Abstract
Aberrant expansion of follicular helper T (TFH) cells occurs in lupus patients. An unanswered question is whether an altered T cell receptor (TCR) repertoire is associated with this expansion. Here, we demonstrate that Blimp-1 repressed expression of the cathepsin S gene (Ctss) which encodes a cysteine protease that cleaves invariant chain and produces antigenic peptides for MHC class II loading. The increased CTSS in dendritic cells (DCs) of female Prdm1 conditional knockout (CKO) mice altered antigen presentation to CD4+ T cells. Analysis of Vβ CDR3s demonstrated a more diverse repertoire of TFH from female CKO mice. In vivo treatment of CKO mice with a CTSS inhibitor abrogated lupus-related phenotypes and reduced the diversity of the TFH TCR repertoire. Thus, Blimp-1 deficiency in DCs leads to a loss of appropriate regulation of Ctss expression in female mice thereby modulating antigen presentation and TFH repertoire to contribute to autoimmunity.
Introduction
The T cell receptor (TCR) repertoire is determined through positive and negative selection of T cells based on recognition by the TCR of peptide–major histocompatibility (MHC) complexes presented by antigen-presenting cells (APCs). In the periphery, CD11chi classical dendritic cells (cDCs) are the primary APCs playing a critical role in both innate and adaptive immune responses1, 2. DCs activate natural killer (NK), NK T, and innate lymphocytes at the site of infection or sterile inflammation. They also process antigens and migrate to local lymphoid organs where they activate naïve T cells3. T cells require signals from a peptide-MHC (MHCII) complex, co-stimulatory molecules and cytokines provided by DCs for differentiation to various subsets of CD4+ T effector cells or CD4+ regulatory cells with each CD4+ T effector cell subset executing unique functions and secreting different cytokines4.
The cytokine milieu is critical to CD4+ T cell differentiation. A dominant cytokine helps establish CD4+ T helper (TH) cell initial polarization; interleukin 12 (IL-12) for TH1, IL-4 for TH2, IL-6 and transforming growth factor-β (TGF-β) for TH17, IL-6 for follicular helper (TFH) and TGF-β and IL-10 for regulatory T (Treg) cells. CD4+ T cell differentiation can be modulated by several other factors such as the type of antigen and dose of exposure, affinity of the TCR for the MHCII complex and the duration of stimulation5, 6.
Antigen-processing pathways have been extensively investigated in mouse DCs. After uptake, antigens are transported into the endolysosomal compartment where they are cleaved and some of the fragments that are generated enter the groove of the MHCII molecule for presentation to CD4+ T cells7. This process is dependent on the action of endocytic proteases in endosomal–lysosomal compartments8 that fall into three main classes: cysteine (cathepsins B, F, H, L, S, Z), aspartate (cathepsins D, E), and serine (cathepsins A, G). While all cathepsins can function in antigen processing and many show an overlapping expression pattern, cathepsin S (CTSS) has been shown to be expressed primarily in professional APCs including B cells and DCs where it plays a critical role in the cleavage of invariant chain (Ii) to permit loading of peptide into MHCII9. CTSS also contributes to antigen processing through degradation of antigen in the endolysosome, helping to establish the pool of peptides that is available for presentation by MHCII10, 11. Appropriate expression of CTSS is critical for establishing the repertoire of immunocompetent cells. Modulation of CTSS and CTSL expression can change the pool of peptides which are presented in vitro to CD4+ T cells10. Overexpression of CTSS in DCs and medullary epithelial cells in the thymus has been shown to permit autoreactive T cells to escape negative selection, presumably through too exuberant degradation of autoantigen12. Whether negative regulation in the periphery is also affected by CTSS has not been addressed.
PRDM1, the gene encoding Blimp-1, was identified as a systemic lupus erythematosus (SLE) risk allele in a genome-wide association studies (GWAS) of Chinese Han and Northern European populations13, 14. We previously demonstrated that CD14+ monocyte-derived DCs (MO-DCs) derived from risk single nucleotide polymorphism (SNP) (C/C) carriers express less PRDM1 transcripts compared to MO-DCs from control SNP (T/T) carriers15. To investigate the pathologic function of Blimp-1 in SLE, we generated mice with a DC-specific deletion of Prdm1 by mating CD11c-Cre to mice with floxed Prdm1 (CKO mice). In female CKO mice, DCs that lack Blimp-1 exhibit an activated phenotype with enhanced MHCII expression and increased production of pro-inflammatory cytokines following Toll-like receptor (TLR) stimulation. These DCs resemble DCs from individuals with the SLE-associated PRDM1 risk allele, which are characterized by increased MHCII expression and hyper-responsiveness to TLR stimulation15. The frequency of TFH cells is increased in the blood of lupus patients16, which correlates with disease activity17, 18. We reported an expansion of TFH cells in female CKO mice which is associated with an increased number of germinal center B cells that produce autoantibodies and cause antibody-mediated glomerulonephritis. We also demonstrated that this phenotype is critically dependent on increased IL-6 produced by Blimp-1 deficient DCs as all autoimmune features are absent in CKO Il6+/− female mice19. In the present study, we show that Blimp-1 also regulates Ctss expression in DCs and that Blimp-1 deficient splenic and bone marrow-derived DCs (BM-DCs) exhibit increased Ctss expression and alterations in antigen presentation compared to wild-type DCs. These alterations lead to a more diverse TCR repertoire in TFH cells of CKO mice. Finally, inhibition of CTSS in CKO mice suppresses the development of the lupus-like phenotype, and is associated with a reduced number and diversity in the TFH TCR repertoire.
Results
Increased cathepsin S expression in Blimp-1-deficient DCs
Blimp-1 has previously been suggested to participate in antigen presentation through downregulation of MHCII transactivator (Ciita) in B cells and CD11c+ DCs20, 21. Consistent with these studies, we had reported increased MHCII expression in Blimp-1 deficient DCs15. To test whether Blimp-1 deficiency in DCs also leads to altered antigen processing, we assayed Blimp-1 deficient DCs from female C57BL/6 mice for expression of H2-Aa, Ciita, H2dmb, and Ctss and other genes involved in antigen presentation.. The expression of H2-Aa, Ciita, H2dmb, and Ctss was significantly higher in Blimp-1 deficient than wild-type DCs whereas other genes showed no difference in expression (Fig. 1a).
Figure 1. Increased Ctss in DCs from female Prdm1 CKO mice.
(a) Splenic CD11c+ cDCs were purified from 6–8 week old female wild-type or CKO mice and gene expression was measured by qPCR using specific primers and relative expression of each gene was calculated by normalization to Polr2a. Mean ± SEM (n=6 from 3 independent experiments). (b) mRNA levels of Ctss and Ctsl were measured in splenic cDCs or BM-DCs from the age-matched female wild-type (open circle) or CKO mice (closed circle) by qPCR. The relative expression was normalized to the level of Hprt. Each dot represents an individual mouse and the bar represents the mean ± SEM (n=9 for splenic DCs and n=6 for BM-DCs from 3 independent experiments). (c) mRNA amount of Ctss in blood cDCs (a representative flow image, top) from age-matched female wild-type or Prdm1 CKO mice was measured by qPCR and the relative expression was normalized to the level of Hprt. Each dot represents an individual mouse and the bar represents the mean± SEM (n=4 from 2 individual experiments). (d) Enzyme activity of CTSS in cDCs was measured in vitro. Fold-increase in CTSS activity was determined by comparing the fluorescence units with the fluorescence of the unincubated control. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=6 from 2 independent experiments). (e) mRNA level of Ctss was measured in splenic DCs of male wild-type and CKO mice. Relative expression was calculated by normalization to Hprt. Mean ± SEM (n=3 from 2 individual experiments). (f) mRNA levels of PRDM1 and CTSS were measured in MO-DCs from control SNP carriers (open box) or from risk SNP carriers (grey box). Relative expression was calculated by normalization to POLR2A. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (female control carriers (n=19), female risk carriers (n=15), male control carriers (n=7) and male risk carriers (n=11)).
To investigate whether Blimp-1 can directly regulate cathepsin gene expression, we searched for the Blimp-1 consensus core binding sequence, 5′-GAAAGT-3′, in a region 1500 bp upstream of the transcriptional start site (TSS) of the first exon of each mouse cathepsin genes22. Ctsc, Ctsd, Ctse, Ctsg, Ctsk, Ctsl1, Ctso, Ctss, and Ctsw contain one or more Blimp-1 consensus sequences (Supplementary Table 1). We, therefore, measured expression of each cathepsin gene in DCs from wild-type and CKO female mice. Expression of Ctss and Ctsl, but not other cathepsins, was increased in splenic DCs and bone marrow-derived DCs (BM-DCs) of CKO female mice (Fig. 1b and Supplementary Fig. 1). Expression of Ctss was also increased in blood DCs of female CKO mice (Fig. 1c). To confirm that the increased expression of Ctss in DCs correlated with increased CTSS functional activity, we assayed CTSS enzymatic activity in DCs purified from spleens of female CKO mice. In vitro enzymatic activity was measured by assaying fluorescence generated upon cleavage of a CTSS substrate. Blimp-1 deficient DCs exhibited increased catalytic activity compared to wild-type DCs (Fig. 1d). We previously observed that CKO male mice do not exhibit altered DC function and do not develop lupus-like disease19. Both Ctss transcript abundance and CTSS catalytic activity were similar in DCs derived from wild-type and CKO male mice (Fig. 1d, e), suggesting that Blimp-1 deficiency in combination with gender-specific alterations in DCs alters Ctss expression.
To address whether CTSS expression is associated with BLIMP-1 expression in human MO-DCs, we measured CTSS expression in MO-DCs of risk and control SNP carriers. MO-DCs from female risk SNP carriers exhibit decreased PRDM1 expression, as reported, and increased CTSS expression compared to MO-DCs from non-risk individuals (Fig. 1f).
Given the presence of a Blimp-1 consensus binding site within the Ctss promoter and the enhanced expression of CTSS in DCs of female CKO mice, we performed chromatin immunoprecipitation (ChIP) assays to determine whether Blimp-1 binds to the Ctss promoter region in splenic CD11c+ cDCs from wild-type mice. As expected, the Ctss promoter region was immunoprecipitated with Blimp-1, indicating Blimp-1 binds in vivo to the Ctss promoter in wild-type, but not CKO mice (Fig. 2a). To test whether Blimp-1 binding negatively regulates transcription of Ctss, assays were performed with a luciferase expression reporter driven by the Ctss promoter (−1300 to +10). There was a significant suppression of luciferase when Blimp-1 was expressed, demonstrating that Blimp-1 downregulates Ctss promoter activity (Fig. 2b). Moreover, Prdm1 siRNA, but not control siRNA, in wild-type DCs led to increased Ctss expression (Fig. 2c) and restoration of Blimp-1 expression suppressed the abundance of Ctss transcripts and CTSS protein in Blimp-1–deficient DCs (Fig. 2d). These data demonstrate that Ctss is a target gene of Blimp-1 in DCs, and that the binding of Blimp-1 to the promoter region of Ctss suppresses transcription.
Figure 2. Blimp-1 regulates Ctss expression in DCs.
(a) Blimp-1 binds to the promoter of Ctss in vivo. DNA was prepared from purified cDCs prepared from spleens of female wild-type or CKO mice, and immunoprecipitated with antibody to Blimp-1 or goat polyclonal IgG. Specific sequences were amplified by Ctss promoter specific primers and visualized in agarose gel (upper panel, one representative image from 4 independent experiments) and quantified by qPCR (lower panel). In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=4 from 4 individual experiments). (b) Increased Blimp-1 suppresses Ctss promoter activity. A Ctss promoter/luciferase plasmid was transfected alone or together with a GFP control vector or a Prdm1 GFP vector at a 2:1 ratio into BM-DCs. 24 hours later, the transfection efficiency was measured by the expression of GFP by flow cytometry (one representative GFP flow image from 4 individual experiments is in the upper panel and quantitation is in the bottom left graph). Ctss-promoter (Luciferase) activity and control luciferase (Renilla) activity was measured immediately after. Luciferase activity was normalized to the level of renilla luciferas. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=4). (c) Knockdown of endogenous Prdm1 increases Ctss expression in BM-DCs. Prdm1 siRNA or scrambled (control) siRNA was transfected into BM-DCs. 48 hours later, Prdm1 and Ctss mRNA were measured by qPCR. Each dot represents an individual experiment and the bar represents the mean ± SEM (n=4). (d) 2 µg of Prdm1 (Blimp-1/IRES/GFP) or control (Luc/IRES/GFP) plasmid was transfected into day 5 Blimp-1 deficient BM-DCs. Two days after transfection, the GFP-positive population of each transfection (upper panel, red rectangle with asterisks, a representative image of each transfection from 3 individual experiments) or unmanipulated cells were purified, and Prdm1 mRNA Blimp-1 and Ctss mRNA and CTSS were measured by qPCR and by immunoblotting. Ctsd mRNA was measured as an irrelevant control. Each dot represents an individual experiment and the bar represents the mean ± SEM (n=3). Immunoblotting image is a representative image from 3 individual experiments.
Gender specificity of altered Ctss expression
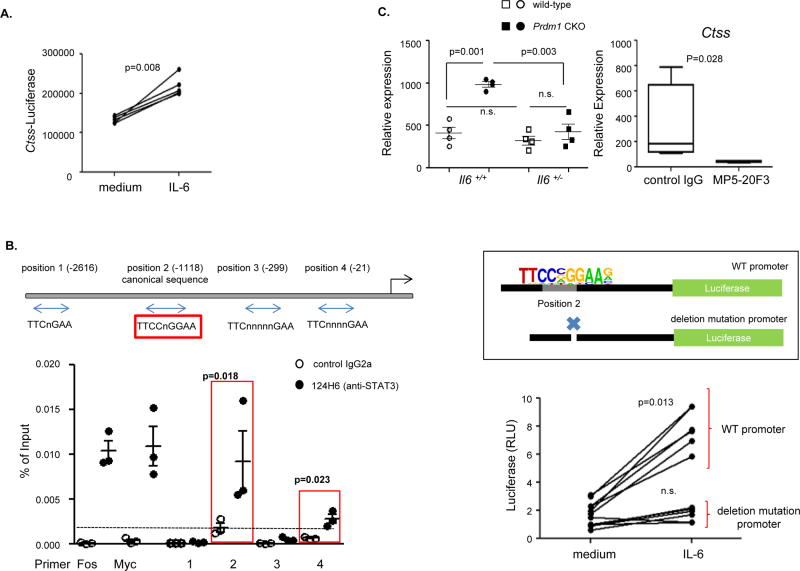
Differential Ctss expression in Blimp-1 deficient DCs from male and female mice suggests that additional regulators are present in cells of female mice that contribute to Blimp-1-mediated Ctss regulation. Our previous study showed that DCs from female CKO mice exhibit increased expression of IL-6 following TLR activation compared to DCs from female wild-type mice, while DCs from male CKO and wild-type mice exhibit similar IL-6 expression19. This observation was not surprising as IL-6 has been shown to be positively regulated by estrogen in BM-DCs23, which we confirmed (Supplementary Fig. 2). The IL-6 signaling pathway activates the transcription factor signal transducer and activator of transcription 3 (STAT3), which has been reported to be involved in the regulation of CTSS activity24 We first asked whether IL-6 exerts a direct effect on Ctss expression. Exogenous IL-6 enhanced Ctss promoter activity in BM-DCs using the luciferase reporter construct described above (Fig. 3a, Supplementary Fig. 3). We also investigated whether STAT3 binds the Ctss promoter. We identified a canonical consensus sequence for STAT3 binding, TTCCnGGAA25, at position −1118, and three additional candidate binding sequences upstream of the transcription initiation site of the Ctss (Fig. 3b)26. STAT3 binding to each region was assessed by ChIP in wild-type BM-DCs. STAT3 bound at −1118 and −21 but not at −2616 and −299, suggesting direct up-regulation of Ctss by IL-6–STAT3 signaling pathway in DCs. To confirm the binding of STAT3 to −1118, we deleted this binding site in the Ctss promoter region of the luciferase construct. IL-6 did not enhance luciferase expression from the construct with the deletion of TTCCnGGAA, while it significantly enhanced luciferase expression from the construct with the intact Ctss promoter (Fig. 3b right).
Figure 3. IL-6 plays a role in Ctss expression in DCs.
(a) Stimulation with IL-6 increases Ctss promoter activity. 2 µg of Ctss promoter/luciferase plasmid was transfected into BM-DCs. Transfected BM-DCs were further cultured with or without IL-6 (100 ng/ml) for 2 hours. Luciferase activity was measured by luminometer. Each dot represents an individual experiment. (b) In vivo binding of STAT3 in Ctss promoter. Putative STAT3 binding sites are depicted and primers are labeled (double arrow) as position 1 through 4. ChIP assay was performed in IL-6 (100 ng/ml) stimulated BM-DCs. After stimulation, BM-DCs were cross-linked and DNA was incubated with either control IgG or anti-STAT3 antibody (124H6) overnight. After precipitation, qPCR was performed with each primer set. Fos and Myc genes were amplified as positive controls. % of input was calculated relative level to the total input. Each dot represents an individual experiment and the bar represents the mean ± SEM (n=3). A mutant luciferase construct was made as depicted in the diagram (box), and the promoter activity was measured by the luciferase assay. Each dot represents an individual sample (n=3). (c) Increased expression of Ctss is abrogated in DCs from Il6+/− Prdm1 CKO mice (left panel) or from anti-IL-6 antibody (MP5-20F3) treated mice (right panel). To inhibit IL-6, 50 µg of anti-IL-6 or control IgG was injected into mice i.p. for 2 weeks. Splenic DCs were prepared from female wild-type or CKO mice (either from Il6+/+ or Il6+/− mice) or from antibody treated mice. Ctss expression was measured by qPCR and the relative expression was calculated by normalization to Polr2a. Each dot represents an individual mouse and the bar represents mean ± SEM (n=4), and in the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=4).
To address the regulation of Ctss expression by IL-6 in vivo, we employed two approaches. First, we analyzed Ctss expression in DCs of CKO female mice that are haplosufficient for IL-6 (Il6+/−). Equivalent Ctss expression was observed in DCs from female Prdm1 CKO Il6+/− mice and wild-type mice (Fig. 3c, left). We also administered neutralizing antibody to female CKO mice. There was a significant decrease in Ctss expression in DCs from mice treated with neutralizing antibody (MP5-20F3) compared to control IgG treated mice (Fig. 3c. right). Thus, IL-6 participates in positive regulation of Ctss in DCs and likely contributes to the increased level of Ctss transcript seen in Blimp-1 deficient female DCs.
Blimp-1 deficient DCs preferentially induce IL-21 producing T cells
The increased numbers of TFH cells in female CKO mice might result from skewed differentiation to TFH induced by Blimp-1 deficient DCs as previously reported19 or from DC-induced changes in gene expression in TFH cells that lead to enhanced survival of TFH cells of CKO mice. Genes encoding transcription factors that positively influence TFH cell numbers include Irf4, Maf, Batf and Bcl627. TFH cells were isolated from female wild-type and CKO mice and expression of these genes was assessed. Only Bcl6 was significantly increased in TFH cells of female CKO mice (Supplementary Fig. 4), suggesting the possibility of enhanced survival of TFH cells.
We reasoned that the increased expression of Ctss in Blimp-1 deficient DCs might affect antigen presentation by MHCII molecules, influencing the TCR repertoire in a manner favorable for the development of autoreactive TFH cells. We, therefore, assayed activation of OT-II CD4+ cells that recognize an ovalbumin (OVA) peptide, amino acids 323–339 (OVA323–339), MHCII complex. CD4+ T cells purified from OT-II mice were co-cultured for 3 days with CD11c+ cDCs isolated from spleens of female CKO or wild-type mice and either intact OVA protein or OVA peptide. T cell proliferation, measured by CFSE dilution, was vigorous in the presence of both antigen preparations, regardless of the source of DCs (Fig. 4a).
Figure 4. Preferential differentiation of IL-21 producing T cells by co-culture with Blimp-1 deficient DCs in vitro.
DCs (spleens of female wild-type (open symbol) or CKO (closed symbol) mice) and OT-II CD4+ T cells were co-cultured for 3 days at various ratios of DCs to T cells with OVA protein (10 µg/ml). (a) To assess proliferation, OT-II T cells were labeled with CFSE (10 µM) before co-culture with DCs. Proliferation was measured by calculation of division index (total number of divisions/number of starting cells). A representative flow image is presented on the left (red line represents wild-type DCs and filled blue represents Blimp-1 deficient DCs). The box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=4). (b) To assess cytokine expression, T cells were harvested after co-culture with DCs (either unfixed DCs or fixed DCs as designated in figure) and total RNA was purified. Cytokine mRNA was measured by qPCR and the relative expression was calculated by normalization to Polr2a. In the box-and-whisker plots, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (unfixed DCs (n=12) and fixed DCs (n=10)). (c) Protein level of cytokines was measured by intracellular staining of T cells. After co-culture, T cells were stimulated with PMA (100 ng/ml)/ionomycin (1 µg/ml) for 6 hours. BFA (20 µg/ml) was added during the last 4 hours of stimulation. Cytokine-positive cells were identified by flow cytometry and their frequency was calculated. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=8). For both (b) and (c), 1 nM CTSS inhibitor was added during the OVA protein incubation with DCs or cells were cultured in transwell plates. Flow images are from one representative set from 4 individual experiments. (d) DQ-OVA protein was incubated with DCs from wild-type (open circle) or CKO (closed circle) mice overnight. After incubation, DCs were extensively washed and OVA uptake was measured by flow cytometry (upper panel, a representative image from 3 individual experiments). The Geo MFI is presented as a dot plot graph (bottom graph). Each do represents an individual experiment and the bar represents the mean ± SEM (n=3).
CD4+ T cell activation is characterized also by cytokine production. Splenic DCs pre-incubated with OVA protein were cultured with OT-II cells and cytokine mRNA expression and the percentage of cells staining positive for cytokine protein were measured by qPCR and flow cytometry, respectively (Fig. 4b, c). Blimp-1 deficient DCs led to a modest but significant increase in IL-2 mRNA, however, the intracellular abundance of IL-2 did not differ between T cells co-cultured with wild-type DCs or Blimp-1 deficient DCs. Secreted IL-2 concentrations were also not different (Supplementary Fig. 5), explaining the lack of difference in proliferation index when T cells were stimulated with either wild-type or Blimp-1 deficient DCs.
Many cytokines that are preferentially expressed by TH1, TH2, TH17 or Treg cells were not significantly different between cultures with Blimp-1 deficient DCs or wild-type DCs (Supplementary Fig. 6). While the expression of IL-21, produced by TFH cells, was relatively low compared to other cytokines, IL-21 transcription and protein production were greater in cultures with Blimp-1 deficient DCs and intact ovalbumin (Fig. 4b, c and Supplementary Fig. 5). The increased IL-21 was dependent on antigen-processing, since it was abrogated in cultures in which OVA323–339 peptide was used as antigen (Supplementary Fig. 7) and on T-DC contact as it was also abrogated in transwell cultures (Fig. 4b,c). Fixation of DCs after incubation with OVA protein also revealed increased expression of IL-21 in cultures with Blimp-1 deficient DCs compared to wild-type DCs, although IL-21 production by fixed DCs of both strains was less than is seen in unfixed cells (Fig. 4b), presumably due to both the absence of cytokine secretion and the absence of membrane fluidity.
The activation of OT-II T cells by OVA protein represented a cognate interaction between T cells and DCs, since the TCR-transgenic T cells were not activated with an irrelevant antigen (hen egg lysozyme, HEL) (Supplementary Fig. 8). To confirm that the enhanced T cell activation mediated by Blimp-1 deficient DCs resulted from increased CTSS, we treated cultures with a CTSS inhibitor, 219393 (~ 400 fold greater selectivity over CTSB)28 or a CTSB inhibitor as a control. When the CTSS inhibitor (Fig. 4b, c), but not the CTSB inhibitor (Supplementary Fig. 8), was added during OVA processing, there was no increased IL-21 production seen in cultures with Blimp-1 deficient DCs. To confirm that CTSS functions to alter antigen processing rather than antigen uptake fluorescence-conjugated DQ-OVA protein was incubated with purified DCs from each strain overnight, and OVA uptake was assessed by flow cytometry. There was no significant difference in the level of OVA internalized by wild-type and Blimp-1 deficient DCs (Fig. 4d). These data strongly suggest that Blimp-1 deficient DCs have altered antigen processing. Thus, increased CTSS alters antigen presentation which induces activated CD4+ T cells differentiates to TFH cells.
Diversity of the TFH TCR repertoire
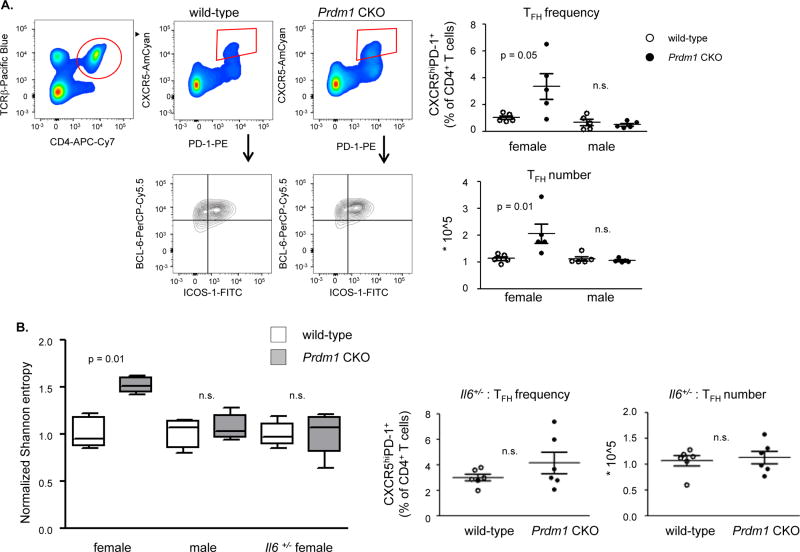
To ask whether the expansion of TFH cells in CKO mice is accompanied by a change in their antigenic specificity, we compared the TCR repertoire of TFH cells in wild-type and CKO mice. TFH cells were purified from the spleens of young (6–10 weeks old) female and male mice. TFH cells were identified by the expression of CXCR5 and PD-1; they also expressed Bcl-6 (~95%) and ICOS-1 (~70%) (Fig. 5a). Although no autoantibodies are detected at this age, there was already an increase in number of TFH cells in female CKO mice compared to wild-type mice (Fig. 5a). We focused on Vβ sequences as these are more diverse than Vα sequences. We analyzed over 100,000 TFH Vβ sequences from each of 5 female and 5 male mice of each strain. TFH cells from female CKO mice exhibited a more diverse repertoire than TFH cells of female wild-type mice. Importantly, TFH cells from male CKO and wild-type mice exhibited similar diversity (Fig. 5b).
Figure 5. TFH cells of female CKO mice harbor diverse CDR3 repertoire.
(a) Flow image and frequency of TFH cells. Spleens were harvested from the age-matched female or male CKO (closed circle) or wild-type (open circle) mice, and TFH cells (red box) were identified by flow cytometry (left panel, a representative set from 3 individual experiments). Percentage of TFH cells was calculated based on the CD4+ T cells and the number of TFH cells was plotted. Each dot represents an individual mouse and the bar represents the mean ± SEM (3 individual experiments). (b) TCR Vβ diversity in TFH cells was analyzed from female or male wild-type or Prdm1 CKO, or from female Il6+− wild-type or Il6+/− CKO. Each dot represents the average true Shannon diversity obtained from 105 sequences per mouse, normalized to the wild-type control group (n=5). Frequency and number of TFH cells in spleens from female Il6+/− wild-type or CKO mice were plotted. Each dot represents an individual mouse and the bar represents the mean ± SEM.
To confirm that the diverse TCR Vβ repertoire in TFH cells was associated with autoimmunity, we examined the Vβ repertoire of wild-type and CKO female mice haplosufficient for Il6. As previously shown, neither of these strains develops disease. No difference in repertoire diversity was seen consistent with their similar TFH cell numbers (Fig. 5b). Thus, the more diverse repertoire was seen only in association with a lupus phenotype.
CTSS inhibitor suppresses the development of lupus
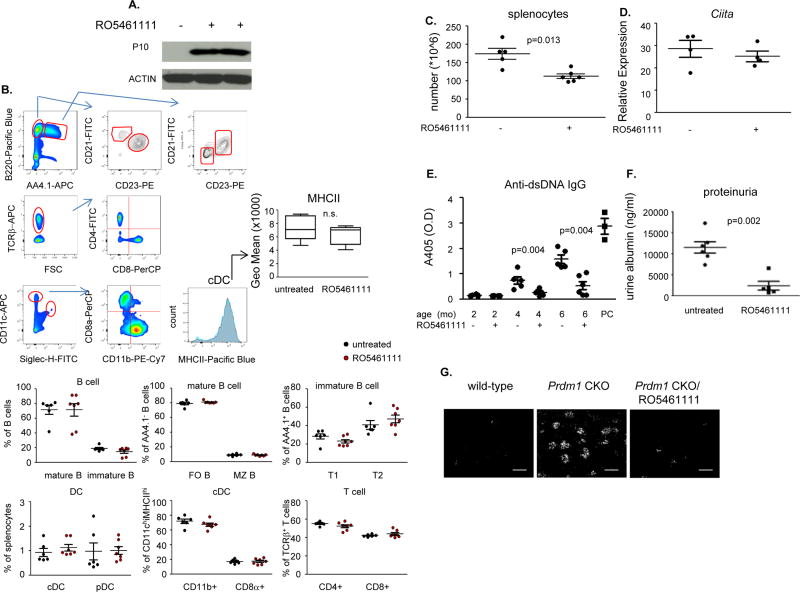
Based on the increased activity of CTSS in Blimp-1 deficient DCs and the promising effects of CTSS inhibition on TFH differentiation in vitro, we investigated whether CTSS inhibition would suppress the development of the lupus-like phenotype in CKO mice as had previously been shown in MRL/lpr mice29. First, we assessed lymphocyte development following long-term treatment with RO5461111. Accumulation of the p10 Ii fragment of invariant chain was seen in splenocytes of RO5461111-treated wild-type and CKO mice but not in splenocytes of mice fed control chow, demonstrating the efficacy of the treatment (Fig. 6a). RO5461111 treatment did not alter the overall frequency of either lymphocytes or DC subsets in wild-type mice (Fig. 6b). DC activation measured by class II expression was also not affected (Fig. 6b). RO5461111 was given to female CKO mice to investigate whether CTSS inhibition prevents the development of lupus-like phenotypes. Splenomegaly was suppressed by CTSS inhibition with a reduction in the number of splenocytes in RO5461111-treated CKO mice (Fig. 6c). Ciita expression was not altered in splenic DCs in RO5461111 treated CKO mice (Fig. 6d). RO5461111-treated CKO mice showed significantly reduced titers of anti-dsDNA antibodies at both 4 and 6 months (Fig. 6e). Proteinuria and IgG deposition in glomeruli was also significantly lower in RO5461111 treated compared to non-treated CKO mice (Fig. 6f, g).
Figure 6. CTSS inhibitor, RO5461111, prevents development of lupus-like phenotypes in female CKO mice.
(a) Efficacy of RO5461111 was measured by accumulation of p10 fragment. Wild-type mice were fed either normal chow or RO5461111 chow for 2 weeks, and p10 from total splenocytes was measured by immunoblot. One representative image is from 3 independent experiments. (b) Lymphocyte and DC subsets and the MHCII expression of DCs was analyzed by flow cytometry (one representative set image from 3 individual experiments). Graph is the mean ± SEM (n=3). (c) Female CKO mice fed normal or RO5461111 chow for 4 months. Mice were sacrificed at 6 months of age and total number of splenocytes was determined. Each dot represents an individual mouse and the bar represents the mean ± SEM (n=5 and 6 from 2 individual experiments). (d) Ciita expression was measured in splenic DCs purified from CKO mice fed with either control chow or RO5461111 treated chow. Expression was normalized to the level of Polr2a. Each dot represents an individual mouse and the bar represents the mean ± SEM. (e) Blood was drawn from 2, 4 and 6 month old mice and anti-dsDNA IgG was measured by ELISA. PC is a positive control serum from sick female CKO mice. Each dot represents an individual mouse and the bar represents mean ± SEM (n=5 and 6 from 2 individual experiments). (f) Urine samples were collected from 6 month old control or RO5461111 treated female CKO mice, and proteinurea was measured by urine albumin ELISA. Each dot represents an individual mouse and the bar represents mean ± SEM (n=6 and 5 from 2 individual experiments). (g) IgG deposition in kidneys was determined from 6 month old wild-type and CKO mice either control chow or RO5461111 chow treated mice. Image was acquired on a fluorescence microscope (10 × magnification; bar is 100 µm). Each image represents from 5 individual samples per group.
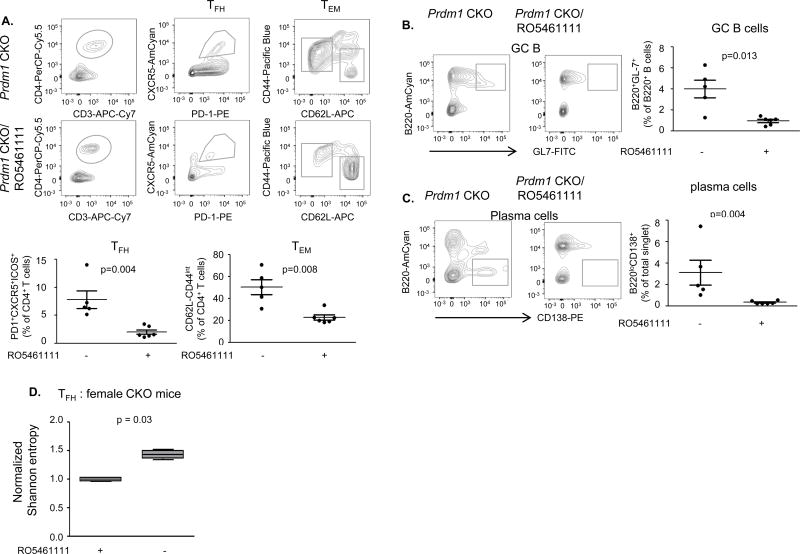
The frequency of TFH and TEM cells was significantly reduced in RO5461111-treated CKO mice (~3.5- and 2-fold respectively) (Fig. 7a) while the frequency of naïve T cells (CD44loCD62Lhi) was slightly higher in RO5461111-treated than in non-treated CKO mice (~ 2 fold). Consistent with the reduction in TFH cells, the frequency of GC B cells and plasma cells was also severely decreased (~ 10 fold) in RO5461111-treated CKO mice (Fig. 7b, c). These data demonstrate increased CTSS expression promotes the development of a lupus-like phenotype in CKO mice, and that inhibition of CTSS activity can suppress the development of disease.
Figure 7. RO5461111 suppresses expansion of TFH cells and GC B cells in CKO mice.
6 month old female CKO mice or RO5461111 treated female CKO mice were sacrificed and lymphocytes were investigated by flow cytometry. (a) TFH and TEM cells were identified (upper panel represents the staining panel and population, a representative image is from 5 individual experiments) and the frequency was calculated (bottom panel). (b–c) GC B cells and plasma cells were stained as described in the flow panel and the frequency of each population was calculated. Each dot represents an individual mouse and the bar represents mean ± SEM (n=3). A representative image is from 5 individual experiments. (d) CTSS inhibitor treatment reduces the TCR diversity of TFH cells in female CKO. Each dot represents the average true Shannon diversity of subsamples of 105 sequences per mouse, normalized to the CKO control group. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=4).
Finally, the Vβ repertoire was significantly less diverse in TFH from RO5461111-treated CKO mice than untreated CKO mice (Fig. 7d). Thus, higher CTSS expression and activity in DCs leads to a TFH population with greater TCR diversity.
Discussion
Alterations in antigen presentation can lead to autoimmune or inflammatory diseases30. Human genetic studies have shown that HLAII genes represent prominent risk alleles for autoimmune diseases, including SLE31, 32. As HLA molecules are critical in establishing thresholds for T cell selection and activation, the TCR repertoire has been presumed to be a key contributor to many autoimmune diseases33.
A role for Blimp-1 in antigen presentation was initially suggested in a study showing that Blimp-1 regulates expression of Ciita, a positive regulator of MHCII expression in B cells20. More recently, Blimp-1 was shown to regulate MHCII in DCs, thereby affecting the threshold for CD4+ TH cell activation in a mouse model of experimental autoimmune encephalomyelitis (EAE)34.
The importance of antigen presentation prompted us to investigate whether Blimp-1 alters antigen presentation to T cells and, thus, the TCR repertoire. Blimp-1 deficient DCs show altered expression of several genes involved in antigen presentation including Ctss. Blimp-1 deletion led to increased Ctss transcription in DCs because Blimp-1 is a transcriptional repressor of Ctss and of Il6. IL-6 is highly expressed in Blimp-1 deficient DCs of female mice leading to increased Ctss expression. Thus, Bimp-1 directly and indirectly regulates Ctss expression. As described in a previous study, increased IL-6 and STAT3 activation also decreases Cst3 expression, subsequently enhancing CTSS activity24. The increased CTSS activity in DCs of female CKO mice led to increased diversity in the TCR repertoire of TFH cells which was abrogated by a CTSS-specific inhibitor. These findings suggest that an increase in CTSS expression associated with absence or low expression of Blimp-1 generates a repertoire more skewed to self- reactivity. While it is likely that the PRDM1 risk allele for SLE leads to a more diverse TFH repertoire also, this requires confirmation in risk allele and non-risk allele human subjects.
The importance of appropriate expression of CTSS in antigen presentation is now widely accepted and its molecular mechanism has been investigated in other studies.. Modulation of expression of CTSS and CTSL led to alterations in the pool of peptides which are presented in class II molecules in an in vitro study10, 35. Thymic DCs express CTSS and efficiently cleave a number of known autoantigens; when CTSS concentrations are increased, it is presumed those T cell epitopes that mediate negative selection are destroyed, allowing escape of autoreactive T cells to the periphery12. Conversely, CTSS deficient mice are markedly resistant to the development of experimental autoimmune myasthenia gravis (EAMG)36. Our current study definitively shows that enhanced CTSS in DCs can lead to an alteration of T cell repertoire in the TFH compartment. Notably, increased CTSS was observed in serum from SLE and lupus nephritis patients37
An increased frequency of TFH cells in peripheral lymphoid organs is a commonly observed phenotype in animal models of SLE38,39. However, it has not been clearly understood whether an increased number of TFH cells predisposes to autoimmunity or whether the antigenic specificity of the TFH cells in these models is also altered. The antigenic specificity of activated CD4+ T cells is largely determined by encounter with peptide: MHCII complexes on DCs40, 41. While TCR selection occurs in the thymus, differentiation of effector T cells occurs in the periphery when CD4+ T cells encounter APCs presenting specific peptide: MHCII complexes42. We do not yet know whether the differential TCR repertoire of TFH cells observed in our study is determined by selection in the thymus or activation in the periphery. Nonetheless, this study demonstrates the importance of precise regulation of CTSS in APCs in determining the T cell repertoire and the fate of autoreactive T cells.
Recently, a CTSS inhibitor was shown to prevent disease onset in the MRL/lpr lupus mouse model29. Our study also demonstrated that CTSS inhibition can prevent disease onset and lupus related phenotypes in CKO mice. More interestingly, inhibitor treatment made the TCR repertoire of TFH cells less diverse. Because long-term treatment with inhibitor did not reduce MHCII expression on DCs, we speculate that the major function of the inhibitor is to change the pool of peptides presented on class II in vivo.
These data help explain how the SLE-associated PRDM1 risk allele with low expression of Blimp-1 and high expression of IL-6 in DCs contributes to the risk of developing SLE.
Online Methods
Mice
Prdm1 CKO mice (Prdm1fl/fCd11c-Cre+) and wild-type (littermate control Prdm1fl/flCd11c-Cre−) mice on a C57BL/6 background are bred and maintained in a specific pathogen-free facility at the Feinstein Institute for Medical Research. OT-II mice were purchased from Jackson laboratory (B6.Cg-Tg (TcraTcrb) 425Cbn/J).
All the experiments conducted in this study strictly followed the guidance in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the committee on the Ethics of Animal Welfare of The Feinstein Institute for Medical Research (protocol number 2009-048). All the animals were euthanized at the end time point of experiments by CO2 instillation.
Sample size to achieve adequate power was chosen based on our previous studies with similar methods. We randomized the female or male mice from different cages and different time points to exclude cage or batch variation. Experiments and data analysis was performed without afore-mentioned genotype or treatment information.
Preparation of PBMCs, blood DCs and in vitro differentiation of MO-DCs
The protocol for study of human samples was approved by the IRB of the Feinstein Institute for Medical Research (FIMR) (approval number: 09-081A). Healthy PRDM1 rs548234 risk allele carriers and non-risk allele controls were identified from the Genotype and Phenotype (GAP) registry at the FIMR and obtained the informed consent from all participants. Both cohorts consisted of hormonally active females under 55 years old and were of various race and ethnicity. Participants consented for the study prior to their participation. Total PBMCs were collected by Ficoll-Paque gradient centrifugation. Briefly, whole blood or leukopack was diluted with HBSS (Life Technologies) and layered on the Ficoll (GE Healthcare Life Sciences). Cells were centrifuged at 750g for 20 min without a break at 20 °C. PBMCs were collected from the intermediate layer and washed three times with HBSS.
To generate MO-DCs, CD14+ monocytes were purified by EasySep kit (Stem Cell Technologies) according to the manufacturer’s protocol. The purity of CD14+ cells was determined by flow cytometry with an LRSII (BD Biosciences). After purification, CD14+ monocytes were cultured with RPMI 1640 supplemented with 10% heat-inactivated FBS, 1% Penicillin-Streptomycin (P/S), 1% L-glutamine, 100 ng/ml of recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) (Peprotech) and 100 ng/ml of recombinant human IL-4 (Peprotech) for 7 days.
CTSS inhibitor treatment
The protocol was adapted from a published study29 with modifications. Briefly, three-week-old female Prdm1 CKO mice were randomized to receive either a medicated diet formulated by mixing the CTSS inhibitor (RO5461111 (provided by F.Hoffman-La Roche), 262.5 mg/kg chow) or standard diet (control) (n = 20 in each group). When the mice became 7–8 weeks old (duration of inhibitor treatment is a minimum of 4 weeks), mice were sacrificed and spleens were collected for TFH cell TCR repertoire experiments (n = 5 in each group). Additional mice were sacrificed at age 6 months or 10 months to investigate lupus-like phenotypes (n = 10 in each group) and lupus nephritis phenotype (n = 5 in each group), respectively.
To confirm the CTSS inhibitor activity in vivo, p10 of Ii accumulation was measured in total splenocytes by immunoblotting. Spleens were collected from control mice and RO5461111 treated mice and lysed in RIPA buffer (Thermo Fisher Scientific), containing protease inhibitor (Roche). Total protein was transferred to polyvinylidene fluoride (PVDF) and the membrane was incubated with anti-CD74 polyclonal antibodies (R&D systems). The membrane was developed with ECL (Thermo Fisher Scientific).
Antibodies and reagents
Taqman primers were purchased from Applied Biosystems: Mm00515580_m1 (Ctsc), Mm00515586_m1 (Ctsd), Mm00456010_m1 (Ctse), Mm00456011_m1 (Ctsg), Mm00484039_m1 (Ctsk), Mm00617413_m1 (Ctso), Mm00515599_m1 (Ctsw), Mm01255859_m1 (CtsS), Mm00515597_m1 (Ctsl), Mm00613524_m1 (March1), Mm00439211_m1 (H2-Aa), Mm00482914_m1 (Ciita), Mm00783707_s1 (H2-dmb2), Mm00511327_m1 (Zbtb46), Mm00658576_m1 (CD74), Mm00468476_m1 (H2-oa), Mm01325350_m1 (Lgmn), Mm00434455_m1 (Itgam), Mm00438347_m1 (Cst3), Mm00499585_m1 (CD209b), Mm00516431_m1 (Irf4), Mm00477633_m1 (Bcl6), Mm00581355_s1 (Maf), Mm00479410_m1 (Batf), Mm00839502_m1 (Polr2a), and Mm03024075_m1 (Hprt). Light cycler 2× master mix was purchased from Roche. OVA protein and OVA323–339 peptide were purchased from Invivogen, and Fluorescence labeled OVA protein was purchased from Molecular Probes.
For the ChIP assay, goat polyclonal anti-Blimp-1 antibody (sc-13206x) and goat control antibody were purchased from Santa Cruz. For immunoblotting, anti-Blimp-1 (sc-13026x, Santa Cruz), anti-CTSS (ab18822, Abcam), anti-CD74 (R&D Systems) and anti-actin (ab6276, Abcam) were used. Anti-IL-6 neutralizing antibody (MP5-20F3) and control antibody were purchased from eBioscience. To detect intracellular cytokines, anti-IL-2 (JES6-5H4, BioLegend) and IL-21R-Fc (R&D Systems) were purchased. For flow cytometry analysis, antibodies were purchased from BioLegend (anti-TCRβ (H57-597), anti-CXCR5 (J252D4), anti-ICOS-1 (C398.4A), anti-CD8 (53-6.7), anti-CD11b (M1/70), anti-CD44 (IM7) and anti-CD62L (MEL-14)), eBioscience (anti-Bcl-6 (BCL-DWN), anti-CD4 (GK1.5), anti-PD-1 (J43), anti-B220 (RA3-6B2), anti-AA4.1 (AA4.1), anti-CD21 (7G6), anti-CD23 (B3B4), anti-CD11c (N418) and anti-MHC II (M5/114.15.2)) or BD Bioscience (anti-GL7 (GL7) and anti-CD138 (281-2).
CTSS inhibitor and CTSB inhibitor, used for in vitro inhibition assays, were purchased from Calbiochem.
Purification of TFH cells for TCRβ sequencing
Age- and gender-matched control and Prdm1 CKO mice, or Il6+/− female Prdm1 CKO mice were sacrificed at 6–8 weeks old and spleens were collected for TFH cell purification. CD4+ TFH cells (defined by TCRβ+CD4+CXCR5+PD-1+ICOS-1+) were sorted on the FACSAria (BD Biosciences). Sorted TFH cells were snap-frozen in liquid nitrogen and kept at −80 °C until sequencing.
Amplification and analysis of the TFH TCR repertoire diversity
For each FACS-sorted TFH cell population, the cells were treated with 1 ml of TRI reagent (Life Technologies) and total RNA was isolated according to the manufacturer’s protocol of the RNeasy Micro Kit (Qiagen). First-strand cDNA was generated from total RNA using a SuperScript RT II kit (Invitrogen) and oligo-dT primer. The resulting cDNA was used as template for FastStart High Fidelity PCR amplification (Roche) using mouse-specific barcoded constant region primer (5’-GCACTGATGTTCTGTGTGACAG-3′) and 23 Vβ-specific primers43. The PCR products were gel-purified to isolate the amplified TCR Vβ DNA and total quantities were determined with a 2100 Bioanalyzer (Agilent). 200 ng of DNA per mouse was processed for Illumina MiSeq DNA sequencing according to the manufacturer’s protocol and sequenced, yielding 2–5 × 105 raw sequences per mouse.
All sequencing data was first processed using the sequence quality and signal filters of the Illumina MiSeq pipeline and then subjected to bioinformatics analysis that relied on homologies to conserved framework regions using the MiXCR software package. Vβ CDR3 sequences were then clustered using the CD-HIT software package with a 90% amino acid sequence similarity cut-off. Repertoire diversity for each mouse was determined by calculating the true Shannon diversity of 3 or more subsamples of 105 sequences and the results were averaged for each mouse and the diversity indices normalized to the wild-type mice.
In vitro OT-II T cell activation with DCs
CD11chiMHCIIhi cDCs were sorted from spleens of age-matched female control or Blimp-1 CKO mice (6–8 weeks old) by FACSAria. cDCs were resuspended at 106/ml with recombinant mouse GM-CSF (20 ng/ml) and 100 µl (105 cDCs) was plated at 96-well flat-bottom plate (Thermo Fisher Scientific). CD4+ T cells were sorted from lymph nodes of female OT-II mice and mixed with cDCs at ratio of 1:10 (DC:T). For OVA protein-derived antigen presentation, 10 µg/ml of OVA protein (InvivoGen) was added to cDCs and incubated for overnight before T cell co-culture. OVA323–339 peptide (InvivoGen) (0.1–10 ng/ml) or HEL (10 µg/ml) (Sigma-Aldrich) was added to cDCs. CTSS inhibitor (1 nM) or CTSB (10 nM), was added with OVA protein incubation in certain experiments. To measure T cell proliferation, OT-II CD4+ T cells were labeled with 10 µM CFSE (Invitrogen). Transwell experiments were performed in 0.4 µm plates (Corning). cDCs purified from wild-type or CKO mice were plated on upper wells and CD4+ OT-II T cells were cultured with OVA-presenting wild-type DCs on the bottom wells. Purified DCs from wild-type or CKO mice were cultured with OVA protein for 6 hour and fixed with 0.001% glutaraldehyde for 30 sec. Fixation was terminated by 0.2 M glycine treatment followed by washing three times with PBS.
Measurement of CTSS activity
In vitro CTSS enzyme activity was measured by fluorometric analysis kit (Abcam). DCs were purified by cell sorter and equal number of DCs (1 × 106) from wild-type or Prdm1 CKO mice were lysed in cell lysis buffer. An equal volume of reaction buffer was added into the cell lysis and amino-4-trifluoromethyl coumarin (AFC)-labeled CTSS substrate peptide (Ac-VVR-AFC) (200 µM) was added and incubated for 2 hour at 37 °C. For negative control, CTSS inhibitor was added to the substrate mixture or a reaction mixture that does not contain cell lysate was prepared. After incubation, samples were read in a fluorometer with a 400 nm excitation and 505 nm emission. Fold change in CTSS activity was determined by comparing the relative fluorescence units (RFU) to the level in the negative control sample.
ChIP assay
5 µg of anti-BLIMP-1 polyclonal antibody (sc-13206, Santa Cruz Biotech) or normal goat IgG were conjugated with 50 µl of Protein G-magnetic beads (Invitrogen) by overnight incubation. CD11chiMHCIIhi cDCs were purified from spleens of wild-type or Prdm1 CKO mice. Purified DCs (5–10 × 106 cells per experiment) were cross-linked in 1% formaldehyde (final concentration) for 10 min at 20 °C. To quench cross-linking, 1.375 M Glycine (100 µl/ml) was added and cells were washed three times with ice-cold PBS. Cells were lysed in cell lysis buffer and subjected to sonication. Antibody-conjugated magnetic beads were incubated with sonicated DNA at 4 °C with rotation. The next day, unbound DNA was washed away and the antibody bound complex was eluted in elution buffer. The protein and DNA complex was de-crosslinked at 65 °C overnight. DNA was further purified by RNase treatment and DNA cleanup using PCR cleanup kit (QIAGEN). Purified DNA was used for PCR.
Kidney histology
Kidneys collected from animals at the end of experiments (6 month-old) were cut into two pieces longitudinally. One half was frozen with tissue freezing, Tissue-Tek O.C.T. compound medium (Fisher) and the other half was immediately fixed with 4% formaldehyde (Sigma-Aldrich) and kept in 70 % alcohol at 4 °C. Frozen or fixed sections were sliced to 7 mm thickness and stored until staining. To detect immunoglobulin deposition, frozen kidney section was fixed with Acetone (−20 °C for 5 min) and blocked with blocking solution (2 % BSA, 0.5 % TritonX-100, 2 % normal goat serum (Invitrogen) in PBS) for 1 hour at 20 °C. Alexafluoro 488-conjugated anti-mouse IgG (1:200 in blocking solution) was incubated with tissue for 2 hour at 20 °C. Tissue was washed with PBS three times and analyzed by microscope.
Anti-dsDNA and albumin ELISA
To measure the titers of anti-dsDNA antibodies, Prdm1 CKO mice or RO5461111 fed Prdm1 CKO mice were bled at indicated ages (2 months, 4 months and 6 months old). Mouse serum was prepared from blood and frozen at −20 °C until assayed. 96-well plates (Costar) were coated with 100 µg/ml of sonicated and filtered calf thymus DNAs (Sigma-Aldrich). The plate was blocked with 3% FCS for 1 hour at 20 °C, and diluted samples from experimental mice or positive control (old NZB/W F1) mice were incubated for 2 hours. The plate was washed and alkaline phosphatase-conjugated anti-mouse IgG were applied. The reaction was developed with p-nitrophenyl phosphate (Sigma-Aldrich). OD was monitored at 405 nm.
Urine samples were collected from 6 month old mice maintained in metabolic cages. Albumin concentrations in urine samples were measured by mouse albumin ELISA quantitation set (Bethyl Laboratories, Inc.) following the manufacturer’s protocol.
Luciferase assays
BM-DCs (2 × 106 cells) were transfected with a wild-type or mutated Ctss promoter construct by Nucleofector kit (Lonza). Luciferase plasmid and Tk-Renilla luciferase plasmid was added to the cells at a 10:1 ratio. Relative light units were measured 6–12 hours post transfection using Dual-Luciferase reporter assay system (Promega).
Transfection of siRNA and Blimp-1 expression vector
SiRNAs (SR420429) targeting Blimp-1 was purchased from Origene and 200 nM of each siRNA or control scrambled siRNA were used for each transfection. Blimp-1 plasmid and control plasmid were purchased from Genecopoeia. siRNAs or plasmid were mixed with DCs in Nucleofector solution and transfected by Amaxa electroporation (program Y-01). Transfected DCs were immediately transferred to culture media (supplemented with GM-CSF) and further cultured for 2 days.
Bio-Plex
Multiple cytokines were measured by Bio-Plex pro mouse cytokine standard 23-plex, group I and group III (Bio-Rad) followed by manufacturer’s protocol. The assay was measured by Bio-Plex suspension array system and the data was analyzed by Bio-Plex manager software.
Statistics
Statistical significance was calculated and determined by a nonparametric, Mann-Whitney test, and P- values less than 0.05 was considered significant. No exclusion of sample was done.
Supplementary Material
Supplemental figure 1: Expression of cathepsin genes in splenic DCs from wild-type or CKO mice. DCs were purified from the spleens of age matched female wild-type or CKO mice, and RNA was purified for qPCR. Gene specific primers were used for each cathepsin gene and the relative expression of each was calculated by normalization to the level of Hprt. Each dot represents an individual mouse and the bar represents the mean ± SEM (n=5).
Supplemental figure 2: Increased expression of Il6 by estrogen in BM-DCs. BM-DCs were differentiated with GM-CSF (200 ng/ml) in hormone-free culture system without or with E2, ICI or E2 and ICI together for 7 days. On day 7, BM-DCs were counted and further cultured with or without LPS (1 µg/ml) for overnight. Total RNA was purified and Il6 transcript was assessed by qPCR. Relative level of Il6 was normalized to housekeeping gene Polr2a. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=7).
Supplemental figure 3: Diagram of Ctss reporter construct.
Supplemental figure 4: Gene expression in TFH and non-TFH cells. Spleens were collected from age-matched female wild-type (open circle) or CKO (closed circle) mice. TFH cells and non-TFH cells were identified as represented in the flow image (left panel), and each population was purified. Total RNA was purified and gene expression was measured by qPCR. Relative level of each gene was normalized to housekeeping gene Polr2a. Each dot represents an individual mouse and the bar represents the mean ± SEM (n=3).
Supplemental figure 5: Increased IL-21 protein in the supernatant from co-culture of OT-II CD4+ T cells and Blimp-1 deficient DCs. CD4+ T cells from OT-II mice and splenic DCs (open box is wild-type mice and grey box is CKO mice) were co-cultured in various conditions for 4 days. To measure the secreted cytokines, IL-2 and IL-21, supernatant was collected and cytokines were detected by a multiplex assay. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=6).
Supplemental figure 6: Cytokine production in the supernatant from day 3 co-cultures of OT-II T cells and DCs. DCs were purified from the spleens of female wild-type (open box) or CKO (grey box) mice, and CD4+ T cells were purified from female OT-II mice. DCs and T cells were co-cultured for 3 days at a 1:10 ratio (DC:T) with either OVA323–339 peptide (100 ng/ml) or OVA protein (10 µg/ml). Cytokine levels were measured in the supernatant by multiplex assay. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=6).
Supplementary figure 7: Proliferation and IL-21 production of OT-II T cells by OVA323–339 presenting DCs. OT-II T cells were co-cultured with splenic DCs for 3 days at 1:10 ratio (DC:T cells) in the presence of OVA323–339 peptide. To assess proliferation, T cells were labeled with CFSE (10 mM) before the culture and proliferation was measured by calculation of the division index (left panel). In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=3). To assess IL-21 production, culture supernatant was harvested and measured by Multiplex assay (right panel). In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=3).
Supplemental figure 8: Protein level of cytokines, IL-2 and IL-21, was measured by intracellular staining of T cells. OT-II T cells were co-cultured with splenic DCs that had been incubated with HEL (10 µ/ml) or OVA (10 µg/ml). CTSB inhibitor (1 nM) was added during processing of OVA protein antigen. After co-culture for 4 days, T cells were stimulated with PMA (100 ng/ml)/ionomycin (1 µg/ml) for 6 hours. BFA (20 µg/ml) was added during the last 4 hours of stimulation. Cytokine-positive cells were identified by flow cytometry and frequency was calculated and presented in the graph. Each dot represents an individual experiment and the bar represents the mean ± SEM (n=4).
Supplementary Table 1: Position of Blimp-1 binding consensus sequence in cathepsin genes and expression of each gene in Blimp-1 ko DCs. Blimp-1 binding sequence was investigated in the upstream of (−1500 to +10 from transcription initiation site (TSS)) each cathepsin gene. Amount of transcript was measured by qPCR from wild-type or Blimp-1 ko DCs. If there is difference more than 2-fold, it is indicated as “increased”. N.D (not detected).
Acknowledgments
We thank H. Borrero and C. Colon for assistance with the flow cytometry; G. Klein and M. DeFranco RN for recruiting PRDM1 genotyped subjects; C. Chrysostomou for helpful discussion and assistance in bioinformatic analysis. The study was supported by NIAMS, US National Institutes of Health (R01 AR065209 to S.J.K. and B.D.), Alliance of Lupus Research grant for B.D., and DTRA contract HDTRA 1-12-C-0105 for G.G.
Footnotes
Author contributions
All contributing authors have agreed to the submission of this manuscript for publications. B.D. designed the study, and analyzed and interpreted results. S.J.K. designed and performed experiments, analyzed data and interpreted results. S.S. performed the sequencing experiments and analyzed data. G.G. designed the sequencing experiments and interpreted data. S.H.J performed the ChIP experiments and Il6 promoter assay. S.S.A and W.H. provided study material, RO5461111, and critically reviewed the manuscript and agreed to the submission of this manuscript for publication. P.K.G. provided the samples for human study. B.D., S.J.K., S.S., and G.G. participated in the interpretation of the study and wrote the manuscript, as well as providing critical review of the paper.
Competing financial interests
The authors S.J.K, S.H.J., S.S., G.G. and B.D declare no competing financial interests. The authors S.S.A. and W.H. are employed by Roche Pharmaceuticals and hold stocks in Roche Pharmaceuticals.
Data availability.
All TCRβ of TFH cells sequences war files that support the findings of this study have been deposited at BioProject under the accession numbers SAMN-7927868-77. The other data are available from the corresponding author upon request.
References
- 1.Pavli P, Hume DA, Van De Pol E, Doe WF. Dendritic cells, the major antigen-presenting cells of the human colonic lamina propria. Immunology. 1993;78:132–141. [PMC free article] [PubMed] [Google Scholar]
- 2.Guilliams M, et al. Skin-draining lymph nodes contain dermis-derived CD103(−) dendritic cells that constitutively produce retinoic acid and induce Foxp3(+) regulatory T cells. Blood. 2010;115:1958–1968. doi: 10.1182/blood-2009-09-245274. [DOI] [PubMed] [Google Scholar]
- 3.Bousso P. T-cell activation by dendritic cells in the lymph node: lessons from the movies. Nat Rev Immunol. 2008;8:675–684. doi: 10.1038/nri2379. [DOI] [PubMed] [Google Scholar]
- 4.Zhu J, Paul WE. CD4 T cells: fates, functions, and faults. Blood. 2008;112:1557–1569. doi: 10.1182/blood-2008-05-078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers PR, Croft M. Peptide dose, affinity, and time of differentiation can contribute to the Th1/Th2 cytokine balance. J Immunol. 1999;163:1205–1213. [PubMed] [Google Scholar]
- 6.Rogers PR, Dubey C, Swain SL. Qualitative changes accompany memory T cell generation: faster, more effective responses at lower doses of antigen. J Immunol. 2000;164:2338–2346. doi: 10.4049/jimmunol.164.5.2338. [DOI] [PubMed] [Google Scholar]
- 7.Thery C, Amigorena S. The cell biology of antigen presentation in dendritic cells. Curr Opin Immunol. 2001;13:45–51. doi: 10.1016/s0952-7915(00)00180-1. [DOI] [PubMed] [Google Scholar]
- 8.Villadangos JA, et al. Proteases involved in MHC class II antigen presentation. Immunol Rev. 1999;172:109–120. doi: 10.1111/j.1600-065x.1999.tb01360.x. [DOI] [PubMed] [Google Scholar]
- 9.Shi GP, et al. Human cathepsin S: chromosomal localization, gene structure, and tissue distribution. J Biol Chem. 1994;269:11530–11536. [PubMed] [Google Scholar]
- 10.Hsieh CS, deRoos P, Honey K, Beers C, Rudensky AY. A role for cathepsin L and cathepsin S in peptide generation for MHC class II presentation. J Immunol. 2002;168:2618–2625. doi: 10.4049/jimmunol.168.6.2618. [DOI] [PubMed] [Google Scholar]
- 11.Beers C, et al. Cathepsin S controls MHC class II-mediated antigen presentation by epithelial cells in vivo. J Immunol. 2005;174:1205–1212. doi: 10.4049/jimmunol.174.3.1205. [DOI] [PubMed] [Google Scholar]
- 12.Stoeckle C, et al. Cathepsin S dominates autoantigen processing in human thymic dendritic cells. J Autoimmun. 2012;38:332–343. doi: 10.1016/j.jaut.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Han JW, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 14.Gateva V, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nature genetics. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SJ, Gregersen PK, Diamond B. Regulation of dendritic cell activation by microRNA let-7c and BLIMP1. J Clin Invest. 2013;123:823–833. doi: 10.1172/JCI64712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu H, et al. Increased frequency of circulating follicular helper T cells in lupus patients is associated with autoantibody production in a CD40L-dependent manner. Cell Immunol. 2015;295:46–51. doi: 10.1016/j.cellimm.2015.01.014. [DOI] [PubMed] [Google Scholar]
- 17.Le Coz C, et al. Circulating TFH subset distribution is strongly affected in lupus patients with an active disease. PLoS One. 2013;8:e75319. doi: 10.1371/journal.pone.0075319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi JY, et al. Circulating follicular helper-like T cells in systemic lupus erythematosus: association with disease activity. Arthritis Rheumatol. 2015;67:988–999. doi: 10.1002/art.39020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim SJ, Zou YR, Goldstein J, Reizis B, Diamond B. Tolerogenic function of Blimp-1 in dendritic cells. J Exp Med. 2011;208:2193–2199. doi: 10.1084/jem.20110658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piskurich JF, et al. BLIMP-I mediates extinction of major histocompatibility class II transactivator expression in plasma cells. Nat Immunol. 2000;1:526–532. doi: 10.1038/82788. [DOI] [PubMed] [Google Scholar]
- 21.Vander Lugt B, et al. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat Immunol. 2014;15:161–167. doi: 10.1038/ni.2795. [DOI] [PubMed] [Google Scholar]
- 22.Doody GM, et al. An extended set of PRDM1/BLIMP1 target genes links binding motif type to dynamic repression. Nucleic Acids Res. 2010;38:5336–5350. doi: 10.1093/nar/gkq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seillet C, et al. Estradiol promotes functional responses in inflammatory and steady-state dendritic cells through differential requirement for activation function-1 of estrogen receptor alpha. J Immunol. 2013;190:5459–5470. doi: 10.4049/jimmunol.1203312. [DOI] [PubMed] [Google Scholar]
- 24.Kitamura H, et al. IL-6-STAT3 controls intracellular MHC class II alphabeta dimer level through cathepsin S activity in dendritic cells. Immunity. 2005;23:491–502. doi: 10.1016/j.immuni.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 25.Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 26.Hutchins AP, Diez D, Miranda-Saavedra D. The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges. Brief Funct Genomics. 2013;12:489–498. doi: 10.1093/bfgp/elt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pratama A, Vinuesa CG. Control of TFH cell numbers: why and how? Immunol Cell Biol. 2014;92:40–48. doi: 10.1038/icb.2013.69. [DOI] [PubMed] [Google Scholar]
- 28.Walker B, Lynas JF, Meighan MA, Bromme D. Evaluation of dipeptide alpha-keto-beta-aldehydes as new inhibitors of cathepsin S. Biochem Biophys Res Commun. 2000;275:401–405. doi: 10.1006/bbrc.2000.3311. [DOI] [PubMed] [Google Scholar]
- 29.Rupanagudi KV, et al. Cathepsin S inhibition suppresses systemic lupus erythematosus and lupus nephritis because cathepsin S is essential for MHC class II-mediated CD4 T cell and B cell priming. Annals of the rheumatic diseases. 2015;74:452–463. doi: 10.1136/annrheumdis-2013-203717. [DOI] [PubMed] [Google Scholar]
- 30.Klein L, Hinterberger M, Wirnsberger G, Kyewski B. Antigen presentation in the thymus for positive selection and central tolerance induction. Nat Rev Immunol. 2009;9:833–844. doi: 10.1038/nri2669. [DOI] [PubMed] [Google Scholar]
- 31.Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6:683–692. doi: 10.1038/nrrheum.2010.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raj P, et al. Regulatory polymorphisms modulate the expression of HLA class II molecules and promote autoimmunity. Elife. 2016;5 doi: 10.7554/eLife.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang H, Rittner H, Weyand CM, Goronzy JJ. Aberrations in the primary T-cell receptor repertoire as a predisposition for synovial inflammation in rheumatoid arthritis. J Investig Med. 1999;47:236–245. [PubMed] [Google Scholar]
- 34.Yang H, et al. Hrd1-mediated BLIMP-1 ubiquitination promotes dendritic cell MHCII expression for CD4 T cell priming during inflammation. J Exp Med. 2014;211:2467–2479. doi: 10.1084/jem.20140283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Honey K, Rudensky AY. Lysosomal cysteine proteases regulate antigen presentation. Nat Rev Immunol. 2003;3:472–482. doi: 10.1038/nri1110. [DOI] [PubMed] [Google Scholar]
- 36.Yang H, et al. Cathepsin S is required for murine autoimmune myasthenia gravis pathogenesis. J Immunol. 2005;174:1729–1737. doi: 10.4049/jimmunol.174.3.1729. [DOI] [PubMed] [Google Scholar]
- 37.Tato M, et al. Cathepsin S inhibition combines control of systemic and peripheral pathomechanisms of autoimmune tissue injury. Sci Rep. 2017;7:2775. doi: 10.1038/s41598-017-01894-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu D, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 39.Kim SJ, et al. Increased IL-12 inhibits B cells' differentiation to germinal center cells and promotes differentiation to short-lived plasmablasts. J Exp Med. 2008;205:2437–2448. doi: 10.1084/jem.20070731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brocker T. Survival of mature CD4 T lymphocytes is dependent on major histocompatibility complex class II-expressing dendritic cells. J Exp Med. 1997;186:1223–1232. doi: 10.1084/jem.186.8.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goldrath AW, Bevan MJ. Selecting and maintaining a diverse T-cell repertoire. Nature. 1999;402:255–262. doi: 10.1038/46218. [DOI] [PubMed] [Google Scholar]
- 42.Rocha B, von Boehmer H. Peripheral selection of the T cell repertoire. Science. 1991;251:1225–1228. doi: 10.1126/science.1900951. [DOI] [PubMed] [Google Scholar]
- 43.Ndifon W, et al. Chromatin conformation governs T-cell receptor Jbeta gene segment usage. Proc Natl Acad Sci U S A. 2012;109:15865–15870. doi: 10.1073/pnas.1203916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1: Expression of cathepsin genes in splenic DCs from wild-type or CKO mice. DCs were purified from the spleens of age matched female wild-type or CKO mice, and RNA was purified for qPCR. Gene specific primers were used for each cathepsin gene and the relative expression of each was calculated by normalization to the level of Hprt. Each dot represents an individual mouse and the bar represents the mean ± SEM (n=5).
Supplemental figure 2: Increased expression of Il6 by estrogen in BM-DCs. BM-DCs were differentiated with GM-CSF (200 ng/ml) in hormone-free culture system without or with E2, ICI or E2 and ICI together for 7 days. On day 7, BM-DCs were counted and further cultured with or without LPS (1 µg/ml) for overnight. Total RNA was purified and Il6 transcript was assessed by qPCR. Relative level of Il6 was normalized to housekeeping gene Polr2a. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=7).
Supplemental figure 3: Diagram of Ctss reporter construct.
Supplemental figure 4: Gene expression in TFH and non-TFH cells. Spleens were collected from age-matched female wild-type (open circle) or CKO (closed circle) mice. TFH cells and non-TFH cells were identified as represented in the flow image (left panel), and each population was purified. Total RNA was purified and gene expression was measured by qPCR. Relative level of each gene was normalized to housekeeping gene Polr2a. Each dot represents an individual mouse and the bar represents the mean ± SEM (n=3).
Supplemental figure 5: Increased IL-21 protein in the supernatant from co-culture of OT-II CD4+ T cells and Blimp-1 deficient DCs. CD4+ T cells from OT-II mice and splenic DCs (open box is wild-type mice and grey box is CKO mice) were co-cultured in various conditions for 4 days. To measure the secreted cytokines, IL-2 and IL-21, supernatant was collected and cytokines were detected by a multiplex assay. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=6).
Supplemental figure 6: Cytokine production in the supernatant from day 3 co-cultures of OT-II T cells and DCs. DCs were purified from the spleens of female wild-type (open box) or CKO (grey box) mice, and CD4+ T cells were purified from female OT-II mice. DCs and T cells were co-cultured for 3 days at a 1:10 ratio (DC:T) with either OVA323–339 peptide (100 ng/ml) or OVA protein (10 µg/ml). Cytokine levels were measured in the supernatant by multiplex assay. In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=6).
Supplementary figure 7: Proliferation and IL-21 production of OT-II T cells by OVA323–339 presenting DCs. OT-II T cells were co-cultured with splenic DCs for 3 days at 1:10 ratio (DC:T cells) in the presence of OVA323–339 peptide. To assess proliferation, T cells were labeled with CFSE (10 mM) before the culture and proliferation was measured by calculation of the division index (left panel). In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=3). To assess IL-21 production, culture supernatant was harvested and measured by Multiplex assay (right panel). In the box-and-whisker plot, horizontal bars indicate the median, boxes indicate 25th to 75th percentile, and whiskers indicate 10th and 90th percentile (n=3).
Supplemental figure 8: Protein level of cytokines, IL-2 and IL-21, was measured by intracellular staining of T cells. OT-II T cells were co-cultured with splenic DCs that had been incubated with HEL (10 µ/ml) or OVA (10 µg/ml). CTSB inhibitor (1 nM) was added during processing of OVA protein antigen. After co-culture for 4 days, T cells were stimulated with PMA (100 ng/ml)/ionomycin (1 µg/ml) for 6 hours. BFA (20 µg/ml) was added during the last 4 hours of stimulation. Cytokine-positive cells were identified by flow cytometry and frequency was calculated and presented in the graph. Each dot represents an individual experiment and the bar represents the mean ± SEM (n=4).
Supplementary Table 1: Position of Blimp-1 binding consensus sequence in cathepsin genes and expression of each gene in Blimp-1 ko DCs. Blimp-1 binding sequence was investigated in the upstream of (−1500 to +10 from transcription initiation site (TSS)) each cathepsin gene. Amount of transcript was measured by qPCR from wild-type or Blimp-1 ko DCs. If there is difference more than 2-fold, it is indicated as “increased”. N.D (not detected).