Drug repurposing, the application of an existing therapeutic to a new disease indication, holds the promise of rapid clinical impact at a lower cost than de novo drug development. To date there has not been a systematic effort to identify such opportunities, limited in part by the lack of a comprehensive library of clinical compounds suitable for testing. To address this challenge, we hand-curated a collection of 4,707 compounds, experimentally confirmed their identity, and annotated them with literature-reported targets. The collection includes 3,422 drugs that are marketed around the world or that have been tested in human clinical trials. Compounds were obtained from more than 50 chemical vendors and the purity of each sample was established. We have thus established a blueprint for others to easily assemble such a repurposing library, and we have created an online Drug Repurposing Hub (www.broadinstitute.org/repurposing) containing detailed annotation for each of the compounds.
Repurposing is attractive and pragmatic given the substantial cost and time requirements— on average, a decade or more— for drug development1. In addition, a large number of potential drugs never reach clinical testing. Moreover, fewer than 15% of compounds that enter clinical development ultimately receive approval despite the majority of them being deemed safe2. For either approved or failed drugs for which safety has already been established, finding new indications can rapidly bring benefits to patients. Prior drug repurposing successes span disease areas; examples include the cyclooxygenase inhibitor aspirin to treat coronary artery disease, the phosphodiesterase inhibitor sildenafil to treat erectile dysfunction, and the antibiotic erythromycin for impaired gastric motility (Supplementary Table 1)3. Even drugs with troubling side effects merit reconsideration, as evidenced by the successful repurposing of the antiemetic thalidomide to treat multiple myeloma4. Risk-mediating measures to avoid the potential teratogenicity of thalidomide and its derivatives are reasonable in patients with life-threatening cancer, whereas use of these drugs to treat nausea remains unacceptable.
While the benefits of repurposing are clear, successes to date have been mostly serendipitous. Systematic, large-scale repurposing efforts have not been possible due to the lack of a definitive physical drug collection, the low quality of drug annotations, and insufficient readouts of drug activity from which new indications can be predicted. Recent technological advances have enabled a step change in our ability to comprehensively assess drug activities. For example, perturbational gene expression profiles can now be obtained at high throughput across multiple cell types5. Gene expression profiling has enabled recent repurposing discoveries, including sirolimus for glucocorticoid-resistant acute lymphocytic leukemia, topiramate for inflammatory bowel disease, and imipramine for small cell lung cancer. For cancer therapeutics, a recently developed assay using barcoded cell lines known as PRISM enables rapid testing of many drugs against a large number of cancer cell lines in pools6. Molecular features of the cell lines (e.g., gene expression, mutation, or copy number variation) can then be used to identify predictive biomarkers of drug sensitivity (Supplementary Table 2). Finally, morphologic changes in cells can be assessed using high-throughput microscopy and machine learning approaches. Such imaging-based screening unexpectedly identified the cholesterol drug lovastatin as a potent inhibitor of leukemia stem cells.
To take advantage of these advances in experimental methods, we sought to assemble a comprehensive library of drugs that have reached the clinic. Surprisingly, we found that no such chemical library of approved and clinical trial drugs is available for purchase. In particular, drugs that have been tested in clinical trials but did not reach approval are not readily accessible. Even obtaining a complete list of such drugs and their annotations is challenging. A prior effort led by the National Institutes of Health (NIH) focused on drugs approved by the Food and Drug Administration (FDA), but the library has few compounds that have yet to achieve FDA approval7. Some chemical vendors offer a subset of approved drugs, but most of these commercial libraries overlap in their content and include only a small fraction of the approximately 10,000 drugs that have reached the clinic in the United States and Europe. Given that no complete collection exists, we launched a three-step effort to create the Repurposing Library by 1) identifying and purchasing compounds, 2) comprehensively annotating their known activities and clinical indications, and 3) experimentally confirming drug identity and purity.
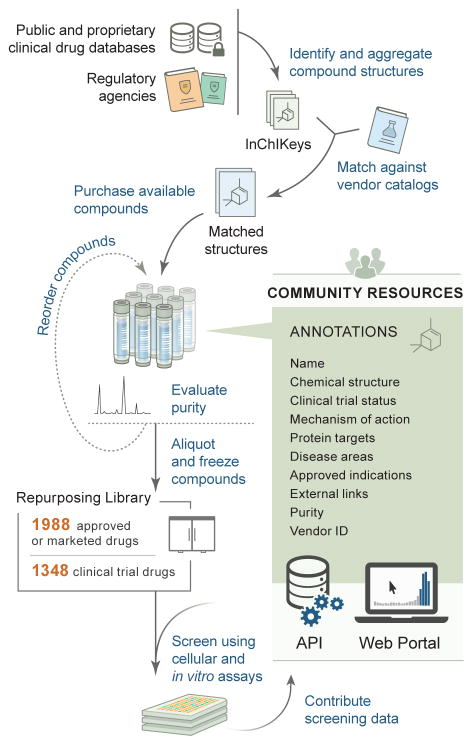
We employed two approaches to identify clinical drug structures for the Repurposing Library. First, existing databases, both publicly accessible and proprietary, were searched for clinically tested drugs and then manually integrated in order to ensure sufficient drug coverage and chemical structure reliability (Supplementary Table 3). Sources included DrugBank, NCATS NCGC Pharmaceutical Collection (NPC), Thomson Reuters IntegritySM, Thomson Reuters Cortellis™, and Citeline Pharmaprojects®7–9. Second, we located marketed or approved ingredient lists from regulatory agencies worldwide, including the FDA. Following structure standardization and removal of duplicates, approximately 10,000 small molecule drugs with disclosed structures were found to have reached clinical development. Most of these drugs are not widely available in commercial screening libraries. Through structure-matching (as opposed to relying on compound names), chemical suppliers were identified for 5,691 compounds (Fig. 1). Controlled substances, non-pharmaceutical substances, and redundant elemental formulations were not pursued further. To assemble the collection, we ultimately purchased 8,584 samples (representing 5,691 unique compounds) from 75 chemical vendors, at an average cost of $29 per sample.
Figure 1. The Repurposing Hub workflow for drug library creation.
Drugs were identified, purchased, confirmed, and fully annotated. The final QC-confirmed library contains 1,988 drugs that are approved or marketed for clinical use around the world and 1,348 drugs that reached Phase 1–3 in clinical development.
We performed chemical structure analysis on all clinical drug structures (whether commercially available or not) in order to assess the extent of structural diversity in the Library. Compounds were clustered into 256 groups using a self-organizing map algorithm. As expected, compounds with known shared features generally clustered together (Supplementary Fig. 1a). All but six clusters had at least one compound that was represented in the Repurposing Library, reflecting the high level of diversity of the Library overall. Poorly represented clusters mostly consisted of large macromolecules and polypeptide-derived drugs with a median molecular weight of 3,200 grams per mole (Supplementary Fig. 1b). We conclude from this analysis that the Repurposing Library is sufficiently representative of the majority of chemotypes that have reached clinical development.
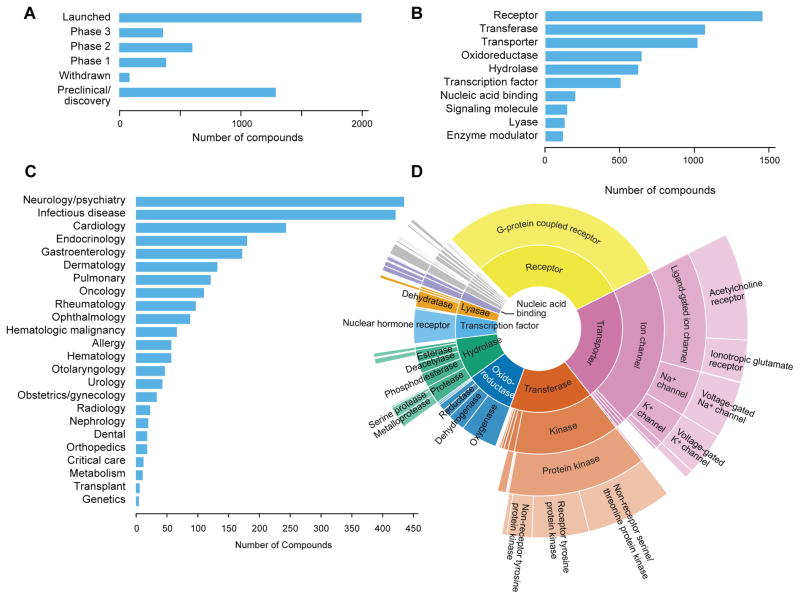
With compounds in hand, the next challenge was to comprehensively annotate known drug functions and clinical development status. This information was often inconsistent or contradictory across databases; manual curation from primary sources was therefore required. The FDA Orange Book, prescribing labels, ClinicalTrials.gov, PubMed, and other Internet resources were manually searched for clinical status information. Based on this analysis, the QC-confirmed Repurposing Library contains 1,988 launched drugs and 1,347 drugs that reached Phase 1–3 clinical development (Fig. 2a). The Library also includes 86 compounds that were previously approved but later withdrawn from use and 1,285 preclinical or tool compounds (tool compounds are valuable to achieve the target redundancy needed to differentiate on-target from off-target activities of clinical drugs being considered for repurposing). In certain cases, annotation of drugs in clinical development was achievable only by searching the patent literature or news releases. Within the launched drug category, 86 are exclusively used in veterinary medicine. In comparison, the published NCATS NPC drug repurposing library contains only 327 clinical trial compounds, based on our annotations. Whereas the clinical trial component of the Repurposing Library remains incomplete (structures and/or vendors are available for fewer than 20% of drugs reaching clinical development but not approved by the FDA), approximately 90% of FDA-approved small-molecule drugs were obtained. Of the 10% of unavailable FDA-approved drugs, one-third are controlled substances that are restricted for purchase and are less amenable to repurposing applications.
Figure 2. Repurposing Library contents.
A) Highest clinical phase achieved by each compound. 3,422 drugs in the Library have reached clinical use as of November 2016. B) Number of compounds per target category. Compounds with multiple targets may be indicated in more than one category. C) Number of approved drugs for indications within each listed disease area. D) Classification of proteins targeted by Library drugs. Protein function hierarchy is shown with increasing specificity of function corresponding to distance from the figure center. The relative area of each segment is proportional to the fraction of the Repurposing Library targeting each protein class. As expected, the Library is enriched in drugs targeting kinases, GPCRs, and ion channels.
Standardization of drug mechanism and protein target information enables experimental interpretation of repurposing efforts. Using a combination of publicly available databases and extensive manual curation, a total of 2,006 human proteins were found to be targeted by compounds in the Repurposing Library. Protein target classification using a published database revealed that the most common drug target categories are G protein-coupled receptors, acetylcholine receptors, non-receptor serine/threonine protein kinases, oxygenases, voltage-gated sodium channels, and nuclear hormone receptors (Fig. 2b,d)10. Overall, 92% of compounds were successfully mapped to a human protein target or assigned a mechanism of action label (useful for anti-infectives and compounds without known human targets).
Surprisingly, we found that it was even difficult to determine the approved clinical indications for existing drugs. The availability of such information in public databases is highly variable, and in many cases terminology is inconsistent. Using the NIH’s DailyMed repository of FDA-approved drug prescribing labels and other resources, a dictionary of 664 unique drug indication terms was manually curated and assigned to 1,918 launched drugs (Supplementary Fig. 2). The most common disease areas covered are neurology/psychiatry, infectious disease, cardiology, and endocrinology (Fig. 2c). While future repurposing opportunities will likely transcend these disease areas, knowledge of the existing drug indications is likely to prove useful.
Knowledge of drug patent and exclusivity status is necessary to identify commercial opportunities and funding mechanisms to conduct clinical trials for new indications. In the United States, the FDA Orange Book serves as an official reference for approved ingredients, therapeutic equivalency, and drug patents, as well as being the repository of exclusivity periods granted by the FDA. To harness these valuable annotations, we manually curated a mapping of compounds to the Orange Book, matching 1,412 of the obtained compounds (1,224 QC-confirmed) to the overall list of 1,576 unique ingredients reported by the FDA (excluding macromolecules, mixtures, radioactive substances, and redundant formulations). Among approved Repurposing Library drugs currently protected by substance patents, we observed an expected correlation between first approval date, exclusivity end date, and patent expiration date (Supplementary Fig. 3). Interestingly, there are more than 50 drugs with patents and/or exclusivity expiring more than 20 years after original launch, largely reflecting new indications and formulations that could be available for further repurposing. In cases where multiple drugs exist against the same target, such patent information can also aid in the selection of individual drugs for repurposing.
The final step in creating a drug screening library is experimental confirmation of compound identity and purity. We therefore tested all compound samples in the Repurposing Library by ultra-performance liquid chromatography-mass spectrometry (UPLC-MS), following receipt of the compound from the vendor. Surprisingly, 2,482 of 8,584 samples (29%) failed quality control (QC), defined as a purity of less than 85% as measured by UPLC absorbance peak area at 210 nm or by an evaporative light scattering detector (ELSD) for peaks containing the expected compound mass (Supplementary Fig. 4a). The majority of QC failures were subsequently confirmed by the vendors of the compounds upon checking of the source stocks. Repeat sourcing of 527 failed compounds (purity less than 85%) from one original vendor resulted in rescue of 55% of those compounds (Supplementary Fig. 4b). The 984 unique compounds that still did not pass QC were excluded from the Repurposing Library. The majority of QC failures appear to have resulted from vendors storing compounds in DMSO, which has been well-documented to result in low stability over time11.
To facilitate use of the resource by the scientific community, we have created an interactive Drug Repurposing Hub website, available at www.broadinstitute.org/repurposing (Supplementary Fig. 5). Users can search and view Repurposing Library compounds by clinical status, drug indication, disease areas, mechanism of action, drug target, purity, and vendor. Database exports are available as text files, and the underlying data can be accessed programmatically via a documented application programming interface (API). We are unaware of such data being systematically available elsewhere. Screening assay results at the Broad Institute will be made available via the website, and we encourage others who replicate the library to share their findings. Experimental results will be collected following the principles developed for the NIH-funded BioAssay Research Database (BARD), which includes descriptions of experimental protocols using controlled vocabulary terms as well as the ability to display compound activity results from multiple assay protocols.
The Drug Repurposing Hub is designed to rapidly identify drugs for evaluation in disease models. To date, there have been few resources available to connect the findings of disease genetics studies to drugs available for preclinical and clinical testing. To illustrate the utility of the Drug Repurposing Hub in identifying drugs targeting proteins encoded by recurrently mutated cancer gene products, we queried the Hub database with the 224 unique genes found to be statistically significantly mutated in a recent analysis of The Cancer Genome Atlas (TCGA)12. We found that 47% of significantly mutated cancer gene products have a corresponding drug in the Repurposing Library that binds to the protein itself or a protein in a related pathway, or recapitulates an established synthetic lethal interaction (Supplementary Fig. 6). Such mapping of gene-drug interactions will help to prioritize repurposing hypotheses for further testing.
Drug repurposing has enormous potential for rapid clinical impact, but there has not yet been a systematic effort to identify such opportunities. We have taken a foundational step forward by creating a robust information resource and comprehensive drug screening library. As outlined by our results using the Drug Repurposing Hub, standardized and computable drug annotations will enable both global analysis as well as rapid individual drug lookups against specific targets, existing indications, or clinical development status.
Integration of disparate resources, coupled with substantial manual curation, was necessary to identify clinical drugs, locate chemical suppliers, and standardize drug annotations. Through this process, we made two important observations that will affect the community’s use of the Repurposing Hub. First, drug targets listed in public resources are often inconsistent and unreliable. To assist users in navigating discrepancies among literature-reported targets, we provide the provenance of the information for each target annotation, allowing users to investigate the evidence in the primary literature underlying each claim and to judge the level of consensus among different data sources. Second, our finding that 29% of purchased compound samples failed quality control should serve as a reminder that experimental verification of compound identity and purity is essential, even when sourcing from established vendors.
We believe that the Repurposing Library and Drug Repurposing Hub will serve as valuable resources for the scientific community, accelerating the search for new indications for existing drugs. We provide detailed sourcing information that makes it possible for others to readily generate the same physical library, thereby facilitating the comparison of results across multiple screening assays performed at different institutions. While the Repurposing Library is the largest of its kind, it is not yet complete. Future work will focus on identifying sources of missing compounds and on developing systematic assays suitable for new indication discovery.
Online Methods
Locating clinical drug names and structures
Clinical drug databases were initially searched for entries with a global clinical development or regulatory approval status of Phase 1 or higher. Structures of FDA-approved drugs were obtained from the DrugBank and NCATS databases (Supplementary Table 3). Additional structures of approved drugs and drugs that entered Phase 1–3 clinical trial drugs were identified from Thomson Reuters IntegritySM, Thomson Reuters Cortellis™, and Citeline Pharmaprojects®. Structures were obtained as SD/MOL file coordinates (where available) or SMILES strings.
Processing chemical structures
Chemical structures were processed using BIOVIA Pipeline Pilot 8.5. After excluding inorganic complexes, the largest molecular fragment was kept in order to remove salts and to separate drugs with multiple active ingredients. Next, structures were converted to standardized InChIKeys to facilitate text matching13.
Matching to vendor libraries
Chemical vendor catalogs were obtained from Selleck Chemicals, Tocris Bioscience, Enzo Life Sciences, EMD Millipore, Prestwick Chemical, MedChem Express, Microsource Discovery, eMolecules, and Aldrich Market Select. Compounds were annotated with vendor catalog IDs and converted to InChIKeys. These InChIKeys were stored in a relational database, where they were matched to desired clinical compound InChIKeys using the first 14 characters (which encode the molecular connectivity but not the stereochemistry). This approach was used because numerous input drug structures were found to lack the correct stereochemistry on initial review. The resulting list of matches was manually reviewed to restore salt forms or stereochemistry where necessary, to avoid inaccurate drug matches.
Compound purchasing
Compounds were purchased in standard screening format wherever possible to minimize cost. Barcoded Matrix tubes (Thermo Fisher Scientific #3734) were shipped in advance to chemical suppliers. Compounds were generally provided in DMSO at 10 millimolar concentration. A subset of compounds were obtained via vendor aggregators eMolecules or Aldrich Market Select. Upon receipt, the barcoded tubes and chemical structure files provided by the vendors were registered in the Broad Institute Compound Management system using ChemAxon JChem version 16.5 for structure normalization. Compounds were assigned a structure identifier and a batch sample number. Annotated tool compounds (non-clinical drugs) were accepted as part of library-wide purchases.
External links
Purchased drug structures were mapped to the publicly available DrugBank, ChEMBL, TTD, and IUPHAR databases by InChIKey or name, followed by manual review and removal of incorrect name matches. Pubchem CID identifier was matched by exact structure searches only.
Drug annotations and curation
Unique protein targets from source databases were mapped to the Human Genome Organization (HUGO) gene symbol by Entrez gene ID. Given the incomplete coverage of annotated drug targets in public databases, more than 2,000 drug-target pairs were manually curated from literature sources. Drug targets were obtained and de-duplicated using HUGO gene symbols. Primary public sources of compound clinical trial annotations were manually curated wherever possible (using the FDA Orange Book, PubMed, and ClinicalTrials.gov). The PANTHER database was used to determine protein target classification, with additional manual curation to add proteins missing from the available hierarchy10. Drug indications were curated using prescription drug labels available on the NIH DailyMed website. A centralized vocabulary of 644 disease indications was manually curated to minimize redundant indication terms. Indications were manually mapped to medical disease areas. Approved drug patent and exclusivity information were obtained from the FDA Orange Book publication (May 2016 edition).
Assessment of compound structure diversity
The chemical space of clinical compounds was modeled and visualized as a self-organizing map with 16×16 hexagonal cells using the batchSOM algorithm (implemented in the R ‘class’ library). The model was trained on ECFP_6 fingerprints folded into 256 count features.
Analysis of mutated cancer genes
The list of statistically significantly mutated genes from The Cancer Genome Atlas was obtained from the published Pan-Cancer analysis12. Each gene was searched using the Repurposing Hub website, and a representative drug was selected among those with the highest clinical development status. For gene products not targeted directly by a small molecule, additional curation was performed from the literature to identify relevant downstream or synthetic lethal targets (Supplementary Table 4). These targets were then searched on the Hub using the same process.
Purity evaluation
Compounds were evaluated by UPLC-MS according to standard compound management practices. Upon receipt of the compounds, aliquots were removed for compound identity and purity assessments, and the remaining stock frozen for future use. To verify structural accuracy, drug names were searched in the Chemical Abstracts Service SciFinder® tool. This allowed confirmation of the expected exact mass prior to identity verification by UPLC-MS. Numerous drawing errors were detected in vendor provided files, as well as mixtures, inorganic compounds, and salts (e.g., pamoate or besylate) not originally contained in the registration database. After detection of errors, the corrected structures were confirmed by viewing the vendor catalog and original sources. Compound purity and identity were determined by UPLC-MS (Waters). Purity was measured by UV absorbance at 210 nm or by Evaporative Light Scattering Detector (ELSD). Compound identity was confirmed on a SQ mass spectrometer by positive and/or negative electrospray ionization. Mobile phase A consisted of either 0.1% ammonium hydroxide or 0.05% trifluoroacetic acid in water, and mobile phase B consisted of either 0.1% ammonium hydroxide or 0.06% trifluoroacetic acid in acetonitrile. The gradient ran from 5% to 95% mobile phase B over 2.65 minutes at 0.9 mL/minute. An Acquity BEH C18, 1.7 micrometer, 2.1×50 millimeter column was used, and the column temperature was maintained at 65°C. Compounds were dissolved in DMSO at a nominal concentration of 1 millimolar, and 1.0 microliter of this solution was injected. Source chemical vendors were contacted for compound replacement when compound purity was found to be less than 85%. Approximately 200 samples had structures judged to be incompatible with the UPLC-MS method due to very low molecular weight (less than 100 grams per mole with no ring assemblies) or the presence of inorganic complexes. In these special cases, chemical vendors generally provided NMR confirmation of compound identity.
Data availability
Drug annotations were stored in MongoDB. A custom Javascript web application was developed to enable interactive access to drug structure, vendor identifiers, mechanism of action, protein targets, clinical status, approved indications, and other information. Finally, a JSON API was added for programmatic access to the dataset. The Repurposing Hub database is publicly available online at www.broadinstitute.org/repurposing.
Supplementary Material
Acknowledgments
We thank the members of the Broad Institute Compound Management team for their invaluable assistance. We also thank C. Yu, J. Boehm, N. Tolliday, J. Athanasopoulos, J. Rosains, S. Loranger, and A. Burgin for helpful scientific discussions. This work was supported in part by NIH LINCS Program grants 3U54 HG006093 (T.R.G. and A.S), U54 HL127366 (T.R.G. and A.S), and U54 HG008699 (T.R.G. and A.S). Additional support was provided by the Howard Hughes Medical Institute (T.R.G), NIH training grant T32 CA009172 (S.M.C.), KL2/Catalyst Medical Research Investigator Training award from Harvard Catalyst/The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health) Award KL2 TR001100 (S.M.C.), and the Conquer Cancer Foundation of ASCO Young Investigator Award (S.M.C.).
Footnotes
Author contributions
S.M.C. and T.R.G. conceived the project, designed experiments, and wrote the paper with input from co-authors; A.V., S.E.J., P.M. and J.A.B. performed analytical chemistry and compound management activities; P.M., J.A.B., and S.M.C. performed chemical structure analysis; S.M.C., Z.L., J.E.H, R.N., and C.C.M. annotated drug properties; B.W. and M.K. designed the Drug Repurposing Hub web portal interface and assisted with manuscript figures; J.G., J.A. and A.S. designed and implemented the Hub database, interactive web platform, and data API.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Nosengo N. Can you teach old drugs new tricks? Nature. 2016;534:314–316. doi: 10.1038/534314a. [DOI] [PubMed] [Google Scholar]
- 2.Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein I, et al. Oral sildenafil in the treatment of erectile dysfunction. Sildenafil Study Group. N Engl J Med. 1998;338:1397–1404. doi: 10.1056/NEJM199805143382001. [DOI] [PubMed] [Google Scholar]
- 4.Palumbo A, et al. Thalidomide for treatment of multiple myeloma: 10 years later. Blood. 2008;111:3968–3977. doi: 10.1182/blood-2007-10-117457. [DOI] [PubMed] [Google Scholar]
- 5.Lamb J, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 6.Yu C, et al. High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat Biotechnol. 2016;34:419–423. doi: 10.1038/nbt.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang R, et al. The NCGC pharmaceutical collection: a comprehensive resource of clinically approved drugs enabling repurposing and chemical genomics. Sci Transl Med. 2011;3:80ps16–80ps16. doi: 10.1126/scitranslmed.3001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Law V, et al. DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–7. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin C, et al. Therapeutic target database update 2014: a resource for targeted therapeutics. Nucleic Acids Res. 2013;42:D1118–D1123. doi: 10.1093/nar/gkt1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mi H, et al. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–8. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blaxill Z, Holland-Crimmin S, Lifely R. Stability through the ages: the GSK experience. J Biomol Screen. 2009;14:547–556. doi: 10.1177/1087057109335327. [DOI] [PubMed] [Google Scholar]
- 12.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heller SR, McNaught A, Pletnev I, Stein S, Tchekhovskoi D. InChI, the IUPAC International Chemical Identifier. J Cheminf. 2015;7:23. doi: 10.1186/s13321-015-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Drug annotations were stored in MongoDB. A custom Javascript web application was developed to enable interactive access to drug structure, vendor identifiers, mechanism of action, protein targets, clinical status, approved indications, and other information. Finally, a JSON API was added for programmatic access to the dataset. The Repurposing Hub database is publicly available online at www.broadinstitute.org/repurposing.