Abstract
Duchenne and Becker muscular dystrophies are caused by mutations in dystrophin. Cardiac manifestations vary broadly, making prognosis difficult. Current dystrophin genotype–cardiac phenotype correlations are limited. For skeletal muscle, the reading-frame rule suggests in-frame mutations tend to yield milder phenotypes. We performed dystrophin genotype–cardiac phenotype correlations using a protein-effect model and cardiac magnetic resonance imaging. A translational model was applied to patient-specific deletion, indel, and nonsense mutations to predict exons and protein domains present within truncated dystrophin protein. Patients were dichotomized into predicted present and predicted absent groups for exons and protein domains of interest. Development of myocardial fibrosis (represented by late gadolinium enhancement [LGE]) and depressed left ventricular ejection fraction (LVEF) were compared. Patients (n = 274) with predicted present cysteine-rich domain (CRD), C-terminal domain (CTD), and both the N-terminal actin-binding and cysteine-rich domains (ABD1 + CRD) had a decreased risk of LGE and trended toward greater freedom from LGE. Patients with predicted present CTD (exactly the same as those with in-frame mutations) and ABD1 + CRD trended toward decreased risk of and greater freedom from depressed LVEF. In conclusion, genotypes previously implicated in altering the dystrophinopathic cardiac phenotype were not significantly related to LGE and depressed LVEF. Patients with predicted present CRD, CTD/in-frame mutations, and ABD1 + CRD trended toward milder cardiac phenotypes, suggesting that the reading-frame rule may be applicable to the cardiac phenotype. Genotype–phenotype correlations may help predict the cardiac phenotype for dystrophinopathic patients and guide future therapies.
Mutations in dystrophin (DMD gene) cause Duchenne and Becker muscular dystrophies (DMD and BMD, respectively).1 The onset and progression of cardiac involvement are quite variable in DMD and/or BMD,2–4 making prognosis and therapy difficult. However, differences in dystrophin genotypes may explain some of the cardiac phenotype variability. Dystrophin genotype–skeletal muscle phenotype correlations suggest that in-frame mutations resulting in a semifunctional dystrophin protein result in less-severe skeletal muscle phenotype (BMD) than frameshift or early truncating mutations resulting in nonfunctional dystrophin (DMD).1,5–7 Previous investigations of dystrophin genotype–cardiac phenotype correlations have found dystrophin exons 45, 48 to 49, and 51 to 52,2,4,8–10 and the N-terminal actin-binding domain (ABD1), rod, hinge-III, cysteine-rich domain (CRD), and C-terminal domains (CTD) correlated with earlier onset of cardiac dysfunction.4,8,9,11–14 These studies focused on mutation type and location without regard to the predicted dystrophin structure and used age of onset of depressed left ventricular ejection fraction (LVEF) by echocardiogram.4,8,10,12,14–16 We aimed to perform a large-scale dystrophin genotype–cardiac phenotype correlation study using a novel direct translation model with phenotyping by cardiac magnetic resonance imaging of myocardial performance and fibrosis as seen on late gadolinium enhancement (LGE) imaging.
Methods
All boys with genetically confirmed DMD or BMD who underwent clinical CMR studies at Cincinnati Children’s Hospital Medical Center (CCHMC) from January 2005 and January 2013 were included. For the evaluation of dystrophin genotype–cardiac phenotype correlations, we included only patients with whole-exon deletion, indel, and nonsense mutations known or predicted to be disease causing (described in the following). The institutional review board approved the study. For each patient with DMD and/or BMD who had undergone a CMR study, we reviewed clinically obtained dystrophin mutation analysis. Several clinical diagnostic laboratories were used during the study period, and methods used for molecular analysis included Southern blot, polymerase chain reaction, single condition amplification/internal primer, comparative genomic hybridization, and/or multiplex ligation-dependent probe amplification. For classification of the mutation data for the cohort, each clinical diagnostic test result was checked using the Leiden reading-frame tool and then analyzed against the Leiden whole exon change database,17 the Leiden point mutation database,18 and/or the Universal Mutation Database7 as appropriate for the specific mutation. Mutations previously described as disease causing, or mutations expected to change the coding sequence of dystrophin, were considered pathogenic. Mutations were defined as nonsense if a base pair change created a premature stop codon at the mutation site, indel if there was an insertion or deletion of 1 to 4 nucleotides that resulted in a premature stop codon, splicing if they occurred at a predicted splice site as reported by the diagnostic laboratory, and intronic if they occurred outside both the coding and splice site sequences, as defined by the diagnostic laboratory.
To predict the presence or absence of critical functional protein domains, we first determined which base pairs were predicted to be present for each patient based on their specific mutation. A direct translation model was then used for whole-exon deletion, indel, and nonsense mutation types. These mutational mechanisms are predicted to result in a truncated protein and/or protein missing specific domains. The model assumed that the mRNA resulting from the patient-specific mutations was stable and not subject to nonsense-mediated decay. The predicted mRNA was then translated to determine the predicted presence or absence of each critical functional protein domain. Exon boundaries were extracted from GenBank (accession NM_004006.2), and protein domain boundaries were extracted from the eDystrophin project19 and GenBank (accession NM_004006.2). For out-of-frame whole-exon deletion mutations, we predicted that exons and protein domains encoded entirely 50′ to the deletion start site would be present. For in-frame whole-exon deletion mutations, we predicted that exons and protein domains coded entirely either 5′ or 3′ of the deleted segment would be present. For indel and nonsense mutations, we predicted that exons and protein domains encoded entirely 5′ to the mutation site would be present. The creation of new 3' protein domains in patients with out-of-frame mutations was not incorporated into the model. For each patient, we then determined whether each exon and protein domain of interest was predicted to be present or absent.
Image acquisition for our DMD/BMD cohort has been described previously.20–22 The CMR studies were conducted on a clinical 3- or 1.5-T scanner, depending solely on schedule availability. At CCHMC, we routinely perform CMR on every DMD and BMD patient annually; only patients who refused or could not tolerate lying in the scanner did not undergo the study, and an annual CMR study was recommended regardless of previous refusal or inability to undergo CMR. No anesthesia or sedation was used for these studies. The LVEF was assessed using standard planimetry techniques (QMASS MR, version 7.5; Medis Medical Imaging Systems, Leiden, Netherlands) by an expert reader (RJF, KNH, JJS, and MDT). The LVEF was defined as depressed if it was <55%. Our interobserver variability was ~4% and intraobserver variability was ~2% for LVEF (unpublished data). LGE was considered positive if any left ventricular segment showed subepicardial or midmyocardial hyperenhancement by visual inspection.
For each region of interest, we compared patients in the predicted present group against those in the predicted absent group. For overall risk of development of LGE and depressed LVEF, we used chi-square analyses; for age of development of LGE and depressed LVEF, we used t tests; and for freedom from LGE and depressed LVEF, we used Kaplan–Meier log-rank analyses (SAS version 9.3, SAS Institute, Cary, North Carolina). All tests were 2-sided, and a p value of <0.05 was considered statistically significant.
Results
The dystrophinopathic cohort contained 322 patients for whom genotype data were available. The mutation distribution was similar to other reported populations6,7: 212 (66%) whole-exon deletions (30 [9.3%] in-frame and 182 [57%] out-of-frame); 39 (12%) whole exon duplications; 39 (12%) nonsense mutations; 23 (7.1%) indel mutations (19 [5.9%] deletion and 4 [1.2%] insertion); 7 (2.2%) splicing mutations; and 2 (0.6%) intronic mutations. Patients ranged in age from 4.9 to 29.7 years (mean 12.3, median 11.4 years) at time of CMR.
We analyzed the dystrophin mutations and cardiac phenotype of patients who met the inclusion criteria for genotype analysis. This subset of patients comprised 274 patients, who ranged in age from 4.9 to 29.4 years (mean 12.2 ± 4.0, median 11.3 years). Of the 274 patients, 11 (4.0%) were patients with BMD. During the study, 15 patients (5.5%) died. Of the 274 in the study, 231 (84%) had been treated with steroids: 138 (50%) with deflazacort only, 38 (14%) with prednisone only, and 55 (20%) with both. In terms of their CMR findings, 118 patients (43%) had at least 1 LGE-positive study; 40 (15%) had at least 1 study with depressed LVEF; 32 (12%) had at least 1 study with both LGE and depressed LVEF; 126 (46%) had 1 study with either LGE or depressed LVEF, and 148 (54%) had all studies with neither LGE nor depressed LVEF. Of the 766 CMR studies examined, 183 (24%) were LGE positive, 539 (70%) were LGE negative, and 44 (5.7%) were LGE indeterminate; 692 (90%) had a normal LVEF; and 74 (10%) had depressed LVEF.
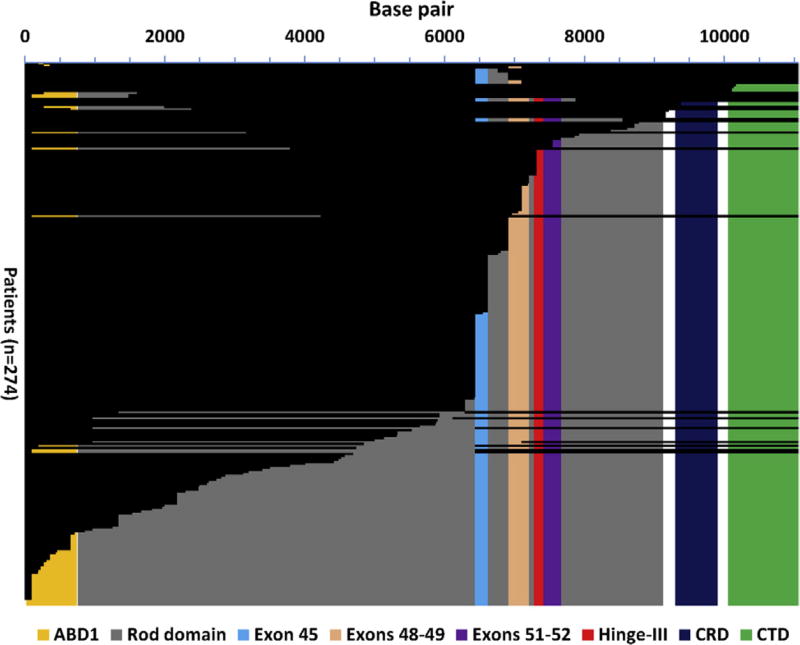
Patients were dichotomized to either the predicted present or predicted absent group for each region of interest based on the process detailed previously. The subset of patients with in-frame deletions overlapped exactly with patients predicted to have the CTD present (Figure 1).
Figure 1.
Graphical representation of predicted present base pairs. The base pairs predicted present for each patient based on mutation data are represented with a horizontal black bar, aligned with the base pair number on the horizontal axis. Exon and protein domain boundaries are marked with a colored background. A patient was predicted to have an exon or protein domain present if all the base pairs that code for that region of interest were predicted to be present.
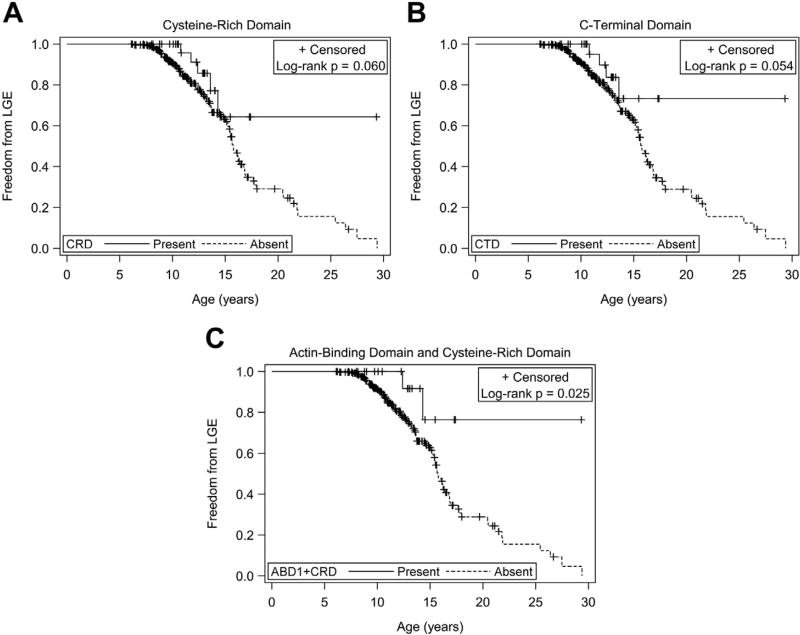
Patients predicted to have exons 45, 48 to 49, 51 to 52, or 1 to 45 present did not show a significant difference in the risk of, age of onset of, or freedom from LGE compared with the respective predicted absent groups. Patients predicted to have the ABD1, hinge-III region, or rod domains present also did not show a significant difference in risk of, age of onset of, or freedom from LGE compared with the respective predicted absent groups. Patients predicted to have the CRD and CTD present did show a decreased risk of LGE (relative risk [RR] 0.40 and 0.37, respectively, Table 1), but there was no significant difference in age of onset of LGE compared with the predicted absent groups. There was a trend toward greater freedom from LGE for patients with predicted present CRD (25% time-to-event 14.3 vs 13.0 years, Figure 2) and CTD (25% time-to-event 13.6 vs 13.0 years, Figure 2). We then considered patients who were predicted to have both the ABD1 and CRD present (ABD1 + CRD).23 Patients predicted to have both ABD1 + CRD present had a lower risk of developing LGE (RR 0.27, Table 1) and greater freedom from LGE (Figure 2) compared with those without both predicted to be present. There was no significant difference in age of onset of LGE.
Table 1.
p Values for relationship of cardiac phenotype markers to dystrophin genotypes
| Exon/domain (n=274) |
Predicted intact (%) |
Decreased risk of LGE |
Freedom from LGE |
Decreased risk of depressed LVEF |
Freedom from depressed LVEF |
|---|---|---|---|---|---|
| CRD | 34 (12%) | 0.012 | 0.060 | 0.191 | 0.194 |
| CTD/in-frame deletions | 30 (11%) | 0.012 | 0.054 | 0.095 | 0.090 |
| ABD1+CRD | 21 (7.7%) | 0.014 | 0.025 | 0.331 | 0.132 |
ABD1 = N-terminal actin-binding domain; CRD = cysteine-rich domain; CTD = C-terminal domain; LGE = late gadolinium enhancement; LVEF = left ventricular ejection fraction.
Figure 2.
(A) Freedom from LGE and CRD intact. Kaplan–Meier freedom from LGE for patients with CRD predicted intact (n = 34) versus predicted disrupted. 25% time-to-event 14.3 versus 13.0 years; log-rank p = 0.060. (B) Freedom from LGE and CTD intact. Kaplan–Meier freedom from LGE for patients with CTD predicted intact (n = 30) versus predicted disrupted. Patients predicted to have the CTD domain intact were exactly those with in-frame deletions. 25% time-to-event 13.6 versus 13.0 years; log-rank p = 0.054. (C) Freedom from LGE and ABD1 + CRD intact. Kaplan–Meier freedom from LGE for patients with both ABD1 + CRD predicted intact (n = 21) versus those predicted to have at least 1 disrupted. Log-rank p = 0.025.
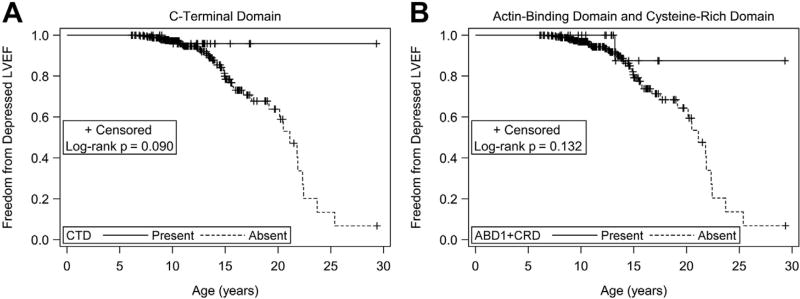
Patients predicted to have exons 45, 48 to 49, 51to 52, or 1 to 45 present did not show a significant difference in the risk of, age of onset of, or freedom from depressed LVEF compared with the respective predicted absent groups. Patients predicted to have the ABD1, hinge-III region, rod domain, or CRD present also did not show a significant difference in the risk of, age of onset of, or freedom from depressed LVEF compared with the respective predicted absent groups. Patients predicted to have the CTD present (same as those with in-frame mutations) trended toward a decreased risk of depressed LVEF (RR 0.21) and greater freedom from depressed LVEF (Table 1 and Figure 3). There was no significant difference in age of onset of depressed LVEF. Patients predicted to have both ABD1 + CRD present trended toward decreased risk of depressed LVEF (RR 0.31) and greater freedom from depressed LVEF (Table 1 and Figure 3). There was no significant difference in the age of onset of depressed LVEF.
Figure 3.
(A) Freedom from depressed LVEF and CTD intact. Kaplan–Meier freedom from depressed LVEF (<55%) for patients with CTD predicted intact (n = 30) versus predicted disrupted. Patients predicted to have the CTD domain intact were exactly those with in-frame deletions. Log-rank p = 0.090. (B) Freedom from depressed LVEF and ABD1 + CRD intact. Kaplan–Meier freedom from depressed LVEF (<55%) for patients with both ABD1 + CRD predicted intact (n = 21) versus those predicted to have at least 1 disrupted. Log-rank p = 0.132.
Discussion
Our data suggest that specific dystrophin mutations affect the severity of the cardiac phenotype in patients with DMD and BMD. Specifically, patients predicted to have the CRD, CTD, and both ABD1 + CRD present and those with in-frame mutations trended toward milder cardiac phenotype. In our cohort, patients with in-frame deletions were likely to have both the CRD (n = 34) and CTD (n = 30) predicted present. These results suggest that the reading-frame rule that applies to skeletal muscle may also apply for the human cardiac phenotype and that this may be due to the CRD or CTD being present. There is evidence that the presence of the CTD can lead to milder skeletal muscle phenotypes in humans.24 Mouse rescue studies suggest that both the ABD1 and CRD are important in restoring skeletal muscle function.23 Our results suggest that this may also be true in human cardiac disease, given the trends toward freedom from depressed LVEF seen in patients predicted to have both ABD1 + CRD present.
This is the first study to use CMR and LGE to examine the correlation of cardiac phenotype with dystrophin genotype. LGE, a marker for myocardial fibrosis, precedes the development of depressed LVEF in dystrophinopathic patients.25,26 We hypothesize that the patterns seen for patients with predicted present CRD, CTD, and both ABD1 + CRD in the development of LGE will be reflected in the development of depressed LVEF as well; studies with a larger cohort and longer longitudinal follow-up will be required to test this hypothesis.
We did not find the same patterns suggested by Kaspar et al,8 Jefferies et al,4 and others who found that mutations in the ABD1 region, around exons 45 to 49, and around exons 51 and 52 were correlated with more severe cardiac phenotype. However, our genotype characterization model takes the predicted effects of the mutation on dystrophin structure into account, whereas previous groups did not. In addition, we use a more standardized and reproducible measurement of cardiac phenotype than previous studies.
Our findings have implications in terms of future gene therapy trials and the cardiac phenotype. Similar to the mouse studies mentioned, therapies that ensure the presence of the ABD1 and CRD may lead to dystrophin proteins with an appropriate three-dimensional structure and can bind actin and appropriately form the dystrophin–glycoprotein complex.
Our study has limitations, many of which are related to its retrospective nature. Genetic data were derived from clinical testing that varied by clinical diagnostic laboratory and evolved over the years covered by the study. Clinical genetic testing does not include evaluation of transcript, protein stability, nonsense-mediated decay, or levels of dystrophin expression in cardiac muscle; in addition, predictions of present regions of interest made in this study assume stability of the message and protein. Given that current diagnostic guidelines for DMD suggest that a genetic diagnosis is necessary although muscle biopsy is optional,27 we believe that basing our analysis on genetic testing alone was reasonable. In terms of phenotypes, we were limited by having data only on those patients able to tolerate a CMR study, which may represent ascertainment bias. Longitudinal follow-up for patients was also not standardized. Our study was not powered to be able to take the effects of cardiac medication usage (e.g., ACE-inhibitors, β-blockers) into account; there is debate in the previous reports as to the effects of these medications on cardiac function.4,28 Given the high percentage of patients who were treated with steroids, the study was not powered to take their effect into account. In terms of statistical analysis, although the sample size is overall large compared with some dystrophinopathic cohorts, when looking at specific mutation groups, the sample size became small at times, limiting power.
Acknowledgments
This publication was supported by Grant 5UL1RR026314, an Institutional Clinical and Translational Science Award, National Center for Research Resources, National Institutes of Health.
Footnotes
All or portions of this report were or will be submitted as a thesis in partial fulfillment of requirements for a Master of Science degree.
The contents of the study are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Disclosures
The authors have no conflicts of interest to disclose.
References
- 1.Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 2.Finsterer J, Stollberger C. The heart in human dystrophinopathies. Cardiology. 2003;99:1–19. doi: 10.1159/000068446. [DOI] [PubMed] [Google Scholar]
- 3.Connuck DM, Sleeper LA, Colan SD, Cox GF, Towbin JA, Lowe AM, Wilkinson JD, Orav EJ, Cuniberti L, Salbert BA, Lipshultz SE. Pediatric Cardiomyopathy Registry Study Group. Characteristics and outcomes of cardiomyopathy in children with Duchenne or Becker muscular dystrophy: a comparative study from the Pediatric Cardiomyopathy Registry. Am Heart J. 2008;155:998–1005. doi: 10.1016/j.ahj.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jefferies JL, Eidem BW, Belmont JW, Craigen WJ, Ware SM, Fernbach SD, Neish SR, Smith EO, Towbin JA. Genetic predictors and remodeling of dilated cardiomyopathy in muscular dystrophy. Circulation. 2005;112:2799–2804. doi: 10.1161/CIRCULATIONAHA.104.528281. [DOI] [PubMed] [Google Scholar]
- 5.Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 6.Flanigan KM, Dunn DM, von Niederhausern A, Soltanzadeh P, Gappmaier E, Howard MT, Sampson JB, Mendell JR, Wall C, King WM, Pestronk A, Florence JM, Connolly AM, Mathews KD, Stephan CM, Laubenthal KS, Wong BL, Morehart PJ, Meyer A, Finkel RS, Bonnemann CG, Medne L, Day JW, Dalton JC, Margolis MK, Hinton VJ, United Dystrophinopathy Project Consortium. Weiss RB. Mutational spectrum of DMD mutations in dystrophinopathy patients: application of modern diagnostic techniques to a large cohort. Hum Mutat. 2009;30:1657–1666. doi: 10.1002/humu.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tuffery-Giraud S, Beroud C, Leturcq F, Yaou RB, Hamroun D, Michel-Calemard L, Moizard MP, Bernard R, Cossee M, Boisseau P, Blayau M, Creveaux I, Guiochon-Mantel A, de Martinville B, Philippe C, Monnier N, Bieth E, Khau Van Kien P, Desmet FO, Humbertclaude V, Kaplan JC, Chelly J, Claustres M. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum Mutat. 2009;30:934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 8.Kaspar RW, Allen HD, Ray WC, Alvarez CE, Kissel JT, Pestronk A, Weiss RB, Flanigan KM, Mendell JR, Montanaro F. Analysis of dystrophin deletion mutations predicts age of cardiomyopathy onset in Becker muscular dystrophy. Circ Cardiovasc Genet. 2009;2:544–551. doi: 10.1161/CIRCGENETICS.109.867242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen N, Muntoni F. Multiple pathogenetic mechanisms in X linked dilated cardiomyopathy. Heart. 2004;90:835–841. doi: 10.1136/hrt.2003.023390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magri F, Govoni A, D’Angelo MG, Del Bo R, Ghezzi S, Sandra G, Turconi AC, Sciacco M, Ciscato P, Bordoni A, Tedeschi S, Fortunato F, Lucchini V, Bonato S, Lamperti C, Coviello D, Torrente Y, Corti S, Moggio M, Bresolin N, Comi GP. Genotype and phenotype characterization in a large dystrophinopathic cohort with extended follow-up. J Neurol. 2011;258:1610–1623. doi: 10.1007/s00415-011-5979-z. [DOI] [PubMed] [Google Scholar]
- 11.Deburgrave N, Daoud F, Llense S, Barbot JC, Recan D, Peccate C, Burghes AH, Beroud C, Garcia L, Kaplan JC, Chelly J, Leturcq F. Protein- and mRNA-based phenotype-genotype correlations in DMD/BMD with point mutations and molecular basis for BMD with nonsense and frameshift mutations in the DMD gene. Hum Mutat. 2007;28:183–195. doi: 10.1002/humu.20422. [DOI] [PubMed] [Google Scholar]
- 12.Diegoli M, Grasso M, Favalli V, Serio A, Gambarin FI, Klersy C, Pasotti M, Agozzino E, Scelsi L, Ferlini A, Febo O, Piccolo G, Tavazzi L, Narula J, Arbustini E. Diagnostic work-up and risk stratification in X-linked dilated cardiomyopathies caused by dystrophin defects. J Am Coll Cardiol. 2011;58:925–934. doi: 10.1016/j.jacc.2011.01.072. [DOI] [PubMed] [Google Scholar]
- 13.Carsana A, Frisso G, Tremolaterra MR, Lanzillo R, Vitale DF, Santoro L, Salvatore F. Analysis of dystrophin gene deletions indicates that the hinge III region of the protein correlates with disease severity. Ann Hum Genet. 2005;69:253–259. doi: 10.1046/j.1529-8817.2005.00160.x. [DOI] [PubMed] [Google Scholar]
- 14.Ferlini A, Sewry C, Melis MA, Mateddu A, Muntoni F. X-linked dilated cardiomyopathy and the dystrophin gene. Neuromuscul Disord. 1999;9:339–346. doi: 10.1016/s0960-8966(99)00015-2. [DOI] [PubMed] [Google Scholar]
- 15.Birnkrant DJ, Ashwath ML, Noritz GH, Merrill MC, Shah TA, Crowe CA, Bahler RC. Cardiac and pulmonary function variability in Duchenne/Becker muscular dystrophy: an initial report. J Child Neurol. 2010;25:1110–1115. doi: 10.1177/0883073810371003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashwath ML, Jacobs IB, Crowe CA, Ashwath RC, Super DM, Bahler RC. Left ventricular dysfunction in Duchenne muscular dystrophy and genotype. Am J Cardiol. 2014;114:284–289. doi: 10.1016/j.amjcard.2014.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White SJ, den Dunnen JT. Copy number variation in the genome; the human DMD gene as an example. Cytogenet Genome Res. 2006;115:240–246. doi: 10.1159/000095920. [DOI] [PubMed] [Google Scholar]
- 18.Aartsma-Rus A, Van Deutekom JC, Fokkema IF, Van Ommen GJ, Den Dunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 19.Nicolas A, Lucchetti-Miganeh C, Yaou RB, Kaplan JC, Chelly J, Leturcq F, Barloy-Hubler F, Le Rumeur E. Assessment of the structural and functional impact of in-frame mutations of the DMD gene, using the tools included in the eDystrophin online database. Orphanet J Rare Dis. 2012;7:45. doi: 10.1186/1750-1172-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hagenbuch SC, Gottliebson WM, Wansapura J, Mazur W, Fleck R, Benson DW, Hor KN. Detection of progressive cardiac dysfunction by serial evaluation of circumferential strain in patients with Duchenne muscular dystrophy. Am J Cardiol. 2010;105:1451–1455. doi: 10.1016/j.amjcard.2009.12.070. [DOI] [PubMed] [Google Scholar]
- 21.Hor KN, Wansapura J, Markham LW, Mazur W, Cripe LH, Fleck R, Benson DW, Gottliebson WM. Circumferential strain analysis identifies strata of cardiomyopathy in Duchenne muscular dystrophy: a cardiac magnetic resonance tagging study. J Am Coll Cardiol. 2009;53:1204–1210. doi: 10.1016/j.jacc.2008.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazur W, Hor KN, Germann JT, Fleck RJ, Al-Khalidi HR, Wansapura JP, Chung ES, Taylor MD, Jefferies JL, Woodrow Benson D, Gottliebson WM. Patterns of left ventricular remodeling in patients with Duchenne muscular dystrophy: a cardiac MRI study of ventricular geometry, global function, and strain. Int J Cardiovasc Imaging. 2012;28:99–107. doi: 10.1007/s10554-010-9781-2. [DOI] [PubMed] [Google Scholar]
- 23.Fairclough RJ, Wood MJ, Davies KE. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat Rev Genet. 2013;14:373–378. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 24.Vainzof M, Takata RI, Passos-Bueno MR, Pavanello RC, Zatz M. Is the maintenance of the C-terminus domain of dystrophin enough to ensure a milder Becker muscular dystrophy phenotype? Hum Mol Genet. 1993;2:39–42. doi: 10.1093/hmg/2.1.39. [DOI] [PubMed] [Google Scholar]
- 25.Puchalski MD, Williams RV, Askovich B, Sower CT, Hor KH, Su JT, Pack N, Dibella E, Gottliebson WM. Late gadolinium enhancement: precursor to cardiomyopathy in Duchenne muscular dystrophy? Int J Cardiovasc Imaging. 2009;25:57–63. doi: 10.1007/s10554-008-9352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walcher T, Steinbach P, Spiess J, Kunze M, Gradinger R, Walcher D, Bernhardt P. Detection of long-term progression of myocardial fibrosis in Duchenne muscular dystrophy in an affected family: a cardiovascular magnetic resonance study. Eur J Radiol. 2011;80:115–119. doi: 10.1016/j.ejrad.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Bushby K, Finkel R, Birnkrant DJ, Case LE, Clemens PR, Cripe L, Kaul A, Kinnett K, McDonald C, Pandya S, Poysky J, Shapiro F, Tomezsko J, Constantin C DMD Care Considerations Working Group. Diagnosis and management of Duchenne muscular dystrophy, part 1: diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 28.Hor KN, Mazur W, Taylor MD, Al-Khalidi HR, Cripe LH, Jefferies JL, Raman SV, Chung ES, Kinnett KJ, Williams K, Gottliebson WM, Benson DW. Effects of steroids and angiotensin converting enzyme inhibition on circumferential strain in boys with Duchenne muscular dystrophy: a cross-sectional and longitudinal study utilizing cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2011;13:60. doi: 10.1186/1532-429X-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]