Abstract
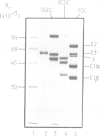
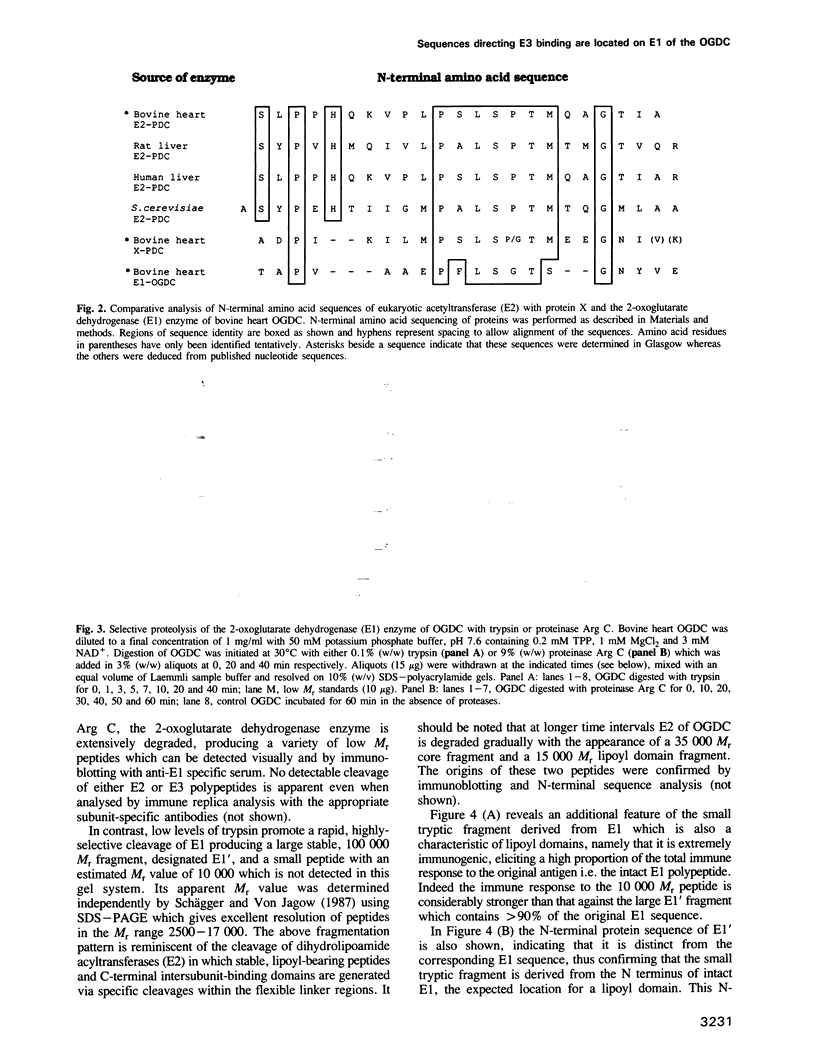
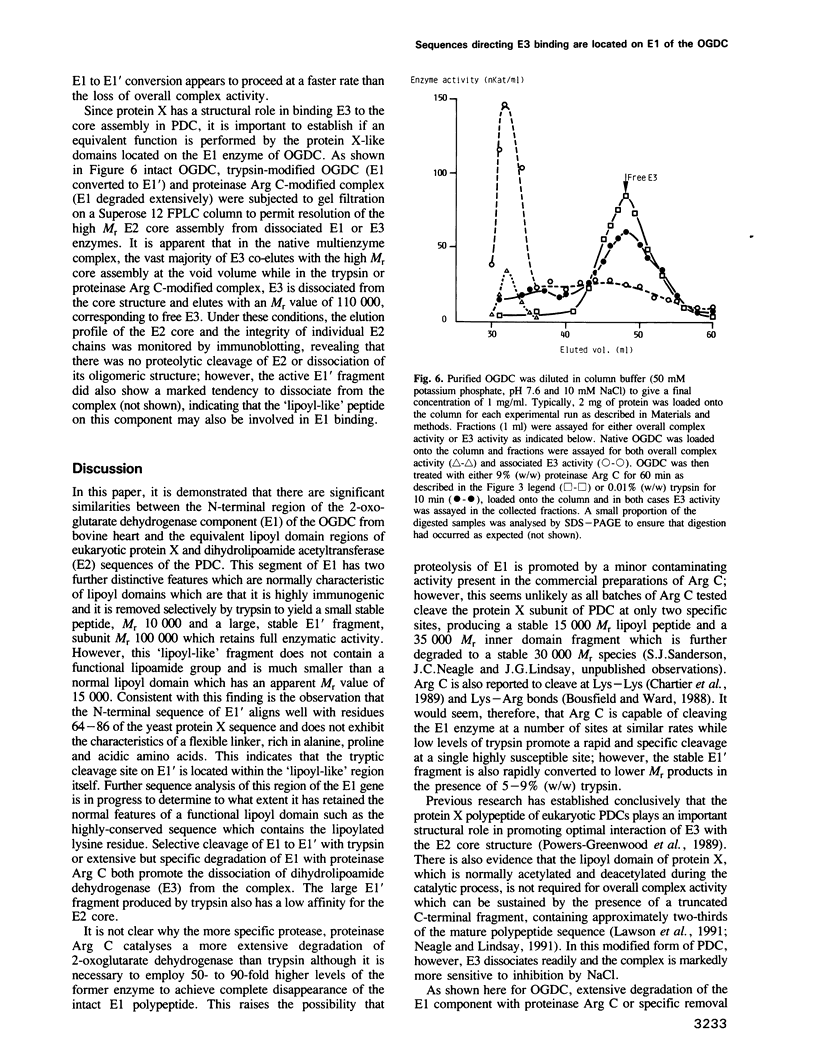
Sequences located in the N-terminal region of the high M(r) 2-oxoglutarate dehydrogenase (E1) enzyme of the mammalian 2-oxoglutarate dehydrogenase multienzyme complex (OGDC) exhibit significant similarity with corresponding sequences from the lipoyl domains of the dihydrolipoamide acetyltransferase (E2) and protein X components of eukaryotic pyruvate dehydrogenase complexes (PDCs). Two additional features of this region of E1 resemble lipoyl domains: (i) it is readily released by trypsin, generating a small N-terminal peptide with an apparent M(r) value of 10,000 and a large stable 100,000 M(r) fragment (E1') and (ii) it is highly immunogenic, inducing the bulk of the antibody response to intact E1. This 'lipoyl-like' domain lacks a functional lipoamide group. Selective but extensive degradation of E1 with proteinase Arg C or specific conversion of E1 to E1' with trypsin both cause loss of overall OGDC function although the E1' fragment retains full catalytic activity. Removal of this small N-terminal peptide promotes the dissociation of dihydrolipoamide dehydrogenase (E3) from the E2 core assembly and also affects the stability of E1 interaction. Thus, structural roles which are mediated by a specific gene product, protein X in PDC and possibly also the E2 subunit, are performed by similar structural elements located on the E1 enzyme of the OGDC.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrera C. R., Namihira G., Hamilton L., Munk P., Eley M. H., Linn T. C., Reed L. J. -Keto acid dehydrogenase complexes. XVI. Studies on the subunit structure of the pyruvate dehydrogenase complexes from bovine kidney and heart. Arch Biochem Biophys. 1972 Feb;148(2):343–358. doi: 10.1016/0003-9861(72)90152-x. [DOI] [PubMed] [Google Scholar]
- Behal R. H., Browning K. S., Hall T. B., Reed L. J. Cloning and nucleotide sequence of the gene for protein X from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8732–8736. doi: 10.1073/pnas.86.22.8732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousfield G. R., Ward D. N. Selective proteolysis of ovine lutropin or its beta subunit by endoproteinase Arg-C. Properties of the Arg beta 43 cleaved hormone. J Biol Chem. 1988 Sep 5;263(25):12602–12607. [PubMed] [Google Scholar]
- Brown J. P., Perham R. N. Selective inactivation of the transacylase components of the 2-oxo acid dehydrogenase multienzyme complexes of Escherichia coli. Biochem J. 1976 May 1;155(2):419–427. doi: 10.1042/bj1550419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier F., Laine B., Bélaïche D., Sautière P. Primary structure of the chromosomal proteins MC1a, MC1b, and MC1c from the archaebacterium Methanothrix soehngenii. J Biol Chem. 1989 Oct 15;264(29):17006–17015. [PubMed] [Google Scholar]
- Clarkson G. H., Lindsay J. G. Immunology, biosynthesis and in vivo assembly of the branched-chain 2-oxoacid dehydrogenase complex from bovine kidney. Eur J Biochem. 1991 Feb 26;196(1):95–100. doi: 10.1111/j.1432-1033.1991.tb15790.x. [DOI] [PubMed] [Google Scholar]
- Dardel F., Laue E. D., Perham R. N. Sequence-specific 1H-NMR assignments and secondary structure of the lipoyl domain of the Bacillus stearothermophilus pyruvate dehydrogenase multienzyme complex. Eur J Biochem. 1991 Oct 1;201(1):203–209. doi: 10.1111/j.1432-1033.1991.tb16275.x. [DOI] [PubMed] [Google Scholar]
- De Marcucci O. G., Hodgson J. A., Lindsay J. G. The Mr-50 000 polypeptide of mammalian pyruvate dehydrogenase complex participates in the acetylation reactions. Eur J Biochem. 1986 Aug 1;158(3):587–594. doi: 10.1111/j.1432-1033.1986.tb09795.x. [DOI] [PubMed] [Google Scholar]
- De Marcucci O. L., Hunter A., Lindsay J. G. Low immunogenicity of the common lipoamide dehydrogenase subunit (E3) of mammalian pyruvate dehydrogenase and 2-oxoglutarate dehydrogenase multienzyme complexes. Biochem J. 1985 Mar 1;226(2):509–517. doi: 10.1042/bj2260509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marcucci O., Lindsay J. G. Component X. An immunologically distinct polypeptide associated with mammalian pyruvate dehydrogenase multi-enzyme complex. Eur J Biochem. 1985 Jun 18;149(3):641–648. doi: 10.1111/j.1432-1033.1985.tb08972.x. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan S., Rahmatullah M., Radke G. A., Powers-Greenwood S., Roche T. E. Role of protein X in the function of the mammalian pyruvate dehydrogenase complex. Biochem Biophys Res Commun. 1989 Apr 28;160(2):715–721. doi: 10.1016/0006-291x(89)92492-3. [DOI] [PubMed] [Google Scholar]
- Hodgson J. A., De Marcucci O. G., Lindsay J. G. Lipoic acid is the site of substrate-dependent acetylation of component X in ox heart pyruvate dehydrogenase multienzyme complex. Eur J Biochem. 1986 Aug 1;158(3):595–600. doi: 10.1111/j.1432-1033.1986.tb09796.x. [DOI] [PubMed] [Google Scholar]
- Hunter A., Lindsay J. G. Immunological and biosynthetic studies on the mammalian 2-oxoglutarate dehydrogenase multienzyme complex. Eur J Biochem. 1986 Feb 17;155(1):103–109. doi: 10.1111/j.1432-1033.1986.tb09464.x. [DOI] [PubMed] [Google Scholar]
- Jackman S. A., Hough D. W., Danson M. J., Stevenson K. J., Opperdoes F. R. Subcellular localisation of dihydrolipoamide dehydrogenase and detection of lipoic acid in bloodstream forms of Trypanosoma brucei. Eur J Biochem. 1990 Oct 5;193(1):91–95. doi: 10.1111/j.1432-1033.1990.tb19308.x. [DOI] [PubMed] [Google Scholar]
- Jilka J. M., Rahmatullah M., Kazemi M., Roche T. E. Properties of a newly characterized protein of the bovine kidney pyruvate dehydrogenase complex. J Biol Chem. 1986 Feb 5;261(4):1858–1867. [PubMed] [Google Scholar]
- Khailova L. S., Bernkhardt R., Khiubner G. Izuchenie kineticheskogo mekhanizma piruvat-2,6-dikhlorfenolindofenolreduktaznoi aktivnosti myshechnoi piruvatdegidrogenazy. Biokhimiia. 1977 Jan;42(1):113–117. [PubMed] [Google Scholar]
- Kochi H., Kikuchi G. Mechanism of reversible glycine cleavage reaction in Arthrobacter globiformis. Function of lipoic acid in the cleavage and synthesis of blycine. Arch Biochem Biophys. 1976 Mar;173(1):71–81. doi: 10.1016/0003-9861(76)90236-8. [DOI] [PubMed] [Google Scholar]
- Lawson J. E., Behal R. H., Reed L. J. Disruption and mutagenesis of the Saccharomyces cerevisiae PDX1 gene encoding the protein X component of the pyruvate dehydrogenase complex. Biochemistry. 1991 Mar 19;30(11):2834–2839. doi: 10.1021/bi00225a015. [DOI] [PubMed] [Google Scholar]
- Nakano K., Matuda S., Yamanaka T., Tsubouchi H., Nakagawa S., Titani K., Ohta S., Miyata T. Purification and molecular cloning of succinyltransferase of the rat alpha-ketoglutarate dehydrogenase complex. Absence of a sequence motif of the putative E3 and/or E1 binding site. J Biol Chem. 1991 Oct 5;266(28):19013–19017. [PubMed] [Google Scholar]
- Neagle J. C., Lindsay J. G. Selective proteolysis of the protein X subunit of the bovine heart pyruvate dehydrogenase complex. Effects on dihydrolipoamide dehydrogenase (E3) affinity and enzymic properties of the complex. Biochem J. 1991 Sep 1;278(Pt 2):423–427. doi: 10.1042/bj2780423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neagle J., De Marcucci O., Dunbar B., Lindsay J. G. Component X of mammalian pyruvate dehydrogenase complex: structural and functional relationship to the lipoate acetyltransferase (E2) component. FEBS Lett. 1989 Aug 14;253(1-2):11–15. doi: 10.1016/0014-5793(89)80919-6. [DOI] [PubMed] [Google Scholar]
- Packman L. C., Borges A., Perham R. N. Amino acid sequence analysis of the lipoyl and peripheral subunit-binding domains in the lipoate acetyltransferase component of the pyruvate dehydrogenase complex from Bacillus stearothermophilus. Biochem J. 1988 May 15;252(1):79–86. doi: 10.1042/bj2520079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packman L. C., Perham R. N. Chain folding in the dihydrolipoyl acyltransferase components of the 2-oxo-acid dehydrogenase complexes from Escherichia coli. Identification of a segment involved in binding the E3 subunit. FEBS Lett. 1986 Oct 6;206(2):193–198. doi: 10.1016/0014-5793(86)80979-6. [DOI] [PubMed] [Google Scholar]
- Patel M. S., Roche T. E. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990 Nov;4(14):3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- Perham R. N. Domains, motifs, and linkers in 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991 Sep 3;30(35):8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- Pettit F. H., Hamilton L., Munk P., Namihira G., Eley M. H., Willms C. R., Reed L. J. Alpha-keto acid dehydrogenase complexes. XIX. Subunit structure of the Escherichia coli alpha-ketoglutarate dehydrogenase complex. J Biol Chem. 1973 Aug 10;248(15):5282–5290. [PubMed] [Google Scholar]
- Powers-Greenwood S. L., Rahmatullah M., Radke G. A., Roche T. E. Separation of protein X from the dihydrolipoyl transacetylase component of the mammalian pyruvate dehydrogenase complex and function of protein X. J Biol Chem. 1989 Mar 5;264(7):3655–3657. [PubMed] [Google Scholar]
- Poyton R. O., Trueblood C. E., Wright R. M., Farrell L. E. Expression and function of cytochrome c oxidase subunit isologues. Modulators of cellular energy production? Ann N Y Acad Sci. 1988;550:289–307. doi: 10.1111/j.1749-6632.1988.tb35344.x. [DOI] [PubMed] [Google Scholar]
- Radford S. E., Laue E. D., Perham R. N., Miles J. S., Guest J. R. Segmental structure and protein domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. Genetic reconstruction in vitro and 1H-n.m.r. spectroscopy. Biochem J. 1987 Nov 1;247(3):641–649. doi: 10.1042/bj2470641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmatullah M., Gopalakrishnan S., Andrews P. C., Chang C. L., Radke G. A., Roche T. E. Subunit associations in the mammalian pyruvate dehydrogenase complex. Structure and role of protein X and the pyruvate dehydrogenase component binding domain of the dihydrolipoyl transacetylase component. J Biol Chem. 1989 Feb 5;264(4):2221–2227. [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Stanley C. J., Perham R. N. Purification of 2-oxo acid dehydrogenase multienzyme complexes from ox heart by a new method. Biochem J. 1980 Oct 1;191(1):147–154. doi: 10.1042/bj1910147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeaman S. J. The 2-oxo acid dehydrogenase complexes: recent advances. Biochem J. 1989 Feb 1;257(3):625–632. doi: 10.1042/bj2570625. [DOI] [PMC free article] [PubMed] [Google Scholar]