp1−/− mice. First, the involvement of OPN derived from donor and recipients was identified in recipient weight changes (Fig. 6a), but the following data indicated more impact of donor OPN than recipient OPN. The increase in colon weight per unit length, a sign of colitis severity, was mediated by WT donor T cells (Fig. 6b, c). Here, not only did WT T cells migrate to the colon more efficiently than Spp1−/− T cells, but WT myeloid cells also did (Fig. 6d). This phenotype entirely depended on OPN in donor T cells (Fig. 6d). With the presence of OPN in donor T cells, proportions of colon-infiltrated T helper subsets were skewed towards Th1 with reduced proportion of Tregs (Fig. 6e). Numbers of total cells and CD4+ T cells in the MLN also shared a similar trend as seen in the colon (Fig. 6f–h), as well as general increase in numbers of most of T helper subsets and myeloid cells in the colon with WT T cell transfer (Supplementary Fig. 6j, k). These results further linked donor T cell-derived OPN and severe pathological features of colitis, such as immune cell infiltration, thickened mucosal and submucosal layers, and disappearance of goblet cells (Fig. 6i). It is of note that OPN in donor T cells and recipients did not impact the expression of integrins, gut homing receptors, and T cell activation markers (Supplementary Fig. 6l, m). Taken together with the cLIP results, T cell-derived sOPN contributes to the T cell increase and significantly exacerbates colitis.
Figure 6. T cell-derived OPN exacerbates T cell-mediated colitis in Rag1−/− mice.
CD62LhiCD4+ T cells (106 per mouse) obtained from WT and Spp1−/− mice were transferred to Rag1−/− or Spp1−/−Rag1−/− recipients. (a–f) Weight change (a)(n=3–4 per group), and a representative image of colons (b) are shown. Colitis severity is shown with the colon weight/length ratios (c), and cell infiltration in the colon (d, e) at 4 weeks after T cell transfer. In (d, e), gating was set as CD4+ T cells (CD3+CD4+), Ly6C+ myeloid cells (CD11b+Ly6C+Ly6G−), neutrophils (CD11b+Ly6G+), cDC (CD11c+B220−), Th1 (CD3+CD4+IFN-γ+), Th17 (CD3+CD4+IL-17+), and Treg (CD3+CD4+Foxp3+). Proportions of the three T helper subsets are relative to total CD4+ T cells. (f–h) Numbers of total MLN cells at 4 weeks after T cell transfer (f), Representative flow panels gated on total leukocytes (g), and frequencies and numbers of CD4+T cells. (i) Histology of colon with histological scores. Scale bars in wide angle and magnified views indicate 1 mm (H&E) or 100 µm (Alcian blue). Datasets in (a–c, f–i) are representatives of successfully repeated three independent experiment. Data in (d, e) was obtained from two independent experiments. Data were analyzed by Student’s t-Test (a, d–f, h, i) and one-way ANOVA (c). * p<0.05. N.S.; not significant.
Ruling out the involvement of iOPN in lymphoid cell expansion
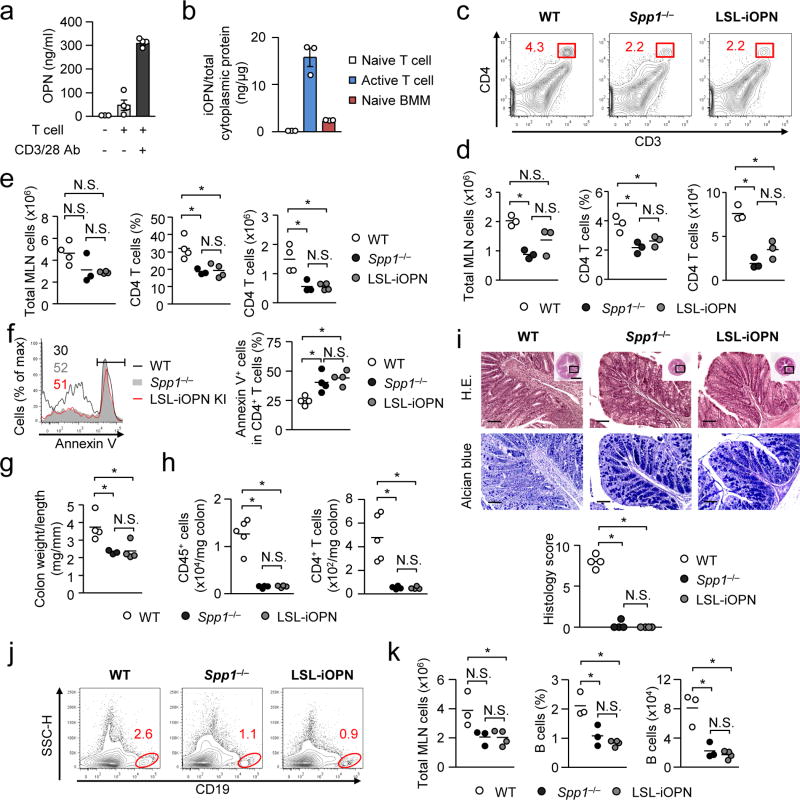
Activation of CD4+ T cells enhanced production of both sOPN and iOPN (Fig. 7a, b). Thus, we evaluated the involvement of iOPN by generating LSL-iOPN (LSLfl/fl-Δ45Spp1;Vav1-cre) mice, which express iOPN but not sOPN, due to the deletion of the signal sequence (45-nt) that target OPN to secretory vesicles (Supplementary Fig. 7a). ES cell clones successfully expressing the target allele were determined by a Southern blot analysis, and the deletion of the signal sequence was confirmed in LSL-iOPN mice (Supplementary Fig. 7b, c). Levels of cytoplasmic iOPN separated by density gradient23 in LSL-iOPN mice were comparable to those in WT mice (Supplementary Fig. 7d). Importantly, serum and cell culture supernatants of LSL-iOPN were free from OPN, i.e., no sOPN (Supplementary Fig. 7e, f).
Figure 7. Ruling out iOPN in exacerbating T cell-mediated colitis.
(a) OPN secretion by T stimulated with anti-CD3/CD28 antibody for 3 days in tissue culture. CD4+ T cells obtained from the MLNs of Spp1−/−Rag1−/− mice 4 weeks after CD62LhiCD4+ T cell transfer. Levels of sOPN were evaluated by ELISA. Error bars denote ± SEM. (b) The amount of iOPN per total cytoplasmic proteins in T cells (naive and activated) and BMMs. Cytoplasmic proteins were separated with the Pierce NE-PER reagent. (c–f) Colitis was induced by adoptively transferring WT, Spp1−/− or LSL-iOPN CD62LhiCD4+ T cells to Spp1−/−Rag1−/− recipients. Total MLN and CD4+ T cells (CD3+CD4+ of total leukocytes) were enumerated on day 6 (c, d) and day 28 (e). Apoptotic CD4+ T cells in MLNs on day 3 were labeled with Annexin-V (f). (g–i) Evaluation of the colon on day 28 after T cell transfer. Colon weight per length (g), CD45+ and CD4+ T cell (CD45+CD3+CD4+) numbers (h), and histology with scores (i) are shown. Scale bars indicate 100 µm (main images) or 1 mm (insets). (j, k) Mature B cells (CD19+B220+CD93−) (2×106 per mouse) from WT, Spp1−/− or LSL-iOPN mice were adoptively transferred to Rag1−/− mice. Representative flow plots (j), and the frequencies and numbers of B cells (k) in the MLNs are shown. All datasets are representatives of two independent experiments. N.S.; Not significant. * p< 0.05. Data was analyzed by one-way ANOVA (d–i and k).
Using CD4+ T cells from the LSL-iOPN mice, we examined cLIP on day 6 (Fig. 7c, d) and day 28 (Fig. 7e). Numbers and proportions of CD4+ T cells were comparable between recipients which received Spp1−/− or LSL-iOPN T cells, but recipients with WT T cells showed more of the values (Fig. 7c–e), suggesting that sOPN enhances T cell expansion during cLIP. Furthermore, apoptosis between LSL-iOPN and Spp1−/− CD4+ T cells was similarly more frequent than WT CD4+ T cell apoptosis (Fig. 7f), again suggesting the involvement of sOPN. This result is also confirmatory to the ex vivo results demonstrating that sOPN, not iOPN, protected T cells from apoptosis (Fig. 5g, h). LSL-iOPN CD4+ T cells induced extremely mild colitis as Spp1−/− T cells did with low numbers of tissue-infiltrated cells and low histology scores, e.g., well preserved goblet cells (Fig. 7g–i). The results suggest that T cell iOPN is dispensable to induce OPN-dependent colitis. We also ruled out B cell iOPN in OPN-mediated B cell increase (Fig. 7j, k). In sum, sOPN is responsible for OPN-mediated lymphocyte survival and the resulting increase of lymphocyte population sizes.
Less myeloid cells by iOPN, more lymphoid cells by sOPN
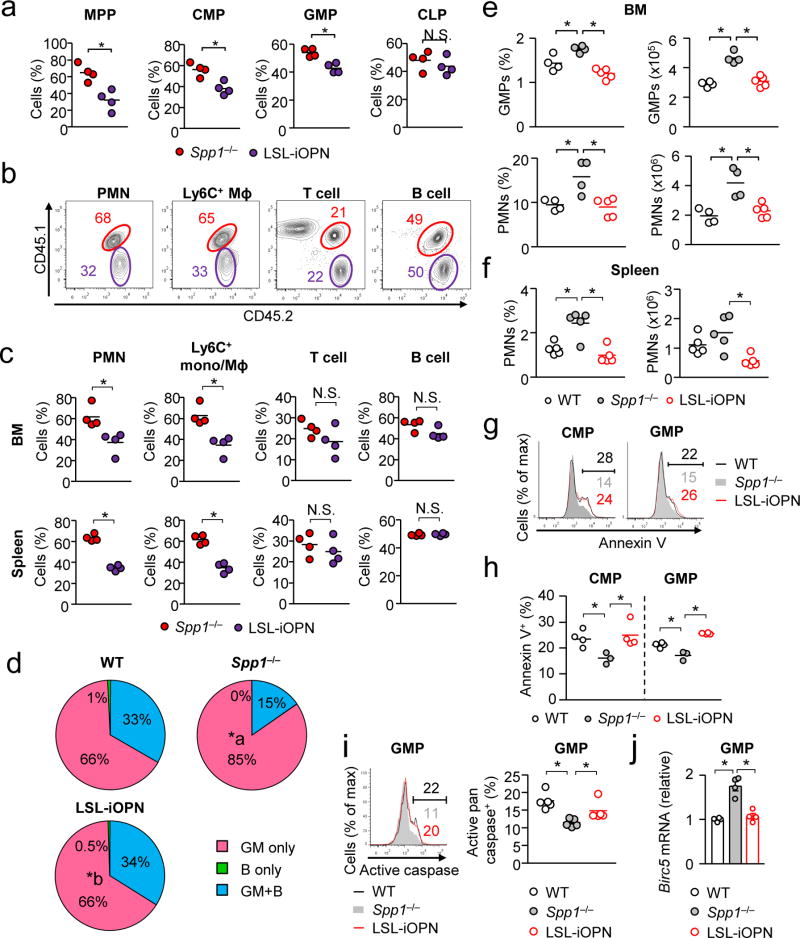
Mixed BM radiation chimera were generated with LSL-iOPN and Spp1−/−donor BM cells (Supplementary Fig. 2f, g). Three weeks after BM transfer, proportions of Spp1−/− MPPs, CMPs, and GMPs predominated over those of LSL-iOPN (Fig. 8a), indicating the involvement of iOPN in restricting the progenitor population sizes. Differentiated leukocytes were examined at 7 weeks after BM transfer. Spp1−/− cells were again predominant over LSL-iOPN cells for neutrophils and Ly6C+ monocytes/macrophages, not for T cells and B cells (Fig. 8b, c). When WT and LSL-iOPN BM cells were used as donors, no difference was observed in myeloid cell population sizes (Supplementary Fig. 8a–c), showing a clear contrast to WT and Spp1−/− mix BM chimera (Supplementary Fig. 8d). These data confirmed that iOPN restricts myeloid cell development by limiting myeloid progenitors, but not differentiated leukocytes, while sOPN expands lymphoid cells.
Figure 8. iOPN inhibits myelopoiesis.
(a–c) Mixed BM chimera mice were generated by transferring BM obtained from Spp1−/− (CD45.1/CD45.2) or LSL-iOPN mice (CD45.2) to irradiated WT mice (CD45.1) as shown in Supplementary Fig. 2f, g. The ratio of Spp1−/−-derived (red) and LSL-iOPN-derived (purple) hematopoietic progenitor (a), myeloid and lymphoid cells (b, c) in the BM were examined at 3 weeks (a) or 7 weeks (b, c) after BM cell transfer. Representative flow images of splenic cells are shown in (b). Data (a, c) is a representative of two independent experiment. Mɸ: macrophage. (d) Ex vivo single-cell MPP differentiation assay, comparing WT, Spp1−/−, and LSL-iOPN MPPs. Development of GM cells between Spp1−/− (indicated with “a”) and LSL-iOPN (indicated with “b”) was significantly different (p < 0.05) by the Chi-square test. Plating efficiencies were 201 of 506 (40%; WT), 246 of 506 (49%; Spp1−/−), and 363 of 960 (38%; LSL-iOPN). Data were pooled from two independent experiments. (e–j) WT, Spp1−/− and LSL-iOPN mice were systemically infected with Candida (2–106 spores/mouse). Shown are frequencies and numbers of GMPs (e), neutrophils (f), Annexin-V+ CMPs and GMPs (g, h), pan-caspase activity (i), and Birc5 expression (j) in GMPs at 15 hrs (g–j) or 24 hrs (e, f) after Candida infection. Each dataset in (e, f, h–j) is a representative of two independent experiments. *; p < 0.05. Data was analyzed by the Student’s t-test (a, c) and ANOVA (e, f, h–j). N.S.; not significantly different.
We further validated the impact of iOPN in myelopoiesis by an ex vivo single-cell progenitor differentiation assay (Fig. 8d). During emergency granulopoiesis induced by systemic Candida infection, iOPN (i.e., in WT and LSL-iOPN mice) decreased proportions and numbers of GMPs and neutrophils in the BM by enhanced apoptosis in CMPs and GMPs (Fig. 8e–h). Furthermore, iOPN enhanced activation of pan-caspases (predominantly caspase-1 and -336) and reduced Birc5 mRNA in GMPs (Fig. 8i, j). In sum, iOPN decreases myeloid cell populations, and sOPN increases lymphoid cell populations.
DISCUSSION
Previous reports showed that OPN decreased the pool size of LSK cells during the homeostatic state and after BM cell transplant19, 20. This study suggested that the function of OPN in GMPs and CMPs is quite distinct from that in LSK cells in the following points: (1) sOPN decreases LSK cells19, 20 but iOPN decreases GMPs and CMPs; (2) the increase of LSK cells by OPN-deficiency does not affect population sizes of differentiated leukocytes19, 20 but the increase of GMPs by iOPN-deficiency results in enlarged population sizes of myeloid cells during emergency myelopoiesis; and (3) non-hematopoietic stroma cells are the source of sOPN that decreases LSK cells but iOPN in the progenitor cells is attributed to the decrease of myeloid progenitors. Thus, although sOPN reduces population sizes of LSK cells, the mechanism by which iOPN reduces myeloid progenitors is quite different.
iOPN was detrimental during early Candida infection by limiting neutrophil supply. This may appear contradictory to a previous study demonstrating that iOPN protects hosts from Pneumocystis fungal infection13. However, major differences in these fungal infection models need to be noted. The previous Pneumocystis study focused on opportunistic and spontaneous infection in lymphopenic hosts and required 10–20 weeks to evaluate host mortality13. Under this long-term infection, emergency myelopoiesis most probably does not apply. Therefore, iOPN appears to be either detrimental or beneficial, depending on timing and condition of a fungal infection.
A series of colitis studies strongly imply multifaceted roles of OPN. Some studies concluded that OPN induces immune responses37, 38, 39, and others demonstrated that OPN suppresses immune responses39, 40, 41. One study provided an insightful suggestion that OPN suppresses immune responses during acute inflammation, but not in chronic inflammation39. In particular, sOPN was previously found to be attributable to chronic colitis37, 39, and our data in this study is congruent with these findings.
sOPN’s anti-apoptotic function was also demonstrated by others42. In contrast, the pro-apoptotic function of iOPN has not been reported in any cells. In this study, we demonstrated that iOPN enhances apoptosis in GMPs. We evaluated a possible involvement of p53 in inducing apoptosis by detecting phosphorylated p53, but no differences were found between WT and Spp1 GMPs after Candida infection (data not shown). Instead, we found that OPN enhances apoptosis through the downregulation of survivin expression and enhancement of pan-caspase activities. Based on our single cell analysis result, it is possible that particular MPP subpopulations were paricularly regulated by iOPN to be advanced into myelopoiesis.
In this study, we dissected roles of sOPN and iOPN in shifting the balance of leukocyte populations during BM cell transplant after irradiation, fungal infection, and autoimmune colitis, and found differential regulation of myeloid vs. lymphoid cell populations by distinct OPN isoforms. Thus, iOPN and sOPN coordinately tip the balance of myeloid vs. lymphoid populations.
ONLINE METHODS
Animals and reagents
All the mice in this study were on the C57BL/6J background. Spp1−/− mice and Vav1-Cre mice were purchased from the Jackson Laboratory. LSL-iOPN mice were created in our laboratory with help from the Duke Transgenic Mouse Facility. Six- to eight-week old sex-matched WT, Spp1−/− and LSL-iOPN mice were used for all experiments. All the experiments were performed as approved by the Institutional Animal Care and Use Committee at Duke University, and all animals were housed under specific pathogen-free conditions. Readouts of in vivo disease model experiments were quantitatively determined by objective measurements; thus, blinding was not performed. The genotype of the ES cells and the mice was examined by Southern blot and PCR by using following probe and primers (Supplementary Fig. 7a–c). Probe for Southern blot: tcatttgcattcaatctgagcttgtcagacaaaattattcactttcattttatgagatttctctcatcaatacgcatataatgtagtctgcatacatgtggtagtctgcacacatgtggtagcctgcacacatagggtcgtatgcacacatggagtagtatcacacatgggataatatacccacatggggtagtgtgcacacatgtgttagtgtgtacacatgaggtagtgtgcacatatgttagtatgcatatgtgttaatatgtatagatggggtagtttgtacacatggggtagtatgcacacatgtgctatatacacacatgtgtgagtgtgcatacatggggtattatgcacatgtgttagtatgtacatatgggatagtgaacatacatggagtagtctacatacatgtgttagtatacacatatgtggaagccagaggacaagtccaggctttgttcctcctgcgttgtctatatatttttttgagacagggtctttcactggcctggaactcagcaaacaggctaaggcagacagagagtcctgagagtacaactgtctgtacctccccagtgctagaatgacaatgctgagcagt. Primers to detect deletion of 45 nucleotides in exon 2 of Spp1 allele, forward: 5’-ACTTGGTGGTGATCTAGTGGTG-3’, reverse: 5’-AGCAACCTTTGCGACGCTGGCGAAC-3’. Antibodies against CD45 (30-F11), CD45.1 (A20), CD45.2 (104), CD4 (GK1.5), CD3 (145-2C11), CD8 (53-6.7), Ly6G (1A8), Ly6C (HK1.4), CD11b (M1/70), CD11c (N418), CD49b (DX5), TER119 (TER-119), Gr-1 (RB6-8C5), B220 (RA3-6B2), CD19 (6D5), FcγR (93), CD34 (MEC14.7), IL-7R (A7R34), Flt3L (A2F10), Sca-1 (D7), c-kit (2B8), CD16/32 (93), E-cadherin (DECMA-1) and BrdU (Bu20a), G-CSF, GM-CSF were purchased from BioLegend. AMD3100 was purchased from Sigma. XMU-MP-1 was purchased from MedChem Express. To check LDH levels in the serum, Pierce™ LDH Cytotoxicity Assay Kit was used.
Tissue-infiltrated cell analyses by flow cytometry
Lung and liver tissues were cut into small pieces and incubated in a 1 mg/ml collagenase D solution at 37 °C for 30 min. Cells were then enriched by Percoll gradient (GE Healthcare). Followed by staining with specific antibodies, cells were analyzed with FACS Canto™ II (BD) and the FlowJo software (Treestar Inc.). For evaluation of apoptosis, FITC Annexin V Apoptosis Detection Kit with 7-AAD was used. Gating strategy is shown in Supplementary Fig. 1a, unless otherwise noted. Data on BM cell enumeration was enumerated per one tibia and one femur from one of the hind legs.
Real-time PCR and ELISA analysis
Total mRNA were reverse-transcribed to cDNA, and gene expression levels were determined by using the −ΔΔCt method of real-time PCR as previously described31, 43 by using primers shown in Supplementary Table 2. Actb expression was used as an internal control. For Fig. 2g, Hsp90ab1 was used as an internal control. Results shown are representatives from multiple independent experiments with similar results. In Supplementary Fig. 3g, 4e, error bars (too short to be identified in Supplementary Fig. 3g) were drawn based on the calculation of RQ-Min = 2−(ΔΔCt + T * SD(ΔCt)) and RQ-Max = 2−(ΔΔCt − T * SD(ΔCt)) from triplicate wells as suggested by a manufacturer of PCR machines (Applied Biosystems). Error bars denote RQ-MIN and RQ-MAX, which constitute the acceptable error for a 95% confidence limit according to Student's t-test. ELISA for OPN was performed as previously described44.
Experiments with Candida
Detailed methods for the experiments with Candida are described in our previous publication45. Candida infection was performed with sex-matched mice of age 6–8 week by intravenous (i.v.) injection of 2 × 106 spores of Candida albicans (ATCC 18804). Fifteen or twenty-four hour after Candida infection, cellularity and apoptotic cell ratio in the BM and spleen were examined by flow cytometry. Host cell phagocytosis of Candida spores was performed as follows. BMMs and thioglycollate-elicited neutrophils were co-cultured with Alexa Fluor 647 (AF647)-labeled Candida spores (host cells vs. spore ratio of 1:1) for 15, 30, 60 or 120 min. Host cells had been stained with antibodies against CD11b, F4/80 (for macrophages) or Ly6G (for neutrophils) before co-culture set up. Flow cytometry was used to determine cells phagocytosed Candida. For evaluating fungicidal activity by host cells, BMMs and thioglycollate-elicited neutrophils were used for the analyses were co-culture with live Candida with the 1:10 ratio (conidia : host)(termed “test samples”) for 12 hours. Negative control culture had Candida spores alone. Host cells were lysed with water, and Candida CFU was determined with YPD plates. Results were obtained with the following formula: Candida Expansion (fold) = [CFU at 12 hrs]/[CFU at 0-h]; Inhibition of Candida expansion (%) = (1 − [CFU of test samples at 12 hrs]/[CFU of negative control at 12 hrs]) × 100.
Treating mice with Thioglycollate or AMD3100
WT or Spp1−/− mice were intraperitoneally injected with 3% thioglycollate (2.5 ml per mouse) or AMD3100 (5 mg/kg mouse weight). GMPs were sorted from BM of the mice at 0, 6, 12, 24 or 48 after the treatment. Spp1 expression in the cells was examined by qPCR. Cell numbers and PMN frequencies were calculated from flow cytometry data.
Ex vivo single-cell MPP differentiation assay
To analyze the differentiation potential of MMPs, single MPP cells were sorted by MoFlo XDP (Beckman Coulter) and plated (one cell/well) in the MEM alpha (Life technology) medium containing 20% ES cell-grade FBS (Millipore), SCF (50 ng/ml: BioLegend), and Flt3 ligand (30 ng/ml: BioLegend) on the layer of OP9 cells in 96-well plates as previously described33. The OP9 cell line was obtained from ATCC and confirmed to be free of mycoplasma by using the MycoAlert PLUS kit (Lonza). Two days after cell plating, IL-3 (10 ng/ml; BioLegend), IL-7 (10 ng/ml; BioLegend), and GM-CSF (10 ng/ml; R&D Systems) were added to each well to induce cell differentiation. “Plating efficiency” denotes cells successfully expanded in culture, determined by microscopic observation. Expanded cells were analyzed by flow cytometry to determine whether they developed into either granulocyte/macrophages (GM) (Gr-1+CD11b+B220−CD19− and Gr-1−CD11b+B220−CD19−) or B cells (Gr-1−CD11b−B220+CD19+).
Separating of cytoplasmic proteins
Separation was performed by two methods. One is by using NE-PER™ Nuclear and Cytoplasmic Extraction Reagents, as instructed by the supplier (Pierce). Another is by the density gradients. In the latter method, tissues from WT, Spp1−/− and LSL-iOPN mice were homogenized in the homogenization medium (0.25 M sucrose, 1mM EDTA 10 mM Hepes-NaOH, pH 7.4), then mixed with iodixanol as the final concentration of 30%. Homogenates in 30% iodixanol solution (1125 µl) were placed at the bottom, then layered with 25% (w/v) of iodixanol solution (2 ml) in the middle and 5% (w/v) iodixanol solution (2 ml) on the top. Using the Beckman Ultracentrifuge L9 and Beckman SW40.1 rotor, samples were centrifuged at 35,000rpm (250,000g) at 4 °C for 3 hr. Eight layers fractions were carefully collected from the top to the bottom by pipette. Cytoplasmic compartments were identified by Western blotting. Separation of the cytoplasmic proteins was confirmed by detecting GAPDH, but not calnexin. The cytoplasmic fractions were used to evaluate the iOPN expression.
Cell proliferation analysis
To analyze cell proliferation in vivo, BrdU (10 ml/kg) (Invitrogen) was intraperitoneally injected into mice. Twenty-four hour after the injection, cells obtained from the spleen or BM. After fixation with 2% PFA, permeabilization with 0.5% Triton X-100 in PBS, cells were treated with RQ1 RNase-Free DNase (Promega). With BrdU antibody, cells were stained, then BrdU incorporation was analyzed by flow cytometry. For experiments shown in Supplementary Fig. 3c, cell proliferation was evaluated by Ki-67 staining. BM cells were harvested and stained for cell surface markers, fixed, and permeabilized by incubating with cold 70% ethanol at −20°C for 1hr. Then, cells were stained with anti-Ki-67 antibody and analyzed by flow cytometry.
Generation of mix BM chimera mice
Mix WT-Spp1−/− BM chimera mice were generated by adoptively transferring 5×106 each of BM cells obtained from naive WT mice (CD45.1/CD45.2) and Spp1−/− mice (CD45.2) to lethally irradiated (900 rad) WT mice (CD45.1), as shown Supplementary Fig. 2a. For LSK cell chimera mice, 104 each of LSK cells isolated from WT (CD45.1) and Spp1−/− mice (CD45.2) were adoptively transferred to lethally irradiated (900 rad) WT mice (CD45.1/CD45.2) with 107 of c-kit− BM cells, as “rescue cells,” obtained from WT mice (CD45.1/CD45.2). Other mix BM chimera mice were also prepared similarly. Ratios between mixed donor cells were analyzed 3, 6, or 7 weeks after BM cell transfer by flow cytometry. The methods were also depicted in Supplementary Fig. 2.
Lymphopenia-induced lymphocyte proliferation and T cell-induced colitis
CD62LhiCD4+ T cells (106/ mouse) were intravenously injected to Rag1−/− or Rag1−/−Spp1−/− mice. Donor T cell apoptosis and population sizes were examined at day 3 and/or 6. The severity of colitis was evaluated by the ratio between weight and length of the colon, as well as histology staining with H&E and Alcian blue, at 4 weeks after T cell transfer. Histological scoring was performed as previously described46. Colon infiltrated cells were isolated by using Lamina Propria Dissociation Kit (Miltenyi Biotec). To evaluate T cell proliferation, donor T cells were labeled with CellTrace Violet (Thermo Fisher Scientific inc.), and evaluated by flow cytometry 6 days after the injection. To examine an impact of OPN in maintaining B cell population, B cells (2× 106/ mouse) were adoptively transferred to Rag1−/− and analyzed the population size by flow cytometry at 4 weeks after the cell transfer.
Ex vivo T cell proliferation assay
Total MLN cells obtained from Rag1−/− or Rag1−/−Spp1−/− mice were co-cultured with WT or Spp1−/− CFSE-labeled CD62LhiCD4+ T cells at the 1:1 ratio in the absence or presence of neutralizing OPN antibody (AF808: 10 µg/ml) for 7 days. Cell proliferation and cell death were evaluated by dilution of CFSE (or CellTrave Violet) and LIVE/DEAD staining, respectively. In some experiment, CD62LhiCD4+ T cells obtained from WT or Spp1−/− mice were labeled with CellTrace Violet and stimulated with Dynabeads T-Activator CD3/CD28 (TermoFisher Scientific). T cell apoptosis was examined by 7-AAD staining.
Single-cell RNA sequencing
MPPs were flow-sorted as Lin−Sca-1+c-kit+CD34+Flt3L+ from BM of WT and Spp1−/− mice 20 hrs after thioglycollate intraperitoneal injection and submitted for single-cell sequencing. For 10x Transcriptome library prep, flow-sorted MPPs were resuspended in a 1× PBS / 0.04% BSA solution, at a concentration of 1000 cells/µl. After size selection (<50 µm), the cell suspensions were washed with a 1× PBS / 0.04% BSA solution to remove debris, clumps, dead cells and contaminants, and a cell counter used to determine the cell concentration and the solution normalized to 1×106 cells/ml. Cell preps were titrated to contain ~3000 cells per library. Cells were then combined with a master mix that contains reverse transcription reagents, and combined on the 10X instrument (10X Genomics, Inc., Pleasanton, CA) with gel beads carrying the Illumina P7 and R2 primers, a 14-bp identifying barcode, a 10-bp randomer for RNA molecule quantitation, and a poly-dT primer, and oil for the emulsion PCR. Cells were partitioned into nanoliter-scale gel beads in emulsion (GEMS) within which reverse-transcription is carried out and all cDNAs within a GEM, i.e. from one cell, share a common barcode. After the RT reaction, the GEMs are/were broken and the full-length cDNAs cleaned with both Silane Dynabeads (Thermo Fisher Scientific, Waltham, MA) and SPRI beads (Beckman Coulter Life Sciences, Indianapolis IN). After purification, the cDNAs were run on the Agilent Bioanalyzer High Sensitivity chip (Agilent Technologies, Santa Clara, CA) for qualitative analysis. cDNA in single cells was sheared to ~200-bp with a Covaris S220 Focused-ultrasonicator (Covaris, Inc., Woburn, MA) and then sequencing libraries constructed for running on an Illumina high-throughput sequencer. This entailed end repair and A-tailing, P5 adapter ligation, SPRI bead clean-up, a sample index PCR, and further SPRI bead clean-ups. The sample index PCR adds a unique sample index for sample multiplexing during sequencing, with P5 and P7 primers used in Illumina bridge amplification. The sequence was generated using 125-bp paired-end sequencing on an Illumina 2500 sequencing platform (Illumina, Inc., San Diego, CA) at the David H Murdock Research Institute (Kannapolis, NC). Supplementary Table 1 shows the cell count and sequencing totals, including unique molecular identifiers which allow for within cell, gene specific counts of RNA molecules.
Single cell sequence cell data analysis
The primary analytical pipeline for the SC data followed the recommended protocols from 10X Genomics. Briefly, raw FASTQ files were processed using Cell Ranger software (v 1.1) to first demultiplex the raw reads and align the reads to the mouse transcriptome. PCR duplicates were flagged, non-cellular reads filtered out, and gene expression matrices created for all single cells in each sample (http://support.10xgenomics.com/single-cell/software/pipelines/latest/what-is-cell-ranger). We performed a secondary analysis using an R statistical methodology, R package Seurat (http://satijalab.org/seurat/). Samples were normalized based on sequencing depth, then combined into a single gene expression matrix in Cell Ranger. In Seurat, the combined expression matrix was normalized on the log scale after basic filtering for minimum gene and cell observance frequency cut-offs (http://satijalab.org/seurat/pbmc-tutorial.html). We then closely examined the data and performed further filtering based a range of metrics in an attempt to identify and exclude possible multiplets (i.e. instances where more than one cell was present and sequenced in an emulsified gel bead). After quality control procedures had completed, we calculated principal components using the most variably expressed genes in the dataset. Significant principal components for downstream analyses were determined through methods mirroring those implemented by Macosko et al47, and these principal components were carried forward to perform cell clustering and enhance visualization (http://satijalab.org/seurat/pbmc-tutorial.html). Cells were grouped into an optimal number of clusters using Seurat’s FindClusters() function, with visualization of cells being achieved through the use of t-SNE, which reduces the information captured in the selected significant principal components to two dimensions. Additional downstream analyses included examining the cellular distribution of a priori genes of interest, and closer examination of genes associated with cell clusters.
Statistical analysis
Statistical analysis was evaluated by using Microsoft Excel or Prism software version 3.02 (GraphPad). The two-tailed Student’s t-test was used for statistical analyses of two-group comparisons. Multigroup comparisons were performed by a one-way analysis of variance (ANOVA) followed by Tukey–Kramer multiple comparisons test. In Fig. 2k, 3d, 3g and 8d, Chi-square test was used to determine if there is a significant relationship between two categorical variables. χ2 statistics were derived using the CHIDIST function of Microsoft Excel. The criterion of significance was set at p<0.05. All results are expressed as mean ± SEM. Blinding or randomization to the groups were not performed and no data points were excluded for analyses. No statistical methods were used to predetermine to justify sample sizes, but our sample sizes are similar to those generally employed in the field.
Supplementary Material
Acknowledgments
We thank Dr. M. Ciofani for and Mr. M. Zuberbuehler (Department of Immunology, Duke University) for helping an experiment with fetal liver, Dr. M. Kondo (Toho University, Japan) for analysis of ex vivo progenitor differentiation assays, and Mr. Gary Kucera (Duke University) for generating the LSL-iOPN BAC construct. We would also like to thank the staff at the David H Murdock Research Institute for generating the sequence data, Ms. Karen Abramson and Ms. Emily Grass (Molecular Genomics Core, Duke Molecular Physiology Institute) for generating the 10X single cell libraries, and Ms. T. Kadota (Duke University) for helping in vitro experiments with BMMs in this study. This work was supported by the National Institutes of Health (R01AI088100 and R21AI103584 to M.L.S.) and U.S. Department of Defense (PC100266 to M.L.S.).
Footnotes
ACCESSION CODES
GSE99735. Comparing single cell sequences between WT vs. Spp1−/− multi-potent progenitor cells (MPP) cells.
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding author upon request.
AUTHOR CONTRIBUTIONS
M.K. performed the majority of experiments; S.X. performed parts of experiments for cell density-mediated OPN upregulation and validating LSL-iOPN mice; K.D. performed parts of BM chimera experiments; M.I. provided experimental help to generate and validate LSL-iOPN mouse line; S.G. and J.G. performed single-cell analysis, analyzed data, and provided editorial help; M.K. and M.L.S. conceived the project and wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no conflict of interests.
References
- 1.Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014;14:302–314. doi: 10.1038/nri3660. [DOI] [PubMed] [Google Scholar]
- 2.Takizawa H, Boettcher S, Manz MG. Demand-adapted regulation of early hematopoiesis in infection and inflammation. Blood. 2012;119:2991–3002. doi: 10.1182/blood-2011-12-380113. [DOI] [PubMed] [Google Scholar]
- 3.Patchen ML, MacVittie TJ, Williams JL, Schwartz GN, Souza LM. Administration of interleukin-6 stimulates multilineage hematopoiesis and accelerates recovery from radiation-induced hematopoietic depression. Blood. 1991;77:472–480. [PubMed] [Google Scholar]
- 4.Boettcher S, Manz MG. Regulation of Inflammation- and Infection-Driven Hematopoiesis. Trends Immunol. 2017 doi: 10.1016/j.it.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Lieschke GJ, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- 6.Zhan Y, Cheers C. Haemopoiesis in mice genetically lacking granulocyte-macrophage colony stimulating factor during chronic infection with Mycobacterium avium. Immunol Cell Biol. 2000;78:118–123. doi: 10.1046/j.1440-1711.2000.00891.x. [DOI] [PubMed] [Google Scholar]
- 7.Bernad A, et al. Interleukin-6 is required in vivo for the regulation of stem cells and committed progenitors of the hematopoietic system. Immunity. 1994;1:725–731. doi: 10.1016/s1074-7613(94)80014-6. [DOI] [PubMed] [Google Scholar]
- 8.Zhan Y, Lieschke GJ, Grail D, Dunn AR, Cheers C. Essential roles for granulocyte-macrophage colony-stimulating factor (GM-CSF) and G-CSF in the sustained hematopoietic response of Listeria monocytogenes-infected mice. Blood. 1998;91:863–869. [PubMed] [Google Scholar]
- 9.Bugl S, et al. Steady-state neutrophil homeostasis is dependent on TLR4/TRIF signaling. Blood. 2013;121:723–733. doi: 10.1182/blood-2012-05-429589. [DOI] [PubMed] [Google Scholar]
- 10.Cain DW, Snowden PB, Sempowski GD, Kelsoe G. Inflammation triggers emergency granulopoiesis through a density-dependent feedback mechanism. PLoS One. 2011;6:e19957. doi: 10.1371/journal.pone.0019957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue M, et al. T cells down-regulate macrophage TNF production by IRAK1-mediated IL-10 expression and control innate hyperinflammation. Proc Natl Acad Sci U S A. 2014;111:5295–5300. doi: 10.1073/pnas.1321427111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinohara ML, Kim JH, Garcia VA, Cantor H. Engagement of the type I interferon receptor on dendritic cells inhibits T helper 17 cell development: role of intracellular osteopontin. Immunity. 2008;29:68–78. doi: 10.1016/j.immuni.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inoue M, et al. Cutting edge: critical role of intracellular osteopontin in antifungal innate immune responses. J Immunol. 2011;186:19–23. doi: 10.4049/jimmunol.1002735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leavenworth JW, Verbinnen B, Wang Q, Shen E, Cantor H. Intracellular osteopontin regulates homeostasis and function of natural killer cells. Proc Natl Acad Sci U S A. 2015;112:494–499. doi: 10.1073/pnas.1423011112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rittling SR, Singh R. Osteopontin in Immune-mediated Diseases. J Dent Res. 2015;94:1638–1645. doi: 10.1177/0022034515605270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uede T. Osteopontin, intrinsic tissue regulator of intractable inflammatory diseases. Pathol Int. 2011;61:265–280. doi: 10.1111/j.1440-1827.2011.02649.x. [DOI] [PubMed] [Google Scholar]
- 17.Shevde LA, Samant RS. Role of osteopontin in the pathophysiology of cancer. Matrix Biol. 2014;37:131–141. doi: 10.1016/j.matbio.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreke MR, Huckle WR, Goldstein AS. Fluid flow stimulates expression of osteopontin and bone sialoprotein by bone marrow stromal cells in a temporally dependent manner. Bone. 2005;36:1047–1055. doi: 10.1016/j.bone.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 19.Stier S, et al. Osteopontin is a hematopoietic stem cell niche component that negatively regulates stem cell pool size. J Exp Med. 2005;201:1781–1791. doi: 10.1084/jem.20041992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nilsson SK, et al. Osteopontin, a key component of the hematopoietic stem cell niche and regulator of primitive hematopoietic progenitor cells. Blood. 2005;106:1232–1239. doi: 10.1182/blood-2004-11-4422. [DOI] [PubMed] [Google Scholar]
- 21.Grassinger J, et al. Thrombin-cleaved osteopontin regulates hemopoietic stem and progenitor cell functions through interactions with alpha9beta1 and alpha4beta1 integrins. Blood. 2009;114:49–59. doi: 10.1182/blood-2009-01-197988. [DOI] [PubMed] [Google Scholar]
- 22.Inoue M, Shinohara ML. Intracellular osteopontin (iOPN) and immunity. Immunol Res. 2011;49:160–172. doi: 10.1007/s12026-010-8179-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shinohara ML, Kim HJ, Kim JH, Garcia VA, Cantor H. Alternative translation of osteopontin generates intracellular and secreted isoforms that mediate distinct biological activities in dendritic cells. Proc Natl Acad Sci U S A. 2008;105:7235–7239. doi: 10.1073/pnas.0802301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leavenworth JW, Verbinnen B, Yin J, Huang H, Cantor H. A p85alpha-osteopontin axis couples the receptor ICOS to sustained Bcl-6 expression by follicular helper and regulatory T cells. Nat Immunol. 2015;16:96–106. doi: 10.1038/ni.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shinohara ML, et al. Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat Immunol. 2006;7:498–506. doi: 10.1038/ni1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashkar S, et al. Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- 27.Sekido Y. Molecular pathogenesis of malignant mesothelioma. Carcinogenesis. 2013;34:1413–1419. doi: 10.1093/carcin/bgt166. [DOI] [PubMed] [Google Scholar]
- 28.Robinson BS, Moberg KH. Cell-cell junctions: alpha-catenin and E-cadherin help fence in Yap1. Curr Biol. 2011;21:R890–892. doi: 10.1016/j.cub.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci U S A. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin F, Tian J, Zhou D, Chen L. Mst1 and Mst2 kinases: regulations and diseases. Cell Biosci. 2013;3:31. doi: 10.1186/2045-3701-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanayama M, et al. Autophagy enhances NFkappaB activity in specific tissue macrophages by sequestering A20 to boost antifungal immunity. Nat Commun. 2015;6:5779. doi: 10.1038/ncomms6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basu S, et al. "Emergency" granulopoiesis in G-CSF-deficient mice in response to Candida albicans infection. Blood. 2000;95:3725–3733. [PubMed] [Google Scholar]
- 33.Lai AY, Lin SM, Kondo M. Heterogeneity of Flt3-expressing multipotent progenitors in mouse bone marrow. J Immunol. 2005;175:5016–5023. doi: 10.4049/jimmunol.175.8.5016. [DOI] [PubMed] [Google Scholar]
- 34.Surh CD, Sprent J. Homeostasis of naive and memory T cells. Immunity. 2008;29:848–862. doi: 10.1016/j.immuni.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Kieper WC, et al. Recent immune status determines the source of antigens that drive homeostatic T cell expansion. J Immunol. 2005;174:3158–3163. doi: 10.4049/jimmunol.174.6.3158. [DOI] [PubMed] [Google Scholar]
- 36.Yang B, et al. Inhibitors directed towards caspase-1 and-3 are less effective than pan caspase inhibition in preventing renal proximal tubular cell apoptosis. Nephron Exp Nephrol. 2004;96:e39–51. doi: 10.1159/000076403. [DOI] [PubMed] [Google Scholar]
- 37.Kourepini E, et al. Osteopontin expression by CD103- dendritic cells drives intestinal inflammation. Proc Natl Acad Sci U S A. 2014;111:E856–865. doi: 10.1073/pnas.1316447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oz HS, Zhong J, de Villiers WJ. Osteopontin ablation attenuates progression of colitis in TNBS model. Dig Dis Sci. 2012;57:1554–1561. doi: 10.1007/s10620-011-2009-z. [DOI] [PubMed] [Google Scholar]
- 39.Heilmann K, et al. Osteopontin as two-sided mediator of intestinal inflammation. J Cell Mol Med. 2009;13:1162–1174. doi: 10.1111/j.1582-4934.2008.00428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toyonaga T, et al. Osteopontin Deficiency Accelerates Spontaneous Colitis in Mice with Disrupted Gut Microbiota and Macrophage Phagocytic Activity. PLoS One. 2015;10:e0135552. doi: 10.1371/journal.pone.0135552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Da Silva AP, et al. Exacerbated tissue destruction in DSS-induced acute colitis of OPN-null mice is associated with downregulation of TNF-alpha expression and non-programmed cell death. J Cell Physiol. 2006;208:629–639. doi: 10.1002/jcp.20701. [DOI] [PubMed] [Google Scholar]
- 42.Hur EM, et al. Osteopontin-induced relapse and progression of autoimmune brain disease through enhanced survival of activated T cells. Nat Immunol. 2007;8:74–83. doi: 10.1038/ni1415. [DOI] [PubMed] [Google Scholar]
- 43.Kanayama M, He YW, Shinohara ML. The lung is protected from spontaneous inflammation by autophagy in myeloid cells. J Immunol. 2015;194:5465–5471. doi: 10.4049/jimmunol.1403249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shinohara ML, et al. T-bet-dependent expression of osteopontin contributes to T cell polarization. Proc Natl Acad Sci U S A. 2005;102:17101–17106. doi: 10.1073/pnas.0508666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kanayama M, et al. Autophagy enhances NFkappaB activity in specific tissue macrophages by sequestering A20 to boost antifungal immunity. Nat Commun. 2015;6:5779. doi: 10.1038/ncomms6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chinen T, et al. Prostaglandin E2 and SOCS1 have a role in intestinal immune tolerance. Nat Commun. 2011;2:190. doi: 10.1038/ncomms1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macosko EZ, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.