Abstract
Dimerization reactions in membranes underlie membrane protein folding, conformational stability and cell receptor signaling. Here we summarize a method that allows for measurement of equilibrium dimerization reactions of membrane proteins in lipid bilayers, using subunit capture into liposomes followed by single-molecule photobleaching analysis. This strategy is grounded in the fact that given a comparable labeling efficiency, monomeric or dimeric forms of a membrane protein will give rise to distinctly different photobleaching probability distributions. These methods have been used to measure the dimer stoichiometry of the Fluc F− ion channel, and the dimerization equilibrium constant of the ClC-ec1 Cl−/H+ antiporter in lipid bilayers. This approach can be applied to any membrane protein system provided it can be purified, fluorescently labeled in a quantitative manner, and verified to be correctly folded by functional assays, even if the structure is not yet known.
1. Introduction
Studies of membrane protein dimerization have been of keen interest in biological research for two reasons. First, there are situations where protein dimerization is physiologically significant, for instance, in the case of receptor activation (Ferré et al., 2014; Kovacs, Zorn, Huang, Barros, & Kuriyan, 2015). Second, dimerization serves as an equilibrium model for studying the physical forces underlying protein folding. In fact, for multi-helix membrane proteins, where reversible assembly is practically intractable, dimerization offers the simplest strategy to obtain a thermodynamic understanding of how protein interactions are stabilized in the non-polar solvent environment of the lipid bilayer. While equilibrium dimerization reactions have been measured in detergent micelles (Fleming, Ackerman, & Engelman, 1997; MacKenzie & Fleming, 2008), numerous technical challenges have limited our ability to study these reactions in lipid bilayers.
One reason is that traditional dilution in membranes is much more challenging than in aqueous solutions. The membrane is a 2-dimensional solvent that adopts a lipid bilayer structure that does not readily exchange with other membranes. Therefore, it is not possible to dilute the system by simply adding more lipids. Second, we work with concentrations of lipids ~35 mM during reconstitution, a ~2000-fold reduction in the solvent phase compared to 55.5 M water, resulting in a proportional decrease in protein signal. For example, in order to measure Glycophorin A dimerization by dilution methods (Keq ~109 POPC lipid/subunit (Hong, Blois, Cao, & Bowie, 2010)) one would have to detect the protein at picomolar concentrations. Other methods have been developed to extract the equilibrium constant while examining the protein at high densities in liposomes (Hong et al., 2010; Mathiasen et al., 2014), however this introduces other technical challenges, as many membrane proteins undergo aggregation at high densities.
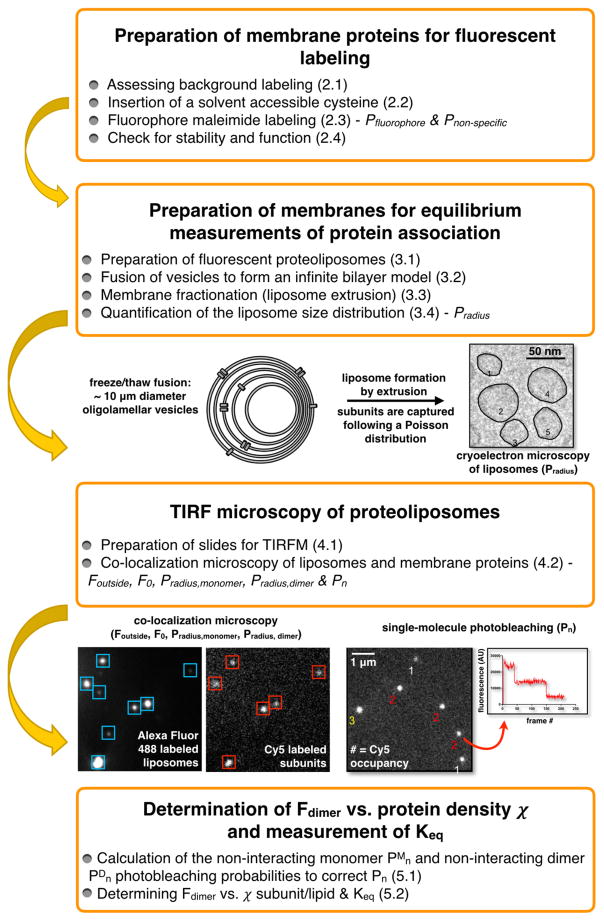
Therefore, macroscopic approaches for measuring dimerization are inherently challenged as high affinity dimers must be diluted to ridiculously low concentrations in order to observe the dissociate state. Single-molecule fluorescence microscopy offers a solution to this problem as it can detect single molecules within the membrane with high sensitivity. Furthermore, photobleaching analysis is a method that allows for rigorous determination of protein stoichiometry on the single particle level (Stockbridge, Robertson, Kolmakova-Partensky, & Miller, 2013). To solve the practical problem of diluting the protein in the membrane, we fuse lipid bilayers through multiple freeze/thaw cycles creating large ~10 μm diameter oligolamellar vesicles (Pozo Navas et al., 2005), our model of an infinite membrane. To prepare this system for single-molecule TIRF imaging, the membranes are fractioned into liposomes, each carrying a small area of the membrane along with the protein subunits embedded in that patch. Membrane protein encapsulation into liposomes follows a Poisson distribution (Maduke, Pheasant, & Miller, 1999; Walden et al., 2007), wherein the number of protein particles per liposome depends on the protein density and stoichiometry. Therefore, measurement of liposome occupancy by single-molecule photobleaching analysis allows us to follow the changing monomer-dimer population in the membrane and extract the equilibrium constant that pertains to the statistical mechanical free energy of dimerization. This method (Figure 1 & Table 1) requires knowledge of the extruded liposome size distribution and experimental determination of subunit fluorescent labeling yields. Protein/liposome co-localization microscopy allows for quality checks on reconstitution, inspection of protein aggregation or non-specific reactions, determination of differences in liposome accessibility and finally, the measurement of the photobleaching probability distribution from which the monomer-dimer equilibrium constant can be obtained. This method has been used to measure Keq of the ClC-ec1 Cl−/H+ antiporter in 2:1 POPE/POPG lipids bilayers (Chadda, R., Krishnamani, V., Mersch, K., Wong, J., Brimberry, M., Chadda, A. 2016) and can reasonably be adapted to study new types of membrane proteins systems using a similar approach.
Figure 1. Flow chart outlining the steps involved in measuring equilibrium membrane protein dimerization in lipid bilayers by subunit capture followed by single-molecule photobleaching analysis.
See Table 1 for definition of experimental parameters.
Table 1.
List of required experimental parameters for determining the fraction of dimerization of a protein in the lipid bilayer as a function of the subunit/lipid mole fraction χ.
| Method/Parameter | Description | Section |
|---|---|---|
| UV-Vis Spectrophotometry | ||
| Pfluorophore | Overall labeling yield (fluorophore/subunits) | 2.3 |
| Pnon-specific | Non-specific labeling yield (fluorophore/subunits) | 2.3 |
| Cryo-EM imaging | ||
| Pradius | The liposome radius distribution from cryoelectron imaging of extruded liposomes | 3.4 |
| Co-localization microscopy | ||
| Foutside | The fraction of protein spots that do not co-localize with liposomes. Measures fidelity of reconstitution. | 4.2.1 |
| F0 | Fraction of un-occupied vesicles, i.e. liposomes that do not co-localize with protein | 4.2.2 |
| Pradius,monomer | Re-normalized liposome size distribution for monomer accessible liposomes | 4.2.2 |
| Pradius,dimer | Re-normalized liposome size distribution for dimer accessible liposomes | 4.2.2 |
| Pexpt(n=1,…,5+) | The experimental photobleaching probabilities | 4.2.3 |
| Simulation | ||
| PM(n=1,…5+) | The calculated photobleaching probabilities for non-interacting monomers. Requires experimental determination of Pradius, Pradius,monomer, Pfluorophore and Pnon-specific. | 5.1.1 |
| PD(n=1,…,5+) | The calculated photobleaching probabilities for non-interacting dimers. Requires experimental determination of Pradius, Pradius,dimer, Pfluorophore and Pnon- specific. | 5.1.2 |
| Fdimer | The fraction of dimer determined by least-squares fitting of the Pexpt to (1-Fdimer)PM+FdimerPD | 5.2 |
2. Preparation of membrane proteins for fluorescent studies
In this section, we will discuss the strategy for subunit specific labeling of a membrane protein for single-molecule fluorescence microscopy experiments in lipid bilayers. We will assume that purification of stable histidine-tagged protein in detergent micelles has already been established, along with reconstitution methods into liposomes. We recommend that there be a quantitative method for assaying functional activity (Stockbridge & Tsai, 2015) as this is the most rigorous measure of whether the protein is properly folded.
In order to measure a dimerization reaction in the membrane using single-molecule photobleaching, each subunit should be labeled at a unique position with a single fluorophore. Complete labeling is desirable for photobleaching experiments but is unrealistic in a laboratory setting. In addition, fluorophores can switch to a dark state (Ha & Tinnefeld, 2012) lowering the effective labeling yield. Increasing the reaction time or rate may improve the yield, but it also increases the chance of non-specific labeling. In reality, both of these situations can be tolerated within reason and corrected for if the overall labeling yield, Pfluorophore, and the non-specific labeling yield, Pnon-specific, are experimentally determined. Using the following procedure, we obtained labeling yields for ClC-ec1 PCy5 = 0.72 and Pnon-specific = 0.14, which allowed for adequate discrimination between the monomer and dimer photobleaching probabilities.
2.1 Assessing the protein for background labeling
We use the standard method of cysteine-maleimide conjugation because it is rapid and fairly specific, but there are other means to attach fluorophores to proteins (Hermanson, 2013). It is important to have only one fluorophore per subunit, so native cysteines must be removed and replaced with a single reactive cysteine. If removal of all cysteines affects the stability of the protein, then it is possible to screen and only remove the ones that are reactive when the protein is in a folded state. For example, ClC-ec1 has three endogenous cysteines, but only one is reactive in the folded structure. The cysteine accessibility Ellman’s reagent assay (Riddles, Blakeley, & Zerner, 1983) measures the amount of reactive thiol groups present, and then step-by-step mutagenesis will identify the cysteines that must be removed. Note that the sulfhydryl groups can be oxidized in vivo during the expression of the protein, so TCEP-HCl is added to maintain the reduced state of the thiol (5 mM in the initial cell suspension, followed by 1 mM in metal affinity or ion affinity buffers). The pH of the buffers must be adjusted after addition of TCEP-HCl. The final purification step (size exclusion chromatography) does not contain TCEP as it can interfere with the maleimide reaction. We find that the cysteines on the purified protein remain reactive for weeks, however a slow loss of reactivity is observed, possibly due to irreversible oxidation (Reddie & Carroll 2008).
2.1.1 Measurement of cysteine accessibility by Ellman’s reagent
In this reaction, the thiolate form of the cysteine sulfhydryl group reacts with Ellman’s reagent (DNTB, 5,5′-Dithiobis(2-nitrobenzoic acid)) to form TNB2−, which absorbs light at 412 nm, A412. Following the absorbance in a UV-Visible light spectrophotometer (Nanodrop 2000c, Thermo-Fisher Scientific) allows for direct monitoring of the reactivity of the cysteines. Baseline absorbance at 750 nm should also be measured to correct for drift during the acquisition.
-
Prepare a fresh stock of Ellman’s reagent in reaction buffer (0.1 M sodium phosphate, 1 mM EDTA, pH 8.0) at 10 mM concentration. This master stock can be stored in the dark at RT for a day. Dilute the master stock to a 5 mM working solution with purification buffer (e.g. 150 mM NaCl, 20 mM MOPS, 5 mM n-Decyl β-D-maltopyranoside (DM), pH 7.0 for ClC-ec1).
Note. The purification buffer can be changed to suit the protein of interest, but it should not contain KCl if SDS is to be added at a later stage.
Take 300 μL of 10 μM protein in a clean quartz cuvette and monitor A412 for a period of 5 minutes to establish the baseline.
Add 20 μL of Ellman’s reagent working solution to the cuvette, mix well by pipetting, and monitor A412 for 20 minutes. Surface exposed cysteines reacts with DTNB in a rapid fashion (i.e. an immediate step up in A412), whereas partially accessible cysteines show a slow creep in the absorbance over time.
Add 40 μL of denaturing buffer (e.g. 20% SDS stock prepared in purification buffer for ClC-ec1) to unfold the protein and expose internal cysteines to react with DTNB. Allow the reaction to proceed for 45 minutes, or until a steady absorbance is reached. For ClC-ec1, we observe this as a slow exponential saturation of A412, presumably due to the slow kinetics of the reaction since unfolding is expected to be fast.
Include a negative control of purification buffer alone to allow measurement of (A412 –A750)control in the presence of Ellman’s reagent and with the addition of denaturing buffer. This is necessary as the spectrum of DNTB bleeds into 412 nm.
-
The TNB2− specific absorbance is calculated using A412,TNB2− = (A412 –A750)protein−(A412–A750)control where the control represents either the purification buffer + Ellman’s reagent, or the addition of denaturing buffer. The concentration of [TNB2−] is calculated using the Beer-Lambert law:
(1) Where ℓ is the path length in cm, and ε is the molar extinction coefficient. Note, this value has been reported in the literature from 11,400–14,140 M−1 cm−1 (Riddles et al., 1983). It is recommended that a standard curve be generated with A412–A750 as a function of cysteine concentration, freshly prepared in the desired buffer condition. The number of reactive cysteines per subunit is calculated as [TNB2−]/([protein]*D), where D is the dilution factor.
2.2 Insertion of a solvent accessible cysteine for subunit specific labeling
Once the protein has been wiped clean of all the reactive endogenous cysteines then a new cysteine residue can be inserted that allows for rapid conjugation with maleimides. The position of the new cysteine should be optimized to obtain high-quality photobleaching data. Since photobleaching relies on observing step-wise changes in fluorescent intensity, any fluctutations can make it difficult to interpret the traces. Fluorescent intensity depends on the solvent environment around a fluorophore (Lakowicz 2007), so it is advisable to place the cysteine in a structured part of the protein far away from the protein-membrane interface. Inserting the cysteine at the end of a helix, in the center of a multi-helix protein may yield better quality data than placing the cysteine in a loop or near one of the termini. It is important that the site be exposed to the aqueous solution so that reaction time can be minimized to prevent non-specific labeling at lysines or the N-terminus (Brewer & Riehm 1967). Note that other factors outside of aqueous exposure, such as steric constraints or the surrounding electrostatic environment can also play a role in the reactivity of the cysteine with maleimides. While knowledge of the structure is useful in this situation, it is not necessary as information from hydropathy analysis can serve as a guide. We recommend preparing 5 cysteine mutants to start, comparing the reactivity of each using Ellman’s reagent and choosing the site that yields an instantaneous reaction of a single cysteine.
2.3 Fluorophore-maleimide labeling of membrane protein subunits
The following procedure has been optimized for labeling of ClC-ec1 with Cy5-maleimide, maximizing the overall labeling yield (Pfluorophore), while minimizing non-specific labeling (Pnon-specific). The procedure here should suffice as a starting point for any new protein system, but optimization is recommended.
-
1
Dissolve lyophilized Cy5-maleimide in anhydrous DMSO (Molecular Probes) to make 10 mM master stocks. This stock should be divided into smaller, workable aliquots to prevent exposure to light, and stored at −80°C in a box with anhydrous CaSO4 crystals (Drierite, USA) to prevent hydrolysis of the maleimide.
-
2
Dilute protein with purification buffer to a final concentration of 10 μM. In the dark, thaw a vial of frozen 10 mM Cy5-maleimide working stock, and add the appropriate volume for a final concentration of 50 μM. Mix well by pipetting, then incubate for 5 minutes at room temperature, in the dark. Stop the reaction with 1 mM cysteine, from a 100 mM stock prepared freshly in purification buffer, and pH adjusted to ~7.0.
-
3
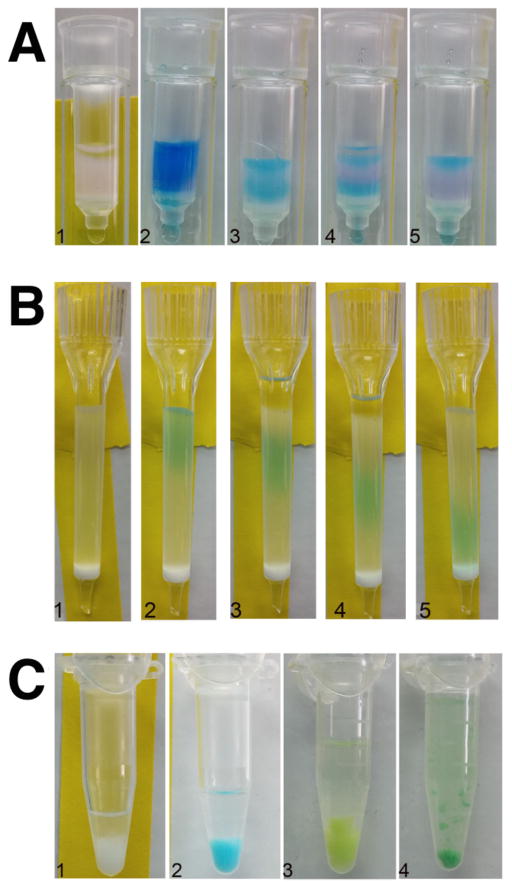
For a 1 mL reaction (~500 μg ClC-ec1), we make a 250 μL cobalt resin column (Talon, Clontech) in a micro-bio spin column (Bio-Rad), and equilibrate the resin with 15 column volumes (CVs) of cobalt wash buffer (100 mM NaCl, 20 mM Tris, 5 mM DM, pH 7.5). Bind the protein to the column, and then wash extensively with 30 CVs of purification buffer, making sure that all inner surfaces of the column are washed to remove any lingering free dye. Since the protein is fluorescently labeled, it is visible as a colored band at the top of the column. The protein is then eluted with 4 CV of 400 mM Imidazole in purification buffer, with only the fluorophore labeled protein collected by eye (Figure 2A).
Note. Imidazole absorbs light at 280 nm, and it is difficult to control for this as the amount of absorbance depends on the preparation, and the final concentration in the elution fraction, which varies. Therefore, all imidazole should be removed from the elution fraction to enable proper quantification of the protein labeling yield.
-
5
Swell Sephadex G50 resin in ddH2O. This can be carried out in advance and the resin stored at 4°C, however, it must be warmed to RT before pouring to prevent de-gassing inside the column. Set up a 5 mL disposable column (Pierce) and pour the pre-swelled G50 resin to the top, letting it drip dry, for a final CV ~2.5 mL. Equilibrate the resin with 15 CV of cobalt wash buffer and let the column drip dry. Add the protein elution fraction from the previous step and then follow with cobalt wash buffer. The blue Cy5 labeled protein can be visualized as it moves through the column, and collected as soon as it reaches the end of the column (Figure 2B). The smaller imidazole will elute at a later volume and be separated from the protein.
-
6Place a sample of the eluted protein in a quartz cuvette and measure the UV-Visible light absorbance spectrum from 260–750 nm. A peak will be observed at 280 nm, representing the protein, and 655 nm for Cy5. The concentration of protein, fluorophore, and the fluorophore/protein labeling yield Pfluorophore, is determined as follows:
(2) (3) Where ℓ is the path length in cm, Afluorophore is the peak absorbance of the fluorophore, CFfluorophore is the correction factor for the absorbance of the fluorophore at 280 nm, εprotein is the molar extinction coefficient of the individual subunit of the protein, εfluorophore is the molar extinction coefficient of the fluorophore at the peak absorbance. For Cy5, ACy5 is typically measured at 655 nm, with εCy5 = 2.5 × 105 at 655 nm and CFCy5=0.02.
Note: Maleimides will react with primary amines on lysines or the N-terminus, albeit at a much slower rate. Therefore, it is expected that there is a small population of subunits that will have more than one label, and thus artificially appear dimeric. The minimization of the reaction time to 5 minutes and the reduction of the reaction pH to 7.0 reduces non-specific labeling. Still, the non-specific labeling yield, Pnon-specific, must be measured experimentally to properly correct the photobleaching data. This can be done by carrying out the same reaction with a construct that lacks the reactive cysteine residue.
Note: We chose Cy5 because it is a far-red fluorophore that provides excellent signal to noise for photobleaching studies. It is also sufficiently far from Alexa Fluor 488 fluorescence that we use for lipid labeling. Other fluorophores have their advantages, for instance Cy3 and Cy3B are far more stable allowing for long-lived photobleaching traces, however liposome-only controls must be imaged to ensure that the signal from large liposomes do not bleed into this channel.
Figure 2. Stages in preparation of fluorescently labeled protein and liposomes. (A.1) Equilibrated Bio-Rad mini-column with Talon His-tag purification resin.
(A.2) Reaction containing 10 μM ClC-ec1 + 50 μM Cy5-maleimide from loaded onto mini-column. (A.3) ClC-ec1-Cy5 bound to the cobalt resin after free dye is washed off. (A.4) Appearance of column during elution of ClC-ec1-Cy5 with 400 mM imidazole. The eluted protein emerges at the bottom of mini-column for collection in an eppendorf. (A.5) Appearance of column near the end of elution. (B.1) 5 mL disposable column filled with 2.5 ml G50 Sephadex resin equilibrated in buffer containing 5 mM DM for separation of imidazole from the eluted protein. Note that the tip of the column is cut at an angle using razor blade to increase the flow rate. (B.2) ClC-ec1-Cy5 eluted in (A.5) is applied to the column. (B.3–B.5) The movement of ClC-ec1-Cy5 in the column can be monitored visually. (C.1) Appearance of 7x freeze/thawed 1 μg/mg ClC-ec1-Cy5 liposomes after storage at room temperature. (C.2) Appearance of 7x freeze/thawed 10 μg/mg ClC-ec1-Cy5 liposomes after storage at room temperature. (C.3) Appearance of 7x freeze/thawed 1 μg/mg ClC-ec1-Cy5 in AF488 labeled liposomes after storage at room temperature. (C.4) Appearance of 7x freeze/thawed 10 μg/mg ClC-ec1-Cy5 AF488 labeled liposomes after storage at room temperature
2.4 Check for stability and folding of the fluorescently labeled protein
Adding a fluorophore to a membrane protein can alter its properties, as fluorophores are hydrophobic, and membrane proteins are situated in a hydrophobic solvent environment. Therefore, it is possible that the fluorophore will preferentially partition into the membrane and alter the structure and stability of the protein. It is imperative to confirm that fluorescent labeling does not affect the folding of the protein in both detergent micelles and in liposomes, preferably by some type of functional assay (Stockbridge & Tsai, 2015).
3. Preparation of membranes for equilibrium measurements of dimerization
Once the protein is fluorescently labeled, and verified to be functionally folded, the protein is reconstituted into lipid bilayers by standard dialysis methods (Stockbridge & Tsai, 2015). Reconstitution into membranes containing fluorescently labeled lipids allows for checks on the fidelity of reconstitution, and measurement of the liposome accessible population for both the monomer and dimer states. In the following steps, we describe how to prepare fluorescent liposomes, how to create the infinite model of the bilayer where equilibrium exchange can take place, and how to fractionate the membrane to quantify the monomer and dimer population by co-localization TIRF microscopy.
3.1 Preparation of fluorescent liposomes
It is prudent to check the fidelity of membrane protein reconstitution. Co-localization microscopy of Alexa Fluor 488 (AF488) labeled liposomes and Cy5 labeled protein can readily provide this information. POPE lipids contain a primary amine on its head-group, which allows for covalent conjugation with NHS ester linked fluorophores. We choose AF488 as it has been shown to exhibit significantly less non-specific membrane incorporation (Hughes, Rawle, & Boxer, 2014). Since the NHS esters are rapidly hydrolyzed in water, we use the more stable SDP ester form that is also available. The following protocol is for 1 mL of lipids at 20 mg/mL but the volumes can be scaled up as needed.
-
1
Under a fume hood, aliquot 18 mg POPE and 9 mg POPG in chloroform into a glass vial. Note that chloroform has a high vapor pressure and will drip from the pipette, leading to inaccuracies in lipid amounts. Make sure to gently pipette the solution up and down 5 times for accurate pipetting. Evaporate the chloroform from the 2:1 POPE/POPG lipids under N2, to form a dried film at the bottom of the vial. To remove trace amounts of chloroform, resuspend the lipids in 500 μL of pentane, then dry the lipids again to obtain a thin film.
-
2
Immediately add 1.5 mL of lipid labeling buffer: 300 mM KCl, 100 mM NaHCO3 pH 8.3, to the lipid film in the glass vial. Sonicate briefly for 10 minutes to obtain a cloudy solution of lipid vesicles.
-
3
Add 33 mg of sol-grade CHAPS and continue sonication until the solution becomes translucent. Adjust the volume to 2 mL with labeling buffer, for a final concentration of 20 mg/mL 2:1 POPE/POPG and 35 mM CHAPS.
Note. CHAPS detergent was chosen as it has a high critical micelle concentration (CMC) and is thus readily removed during dialysis (Stockbridge & Tsai, 2015).
Note. Different sonication methods have been used for solubilization of lipids, including vortexing, probe sonication, and cylindrical bath sonication. Recently, we have turned to sonication using a cup horn sonicator (Qsonica) that allows for rapid solubilization of the lipids producing a transparent solution in 15 minutes.
-
4
Prepare 10 mM AF488 5-SDP ester stocks in anhydrous DMSO as described in section 2.3. Aliquot the stock into single-use 16 μL volumes and store at −80 °C to prevent exposure to light and hydrolysis of the reactive group during freeze/thaw.
-
5
When labeling, thaw an aliquot of 10 mM AF488 and add 15 μL to the solubilized lipids. The final mole ratio of AF488 to total lipids is 0.003.
Note. A liposome with radius 25 nm will contain ~104 lipids, assuming a surface area per lipid of 0.6 nm2 (Murzyn, Róg, & Pasenkiewicz-Gierula, 2005). Even if the reaction is only 50% successful, then this would predict ~15 fluorophores per liposome of that size. We confirmed that this saturated the liposomes by co-localization microscopy, examining the reconstitution as a function of the AF488/lipid mole fraction. If some liposomes were not labeled, then we would expect to see protein spots that do not co-localize with liposomes concomitant with a decrease in the fraction of unoccupied liposomes, F0. In fact, we do not observe protein outside of the liposomes and there is no change in F0 as the amount of AF488 is increased, indicating that there are few ‘dark’ liposomes.
-
4
Incubate the reaction at RT on a cell mixer for 15 minutes, in the dark. The reaction is stopped by addition of 85 μL 1.5 M of Tris-Cl, pH 8.3, which quenches the free, reactive fluorophore with its primary amine.
-
5
At this point, the fluorescently labeled protein can be combined with the lipid/fluorophore mixture for reconstitution of the protein. For ClC-ec1-Cy5, we reconstituted the protein at χ = 1.5 × 10−9 to 7.5 × 10−4 subunit/lipid mole fraction densities. Typically, Cy5 labeled protein is obtained at ~5 μM during the purification, so the protein must be serially diluted in 5 mM DM purification buffer before combining with the lipid mixture, for the low density reconstitutions.
-
6
Liposomes are formed by dialysis using a 10 kDa MWCO dialysis cassette. This step does two things, it reconstitutes the protein in the membrane by dialyzing the CHAPS and DM detergent, and it also removes the free AF488 dye from the lipid labeling. For a 1 mL sample, we dialyze against 1 L of imaging buffer (300 mM KCl, 20 mM Citrate pH 4.5) at 4 °C, with buffer changes every 12 hours for 48–60 hours. When scaling up the sample volumes, we use a ratio of at least 1 sample volume to 500 volumes of dialysis buffer. The dialysis should be performed in the dark.
Note. The imaging buffer was selected to match the functional buffer for ClC-ec1 chloride transport studies (Walden et al., 2007). For the protein of interest, a buffer should be selected to match conditions where function can be measured.
Note. The progress of removal of free dye by dialysis can be followed by taking an aliquot of the spent dialysis buffer and measuring AF488 fluorescence in a fluorometer. We observe that using a 10 kDa MWCO dialysis membrane, all free dye is removed after four buffer changes, i.e. 48 hours.
Note. The MWCO of the dialysis cassette should be selected with the protein of interest in mind. For smaller molecular weight species, a 3 kDa MWCO cassette can be used, but the dialysis should be extended as it will take a longer amount of time and cycles to remove the detergent.
-
7
After dialysis, the liposomes should appear slightly cloudy in the 2:1 POPE/POPG lipid composition (Figure 2C), but this may depend on the protein being reconstituted. The samples should be homogenized in the dialysis cassette by light vortexing, transferred to an eppendorf tube then supplemented with 0.02% NaN3 to inhibit bacterial activity. The samples can be stored at −80 °C until further use. Care must be taken to keep the samples away from light at all times.
Note: To check that the detergent has been fully removed, the samples should be tested by freeze/thaw after removal from the dialysis cassette. They should fuse and become cloudy, indicating the formation of large multilamellar vesicles. If the samples remain clear after several freeze/thaw cycles, then dialysis should be repeated to remove the remaining detergent.
Note. We recommend carrying out the lipid labeling at the beginning of working with a new protein to check the fidelity of reconstitution and the liposome accessibility. However, once these parameters have been measured, the remaining photobleaching experiments can be carried out in un-labeled lipids. For ClC-ec1-Cy5, we do not observe any difference in the photobleaching probabilities from labeled or unlabeled lipid membranes.
3.2 Fusion of vesicles to form an infinite bilayer model
During reconstitution, subunits will become trapped into liposomes with little possibility of exchange between vesicles. Since we are interested in measuring the equilibrium reaction of dimerization in the lipid bilayer, we must first create a large bilayer state where multiple copies of subunits exist and can exchange with one another, even at low densities. 2:1 POPE/POPG liposomes undergo fusion by freeze/thaw, forming large oligolamellar vesicles that are 10 μm in diameter, i.e. ~ 5 × 108 lipids per bilayer (Pozo Navas et al., 2005). Our lowest experimental mole fraction density of χ = 1.5 × 10−9 is expected to have 1 subunit per bilayer, just below the limit where equilibrium behavior can theoretically be observed. This large membrane state therefore creates a model of an infinite bilayer that allows for equilibrium behavior to be observed and measurement of the free energy of dimerization. The freeze/thaw procedure is carried out in the dark as follows:
Transfer dialyzed proteoliposomes to eppendorfs, and place them in a floating tube rack.
Prepare a dry ice-100% ethanol bath in a sturdy ice bucket. Allow 30 minutes for the temperature to equilibrate. Set up a separate styrofoam bucket with 25–30 °C water for thawing.
-
Immerse the tubes containing the proteoliposomes in the dry-ice-ethanol bath for 5 minutes. At the end of incubation transfer tubes to the water bath for thawing for 15 minutes. Repeat this cycle 7 times. The samples should appear opaque, and after sitting for some time, the OLVs will settle to the bottom of the tube. If the sample remains clear after freeze/thaw then it is possible that there is still detergent in the solution that is preventing the formation of bilayers. Dialysis should be repeated or preferably new samples prepared.
Note. The 7 times freeze/thaw procedure was developed by monitoring the FRET signal after freeze/thaw of 2:1 POPE/POPG/PE-RhB and 2:1 POPE/POPG/PE-NBD. The FRET signal saturates after 4–7 cycles.
Note. Care must be taken to ensure that ethanol, or water, does not enter the tubes. In particular, ethanol can creep up the sides of the vial and leach into the tube. The levels of the tubes should be maintained with a tube rack to make sure that this does not occur.
After freeze-thaw the samples should be stored at room temperature and away from ambient light.
The protein is now in the infinite membrane reaction state where subunits can exchange with one another. For ClC-ec1-Cy5, we find that the protein is stable, at least up to 27 days, at room temperature provided 0.02% NaN3 is added to prevent bacterial growth. With this, we can examine the photobleaching probabilities as a function of incubation time after fusion of membranes, and find that the monomer/dimer population comes to equilibrium during the freeze/thaw process, and this is stable over the course of a month. Of course, this could be different for other protein systems so we advise investigating this experimentally.
3.3 Membrane fractionation by liposome formation
To measure the population of monomers and dimers in the large membrane state by TIRF microscopy, we fractionate the membrane by extrusion. This forms a population of liposomes with a defined size distribution, and the encapsulation of protein into these liposomes follows a Poisson distribution (Maduke et al., 1999; Walden et al., 2007). The liposome extrusion is carried out as follows:
Assemble the extruder following manufacturer’s guidance. We recommend using the Avestin LiposoFast-Basic with a stabilizer support.
Using 0.5 mL gas-tight glass syringe pass ddH2O, back and forth, through the extruder 4–5 times. Discard.
-
Now, using the same glass syringe, pass filtered imaging buffer through the extruder 4–5 times. Discard.
Note. Imaging buffer can be prepared in advance and frozen in 10 mL aliquots and stored at −20 °C. The imaging buffer is filtered by passing three times through a 0.22 μm syringe filter. The filtered buffer should be checked on the microscope, with practically no observation of background debris.
Wet a 0.4 μm Nucleopore membrane filter (Sigma Aldrich) in ddH20 and gently place the white membrane between the two O-rings on the Teflon cylinders. Be careful not to touch the membrane directly. Hold it at the edges with forceps, or with clean gloves.
To equilibrate the membrane filter pass 0.4 mL imaging buffer through extruder 4–5 times. Discard.
-
Finally fill the glass syringe with 0.4 mL of the freeze/thawed proteo-liposome sample and pass 21 times through the membrane filter. Apply steady but gentle pressure while moving back and forth between two syringes.
Note: It is imperative that the extruder be properly cleaned in between samples. We found the easiest and efficient way of cleaning the extruder, was to dismantle it completely (including o-rings) and soak in hot water/dilute detergent (0.5% v/v Dial dish soap) followed by sonication in ddH2O. ‘Extruding’ buffer after this cleaning and checking on the microscope shows that there is no cross contamination following this method.
3.4 Quantification of the liposome size distribution
The size distribution of the extruded liposome population must be known in order to correct the photobleaching probability data and extract the monomer-dimer reaction. Extrusion yields reproducible liposome populations, but the radii are far from uniform (Walden et al., 2007). As such, liposome sizes must be measured experimentally, and cryoelectron microscopy (cryo-EM) provides a direct approach. The methods are well established (Almgren et al. 2000; Frederik & Hubert 2005) and any cryo-EM imaging facility should be able to carry out these experiments, but the liposomes should be extruded immediately before freezing to prevent changes in the sample over time. The images can be analyzed in Fiji (Schindelin et al., 2012) using the polygon outline tool to obtain the radius of each liposome, and then a probability distribution, Pradius, can be determined from the normalized histogram of radii. If an internal control is desired for pixel-to-nm conversion, 0.1 mg/mL of purified tobacco mosaic virus (width = 1.8 nm (Namba & Stubbs 1986)) can be added to the liposome sample prior to freezing. We find that experimental distributions reported in the literature are quite reproducible even with small changes in composition, e.g. 0.4 μm extrusion of E. coli polar lipids or 2:1 POPE/POPG, both in 300 mM KCl, 25 mM citrate pH 4.5, yield similar Pradius distributions (Walden et al., 2007), even though cardiolipin is lacking in the latter condition.
4. TIRF microscopy of proteo-liposomes
We use passive adsorption (Johnson, Ha, Chu, & Boxer, 2002) to attach 2:1 POPE/POPG liposomes to the glass slide. This is an extremely effective method that requires little slide preparation, outside of a basic cleaning procedure. While binding to the slide is expected to deform the liposome, we rarely observe rupture or fusion of the membrane in the 2:1 POPE/POPG condition (Nollert, Kiefer, & Jähnig, 1995). This is not expected for all lipid conditions, as we observe POPC liposomes forming blurry spots, indicating rupture and formation of membrane patches on the slide. At high densities of POPC liposomes, we observe formation of a complete bilayer on the glass where single-molecule diffusion of lipids can be observed. Therefore, a liposome tethering method may be used, e.g. via PE-biotin (Diao et al., 2012) or lipid-DNA tethering (M. Chung, Lowe, Chan, Ganesan, & Boxer, 2009) for other types of lipid compositions. Our choice of 2:1 POPE/POPG to study ClC-ec1 was based on the fact that this protein is native to E. coli, and is functional in this membrane composition. When working with a new protein system, the lipid composition should be selected to match the condition where functional activity can be measured. However, for preliminary studies, 2:1 POPE/POPG offers a quick and simple way of carrying out single-vesicle microscopy studies.
4.1 Preparation of slides for TIRFM
The cleaning of glass slides and coverslips serves two purposes. First, it leads to a uniformly low background, and second, KOH treatment renders the surface hydrophilic enabling passive adhesion of liposomes to the surface (Chandradoss et al., 2014).
Place glass slides and coverslips (Gold-Seal slides were 24 × 60 mm no. 1.5 thickness and coverslips were 25 × 25 mm no. 1.0 thickness) in slide mailers 5 at a time. Fill up the container with 0.1% Micro-90 detergent (Cole-Parmer) and sonicate in bath sonicator for 30 minutes. At the end of 30 minutes empty out detergent and rinse 10 times with deionized, distilled grade water (ddH2O).
Next, fill slide mailers with 100% ethanol and sonicate for 30 minutes. At the end of 30 minutes empty out ethanol and rinse 10 times with ddH2O.
-
Add freshly prepared 0.2 M KOH and sonicate for 5 minutes followed by rinsing 10 times with ddH2O.
Note. Clean coverslips and slides can be stored in ddH2O in a laminar flow bench for up to a week.
4.2 Co-localization microscopy of liposomes and membrane proteins
Our experiments were carried out on a multi-wavelength single molecule total internal reflection fluorescence microscope that we built in our lab following the CoSMoS design (Friedman, Chung, & Gelles, 2006; Larson et al., 2014). Imaging is carried out in two channels, using 488 nm and 637 nm excitation lasers while emitted fluorescence is collected and filtered using ET525/50m or ET650LP, respectively and focused onto the CCD chip of an EM/CCD camera. Since liposomes adsorb onto the glass slide, and free liposomes can be washed away, epi-fluorescence or confocal microscopy (Mathiasen et al., 2014) could be used as long as the signal is sufficient to observe unambiguous photobleaching steps.
-
Apply extruded proteo-liposomes to the flow cell for passive binding to the glass coverslip. For high protein densities, the samples must be serially diluted with filtered imaging buffer in low-adhesion tubes. The binding occurs quickly and excess liposomes can be washed off within 1–2 minutes of application. Low protein density liposomes can be extruded and applied directly to the chamber, allowing 20–30 minute incubation time to maximize liposome binding. The density of fluorescent spots on the glass slide was maintained at 0.02 to 0.09 spots/um2 to minimize overloading.
Note. The binding of liposomes to the glass is practically irreversible (Johnson et al., 2002). We confirm this by imaging the same field of liposomes over time, demonstrating little change in the number of spots.
Note. Cy5 triplet-state blinking (Ha & Tinnefeld, 2012)is avoided by using imaging buffers that do not contain oxygen scavengers.
Set EM gain to 300 and acquisition rate to 1 frame per second. The laser intensity depends on the sample, but we typically set the 488 nm laser to 15 μW to image the liposomes and the 637 nm laser to 240 μW for the Cy5 labeled subunits. The latter allows for long photobleaching traces while maintaining good signal to noise, but this must be optimized for different samples.
-
On the same day of imaging, collect a mapping dataset to allow for pixel registration between 488 nm and 637 nm imaging channels, especially necessary for determining co-localization at low densities. This can be done independently with a separate high protein density sample of sonicated liposomes, or co-labeled beads (Friedman & Gelles, 2015).
Note. When carrying out AF488/Cy5 co-localization imaging, image the Cy5 channel first, followed by AF488. We observe Cy5 going to a dark state upon exposure to the 488 nm laser.
4.2.1 Measurement of Foutside, the fraction of protein that does not co-localize with liposomes
Foutside is the fraction of Cy5 protein spots that do not co-localize with AF488 liposomes. This is a measurement of the fidelity of reconstitution, as appearance of protein outside of liposomes indicates aggregation during reconstitution (Mathiasen et al., 2014). If this value is more than 1–2% then the protein does not properly reconstitute into the membrane or the lipid-labeling yield is too low. If increasing the amount of lipid labeling does not reduce Foutside, then it can be concluded that there is significant protein loss during the reconstitution preventing reliable measurement of the dimerization reaction.
4.2.2 Measurement of F0, the fraction of unoccupied liposomes
As reconstitution follows a Poisson distribution, the fraction of unoccupied vesicles, F0, should decrease as the protein density increases. However, it has been shown that incorporation of protein in liposomes can depend on both size and curvature (Mathiasen et al., 2014), and so it is possible that there is sub-population of liposomes that are not accessible to protein, either in the monomeric or dimeric state. Measuring F0 as a function of protein density allows for direct determination of the accessible liposome population. For this, we recommend measuring the protein at high densities, χ = 7.5 × 10−5 to 7.5 × 10−4 subunit/lipid as this will reveal the plateau of the curve. For example, the dimeric wild-type ClC-ec1 exhibits a plateau at F0 = 0.4, whereas the monomeric I201W/I422W plateau is ~10%. This indicates that even at the highest protein density, dimers cannot access 40% of the liposomes, whereas the monomer occupies almost all of them. We assume that this is related to a conflict in protein vs. liposome size, indicating that liposomes with radius < 25 nm are excluded from the dimer population. With this, Pradius,dimer can be calculated as the re-normalized accessible liposome probability distribution, excluding the smaller radii bins. ClC-ec1 is a large protein (10 nm end-to-end), and so this may not be needed for smaller protein systems. It is worth checking but it requires having a positive dimer control, either through mutagenesis or by reconstitution of a cross-linked form of the complex.
4.2.3 Measurement of the photobleaching probabilities, Pn
Here, we describe the manual counting of photobleaching steps. While there are numerous methods that use algorithms for automated counting (McGuire, Aurousseau, Bowie, & Blunck, 2012) (Hines, Bankston, & Aldrich, 2015), we find that these results often have to be checked against manual counting. We have compared the subjectivity of counting between different individuals and find no significant difference as long as the quality of the images have been optimized to maximize signal to noise (by increasing laser power) while maximizing the length of each photobleaching state (by decreasing laser power). This will be different for each protein system due to changes in photostability of the fluorophore on the protein near the membrane. Note, we do not observe Cy5 blinking behavior, as we do not include an oxygen scavenging system in our buffers. Since our goal is to measure a reaction in the membrane, we avoid triplet state quenchers such as Trolox, which partitions into membranes and may affect protein properties (Alejo, Blanchard, & Andersen, 2013).
Image files can be stored as stacked tiff files (or some other binary image format) and analyzed in any image analysis program. We use a Matlab based CoSMoS image analysis program (Friedman & Gelles, 2015) as it has been customized to serve many needs in co-localization imaging analysis such as mapping, drift correction and auto-detection of fluorescent spots. We used raw images directly for analysis, as conditions were optimized for maximum signal to noise and relatively long photobleaching traces. An area of interest (AOI) around each fluorescent spot can be detected manually, or by an auto-detection method. Pixel intensity within an AOI is then intergrated over time to obtain a photobleaching trajectory, where photodestruction steps can be clearly counted.
The photobleaching probability distribution, Pn, is calculated as the number of Cy5 occupied liposomes that bleach in n steps, over the total number of Cy5 occupied liposomes. On our microscope, we can count up to 9 discrete photobleaching steps, while higher occupied liposomes exhibit a macroscopic decay in bulk fluorescence. In general, it is sufficient to calculate probabilities for n = 1, 2, 3, 4 and >4 to follow the monomer-dimer reaction.
5. Calculation of correction factors for determination of Fdimer vs. χ
5.1 Correcting the experimental photobleaching probabilities
The photobleaching probabilities will describe the monomer-dimer equilibrium in the membrane, but to do so, this data must be corrected since liposome occupancy independently depends on the protein density in the membrane. In addition, fluorescence microscopy detects the fluorophores attached to subunits, and so the data must also be corrected for the fluorescent labeling yields, as different amounts of labeling will yield different results. Here, we describe the steps involved for simulating the photobleaching probabilities for non-interacting monomers and non-interacting dimers as a function of the protein density. The procedure is depicted in Figure 3 and described in full detail in Chadda et al. (Chadda, R., Krishnamani, V., Mersch, K., Wong, J., Brimberry, M., Chadda, A. 2016).
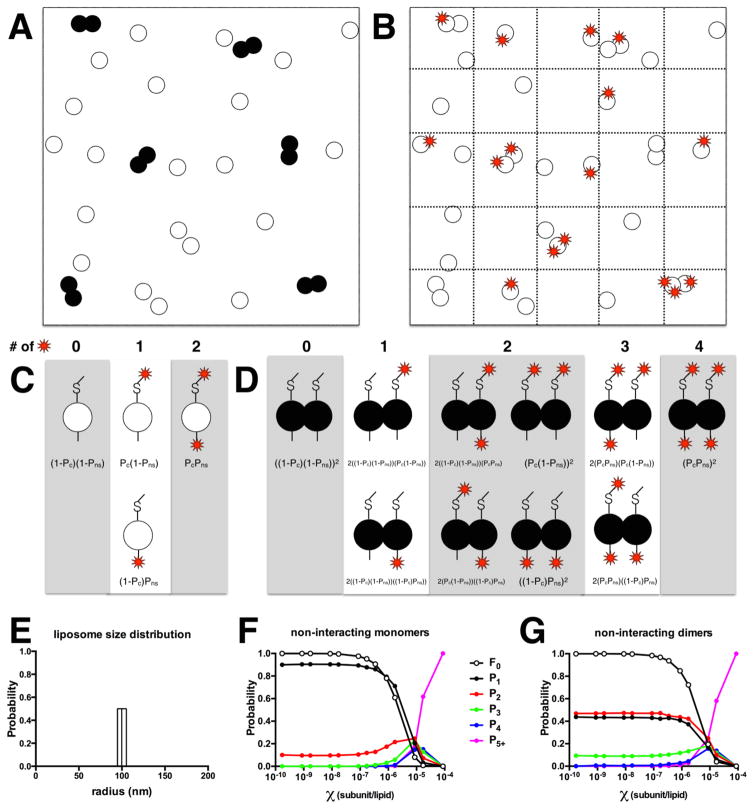
Figure 3. Simulation of subunit capture into liposomes and photobleaching probabilities.
(A) Large membrane (square) containing monomers (white) and dimers (black). (B) In the subunit capture method, the membrane is fractionated (dotted lines) by extrusion that forms liposomes, encapsulating monomers and/or dimers into each area of membrane. In the photobleaching analysis, only the fluorophores (red stars) are detected, and the probabilities Pn=1,…,5+ depend on protein density, χ subunit/lipid, fluorescent labeling yields Pfluorophore & Pnon-specific, and monomer-dimer equilibrium constant Keq. (C) Monomer fluorescent species and association probabilities. Each subunit has two sites for labeling, a high probability cysteine (-S site, Pcysteine abbreviated to Pc) and a low probability non-specific site (- site, Pnon-specific abbreviated to Pns). (D) Dimer fluorescent species with associated probabilities. Simulation of the photobleaching probabilities using a homogeneous liposome size distribution in (E) with Pfluorophore = 0.75 and Pnon-specific = 0.1 (i.e. Pcysteine = 0.65) for (F) non-interacting monomer species PMn=1,…,5+ (F) and (G) non-interacting dimer species PDn=1,…,5+.
5.1.1 The non-interacting monomer distribution
The non-interacting monomer distribution represents the probability of co-encapsulation of membrane proteins that do not associate in the membrane. It has been referred to as “artifactual togetherness” for association reactions in detergent micelles (Fleming et al., 1997; Tanford & Reynolds, 1976). This can be simulated using the experimental parameters obtained so far (Table 1) and the following steps:
-
Set Nmonomers = Nsubunits and Nliposomes. The experimental mole fraction, χ, is calculated as:
(4) Where SAlipid is the surface area per lipid in a bilayer, and Pradius is the liposome size probability distribution measured by cryo-EM (Section 3.4).
Define a matrix for each radius bin in the Pradius distribution. The number of rows in the matrix is equal to Pradius(r)Nliposomes, where each individual row represents a single liposome.
Calculate the number of each fluorescently labeled species. We assume that each subunit has two sites for fluorescent labeling: (1) the reactive cysteine and (2) a non-specific site. The probability of labeling at the cysteine is Pcysteine = Pfluorophore−Pnon-specific, where Pfluorophore and Pnon-specific are the labeling yields measured from the absorbance spectrum (Section 2.3). For monomers, this results in subunits labeled with 0, 1, and 2 fluorophores with associated probabilities listed in Figure 3C.
- The proportion of each monomer species to be inserted into the liposomes is calculated using the fractional surface area from the accessible liposome population, Pradius,monomer(r) determined from the measurements of F0 (Section 4.2.2).
(5) Define three columns in each matrix representing the occupancy of each of the different fluorescent species. Randomly insert the calculated number of the each monomer species into each sub-population of liposomes.
At the end of the simulation, add up the number of fluorophores in each liposome row and generate a histogram that is normalized by Nliposomes. This gives the theoretical fluorophore occupancy probability distribution, P*(k), where k=0,1,2,… represents the number of fluorophores per liposome, and P*(0) is equivalent to the experimentally measured F0 (Section 4.2.2).
The photobleaching distribution is calculated from P*(k) by considering only the fluorophore occupied liposomes, i.e. P*(k)/(1-P*(0)) for k > 1. This gives Pn, where n=1,2,3,… represents the number of photobleaching steps corresponding to the experimental photobleaching probabilities (Section 4.2.3).
The simulation is repeated for different χ subunit/lipid densities by changing Nsubunits and Nliposomes. Note that Nsubunits > 100 and Nliposomes > 1000 should be used to avoid rounding errors.
5.1.2 The non-interacting dimer distribution
The non-interacting dimer distribution must also be calculated. For single dimer occupied liposomes with 100% labeling, only double steps would be observed. However, given typical labeling yields i.e. ~ 75%, single and double steps are observed with equal probabilities. This is because there are now 5 different dimer fluorescent species with 0–4 fluorophores, where the 1 and 2 fluorophore labeling states are almost equal in probability (Figure 3C). The rest of the calculation is almost identical to the monomer simulation described in section 5.1.1, except now the user sets Ndimers = Nsubunits/2, the dimer accessible liposome distribution Pradius,dimer(r) is used, and the simulation involves the 5 fluorescent dimer species.
5.2 Determination of Fdimer vs. χ
With the all-monomer and all-dimer probabilities calculated, the experimental photobleaching data (Pn) can be fit to the non-interacting monomer (PMn) and non-interacting dimer (PDn) distributions by least-squares analysis of the sum of the squared residuals (S) to obtain Fdimer as a function of χ.
| (6) |
The resulting data is fit to an equilibrium isotherm for extrapolation of the equilibrium constant, Keq, of the reaction of the protein in the large membrane state.
| (7) |
Note that the reactive mole fraction density, χ* = χ/2 must be used to account for random orientation of the protein in the membrane and that the reaction only occurs between proteins in the same direction. In order to establish that the reaction is at equilibrium, subunit exchange, reversibility and path independence must be experimentally tested.
6. Summary
The methods in this chapter describe the general procedure for measuring subunit stoichiometry by subunit capture into liposomes followed by co-localization microscopy and photobleaching analysis. This approach can be used to determine protein stoichiometry in the membrane even if the structure is not yet known, as well as reactions of changing stoichiometry, such as equilibrium dimerization. Co-localization microscopy offers an opportunity to check the protein during the reconstitution process, to measure the success of protein incorporation, aggregation, and other higher-order oligomeric states. In principle, the same approach can be used to measure the equilibrium of higher-order oligomeric structures as well as heteromeric complexes. Finally, these methods map out the mole fraction range for single-molecule liposome experiments that can be applied to studies of conformational dynamics by FRET or functional experiments.
References
- Alejo JL, Blanchard SC, Andersen OS. Small-molecule photostabilizing agents are modifiers of lipid bilayer properties. Biophysical Journal. 2013;104(11):2410–2418. doi: 10.1016/j.bpj.2013.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming KG, Ackerman AL, Engelman DM. The effect of point mutations on the free energy of transmembrane alpha-helix dimerization. Journal of molecular biology. 1997;272(2):266–275. doi: 10.1006/jmbi.1997.1236. [DOI] [PubMed] [Google Scholar]
- Friedman LJ, Gelles J. Multi-wavelength single-molecule fluorescence analysis of transcription mechanisms. Methods. 2015 doi: 10.1016/j.ymeth.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman LJ, Chung J, Gelles J. Viewing Dynamic Assembly of Molecular Complexes by Multi-Wavelength Single-Molecule Fluorescence. Biophysical Journal. 2006;91(3):1023–1031. doi: 10.1529/biophysj.106.084004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T, Tinnefeld P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annual review of physical chemistry. 2012;63(1):595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanson GT. Bioconjugate Techniques - Greg T. Hermanson - Google Books. 2013. [Google Scholar]
- Hines KE, Bankston JR, Aldrich RW. Analyzing Single-Molecule Time Series via Nonparametric Bayesian Inference. Biophysical Journal. 2015;108(3):540–556. doi: 10.1016/j.bpj.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong H, Blois TM, Cao Z, Bowie JU. Method to measure strong protein-protein interactions in lipid bilayers using a steric trap. Proc Natl Acad Sci USA. 2010;107(46):19802–19807. doi: 10.1073/pnas.1010348107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes LD, Rawle RJ, Boxer SG. PloS one. 2. Vol. 9. Public Library of Science; 2014. Choose Your Label Wisely: Water-Soluble Fluorophores Often Interact with Lipid Bilayers; p. e87649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Ha T, Chu S, Boxer SG. Early steps of supported bilayer formation probed by single vesicle fluorescence assays. Biophysical Journal. 2002;83(6):3371–3379. doi: 10.1016/S0006-3495(02)75337-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson J, Kirk M, Drier EA, O’Brien W, MacKay JF, Friedman LJ, Hoskins AA. Design and construction of a multiwavelength, micromirror total internal reflectance fluorescence microscope. Nature Protocols. 2014;9(10):2317–2328. doi: 10.1038/nprot.2014.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKenzie K, Fleming KG. Association energetics of membrane spanning α-helices. Current opinion in structural biology. 2008;18(4):412–419. doi: 10.1016/j.sbi.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduke M, Pheasant DJ, Miller C. High-level expression, functional reconstitution, and quaternary structure of a prokaryotic ClC-type chloride channel. The Journal of general physiology. 1999;114(5):713–722. doi: 10.1085/jgp.114.5.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathiasen S, Christensen SM, Fung JJ, Rasmussen SGF, Fay JF, Jorgensen SK, Veshaguri S, et al. Nanoscale high-content analysis using compositional heterogeneities of single proteoliposomes. Nature Methods. 2014;11(9):931–934. doi: 10.1038/nmeth.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire H, Aurousseau MRP, Bowie D, Blunck R. Automating single subunit counting of membrane proteins in mammalian cells. The Journal of biological chemistry. 2012;287(43):35912–35921. doi: 10.1074/jbc.M112.402057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzyn K, Róg T, Pasenkiewicz-Gierula M. Phosphatidylethanolamine-Phosphatidylglycerol Bilayer as a Model of the Inner Bacterial Membrane. Biophysical Journal. 2005;88(2):1091–1103. doi: 10.1529/biophysj.104.048835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollert P, Kiefer H, Jähnig F. Lipid vesicle adsorption versus formation of planar bilayers on solid surfaces. Biophysical Journal. 1995;69(4):1447–1455. doi: 10.1016/S0006-3495(95)80014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo Navas B, Lohner K, Deutsch G, Sevcsik E, Riske KA, Dimova R, Garidel P, et al. Composition dependence of vesicle morphology and mixing properties in a bacterial model membrane system. Biochimica et biophysica acta. 2005;1716(1):40–48. doi: 10.1016/j.bbamem.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Riddles PW, Blakeley RL, Zerner B. Enzyme Structure Part I, Methods in Enzymology. Vol. 91. Elsevier; 1983. [8] Reassessment of Ellman’s reagent; pp. 49–60. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 2012;9(7):676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbridge RB, Tsai MF. Methods in enzymology. Vol. 556. Elsevier; 2015. Lipid reconstitution and recording of recombinant ion channels; pp. 385–404. [DOI] [PubMed] [Google Scholar]
- Stockbridge RB, Robertson JL, Kolmakova-Partensky L, Miller C. eLife. Vol. 2. eLife Sciences Publications Limited; 2013. A family of fluoride-specific ion channels with dual-topology architecture. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanford C, Reynolds JA. Characterization of membrane proteins in detergent solutions. Biochimica et biophysica acta. 1976;457(2):133–170. doi: 10.1016/0304-4157(76)90009-5. [DOI] [PubMed] [Google Scholar]
- Walden M, Accardi A, Wu F, Xu C, Williams C, Miller C. The Journal of general physiology. 4. Vol. 129. Rockefeller Univ Press; 2007. Uncoupling and turnover in a Cl−/H+ exchange transporter; pp. 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]