Abstract
Parathyroid hormone (PTH) and Notch receptors regulate bone formation by governing the function of osteoblastic cells. To determine whether PTH interacts with Notch signaling as a way to control osteoblast function, we tested the effects of PTH on Notch activity in osteoblast- and osteocyte-enriched cultures. Notch signaling was activated in osteoblast-enriched cells from wild-type C57BL/6J mice following exposure to the Notch ligand Delta-like (Dll)1 or by the transient transfection of the Notch intracellular domain (NICD), the transcriptionally active fragment of Notch1. To induce Notch signaling in osteocyte-enriched cultures, a murine model of Notch2 gain-of-function was used. PTH opposed the stimulatory effects of Dll1 on Hey1, Hey2 and HeyL mRNA levels in osteoblast-enriched cells and suppressed the expression of selected Notch target genes in osteocyte-enriched cultures, either under basal conditions or in the context of Notch2 gain-of-function. Induction of Notch signaling in osteocytes did not alter the inhibitory effect of PTH on Sost expression, but reduced the stimulation of Tnfsf11 mRNA levels by PTH. In agreement with these in vitro observations, male mice administered with PTH displayed suppressed Hey1 and HeyL expression in parietal bones. Transactivation experiments with a Notch reporter construct and electrophoretic mobility shift assays in osteoblast-enriched cells suggest that PTH acts by decreasing the capacity of Rbpjκ to bind to DNA. In conclusion, downregulation of Notch in osteoblasts and osteocytes may represent a mechanism contributing to the anabolic effects of PTH in bone.
Keywords: PTH, Notch, Rbpjκ, osteoblast, osteocyte
1. INTRODUCTION
Bone remodeling is the process whereby the coordinated activities of osteoblasts and osteoclasts preserve skeletal integrity and contribute to the regulation of mineral metabolism [1]. Bone-forming osteoblasts derive from mesenchymal precursors and can mature further into quiescent bone lining cells or become embedded in the mineralized matrix as osteocytes. These are terminally differentiated cells that confer the skeleton the ability of sensing and adapting to mechanical stimuli [2]. Osteoblasts and osteocytes secrete receptor activator of nuclear factor-Kappa B-ligand (Rankl), a peptide encoded by Tnfsf11 that stimulates the formation of bone-resorbing osteoclasts. As a consequence, osteoblasts and osteocytes have the capacity to control bone resorption [3]. An attribute that distinguishes osteocytes from osteoblasts is the ability to express high levels of Dmp1 and Sost, the genes encoding for dentin matrix protein 1 and sclerostin, respectively [4–6]. The latter is a secreted molecule that inhibits osteoblast differentiation by suppressing Wnt/β-catenin signaling [7].
Parathyroid hormone (PTH) is an 84-amino acid peptide that regulates the function of osteoblasts and osteocytes, and is secreted by the parathyroid gland in response to changes in the concentration of extracellular calcium [8–10]. PTH enhances bone formation by a variety of mechanisms, including direct effects on the osteoblast, the induction of insulin-like growth factor I and possibly the suppression of Sost [11, 12]. However, neither changes in Sost expression nor the presence of osteocytic/osteoblastic β-catenin are required for the anabolic actions of PTH suggesting the involvement of other mechanisms [13, 14]. PTH also enhances bone resorption by inducing Tnfsf11 in osteoblasts and osteocytes [15, 16]. Direct stimulation of osteoblast function by PTH is mediated primarily by the activation of the cAMP/protein kinase A and the mitogen activated protein kinases (MAPK) [17]. The intracellular mechanisms of PTH action in osteocytes are not well understood, in part due to the lack of a suitable model for the study of osteocyte function in vitro.
The four Notch receptors and their cognate ligands of the Delta-like (Dll) and Jagged (Jag) families are transmembrane proteins that mediate communication between adjacent cells. Activation of Notch receptors occurs following their cleavage and release of the intracellular domain (NICD), which translocates to the nucleus, associates with a DNA binding factor termed recombination signal binding protein for immunoglobulin kappa J region (Rbpj)κ, and induces gene expression [18]. Rbpjκ is the murine ortholog of core binding factor-1/suppressor of hairless/Lag-1 (CSL). Classic target genes of Rbpjκ-mediated Notch signaling are Hes1, Hes5 and Hes7 as well as Hey1, Hey2 and HeyL [19]. Notch1 and Notch2 regulate bone remodeling, and their actions are determined by the degree of osteoblast maturity [20]. In contrast to the bone anabolic effects of PTH, induction of Notch1 in mesenchymal precursor cells and mature osteoblasts suppresses cell differentiation and trabecular bone formation, and Notch2 exerts similar inhibitory effects on osteoblast function [21–26]. Conversely, activation of Notch1 in osteocytes induces osteoprotegerin and suppresses sclerostin, leading to an osteopetrotic phenotype [23, 27].
PTH is known to induce the Notch ligand Jag1 in osteoblasts and potentially activate Notch signaling, and the effects of PTH on osteoblast differentiation were opposed by γ-secretase inhibitors, suggesting that Notch mediates selected actions of PTH in osteoblasts [28, 29]. However, γ-secretase inhibitors have nearly 100 substrates and therefore are not specific to the activation Notch signaling [30]. In addition, PTH exposure and Notch induction have opposite consequences on bone formation and osteoblast activity. These apparently contradictory observations could be explained by additional interactions between PTH and Notch in osteoblastic cells.
In the present study, we tested the effects of PTH on Notch signaling in osteoblasts from wild-type mice and osteocytes from a recently described murine model of Notch2 gain-of-function [25]. These experiments were complemented by in vivo work to determine the impact of PTH on Notch signaling in bone tissue of wild-type mice.
2. MATERIALS AND METHODS
2.1 Mice
Wild-type C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were used as a source of primary osteoblastic cells for in vitro experiments. To test the effects of PTH on Notch signaling in bone in vivo, 7 week old wild-type male mice were injected subcutaneously with 80 μg/Kg of the PTH amino terminal fragment 1–34 (Sigma-Aldrich, St. Louis, MO) dissolved in phosphate buffered saline (PBS). Mice injected with an equal volume of PBS served as controls. Parietal bones were harvested at sacrifice, frozen in liquid nitrogen and stored at −80°C.
To achieve Notch2 gain-of-function, mice harboring a point mutation in exon 34 of the Notch2 locus were generated by homologous recombination, as described [25]. A 6955C>T mutation was introduced creating a stop codon leading to the translation of a truncated protein lacking the proline-, glutamic acid-, serine- and threonine-rich (PEST) domain, making it resistant to degradation and resulting in a Notch2 gain-of-function [20]. The homologous mutation in humans is associated with Hajdu Cheney syndrome (HCS), and the model was named Notch2HCS. Heterozygous Notch2HCS mice were bred to wild-type mice to create experimental heterozygous Notch2HCS mice and wild-type littermate controls in a 129SvJ/C57BL/6J genetic background. The presence of the Notch2HCS allele was determined in tail DNA extracts by polymerase chain reaction (PCR) using forward 5′-CCCTTCTCTCTGTGCGGTAG-3′ and reverse 5′-CTCAGAGCCAAAGCCTCACTG-3′ primers (both from Integrated DNA Technologies; IDT, Coralville, IA). Experimental protocols were approved by the Institutional Animal Care and Use Committees of Saint Francis Hospital and Medical Center (Hartford, CT) and of UConn Health.
2.2 Osteoblast-enriched Cultures
Parietal bones of 3 to 5 day old wild-type mice were digested at 37°C with type-II collagenase from Clostridium histolyticum (Worthington Biochemical Corp., Lakewood, NJ) and treated with N-α-tosyl-L-lysyl-chloromethylketone hydrochloride (TLCK; Calbiochem, La Jolla, CA). Five sequential digestions were carried out and cells extracted from the last 3 digestions were pooled and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with non-essential amino acids (both from Life Technologies, Grand Island, NY), 20 mM HEPES, 100 μg/ml ascorbic acid (both from Sigma-Aldrich) and heat-inactivated 10% fetal bovine serum (FBS, Atlanta Biologicals, Inc., Atlanta, GA), in a humidified 5% CO2 incubator at 37°C, as described [31]. To activate Notch receptors, osteoblast-enriched cells were trypsinized and seeded at a density of 22,000 cells/cm2 on culture plates coated with a Dll1-IgG2A fragment crystallizable (Fc) recombinant fusion protein 125 ng/cm2 (R&D Systems, Minneapolis, MN), and cultured for 24 h. Parallel cultures on an equimolar amount of bovine serum albumin (BSA) served to establish basal levels of Notch signaling, as BSA is considered to be a suitable control for recombinant Fc fusion proteins [32]. To prevent the effects of serum-derived factors, cultures were serum deprived 24 h prior to the addition of PTH at 10 nM for 6 h. No negative consequences are observed on osteoblast viability after a 24 h serum deprivation [33].
2.3 Osteocyte-enriched Cultures
Femurs from 6 or 7 week old wild-type or Notch2HCS mice were collected after sacrifice, surrounding tissues dissected, the proximal epiphysis excised, and the bone marrow removed by centrifugation. The distal epiphysis was removed, and to release the endosteal and periosteal cellular layers, the femoral fragments were sequentially exposed for 20 min periods at 37°C to type-II collagenase pretreated with TLCK 17 μg/ml or EDTA 5 mM (Life Technologies), as stated in figure legends. In an initial experiment, cells were obtained after 5 sequential collagenase/EDTA digestions as described in previous publications [34]. Cells from each digestion step, and the remaining bone fragments were collected and analyzed for gene expression. Subsequently, osteocyte-enriched bone fragments were obtained after a single collagenase and an EDTA digestion, and either analyzed or cultured individually in DMEM supplemented with nonessential amino acids, 100 μg/ml ascorbic acid, and heat-inactivated 10% FBS for 4 days in a humidified 5% CO2 incubator at 37°C [35, 36]. Cultures were serum deprived for 24 h prior to treatment with PTH 10 nM or vehicle for 6 h, and femoral fragments frozen in liquid nitrogen and stored at −80°C.
RNA Integrity and Quantitative Reverse Transcription-PCR (qRT-PCR)
Total RNA was extracted from osteoblast-enriched cells and from cellular fractions obtained after collagenase/EDTA digestions of femoral fragments with an RNeasy kit (Qiagen, Valencia, CA). Osteocyte-enriched femoral fragments or parietal bones were homogenized prior to chloroform/phenol extraction of total RNA with the micro RNeasy kit (Qiagen). Integrity of RNA preparations was assessed by microfluidic electrophoresis on an Experion system (Bio-Rad, Hercules, CA) [37, 38]. Equal amounts of RNA were reverse-transcribed using the iScript RT-PCR kit (Bio-Rad) according to manufacturer’s instructions, and amplified in the presence of specific primers (all from IDT, Table 1) and iQ SYBR Green Supermix (Bio-Rad) at 60°C for 35 cycles. Transcript copy number was estimated by comparing to a serial dilution of cDNA for Alpl, encoding for alkaline phosphatase, and Rpl38 (both from American Type Tissue Culture Collection, Manassas, VA), Bglap, encoding for osteocalcin (J. Lian, Burlington, VT), Dll1 (E. Six, Paris, France), Dll3, Dll4 and Jag2 (GE Healthcare Dharmacon, Lafayette, CO), Dmp1, Sost, Jag1 and Notch2 (Thermo Fisher Scientific, Waltham, MA), Hey1 and Hey2 (T. Iso, Los Angeles, CA), HeyL (D. Srivastava, Dallas, TX), Notch1 (J.S. Nye, Cambridge, MA), Notch4 (Y. Shirayoshi, Tottori, Japan) and Tnfsf11 (Source BioScience, Nottingham, UK) [39–44]. Notch3 copy number was estimated by comparison to a serial dilution of a ~100 base pairs synthetic DNA template (IDT) cloned into pcDNA3.1(–) (Thermo Fisher Scientific) by isothermal single reaction assembly using commercially available reagents (New England BioLabs, Ipswich, MA) [45]. Reactions were conducted in a CFX96 qRT-PCR detection system (Bio-Rad), and fluorescence was monitored during every PCR cycle at the annealing step. Data are expressed as copy number corrected for Rpl38 [46].
Table 1. Primers for qRT-PCR.
Forward (Fwd) and reverse (Rev) primers used for detection of gene expression by qRT-PCR. GenBank accession numbers indicate transcript variants with homologous sequences to the primers.
| Gene | Strand | Primer Sequence | GenBank Accession Numbers |
|---|---|---|---|
| Alpl | Fwd Rev |
5′-TGGTATGGGCGTCTCCACAGTAACC-3′ 5′-CTTGGAGAGGGCCACAAAGG-3′ |
NM_007431 |
| Bglap | Fwd Rev |
5′-GACTCCGGCGCTACCTTGGGTAAG-3′ 5′-CCCAGCACAACTCCTCCCTA-3′ |
NM_001037939 |
| Dll1 | Fwd Rev |
5′-CTCTTCCCCTTGTTCTAAC-3′ 5′-ACAGTCATCCACATTGTC-3′ |
NM_007865 |
| Dll3 | Fwd Rev |
5′-TCTATCTTGTCCCTTCTCTATCA-3′ 5′-AATCATTCAGGCTCCATCTC-3′ |
NM_007866 |
| Dll4 | Fwd Rev |
5′-TGACAAGAGCTTAGGAGAG-3′ 5′-GCTTCTCACTGTGTAACC-3′ |
NM_019454 |
| Dmp1 | Fwd Rev |
5′-CTGTCATTCTCCTTGTGTTC-3′ 5′-TTCAGATTCAGTATTGTGGTATC-3′ |
NM_016779 |
| Hey1 | Fwd Rev |
5′-ATCTCAACAACTACGCATCCCAGC-3′ 5′-GTGTGGGTGATGTCCGAAGG-3′ |
NM_010423 |
| Hey2 | Fwd Rev |
5′-AGCGAGAACAATTACCCTGGGCAC-3′ 5′-GGTAGTTGTCGGTGAATTGGACCT-3′ |
NM_013904 |
| HeyL | Fwd Rev |
5′-CAGTAGCCTTTCTGAATTGCGAC-3′ 5′-CCCAGCACAACTCCTCCCTA-3′ |
NM_013905 |
| Jag1 | Fwd Rev |
5′-TGGGAACTGTTGTGGTGGAGTCCG-3′ 5′-GTGACGCGGGACTGATACTCCT-3′ |
NM_013822 |
| Jag2 | Fwd Rev |
5′-AAGGTGGAAACAGTTGT-3′ 5′-CACGGGCACCAACAG-3′ |
NM_010588 |
| Notch1 | Fwd Rev |
5′-GTCCCACCCATGACCACTACCCAGTTC-3′ 5′-GGGTGTTGTCCACAGGTGA-3′ |
NM_008714 |
| Notch2 | Fwd Rev |
5′-TGACGTTGATGAGTGTATCTCCAAGCC-3′ 5′-GTAGCTGCCCTGAGTGTTGTGG-3′ |
NM_010928 |
| Notch3 | Fwd Rev |
5′-CCGATTCTCCTGTCGTTGTCTCC-3′ 5′-TGAACACAGGGCCTGCTGAC-3′ |
NM_008716 |
| Notch4 | Fwd Rev |
5′-CCAGCAGACAGACTACGGTGGAC-3′ 5′-GCAGCCAGCATCAAAGGTGT-3′ |
NM_010929 |
| Rpl38 | Fwd Rev |
5′-AGAACAAGGATAATGTGAAGTTCAAGGTTC-3′ 5′-CTGCTTCAGCTTCTCTGCCCTTT-3′ |
NM_001048057 NM_023372 NM_001048058 |
| Sost | Fwd Rev |
5′-AGGAATGATGCCACAGAGGTC-3′ 5′-CTGGTTGTTCTCAGGAGGAGGCTC-3′ |
NM_024449 |
| Tnfsf11 | Fwd Rev |
5′-TATAGAATCCTGAGACTCCATGAAAAC-3′ 5′-CCCTGAAAGGCTTGTTTCATCC-3′ |
NM_011613 |
2.4 Transient Transfections
A plasmid containing six dimeric consensus sequences for CSL, the human ortholog of Rbpjκ, upstream of the β-globin basal promoter and Luciferase (12xCSL-Luc; L.J. Strobl, Munich, Germany) was used to detect Rbpjκ-mediated transcriptional activity [47]. To induce Notch1, cells were co-transfected with a 2.4 kilobase DNA fragment containing the coding sequence of the murine Notch1 NICD (J.S. Nye) cloned into pcDNA3.1 downstream of the cytomegalovirus (CMV) promoter (pcDNA-NICD), or with control pcDNA3.1 [43]. Transfections were conducted in sub-confluent cultures of osteoblast-enriched cells with X-tremeGENE 9 (2 μl X-tremeGENE 9/1 μg DNA), according to manufacturer’s instructions (Roche, Indianapolis, IN). A construct where the CMV promoter directs the expression of β-galactosidase (Clontech, Mountain View, CA) was used to correct for transfection efficiency. Cells were exposed to the X-tremeGENE 9/DNA mix for 16 h, transferred to DMEM without serum for 6 h, and exposed to PTH 10 nM or to vehicle for 24 h. Cells were harvested in reporter lysis buffer (Promega, Madison, WI) and lysed by freezing and thawing. Luciferase and β-galactosidase activity were determined with a luciferase assay system kit (Promega) and with galacton plus (Life Technologies), respectively, in accordance with manufacturers’ instructions on an Optocomp II luminometer (MGM Instruments, Hamden, CT).
2.5 Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were obtained from wild-type osteoblast-enriched cells seeded on BSA or Dll1 and exposed either to vehicle or PTH at 10 nM. A double-stranded DNA oligonucleotide containing the CSL consensus sequence found in the Epstein-Barr virus nuclear antigen (EBNA)2 promoter (forward strand sequence: 5′-GGAAACACGCCGTGGGAAAAAATTTGGG-3′) biotinylated on both 5′-termini was synthesized commercially (IDT) [48]. Binding reactions of nuclear extracts with biotinylated DNA at a concentration of 1 μM were carried out with the LightShift Chemiluminescent EMSA Kit, as recommended by the manufacturer (Thermo Fisher Scientific). To determine the specificity of the interactions between the nuclear extracts and the biotinylated oligonucleotides, unlabeled homologous DNA was added in 200 fold excess. Nucleic acid-protein complexes were resolved on non-denaturing, non-reducing 4% polyacrylamide gels for 45 min and subsequently transferred to a nylon membrane with a 0.45 μm pore size (MP Biomedicals, Solon, OH) in 45 mM Tris, 45 mM boric acid, 1 mM EDTA at pH 8.3 (all from Sigma-Aldrich). Transfer was conducted for 30 min at 4°C, and crosslinking of the transferred complexes to the nylon membrane was carried out at 120 mJ/cm2 for 1 min using a CL-1000 UV-light crosslinking instrument (UVP, Upland, CA). The biotinylated DNA was detected with a streptavidin-horseradish peroxidase conjugate following manufacturers’ instructions for the LightShift Chemiluminescent EMSA Kit detection module, and images of chemiluminescence reactions were acquired with a Chemidoc XSR molecular imager (Bio-Rad).
2.6 Statistical Analysis
Data are expressed as means ± SEM. Statistical differences were determined by Student’s t test or analysis of variance (ANOVA) with Holm-Šídák post-hoc analysis for pairwise or multiple comparisons, respectively.
3. RESULTS
3.1 PTH Suppresses Notch Signaling in Osteoblast-enriched Cells
To test the effects of PTH, osteoblast-enriched cells from the parietal bones of C57BL/6J wild-type mice were seeded on Dll1 for 24 h to activate Notch receptors or on BSA, and subsequently exposed to PTH or vehicle. Although Jag1 has a documented skeletal function, Dll1 was chosen because it enhances Notch activation consistently in multiple cell lineages and in osteoblasts, and both ligands induce Notch target genes to a comparable extent (Canalis and Zanotti unpublished observations) [49–51]. A PTH dose of 10 nM, known to have a maximal effect on osteoblast function was used for the present studies [52–55]. Cells cultured on Dll1 exhibited higher expression of the Notch target genes Hey1, Hey2 and HeyL than those cultured on BSA, confirming activation of Notch signaling. Basal levels of Hey1, Hey2 and HeyL mRNA obtained in the absence of Dll1 were not affected by PTH, but PTH opposed the effect of Dll1 on Notch activation reducing the expression of the three Notch target genes to basal levels (Figure 1).
Figure 1. PTH prevents the induction of Notch target genes in osteoblasts.
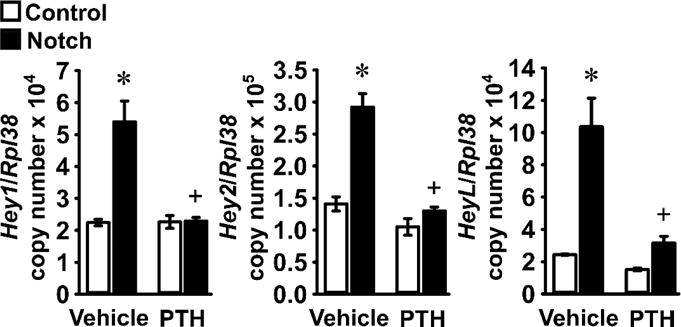
Primary osteoblast-enriched cells were harvested from parietal bones of 3 to 5 day old wild-type C57BL/6J mice and seeded on BSA (Control, white bars) or Dll1 (Notch, black bars) coated culture plates. After confluence, cells were cultured for 24 h in the absence of serum and exposed to vehicle or PTH at 10 nM (PTH) for 6 h. Total RNA was extracted and mRNA levels quantified by qRT-PCR in the presence of specific primers. Data are expressed as Hey1, Hey2 and HeyL copy number corrected for Rpl38 expression. Values are means ± SEM; n = 3 – 4. *Significantly different between Control and Notch, p < 0.05; +significantly different between Vehicle and PTH, p < 0.05; two-way ANOVA with Holm-Šídák post-hoc analysis.
3.2 Characterization of Osteocyte-enriched Cultures
To establish a cell culture system for the study of osteocytes in vitro, we obtained cells from sequential collagenase and EDTA digestions of femurs devoid of cartilage and bone marrow, and collected the resulting bone fragments after their final digestion with EDTA [34]. To identify the cell population with the highest enrichment in osteocytes, levels of transcripts for gene markers preferentially expressed by osteoblasts and osteocytes were determined. Expression of the osteoblastic markers Alpl and Bglap was detected in all cellular fractions and the digested femoral fragments displayed the highest levels of Bglap transcripts. Expression of Dmp1 and Sost is higher in osteocytes than in osteoblasts, and both genes were detected only in bone fragments after their digestion with collagenase and EDTA, and these were considered osteocyte-enriched fractions (Figure 2) [4–6].
Figure 2. Characterization of Notch expression in osteoblast and osteocyte-enriched cell fractions.
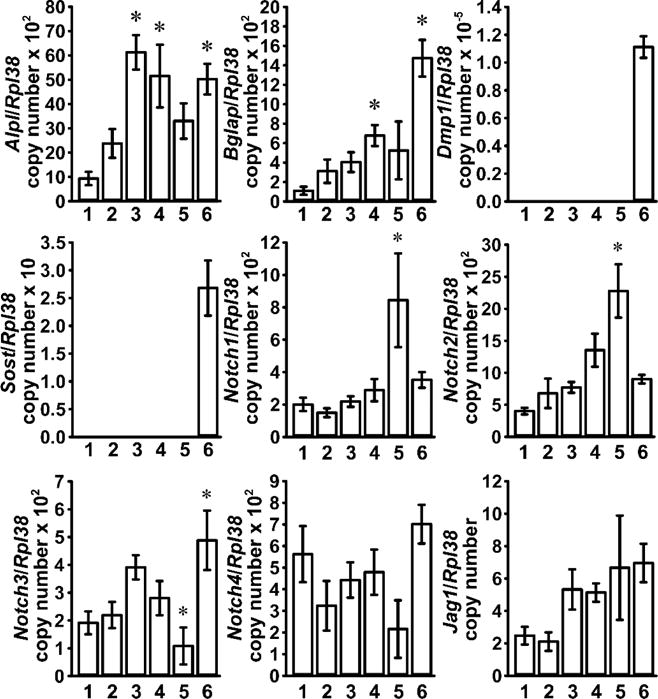
Femoral fragments from 6 week old wild-type C57BL/6J mice were exposed sequentially to type-II collagenase (fraction 1, 3 and 4), or EDTA (fraction 2 and 5) and cells collected after each digestion; femoral diaphyses (6) were frozen immediately after the final exposure to EDTA. Total RNA was extracted from cellular fractions and digested bone remnants, and mRNA quantified by qRT-PCR in the presence of specific primers. Data are expressed as Alpl, Bglap, Dmp1, Sost, Notch1, Notch2, Notch3, Notch4 and Jag1 copy number, corrected for Rpl38 expression. Values are means ± SEM; n = 5. *Significantly different from fraction 1, p < 0.05 by one-way ANOVA with Holm-Šídák post-hoc analysis.
To determine the identity of the Notch paralogs and specific ligands expressed in the cellular fractions, mRNA levels for all Notch receptors, Jag and Dll ligands were measured. Notch1 to Notch4 had comparable expression levels and were detected in all fractions. Expression of Notch1 and Notch2 was more pronounced in fraction 5, whereas Notch3 expression was highest in the osteocyte-enriched fraction. Expression of Jag1 was uniform across all fractions while transcripts for Dll1, Dll3, Dll4 and Jag2 were not detected (Figure 2). Subsequently, femoral fragments were subjected to a single collagenase and an EDTA digestion and their treatment with PTH induced Tnfsf11 and decreased Sost transcripts, confirming the known effects of PTH in osteocytes and documenting that the culture system is enriched in osteocytes (Figure 3) [11, 56].
Figure 3. Characterization of osteocyte-enriched cultures.
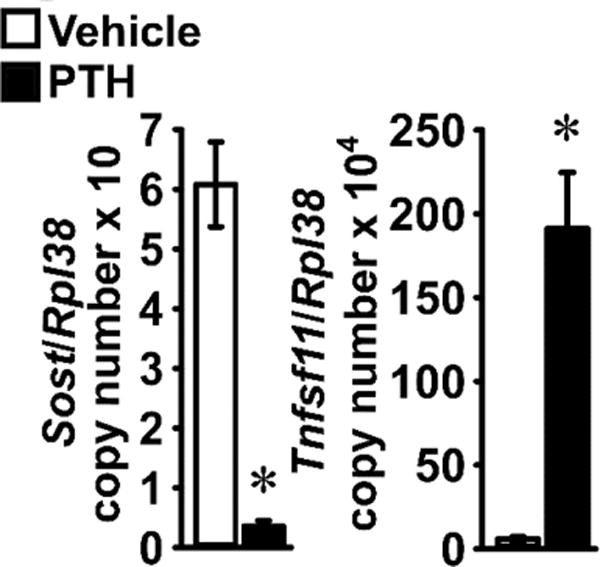
Primary osteocyte-enriched femoral fragments from 6 week old wild-type C57BL/6J mice were cultured for 24 h in the absence of serum and exposed to vehicle (white bars) or to PTH 10 nM (black bars) for 6 h. Total RNA was extracted and mRNA quantified by qRT-PCR in the presence of specific primers. Data are expressed as Sost and Tnfsf11 copy number, corrected for Rpl38 expression. Values are means ± SEM; n = 6. *Significantly different between vehicle and PTH, p < 0.05 by unpaired Student’s t-test.
3.3 PTH Inhibits Notch Signaling in Osteocyte-enriched Cultures
PTH suppressed the basal expression of the Notch target genes Hey1, Hey2 and HeyL in osteocyte-enriched cultures, suggesting that mature osteoblastic cells are sensitized to the inhibitory effects of PTH on Notch signaling (Figure 4).
Figure 4. PTH suppresses Hey1, Hey2 and HeyL expression in osteocytes.
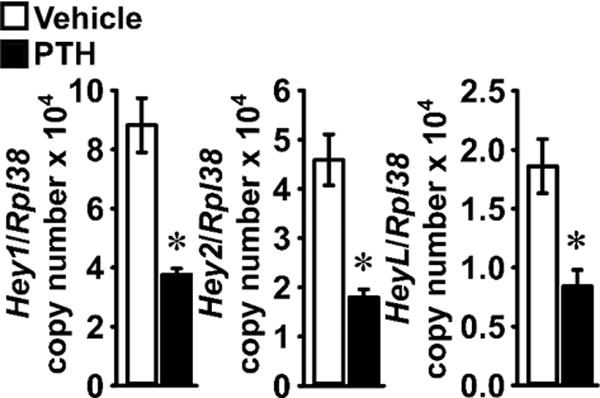
Primary osteocyte-enriched femoral fragments from 6 week old wild-type C57BL/6J mice were cultured for 24 h in the absence of serum and subsequently exposed to vehicle (white bars) or PTH 10 nM (black bars) for 6 h. Total RNA was extracted and mRNA quantified by qRT-PCR in the presence of specific primers. Data are expressed as Hey1, Hey2 and HeyL copy number, corrected for Rpl38 expression. Values are means ± SEM; n = 6. *Significantly different between vehicle and PTH, p < 0.05 by unpaired Student’s t-test.
To determine the effects of PTH in the context of Notch activation in osteocytes, we tested the consequences of PTH exposure in osteocyte-enriched cells from heterozygous Notch2HCS mice, a model of Notch2 gain-of-function [25]. This was necessary because bone fragments cannot be acutely transfected with Notch expression vectors nor successfully cultured on Notch-specific ligands to achieve activation. In the absence of PTH, osteocyte-enriched cells from Notch2HCS mice exhibited higher Hey1, Hey2 and HeyL mRNA levels than those from wild-type littermates, confirming activation of Notch signaling (Figure 5). PTH opposed basal and Notch2-induced expression of Hey1 and Hey2 transcripts. PTH suppressed HeyL in cultures from wild-type mice by ~50%, although the effect did not reach statistical significance, and PTH did not alter HeyL mRNA levels in bone fragments from Notch2HCS mutants (Figure 5). The reason for the sustained expression of HeyL in the presence of PTH remains without an obvious explanation. Confirming previous results, PTH induced Tnfsf11 and suppressed Sost expression in cultures from Notch2HCS mutants and wild-type littermates. Notch2 had a lesser effect than PTH on Tnfsf11 and Sost expression, and no additive or synergetic effects with PTH were observed.
Figure 5. PTH opposes selected effects of Notch2 gain-of-function in osteocytes.
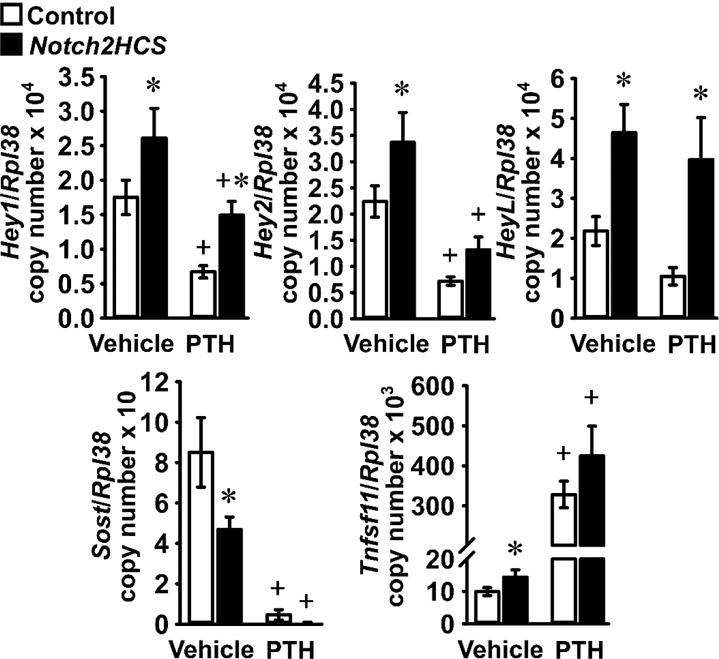
Primary osteocyte-enriched femoral fragments from 7 week old 129SvJ;C57BL/6J wild-type mice (Control, white bars) or Notch2HCS littermates (black bars) were cultured for 24 h in the absence of serum and subsequently exposed to vehicle or PTH 10 nM (PTH) for 6 h. Total RNA was extracted and mRNA quantified by qRT-PCR in the presence of specific primers. Data are expressed as Hey1, Hey2, HeyL, Sost and Tnfsf11 copy number, corrected for Rpl38 expression. Values are means ± SEM; n = 5 – 7. *Significantly different between Control and Notch2HCS, p < 0.05; +significantly different between Vehicle and PTH, p < 0.05; two-way ANOVA with Holm-Šídák post-hoc analysis.
3.4 PTH Suppresses Notch Signaling In Vivo
To establish whether PTH regulates Notch signaling in osteoblastic cells in vivo, 1 month old C57BL/6J wild-type male mice were administered with PTH 80 μg/Kg or vehicle, and the expression of Notch target genes was measured in parietal bones 3 h after injection. PTH suppressed Hey1 and HeyL mRNA levels, but not Hey2 (Figure 6). Confirming previous observations, PTH downregulated Sost mRNA levels [11].
Figure 6. PTH suppresses expression of Notch target genes by bone cells in vivo.
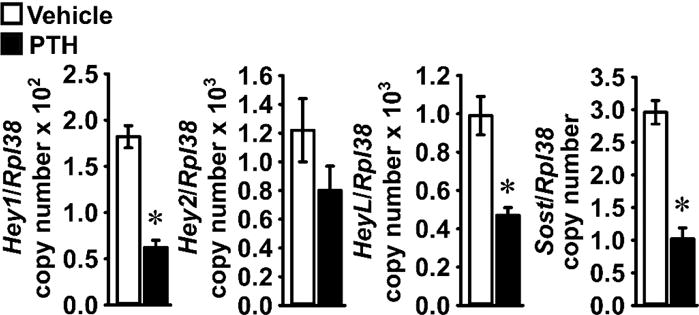
Seven week-old wild-type C57BL/6J male mice were administered vehicle 5 ml/Kg (white bars) or PTH 80 μg/Kg (black bars) subcutaneously. Mice were sacrificed 3 h after the injections and total RNA extracted from parietal bones and mRNA quantified by qRT-PCR in the presence of specific primers. Data are expressed as Hey1, Hey2, HeyL and Sost copy number, corrected for Rpl38 expression. Values are means ± SEM, n = 4 – 5. *Significantly different between Vehicle and PTH, p < 0.05 by unpaired Student’s t-test.
3.5 PTH Inhibits Rbpjκ-mediated Notch Signaling
To determine whether the inhibition of Notch target gene expression by PTH is mediated by an effect on Rbpjκ-mediated signaling, wild-type osteoblasts were transiently transfected with the 12xCSL-Luc reporter, which contains consensus sequences for CSL, the human ortholog for Rbpjk. Cells were co-transfected with pcDNA-NICD to overexpress Notch, or a control vector, and exposed to either PTH or vehicle. Transactivation of the 12xCSL-Luc construct was detected only in cells co-transfected with pcDNA-NICD and PTH reduced luciferase activity by ~70%, indicating that PTH inhibits Rbpjκ-mediated Notch signaling (Figure 7A).
Figure 7. PTH inhibits Rbpjκ-mediated Notch signaling in osteoblasts.
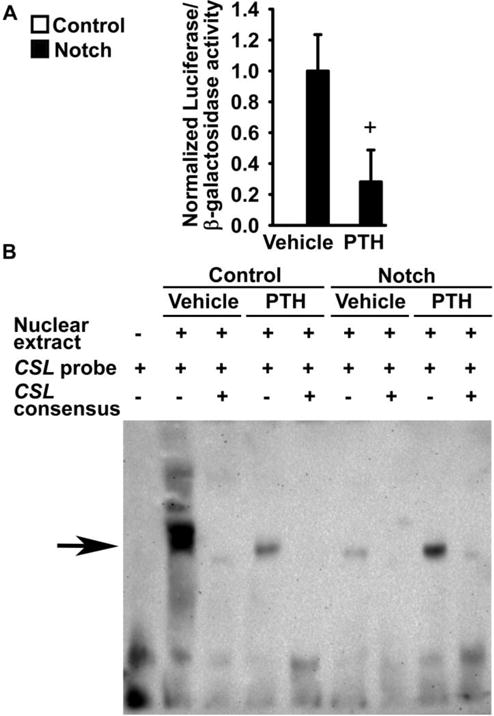
Primary osteoblast-enriched cells were harvested from parietal bones of 3 to 5 day old wild-type C57BL/6J mice. In panel A, cells were transiently transfected with pcDNA3.1 (Control) or with pcDNA-NICD (Notch) and co-transfected with the 12xCSL-Luc reporter and a CMV/β-galactosidase expression vector. Cells were cultured for 6 h in the absence of serum, exposed to vehicle or PTH at 10 nM (PTH), and harvested after 24 h. Data represent luciferase/β-galactosidase activity. Values are means ± SEM, n = 6. +significantly different between Vehicle and PTH, p < 0.05 by unpaired Student’s t-test. In panels B and C, cells were seeded on BSA (Control) or Dll1 (Notch) coated culture plates. After confluence, cells were cultured for 24 h in the absence of serum and exposed to vehicle or PTH at 10 nM (PTH) for 6 h. In panel B, a biotinylated oligonucleotide containing a CSL consensus sequence from the EBNA2 promoter was used. Competition of binding reactions was performed in the presence of unlabeled oligonucleotides containing homologous CSL consensus sequences in 200 fold excess. DNA-nuclear protein complexes were resolved by gel electrophoresis, transferred to a nylon membrane, exposed to a streptavidin-horseradish peroxidase conjugate and visualized by chemiluminescence. The arrow indicates the position of DNA protein complexes.
To explore the mechanisms responsible for the inhibition of Notch signaling by PTH, EMSA were carried out in osteoblast-enriched cells harvested from C57BL/6J wild-type mice and seeded on Dll1, to induce Notch, or on BSA. A biotinylated oligonucleotide containing the consensus sequence for CSL was bound by nuclear protein extracts from cells cultured on BSA, and an excess of unlabeled CSL oligonucleotides prevented this effect, demonstrating specificity of the protein-DNA interaction. In cells cultured on BSA, PTH suppressed the formation of nuclear protein complexes with the biotinylated CSL consensus oligonucleotide, supporting the notion that PTH prevents the interaction of Rbpjκ with DNA (Figure 7B). Previous reports demonstrated that the binding of Rbpjκ to DNA oligonucleotides decreases in the presence of active Notch receptors; similarly, a nuclear protein-CSL consensus oligonucleotide complex was not observed in osteoblasts seeded on Dll1 [57]. The complex was partially restored in cultures exposed to both Dll1 and PTH, suggesting that activation of the Notch receptors interferes with the inhibitory effects of PTH on the ability of Rbpjκ to bind DNA (Figure 7B).
4. DISCUSSION
The present work demonstrates that PTH suppresses Notch signaling in osteoblasts and osteocytes. Expression of the four Notch receptors was documented in both cell types, suggesting that PTH affects a signaling mechanism that may be common to all Notch paralogs. Inhibition of Notch target gene expression was observed at a concentration of PTH that regulates parameters of osteoblastic function, such as Alpl, Col1a1, Igf1 and Bglap expression and collagen type I synthesis, [52–55] (Zanotti and Canalis unpublished). Suppression of Notch signaling is an acute response to PTH that includes inhibition of Rbpjκ/DNA interactions and of Notch transactivating capacity. In contrast, the regulation of osteoblastic function is noted after longer exposure to PTH, suggesting that PTH regulates activation of Notch directly [52–55]. Notch activation during the early stages of the osteoblast differentiation program suppresses bone formation and leads to osteopenia, whereas PTH enhances bone formation in rodents and humans [12, 21–24]. Therefore, suppression of Notch signaling by PTH may be required to achieve an optimal anabolic response to the hormone.
The current work confirmed the effects of PTH on Tnfsf11 expression in osteoblast-enriched cells and those of PTH and Notch on Tnfsf11 and Sost transcripts in cultures of osteocyte-enriched femoral fragments. These results indicate that the osteocyte-enriched cultures used in the present work are a viable model to investigate osteocyte biology. However, studies in this system must be interpreted with caution, since expression of Alpl indicates the possible presence of contaminating osteoblasts in femoral fragments. It is of interest that PTH induced Tnfsf11 expression to a greater extent in osteocyte-enriched cultures (~30 fold) than in primary osteoblasts (~3 fold). This is in accordance with the fundamental role of osteocyte-derived Rankl in bone resorption [58, 59]. The suppression of Sost mRNA levels in vivo is consistent with the bone anabolic response to PTH [1]. The effects of Notch on Tnfsf11 and Sost expression were less pronounced than those of PTH, and Notch activation did not enhance the effect of PTH on either gene.
Transactivation and EMSA experiments conducted in osteoblast-enriched cultures suggest that PTH opposes Notch signaling by decreasing the capacity of Rbpjκ to associate with DNA and also by interfering with the formation of an active NICD/Rbpjκ transcriptional complex. These effects are mediated by intracellular events, since Notch signaling was suppressed in the context of the Notch1 NICD overexpression, which is a model of activation that does not require the presence of ligands or cleavage of the extracellular domain of the receptor [43]. The partial restoration of the complex in the presence of PTH and Dll1 suggests that the nuclear translocation of the NICD may lessen the inhibitory effect of PTH on the ability of Rbpjκ to bind to DNA. This mechanism may explain the residual expression of Notch target genes in the context of PTH exposure. In the present study, known limitations to the study of Rbpjk/DNA interactions by EMSA are documented. Namely, displaced CSL probes are not always visible in the presence of unlabeled competitors, and formation of an active transcriptional complex in the context of Dll1 exposure may lead to a shift of the nuclear protein bound to the CSL probe and attenuation of the signal [57, 60, 61]. Although the identity of the factors that prevent the association of Rbpjκ with DNA is not known, activation of MAPK in response to PTH may be involved in this process. Previous work demonstrated that selected members of the MAPK family are expressed by osteoblasts, regulate bone remodeling, and can prevent the formation of an active Notch transcriptional complex [62–65]. Another alternative is activation of cyclic AMP, a mechanism that has been found to be responsible for the downregulation of Sost in osteocytes expressing a constitutively active form of the PTH receptor [66, 67].
The inhibitory effect of PTH on Notch signaling is analogous to the one reported for nuclear factor of activated T-cells (Nfatc)1 and Nfatc2 [22, 68]. In the present studies, we tested and determined that PTH induces Nfatc1 and Nfatc2 in osteocyte-enriched cultures, but not in calvarial osteoblasts (Zanotti and Canalis unpublished). However, since suppression of Notch signaling was observed in both cell preparations, it is probably not related to an induction of Nfatc1 or Nfatc2. Although the effect of PTH in osteocytes was assessed by examining for the downregulation of Notch target genes and not by transactivation or EMSA experiments, it is probable that PTH acts by similar mechanisms in both osteoblasts and osteocytes [19]. Additional osteocyte-specific mechanisms may be operational, since in contrast to the observations in osteoblast cultures, PTH suppressed Hey expression in osteocytes even in the absence of Notch activation. This suggests a sensitization to the effects of PTH on Notch signaling or higher basal levels of Hey1, Hey2 and HeyL transcripts in osteocytes than in osteoblasts. Moreover, the expression of Notch3 in osteocyte-enriched cells may be accountable for the different response of osteocytes and osteoblasts to PTH, since the Notch3 receptor exhibits higher degree of basal activation than Notch1 and Notch2 [69].
The effect of PTH was observed in osteocyte cultures from Notch2HCS mice expressing a truncated and stable Notch2 NICD that is resistant to proteasomal degradation [70]. This would suggest that PTH is not likely to act by destabilizing the Notch NICD. The relative increase in Hey1 and Hey2 mRNA levels in the context of the Notch2HCS mutation was not affected by PTH. This indicates that suppression of Hey1 and Hey2 expression occurs by mechanisms that do not require inhibition of Notch2 or Rbpjκ function and may be secondary to an effect of PTH on the osteocytic phenotype. PTH did not suppress HeyL expression in the context of the Notch2HCS mutation. The reason is not known, but recent work has shown that the NICD of Notch1 and Notch2 activate transcription by associating with distinct regions of Rbpjκ, and that the structure of the target gene promoter determines differences in the capacity of Notch to induce transcription [71]. It is possible that the association of the Notch2 NICD with Rbpjκ bound to HeyL regulatory sequences was not affected by the mechanisms that mediate the inhibitory effects of PTH on Hey1 and Hey2.
Previous work demonstrated that PTH induces the Notch ligand Jag1 in cells of the osteoblastic lineage and in the bone marrow niche with an apparent increase in Notch signaling [28, 29]. Although we confirmed this effect (Zanotti and Canalis unpublished) and reported that Jag1 is the only Notch specific ligand expressed by osteoblastic cells in femurs, we observed that PTH downregulates Notch signaling in osteoblasts and osteocytes. A possible explanation is that Jag1 induction is not sufficient to overcome the inhibitory effect of PTH on Rbpjκ-mediated signaling that occurs primarily in the osteoblastic lineage but might not take place in the bone marrow niche. Alternatively, Jag1 induced by PTH may sequester and suppress Notch receptors with a mechanism of cis-inhibition that takes place when receptors and ligands are expressed by the same cell and is known to occur in Drosophila and mammalian cells [72].
In conclusion, PTH prevents the association of Rbpjκ with DNA and suppresses Notch signaling in the osteoblastic lineage, effects that may contribute to the anabolic actions of PTH in bone.
HIGHLIGHTS.
PTH and Notch regulate bone formation
PTH suppresses Notch signaling in osteoblasts and osteocytes in vitro
PTH suppresses Notch signaling in bone in vivo
Suppression of Notch may contribute to the bone anabolic effects of PTH
Acknowledgments
The authors thank J. Lian for Bglap cDNA, E. Six for Dll1 cDNA, T. Iso for Hey1 and Hey2 cDNAs, D. Srivastava for HeyL cDNA, Y. Shirayoshi for Notch4 cDNA, L.J. Strobl for 12xCSL-Luc and J.S. Nye for the pcDNA-NICD. The authors thank D. Bridgewater, T. Eller, and L. Schilling for technical assistance and M. Yurczak for secretarial support.
*This work was supported by [Grants AR063049 and AR068160] from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) and [Grant DK045227] from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS
- ANOVA
analysis of variance
- BSA
bovine serum albumin
- CMV
cytomegalovirus
- CSL
core binding factor-1/suppressor of hairless/Lag-1
- Dll
delta-like
- DMEM
Dulbecco’s modified Eagle’s medium
- Dmp
dentin matrix protein
- EBNA
Epstein-Barr virus nuclear antigen
- EMSA
electrophoretic mobility shift assay
- FBS
fetal bovine serum
- GFP
green fluorescent protein
- HCS
Hajdu Cheney syndrome
- IDT
Integrated DNA Technologies
- Jag
Jagged
- MAPK
mitogen activated protein kinase
- Nfatc
nuclear factor of activated T-cells
- NICD
Notch intracellular domain
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- PEST
proline-, glutamic acid-, serine- and threonine-rich
- PTH
parathyroid hormone 1–34 fragment
- qRT-PCR
quantitative reverse transcription-PCR
- Rankl
Receptor activator of nuclear factor kappa B ligand
- Rbpjκ
recombination signal binding protein J kappa
- SEM
standard error of the mean
- TLCK
N-α-tosyl-L-lysyl-chloromethylketone hydrochloride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Canalis E, Giustina A, Bilezikian JP. Mechanisms of anabolic therapies for osteoporosis. The New England journal of medicine. 2007;357(9):905–16. doi: 10.1056/NEJMra067395. [DOI] [PubMed] [Google Scholar]
- 2.Dallas SL, Prideaux M, Bonewald LF. The osteocyte: an endocrine cell … and more, Endocr. Rev. 2013;34(5):658–90. doi: 10.1210/er.2012-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S, Hsu H, Sullivan J, Hawkins N, Davy E, Capparelli C, Eli A, Qian YX, Kaufman S, Sarosi I, Shalhoub V, Senaldi G, Guo J, Delaney J, Boyle WJ. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93(2):165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 4.Brunkow ME, Gardner JC, Van Ness J, Paeper BW, Kovacevich BR, Proll S, Skonier JE, Zhao L, Sabo PJ, Fu Y, Alisch RS, Gillett L, Colbert T, Tacconi P, Galas D, Hamersma H, Beighton P, Mulligan J. Bone dysplasia sclerosteosis results from loss of the SOST gene product, a novel cystine knot-containing protein. Am J Hum Genet. 2001;68(3):577–89. doi: 10.1086/318811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nioi P, Taylor S, Hu R, Pacheco E, He YD, Hamadeh H, Paszty C, Pyrah I, Ominsky MS, Boyce RW. Transcriptional Profiling of Laser Capture Microdissected Subpopulations of the Osteoblast Lineage Provides Insight Into the Early Response to Sclerostin Antibody in Rats. J Bone Miner Res. 2015;30(8):1457–67. doi: 10.1002/jbmr.2482. [DOI] [PubMed] [Google Scholar]
- 6.Xiong J, Piemontese M, Onal M, Campbell J, Goellner JJ, Dusevich V, Bonewald L, Manolagas SC, O’Brien CA. Osteocytes, not Osteoblasts or Lining Cells, are the Main Source of the RANKL Required for Osteoclast Formation in Remodeling Bone. PLoS One. 2015;10(9):e0138189. doi: 10.1371/journal.pone.0138189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bezooijen RLvan, Roelen BA, Visser A, Wee-Pals Lvander, Wilt Ede, Karperien M, Hamersma H, Papapoulos SE, Dijke Pten, Lowik CW. Sclerostin is an osteocyte-expressed negative regulator of bone formation, but not a classical BMP antagonist. J Exp Med. 2004;199(6):805–14. doi: 10.1084/jem.20031454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray TM, Rao LG, Divieti P, Bringhurst FR. Parathyroid hormone secretion and action: evidence for discrete receptors for the carboxyl-terminal region and related biological actions of carboxyl-terminal ligands. Endocrine reviews. 2005;26(1):78–113. doi: 10.1210/er.2003-0024. [DOI] [PubMed] [Google Scholar]
- 9.Rouleau MF, Mitchell J, Goltzman D. In vivo distribution of parathyroid hormone receptors in bone: evidence that a predominant osseous target cell is not the mature osteoblast. Endocrinology. 1988;123(1):187–91. doi: 10.1210/endo-123-1-187. [DOI] [PubMed] [Google Scholar]
- 10.Fermor B, Skerry TM. PTH/PTHrP receptor expression on osteoblasts and osteocytes but not resorbing bone surfaces in growing rats. J Bone Miner Res. 1995;10(12):1935–43. doi: 10.1002/jbmr.5650101213. [DOI] [PubMed] [Google Scholar]
- 11.Bellido T, Ali AA, Gubrij I, Plotkin LI, Fu Q, O’Brien CA, Manolagas SC, Jilka RL. Chronic elevation of parathyroid hormone in mice reduces expression of sclerostin by osteocytes: a novel mechanism for hormonal control of osteoblastogenesis. Endocrinology. 2005;146(11):4577–83. doi: 10.1210/en.2005-0239. [DOI] [PubMed] [Google Scholar]
- 12.Canalis E. Update in new anabolic therapies for osteoporosis. J Clin Endocrinol Metab. 2010;95(4):1496–504. doi: 10.1210/jc.2009-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kedlaya R, Kang KS, Hong JM, Bettagere V, Lim KE, Horan D, Divieti-Pajevic P, Robling AG. Adult-Onset Deletion of beta-Catenin in (10kb)Dmp1-Expressing Cells Prevents Intermittent PTH-Induced Bone Gain. Endocrinology. 2016;157(8):3047–57. doi: 10.1210/en.2015-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robling AG, Kedlaya R, Ellis SN, Childress PJ, Bidwell JP, Bellido T, Turner CH. Anabolic and catabolic regimens of human parathyroid hormone 1–34 elicit bone- and envelope-specific attenuation of skeletal effects in Sost-deficient mice. Endocrinology. 2011;152(8):2963–75. doi: 10.1210/en.2011-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Q, Jilka RL, Manolagas SC, O’Brien CA. Parathyroid hormone stimulates receptor activator of NFkappa B ligand and inhibits osteoprotegerin expression via protein kinase A activation of cAMP-response element-binding protein. J Biol Chem. 2002;277(50):48868–75. doi: 10.1074/jbc.M208494200. [DOI] [PubMed] [Google Scholar]
- 16.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A, Tsuda E, Morinaga T, Higashio K, Udagawa N, Takahashi N, Suda T. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL, Proc. Natl Acad Sci U S A. 1998;95(7):3597–602. doi: 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahalingam CD, Sampathi BR, Sharma S, Datta T, Das V, Abou-Samra AB, Datta NS. MKP1-dependent PTH modulation of bone matrix mineralization in female mice is osteoblast maturation stage specific and involves P-ERK and P-p38 MAPKs. J Endocrinol. 2013;216(3):315–29. doi: 10.1530/joe-12-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(3):237–55. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 20.Zanotti S, Canalis E. Notch Signaling and the Skeleton. Endocr Rev. 2016;37(3):223–53. doi: 10.1210/er.2016-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zanotti S, Smerdel-Ramoya A, Stadmeyer L, Durant D, Radtke F, Canalis E. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology. 2008;149(8):3890–9. doi: 10.1210/en.2008-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zanotti S, Smerdel-Ramoya A, Canalis E. Reciprocal regulation of Notch and nuclear factor of activated T-cells (NFAT) c1 transactivation in osteoblasts. J Biol Chem. 2011;286(6):4576–88. doi: 10.1074/jbc.M110.161893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canalis E, Adams DJ, Boskey A, Parker K, Kranz L, Zanotti S. Notch Signaling in Osteocytes Differentially Regulates Cancellous and Cortical Bone Remodeling. J Biol Chem. 2013;288(35):25614–25625. doi: 10.1074/jbc.M113.470492. M113.470492 [pii];10.1074/jbc.M113.470492 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Canalis E, Parker K, Feng JQ, Zanotti S. Osteoblast Lineage-specific Effects of Notch Activation in the Skeleton. Endocrinology. 2013;154(2):623–634. doi: 10.1210/en.2012-1732. en.2012-1732 [pii];10.1210/en.2012-1732 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canalis E, Schilling L, Yee SP, Lee SK, Zanotti S. Hajdu Cheney Mouse Mutants Exhibit Osteopenia, Increased Osteoclastogenesis and Bone Resorption. J Biol Chem. 2016;291:1538–1551. doi: 10.1074/jbc.M115.685453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yorgan T, Vollersen N, Riedel C, Jeschke A, Peters S, Busse B, Amling M, Schinke T. Osteoblast-specific Notch2 inactivation causes increased trabecular bone mass at specific sites of the appendicular skeleton. Bone. 2016;87:136–146. doi: 10.1016/j.bone.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Canalis E, Bridgewater D, Schilling L, Zanotti S. Canonical Notch Activation in Osteocytes Causes Osteopetrosis, American journal of physiology. Endocrinology and metabolism. 2015;310(2):E171–E182. doi: 10.1152/ajpendo.00395.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber JM, Forsythe SR, Christianson CA, Frisch BJ, Gigliotti BJ, Jordan CT, Milner LA, Guzman ML, Calvi LM. Parathyroid hormone stimulates expression of the Notch ligand Jagged1 in osteoblastic cells. Bone. 2006;39(3):485–93. doi: 10.1016/j.bone.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, Milner LA, Kronenberg HM, Scadden DT. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425(6960):841–6. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 30.Duggan SP, McCarthy JV. Beyond gamma-secretase activity: The multifunctional nature of presenilins in cell signalling pathways. Cell Signal. 2016;28(1):1–11. doi: 10.1016/j.cellsig.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Zanotti S, Kalajzic I, Aguila HL, Canalis E. Sex and genetic factors determine osteoblastic differentiation potential of murine bone marrow stromal cells. PLoS One. 2014;9(1):e86757. doi: 10.1371/journal.pone.0086757. [doi];PONE-D-13-31354 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu H, Lacey DL, Dunstan CR, Solovyev I, Colombero A, Timms E, Tan HL, Elliott G, Kelley MJ, Sarosi I, Wang L, Xia XZ, Elliott R, Chiu L, Black T, Scully S, Capparelli C, Morony S, Shimamoto G, Bass MB, Boyle WJ. Tumor necrosis factor receptor family member RANK mediates osteoclast differentiation and activation induced by osteoprotegerin ligand. Proc Natl Acad Sci U S A. 1999;96(7):3540–5. doi: 10.1073/pnas.96.7.3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Centrella M, McCarthy TL, Canalis E. Parathyroid hormone modulates transforming growth factor beta activity and binding in osteoblast-enriched cell cultures from fetal rat parietal bone. Proc Natl Acad Sci U S A. 1988;85(16):5889–93. doi: 10.1073/pnas.85.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halleux C, Kramer I, Allard C, Kneissel M. Isolation of mouse osteocytes using cell fractionation for gene expression analysis. Methods Mol Biol. 2012;816:55–66. doi: 10.1007/978-1-61779-415-5_5. [doi] [DOI] [PubMed] [Google Scholar]
- 35.Zanotti S, Canalis E. The Dmp1-SOST Transgene Interacts With and Downregulates the Dmp1-Cre Transgene and the Rosa Allele. J Cell Biochem. 2015 doi: 10.1002/jcb.25405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canalis E, Zanotti S. Hairy and Enhancer of Split-related with YRPW Motif-Like (HeyL) is Dispensable for Bone Remodeling in Mice. J Cell Biochem. 2016 doi: 10.1002/jcb.25859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nazarenko I, Lowe B, Darfler M, Ikonomi P, Schuster D, Rashtchian A. Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res. 2002;30(9):e37. doi: 10.1093/nar/30.9.e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nazarenko I, Pires R, Lowe B, Obaidy M, Rashtchian A. Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 2002;30(9):2089–195. doi: 10.1093/nar/30.9.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lian J, Stewart C, Puchacz E, Mackowiak S, Shalhoub V, Collart D, Zambetti G, Stein G. Structure of the rat osteocalcin gene and regulation of vitamin D-dependent expression. Proc Natl Acad Sci USA. 1989;86(4):1143–7. doi: 10.1073/pnas.86.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Six EM, Ndiaye D, Sauer G, Laabi Y, Athman R, Cumano A, Brou C, Israel A, Logeat F. The notch ligand Delta1 recruits Dlg1 at cell-cell contacts and regulates cell migration. J Biol Chem. 2004;279(53):55818–26. doi: 10.1074/jbc.M408022200. [DOI] [PubMed] [Google Scholar]
- 41.Iso T, Sartorelli V, Chung G, Shichinohe T, Kedes L, Hamamori Y. HERP, a new primary target of Notch regulated by ligand binding. Mol Cell Biol. 2001;21(17):6071–9. doi: 10.1128/MCB.21.17.6071-6079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol. 1999;216(1):72–84. doi: 10.1006/dbio.1999.9454. [doi];S0012-1606(99)99454-X [pii] [DOI] [PubMed] [Google Scholar]
- 43.Nye JS, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120(9):2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 44.Shirayoshi Y, Yuasa Y, Suzuki T, Sugaya K, Kawase E, Ikemura T, Nakatsuji N. Proto-oncogene of int-3, a mouse Notch homologue, is expressed in endothelial cells during early embryogenesis. Genes Cells. 1997;2(3):213–224. doi: 10.1046/j.1365-2443.1997.d01-310.x. [DOI] [PubMed] [Google Scholar]
- 45.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6(5):343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 46.Kouadjo KE, Nishida Y, Cadrin-Girard JF, Yoshioka M, St-Amand J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics. 2007;8:127. doi: 10.1186/1471-2164-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strobl LJ, Hofelmayr H, Stein C, Marschall G, Brielmeier M, Laux G, Bornkamm GW, Zimber-Strobl U. Both Epstein-Barr viral nuclear antigen 2 (EBNA2) and activated Notch1 transactivate genes by interacting with the cellular protein RBP-J kappa. Immunobiology. 1997;198(1–3):299–306. doi: 10.1016/s0171-2985(97)80050-2. [DOI] [PubMed] [Google Scholar]
- 48.Henkel T, Ling PD, Hayward SD, Peterson MG. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science (New York, NY) 1994;265(5168):92–5. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 49.Canalis E, Zanotti S, Smerdel-Ramoya A. Connective Tissue Growth Factor is a Target of Notch Signaling in Cells of the Osteoblastic Lineage. Bone. 2014;64:273–280. doi: 10.1016/j.bone.2014.04.028. S8756–3282(14)00163-X [pii];10.1016/j.bone.2014.04.028 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Youngstrom DW, Dishowitz MI, Bales CB, Carr E, Mutyaba PL, Kozloff KM, Shitaye H, Hankenson KD, Loomes KM. Jagged1 expression by osteoblast-lineage cells regulates trabecular bone mass and periosteal expansion in mice. Bone. 2016;91:64–74. doi: 10.1016/j.bone.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawal RA, Zhou X, Batey K, Hoffman CM, Georger MA, Radtke F, Hilton MJ, Xing L, Frisch BJ, Calvi LM. The Notch Ligand Jagged1 Regulates the Osteoblastic Lineage by Maintaining the Osteoprogenitor Pool. J Bone Miner Res. 2017 doi: 10.1002/jbmr.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dietrich JW, Canalis EM, Maina DM, Raisz LG. Hormonal control of bone collagen synthesis in vitro: effects of parathyroid hormone and calcitonin. Endocrinology. 1976;98(4):943–949. doi: 10.1210/endo-98-4-943. [DOI] [PubMed] [Google Scholar]
- 53.Canalis E. Effect of hormones and growth factors on alkaline phosphatase activity and collagen synthesis in cultured rat calvariae. Metabolism. 1983;32(1):14–20. doi: 10.1016/0026-0495(83)90149-x. [DOI] [PubMed] [Google Scholar]
- 54.Noda M, Yoon K, Rodan GA. Cyclic AMP-mediated stabilization of osteocalcin mRNA in rat osteoblast-like cells treated with parathyroid hormone. J Biol Chem. 1988;263(34):18574–7. [PubMed] [Google Scholar]
- 55.McCarthy TL, Centrella M, Canalis E. Parathyroid hormone enhances the transcript and polypeptide levels of insulin-like growth factor I in osteoblast-enriched cultures from fetal rat bone. Endocrinology. 1989;124(3):1247–1253. doi: 10.1210/endo-124-3-1247. [DOI] [PubMed] [Google Scholar]
- 56.Lee SK, Lorenzo JA. Parathyroid hormone stimulates TRANCE and inhibits osteoprotegerin messenger ribonucleic acid expression in murine bone marrow cultures: correlation with osteoclast-like cell formation. Endocrinology. 1999;140(8):3552–61. doi: 10.1210/endo.140.8.6887. [DOI] [PubMed] [Google Scholar]
- 57.Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. p300 acts as a transcriptional coactivator for mammalian Notch-1, Mol. Cell Biol. 2001;21(22):7761–7774. doi: 10.1128/MCB.21.22.7761-7774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakashima T, Hayashi M, Fukunaga T, Kurata K, Oh-Hora M, Feng JQ, Bonewald LF, Kodama T, Wutz A, Wagner EF, Penninger JM, Takayanagi H. Evidence for osteocyte regulation of bone homeostasis through RANKL expression. Nat Med. 2011;17(10):1231–1234. doi: 10.1038/nm.2452. nm.2452 [pii];10.1038/nm.2452 [doi] [DOI] [PubMed] [Google Scholar]
- 59.Xiong J, Onal M, Jilka RL, Weinstein RS, Manolagas SC, O’Brien CA. Matrix-embedded cells control osteoclast formation. Nat Med. 2011;17(10):1235–1241. doi: 10.1038/nm.2448. nm.2448 [pii];10.1038/nm.2448 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang Y, Ganem D. RBP-J (CSL) is essential for activation of the K14/vGPCR promoter of Kaposi’s sarcoma-associated herpesvirus by the lytic switch protein RTA. J Virol. 2004;78(13):6818–26. doi: 10.1128/jvi.78.13.6818-6826.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi M, Hu ZL, Zheng MH, Song NN, Huang Y, Zhao G, Han H, Ding YQ. Notch-Rbpj signaling is required for the development of noradrenergic neurons in the mouse locus coeruleus. J Cell Sci. 2012;125(Pt 18):4320–32. doi: 10.1242/jcs.102152. [DOI] [PubMed] [Google Scholar]
- 62.Canalis E, Kranz L, Zanotti S. Nemo-like kinase regulates postnatal skeletal homeostasis. J Cell Physiol. 2014;229(11):1736–43. doi: 10.1002/jcp.24625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ishitani T, Hirao T, Suzuki M, Isoda M, Ishitani S, Harigaya K, Kitagawa M, Matsumoto K, Itoh M. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat Cell Biol. 2010;12(3):278–85. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- 64.Zanotti S, Canalis E. Nemo-like kinase inhibits osteoblastogenesis by suppressing bone morphogenetic protein and WNT canonical signaling. J Cell Biochem. 2012;113(2):449–456. doi: 10.1002/jcb.23365. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nifuji A, Ideno H, Ohyama Y, Takanabe R, Araki R, Abe M, Noda M, Shibuya H. Nemo-like kinase (NLK) expression in osteoblastic cells and suppression of osteoblastic differentiation. Exp Cell Res. 2010;316(7):1127–36. doi: 10.1016/j.yexcr.2010.01.023. [DOI] [PubMed] [Google Scholar]
- 66.O’Brien CA, Plotkin LI, Galli C, Goellner JJ, Gortazar AR, Allen MR, Robling AG, Bouxsein M, Schipani E, Turner CH, Jilka RL, Weinstein RS, Manolagas SC, Bellido T. Control of bone mass and remodeling by PTH receptor signaling in osteocytes. PLoS One. 2008;3(8):e2942. doi: 10.1371/journal.pone.0002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schipani E, Kruse K, Juppner H. A constitutively active mutant PTH-PTHrP receptor in Jansen-type metaphyseal chondrodysplasia. Science (New York, NY) 1995;268(5207):98–100. doi: 10.1126/science.7701349. [DOI] [PubMed] [Google Scholar]
- 68.Zanotti S, Smerdel-Ramoya A, Canalis E. Nuclear Factor of Activated T-cells (Nfat)c2 Inhibits Notch Signaling in Osteoblasts. J Biol Chem. 2013;288(1):624–632. doi: 10.1074/jbc.M112.340455. M112.340455 [pii];10.1074/jbc.M112.340455 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu X, Choi SH, Hu T, Tiyanont K, Habets R, Groot AJ, Vooijs M, Aster JC, Chopra R, Fryer C, Blacklow SC. Insights into Autoregulation of Notch3 from Structural and Functional Studies of Its Negative Regulatory Region. Structure. 2015;23(7):1227–35. doi: 10.1016/j.str.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oberg C, Li J, Pauley A, Wolf E, Gurney M, Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J Biol Chem. 2001;276(38):35847–53. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- 71.Yuan Z, Friedmann DR, Vanderwielen BD, Collins KJ, Kovall RA. Characterization of CSL (CBF-1, Su(H), Lag-1) Mutants Reveals Differences in Signaling Mediated by Notch1 and Notch2. J Biol Chem. 2012;287(42):34904–34916. doi: 10.1074/jbc.M112.403287. M112.403287 [pii];10.1074/jbc.M112.403287 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.del Alamo D, Rouault H, Schweisguth F. Mechanism and significance of cis-inhibition in Notch signalling. Curr Biol. 2011;21(1):R40–7. doi: 10.1016/j.cub.2010.10.034. [DOI] [PubMed] [Google Scholar]


