Abstract
In arterial tissue engineering, mimicking native structure and mechanical properties is essential because compliance mismatch can lead to graft failure and further disease. With bottom-up tissue engineering approaches, designing tissue components with proper microscale mechanical properties is crucial to achieve the necessary macroscale properties in the final implant. Here, we developed a thermo-responsive cell culture platform for growing aligned vascular smooth muscle cell sheets by photo-grafting N-isopropylacrylamide (NIPAAm) onto micro-patterned PDMS. We experimentally and computationally optimized the grafting process to produce PNIPAAm-PDMS substrates optimal for vascular smooth muscle cell (VSMC) attachment. To allow long-term VSMC sheet culture and increase the rate of VSMC sheet formation, PNIPAAm-PDMS surfaces were further modified with 3-aminopropyltriethoxysilane (APTES) yielding a robust, thermo-responsive cell culture platform for culturing VSMC sheets. VSMC cell sheets cultured on patterned thermo-responsive substrates exhibit cellular and collagen alignment in the direction of the micro-pattern. Mechanical characterization of patterned, single-layer VSMC sheets reveals increased stiffness in the aligned direction compared to the perpendicular direction whereas non-patterned cell sheets exhibit no directional dependence. Structural and mechanical anisotropy of aligned, single-layer VSMC sheets makes this platform an attractive micro-structural building block for engineering a vascular graft to match the in vivo mechanical properties of native arterial tissue.
Keywords: NIPAAm, cell sheet mechanics, vascular tissue engineering, micro-patterning
Graphical abstract
Vascular smooth muscle cell sheets were cultured using a novel, micro-patterned, thermo-responsive cell culture platform. The method was optimized to produce vascular smooth muscle cell sheets with cellular and extracellular alignment. Aligned cell sheets exhibit nonlinear, anisotropic mechanical properties that make them an attractive material for building a vascular graft that mimics the structural and mechanical properties of native vascular tissue.
1. Introduction
A major goal in tissue engineering is to design biomaterials that mimic native tissue structure and mechanical properties. As tissue-engineered constructs become more complex, achieving this goal is essential because incompatibility of biomaterial integration can lead to further disease and possible host rejection.[1] Arterial tissue engineering is especially challenging because mechanical mismatch between native and engineered tissue can cause complications such as anastomotic failure, aneurysm formation[2–4], or intimal hyperplasia[5–7], which may require repeat surgical intervention or lead to death. To avoid these complications, a tissue-engineered artery must have a similar stiffness and stress-strain response to native tissue, while also being strong enough to avoid rupture.[2]
The arterial wall is composed of three distinct layers: the tunica intima, tunica media, and tunica adventitia. The tunica intima, the innermost, blood contacting layer, is composed of a single layer of endothelial cells and is supported by the inner elastic lamina composed of elastin, collagen, and proteoglycans.[8,9] The tunica media, the middle layer, has a well-defined hierarchical structure of cells and extracellular matrix (collagen, elastin, and proteoglycans; ECM) driving arterial non-linear mechanical response.[8,10] Individual layers of the tunica media feature vascular smooth muscle cells (VSMCs) and collagen fibers that align helically down the length of the artery with successive layers aligned in distinct directions.[8,11] The tunica adventitia, the outermost layer, is composed of an ill-defined structure of densely woven collagen, VSMCs, fibroblasts and provides mechanical support to withstand super-physiological loads.[8]
At low strains, the arterial wall is compliant, but as strain increases, collagen fibers gradually begin to bear load and the artery wall stiffens.[4] Designing a tissue that recaptures this non-linear stress-strain response is essential for achieving long-term patency of engineered arterial tissue. Because the mechanical response is dominated by the mechanical properties of the tunica media, we believe that a tissue engineering strategy that recapitulates the micro-structural features of the medial layer is essential for constructing a replacement blood vessel that overcomes the limitations that plague current treatments[12].
Bottom-up tissue engineering techniques allow complex tissues to be constructed with control of the tissue micro-structure.[13,14] One strategy, known as cell sheet engineering, involves building three-dimensional tissues layer-by-layer from monolayers of cells and their integrated extracellular matrix.[15,16] Multiple techniques have been developed to promote the non-destructive detachment of cell sheets from culture including the use of thermo-responsive surfaces[17] and hydrogels[18] as well as enzymatically degradable substrates[19]. Thermo-responsive surfaces are typically modified with poly(N-isopropylacrylamide) (PNIPAAm) as the polymer undergoes a phase transition at its lower critical solution temperature (LCST) at 32 °C.[17,20] Specifically, the substrate surface is thought to be hydrophobic at 37 °C and transitions to hydrophilic as the temperature drops below 32 °C.[17] At 37 °C, cells can attach to PNIPAAm-coated surfaces and proliferate to form cell sheets – confluent monolayers of cells and extracellular matrix – that can be detached as intact sheets by reducing the temperature below the LCST (32 °C).[21] Okano et al. first demonstrated that cell sheets could be grown and harvested on thermo-responsive PNIPAAm-coated substrates after developing a method to graft PNIPAAm onto polystyrene tissue culture dishes.[21] Poly(dimethysiloxane) (PDMS) is an attractive alternative to polystyrene because it can easily be cast using micro-patterned silicon wafers, while remaining optically clear and inexpensive.[22] Moreover, the Young’s modulus of PDMS spans 5 kPa to 1.72 MPa covering a large, physiologically relevant range of elastic moduli.[23] Moreover, previous studies within our lab have studied the effect of PDMS substrate stiffness on VSMC behavior to show that decreased PDMS stiffness increased VSMC growth rate.[24] As a result of these advantages, significant effort has been devoted towards grafting NIPAAm onto PDMS.[25–29]
Many techniques have been developed to control cell sheet microstructure including the use of micro-contact printing[30], topographically patterned surfaces[31,32], and dynamic stretching.[33] Previous work within our lab has demonstrated that micro-patterned PDMS substrates induce VSMC alignment, enhance cellular F-actin alignment, and improve VSMC aspect ratio.[22] Although significant effort has been applied towards controlling cell sheet organization, the relationship between single-layer cell sheet structure and mechanical properties has been unexplored. Previous work within our lab has shown that VSMC sheets with structural alignment (cellular and extracellular) are 50% stiffer in the aligned direction compared to the perpendicular direction.[32] Because these cell sheets were cultured on micro-patterned PDMS with a layer-by-layer polyelectrolyte coating, they could not be selectively detached and had to be cultured until spontaneous detachment (8–10 weeks), resulting in thick (40 μm) cell sheets that were no longer single cell layers. Furthermore, cellular alignment throughout the cell sheet construct decreased in cells further from the micro-patterned PDMS surface. Based upon this result, we hypothesize that culturing cell sheets for shorter durations will result in uniformly organized monolayers that will eventually allow for biomimetic recapitulation of native arterial structure.
In this paper, we developed a robust method to culture vascular smooth muscle cell sheets using thermo-responsive PDMS substrates and then applied this method to produce cell sheets with structural and mechanical anisotropy. First, we computationally and experimentally optimized a method to photo-graft NIPAAm onto micro-patterned PDMS substrates. We further optimized the substrate surface properties to promote VSMC attachment and cell sheet formation by modifying the surface with 3-aminopropyltriethoxysilane (APTES). Moreover, we established a non-destructive method to quantify substrate quality and therefore assess the likelihood of successful tissue formation. Using this thermo-responsive cell sheet culture platform, single-layer, patterned VSMC sheets were cultured so that the cells and extracellular matrix were oriented in a preferential direction. Mechanical characterization of these sheets in the aligned and perpendicular directions revealed the tissue was significantly stiffer in the aligned direction, thus demonstrating mechanical anisotropy in single-layer, patterned cell sheets. This result demonstrates a clear relationship between the structure and mechanical properties of an aligned VSMC sheet, the basic, micro-structural unit of vasculature. Using the methods and findings presented here, researchers can design a biomimetic vascular graft using aligned VSMC sheets to engineer multi-layered vascular tissue with mechanical properties similar to native tissue.
2. Results and Discussion
2.1 Thermo-responsive PDMS Substrate Preparation
Thermo-responsive substrates for vascular smooth muscle cell sheet culture were prepared using the multi-step process shown in Figure 1. First, one side of flat or patterned PDMS substrates was soaked in a solution of benzophenone (BP), a UV photo-initiator (Figure 1a–b). Next, substrates were exposed to UV irradiation while coated in an aqueous solution of N-isopropylacrylamide monomer (Figure 1c). When exposed to UV irradiation, benzophenone abstracts hydrogen from methyl groups on the PDMS surface to create a free radical on the PDMS surface. This free radical initiates the polymerization of NIPAAm onto the PDMS surface.[34] To facilitate cell sheet formation, PNIPAAm-PDMS substrates were coated with 3-aminopropyltriethoxysilane and heated to 80 °C (Figure 1d).
Figure 1.
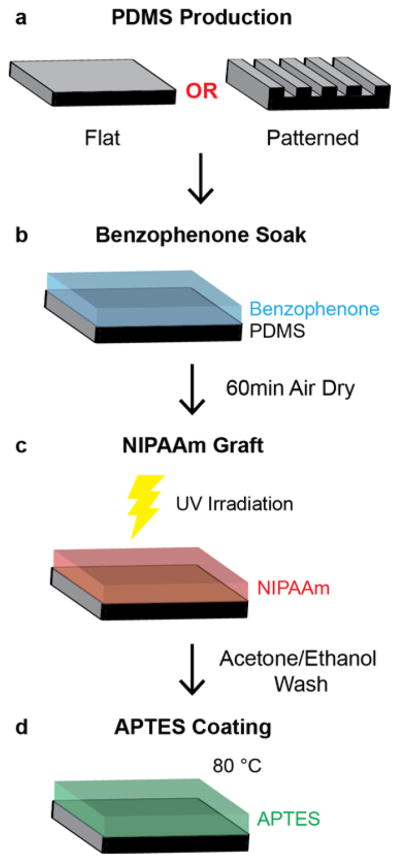
Schematic of thermo-responsive substrate preparation. (a) Flat and patterned PDMS substrates are prepared for subsequent grafting. (b) PDMS substrates are soaked in a 20% (w/v) benzophenone in acetone for 3 min and air dried for 60 min by exposing one side of the PDMS to benzophenone solution. (c) Benzophenone soaked PDMS substrates are then exposed to UV irradiation for 3 min while coated with a 20% aqueous solution of NIPAAm and then washed with acetone and ethanol. (d) PNIPAAm-PDMS substrates are then heated to 80 °C for 2 hours while coated with a 10% (v/v) solution of APTES in ethanol.
Developing a robust method to reliably culture VSMC sheets requires each step of the grafting process to be carefully controlled for optimal reliability. To control the reaction rate, we investigated the relationship between benzophenone soaking conditions and benzophenone concentration at the PDMS surface – where the free radical polymerization occurs. However, the concentration of benzophenone depends not only on the soaking conditions (time, solvent, concentration), but also the drying time because benzophenone diffuses into the PDMS bulk during drying causing the concentration of benzophenone at the surface to decrease over longer drying durations.[34] To understand the evolution of benzophenone concentration at the surface, we combined experimental and computational predictions (finite difference methods).
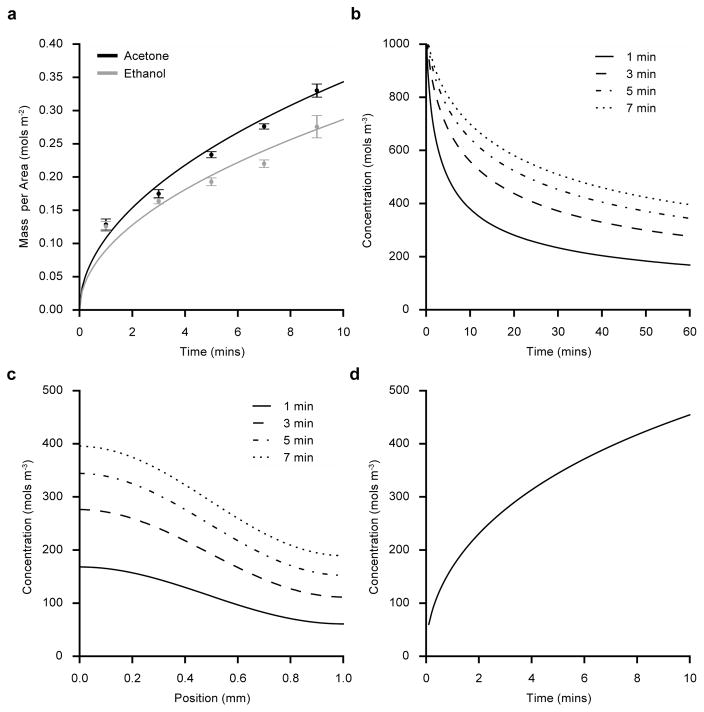
One side of dry PDMS substrates were soaked in 20% (w/v) benzophenone dissolved in either acetone or ethanol for varying durations (1–9 min) and the absorbed benzophenone mass per area was determined by measuring absorbance at 345 nm – the UV absorption peak of benzophenone.[34] In both solvents, PDMS rapidly absorbs benzophenone initially, however the rate of BP uptake slows substantially over time (Figure 2a). Interestingly, PDMS absorbs benzophenone at a faster rate when dissolved in acetone compared to ethanol. Although these experiments relate the benzophenone soaking conditions to the total benzophenone mass absorbed, they do not directly relate to the BP concentration at the PDMS surface. Benzophenone absorption was modeled as a diffusion process using the experimental data of the bulk absorption to predict BP surface concentration. First, an analytical solution to Fick’s law was applied to estimate the diffusion coefficient for benzophenone into PDMS (See Supporting Information for model implementation details). The diffusion model fits the experimental benzophenone absorption data (Figure 2a) using diffusion coefficients of 1.3 10−10 m2 s−1 or 0.9 10−10 m2 s−1 when dissolved in either acetone or ethanol, respectively.
Figure 2.
Benzophenone diffusion into PDMS substrates. (a) Mean experimentally measured benzophenone (BP) mass per area (± s.e.m.) absorbed by PDMS after soaking substrates in 20% BP for varying durations in ethanol (gray circles) or acetone (black circles) (N=4). Analytical prediction for absorbed BP mass when dissolved in ethanol (gray) or acetone (black) and soaked for 1–10 min. (b) Finite difference prediction of benzophenone surface concentration during air drying after soaking the PDMS substrates for 1–7 min in 20% BP (w/v) (acetone). (c) Predicted BP concentration at positions below the soaked surface of the PDMS substrates after soaking in BP (acetone) for 1–7 min and then drying for 60 min. (d) Predicted BP surface concentration after soaking PDMS substrates in BP (acetone) for 1–10 min and then drying for 60 min.
After soaking the substrates in benzophenone, the substrates are dried to evaporate solvent absorbed into the PDMS bulk (acetone or ethanol). During drying, benzophenone continues to diffuse into the PDMS bulk leading to a decrease in benzophenone concentration at the surface. Because benzophenone concentration at the surface dictates photo-grafting efficiency, we modeled the drying process in order to better understand the BP concentration at the surface – a quantity that could not easily be directly measured. To predict benzophenone concentration within the PDMS substrates during drying, the drying process was modeled using Fick’s law. Finite difference methods were employed to solve Fick’s law (see Supporting Information for details about the finite difference solution implementation) and the diffusion coefficient was assumed to be 3.0 10−11 m2 s−1 as reported elsewhere.[34] When PDMS substrates are soaked in 20% (w/v) BP in acetone, the model can predict the evolution of benzophenone surface concentration during drying (Figure 2b). For all soak times, the surface concentration rapidly drops initially as benzophenone diffuses into the PDMS bulk, but slowly approaches a constant surface concentration. After a 60 min dry time, the surface concentration continues to slowly decrease. The benzophenone concentration profile through the thickness of the PDMS substrates after a 60 min dry interval is plotted in Figure 2c. At positions near the PDMS surface, the predicted concentration of benzophenone is relatively constant and eventually drops near the middle. Finally, the model predicts a relationship between benzophenone surface concentration and soak time as shown in Figure 2d. As expected, increased soaking time results in increased surface concentration in a relationship that mirrors the experimental data in Figure 2a. The diffusion model results demonstrate how benzophenone surface concentration can be controlled by modifying the soaking time of benzophenone.
The experimental results reveal that benzophenone diffuses into PDMS faster when dissolved in acetone than when dissolved in ethanol, which is consistent with differences in the diffusion rate of pure ethanol or acetone into PDMS[34]. Schneider et al. also measured the diffusion rate of benzophenone into PDMS and their measurements were similar but smaller (1.0 10−11 m2 s−1 for benzophenone dissolved in acetone compared with our measurement of 1.3 10−10 m2 s−1). However, the difference can be attributed to alterations in experimental conditions; for the experiments in this paper, PDMS substrates were dry before being exposed to benzophenone solution, while Schneider et al. soaked PDMS substrates with acetone before exposing the substrates to benzophenone solution. During drying, benzophenone concentration slowly approaches a constant surface concentration (Figure 2b). However, after drying for 60 min, the benzophenone surface concentration changes slowly, making it ideal; small variations in dry time lead to minimal changes in benzophenone surface concentration. Because oxygen quenches benzophenone initiated reactions, the benzophenone concentration profile shown in Figure 2c is ideal since it buffers the reaction site from oxygen diffused in the PDMS.[34,35] The development and experimental validation of a diffusion model of benzophenone provided insights that allowed robust control of the grafting process to ensure consistent substrates.
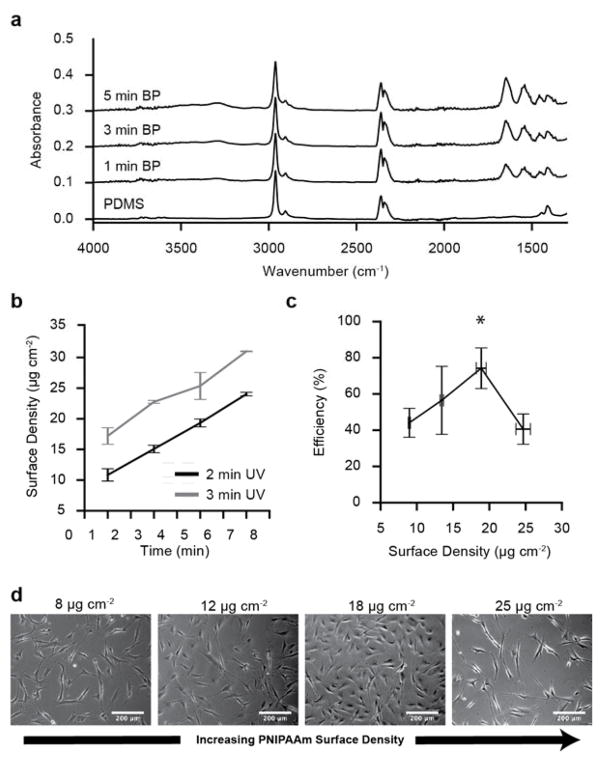
To understand the relationship between benzophenone surface concentration and PNIPAAm graft density (mass per surface area) after UV exposure, PDMS substrates were soaked in benzophenone for varying durations (1–7 min) and then irradiated with UV light for either 2 or 3 min while coated with a 20% N-isopropylacrylamide monomer solution. Attenuated total reflection Fourier transformed infrared (FT-IR) spectroscopy measurements confirmed grafting of NIPAAm onto PDMS (Figure 3a). Bare PDMS and PNIPAAm-PDMS substrates have FT-IR absorption peaks near 2900 cm−1, corresponding to C-H stretching within the methyl groups of the PDMS. The PNIPAAm-PDMS spectra confirms the presence of peaks at 1650 cm−1 and 1540 cm−1, which correspond to the C=O stretching and N-H bending within PNIPAAm that are not present for bare PDMS. Moreover, the broad peak at 3300 cm−1 corresponds to the N-H stretching within the PNIPAAm. FT-IR absorption spectra show a clear increase PNIPAAm absorbance peak heights (1650 cm−1 and 1540 cm−1) as the benzophenone soak time increases, while the Si-CH2 of the PDMS remains unchanged (1019 cm−1). Using the ratio of the 1650 cm−1 PNIPAAm amide peak to the 1019 cm−1 PDMS Si-CH2 peak, PNIPAAm graft density (μg cm−2) on the PDMS was quantified. PNIPAAm graft density increases with benzophenone soak time for UV irradiation times of 2 and 3 min (Figure 3b). The results demonstrate that modulating benzophenone soaking time and UV irradiation time provide significant control over PNIPAAm graft density.
Figure 3.
Fourier transformed infrared spectra and cell attachment efficiency of PNIPAAm-PDMS substrates. (a) FT-IR absorbance spectra for PNIPAAm-PDMS substrates soaked in BP (acetone) for 0–5 min and then grafted with 3 min UV exposure. (b) Mean PNIPAAm surface density (± s.e.m.) for substrates soaked in BP for varying durations (1–7 min) and then PNIPAAm grafted with 2 or 3 min UV exposure (N=5). (c) Mean vascular smooth muscle cell seeding efficiency (± s.d.) for PNIPAAm-PDMS substrates with varying PNIPAAm surface density (N=3). (d) Phase contrast microscopy representative images of vascular smooth muscle cell attachment on PNIPAAm-PDMS with different graft densities taken 18 hours after seeding.
Using the ability to control PNIPAAm surface density, we investigated how grafting density affected vascular smooth muscle cell attachment. VSMCs were seeded onto flat PNIPAAm-PDMS substrates in four graft density ranges (N=3), and cell attachment efficiency was measured at multiple positions on each substrate. Cell attachment quantification reveals that PNIPAAm-PDMS substrates with PNIPAAM surface densities around 18 μg cm−2 have the highest seeding efficiency compared to the other grafting conditions tested (p < 0.01; ANOVA). Phase contrast images of VSMCs adhered to PNIPAAm-PDMS substrates demonstrate that VSMC attachment and cell spreading also depends on surface graft density (Figure 3d). At low (~ 8 μg cm−2) and high graft densities (~ 25 μg cm−2), VSMCs have low adherence to the surface, while near the optimal graft density (~18 μg cm−2) VSMCs adhere to the surface and maintain a more spread out morphology. VSMCs do not adhere to bare PDMS (0 μg cm−2) and PNIPAAm-PDMS substrates with graft densities greater than 25 μg cm−2.
The relationship between PNIPAAm surface density and cell attachment has been extensively studied in other PNIPAAm based cell culture systems.[21,36] These studies demonstrated that increasing PNIPAAm graft density above the optimal level completely inhibits successful cell attachment and that the optimal PNIPAAm surface is cell type dependent.[21,31] Interestingly, the optimal graft density for the PNIPAAm-PDMS substrates presented here is approximately 18 μg cm−2, an order of magnitude higher than optimal grafting densities for photo-grafting PNIPAAm onto polystyrene (~1.4 μg cm−2).[21] Another study where PNIPAAm was grafted onto PDMS found the optimal graft density to be 11.9 μg cm−2 – but this was the highest graft density studied – thus, the optimal graft density could have occurred at a higher surface density.[29] While the differences are significant, they can be attributed to fundamental differences in the polymer architecture of the grafted PNIPAAm surface that are achieved through the different polymerization method for producing PNIPAAm surfaces (electron beam irradiation[17], RAFT polymerization[37], self assembled monolayers[38]). Moreover, surface chemistry of the underlying substrate – PDMS and polystyrene – has been shown to modulate cell attachment and protein adsorption and could possibly explain these differences.[38]
Because the reaction that grafts PNIPAAm onto PDMS is a free-radical polymerization, the process is sensitive to environmental conditions such as temperature, and oxygen concentration.[39] Furthermore, benzophenone surface concentration variability and heterogeneous UV irradiation are sources of processing variability that can be minimized, but not completely eliminated. Therefore, it was essential to design the process to minimize these sources of variability by computationally modeling key components of the grafting process and relating processing parameters to experimental characterization. Moreover, identifying the optimal PNIPAAm surface density allows for rapid quantification of substrate quality (optimal graft density and graft density spatial variability) using FT-IR, a non-destructive method.
2.2 APTES Modification of Thermo-responsive Substrates
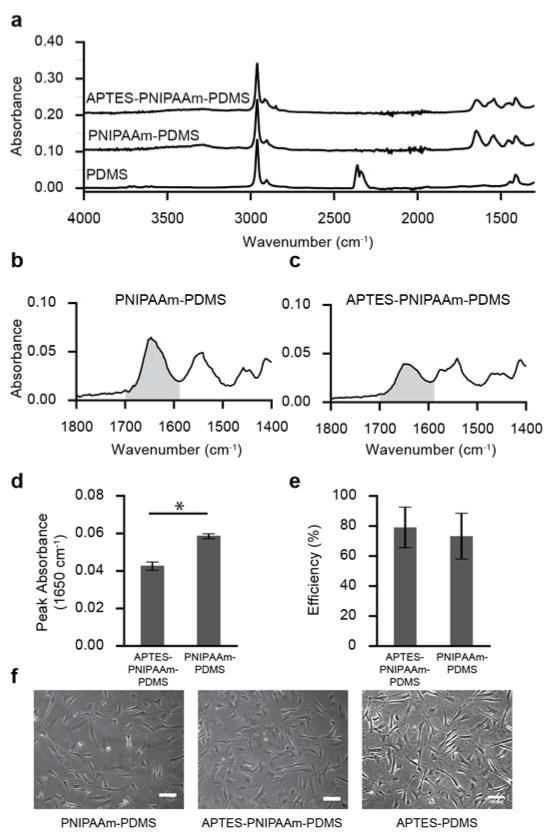
While PNIPAAm graft density was optimized for VSMC attachment, the cell sheet formation yield was considerably low. While possible to grow cell sheets on the PNIPAAm grafted PDMS, VSMC sheets would often spontaneously develop holes, clump or spontaneously detach during longer culture durations (see Figure S2 in the Supporting Information). Surface modification with 3-aminopropyltriethoxysilane has been shown to improve cell attachment by adding extra primary amine groups to the substrate surface thus facilitating cell attachment in other systems.[40,41] To improve the cell sheet yield, we developed a method to incorporate APTES into the surface of the PNIPAAm-PDMS substrate. PNIPAAm-PDMS substrates were baked in 10% (v/v) APTES in ethanol. APTES coated PNIPAAm-PDMS substrates show clear differences in the FT-IR absorption spectra (Figure 4a). FT-IR absorbance for the C=O stretching peak height decreases after APTES treatment, but also becomes broader (Figure 4b,c). The PNIPAAm C=O stretching peak (1650 cm−1) absorbance decreases from 0.058 ± 0.0005 to 0.042 ± 0.001 (p < 0.01; t-test) after APTES modification (Figure 4d).
Figure 4.
Fourier transformed infrared spectroscopy and cell attachment efficiency for APTES-PNIPAAm-PDMS substrates. (a) FT-IR absorption spectra for PDMS, PNIPAAm-PDMS and APTES-PNIPAAm-PDMS substrates. (b) Absorbance spectra of the C=O stretching bond of an APTES-PNIPAAm-PDMS substrate. Grey region corresponds to wavenumbers where the area under the curve was calculated in panel e. (c) Absorbance spectra of the C=O stretching bond of an APTES-PNIPAAm-PDMS substrate. (d) Peak FT-IR absorbance of the 1650 cm−1 peak for PNIPAAm-PDMS and APTES-PNIPAAm-PDMS substrates (N=3). (e) Mean cell seeding efficiency (± s.d.) for VSMCs seeded onto PNIPAAm-PDMS and APTES-PNIPAAm-PDMS surfaces (N=3; p>0.05; t-test). (f) VSMC attachment on PNIPAAm-PDMS APTES-PDMS and PNIPAAm-APTES-PDMS substrates 18 hours after seeding substrates with 15k cells cm−2.
Both PNIPAAm-PDMS and APTES-PNIPAAm-PDMS substrates were seeded with VSMCs to determine whether APTES modification improved cell attachment. Interestingly, mean VSMC cell attachment efficiency is slightly higher for APTES-PNIPAAm substrates, although the difference is not significant (p=0.36; t-test) (Figure 4f). However, VSMCs cultured on APTES-PNIPAAm-PDMS (Figure 4h) have a more spread out morphology compared to VSMCs seeded on PNIPAAm only (Figure 4g). Most importantly, VSMCs seeded on APTES-PNIPAAm-PDMS substrates form cell sheets at a higher rate than PNIPAAm-PDMS substrates. For example, when directly comparing substrates, VSMCs seeded on APTES-PNIPAAm-PDMS substrates for cell sheets 81% of the time while PNIPAAm-PDMS substrates failed to produce an intact, confluent sheet (p < 0.01; Fisher’s Exact Test; N=16).
Sessile drop water contact angle measurements on both PNIPAAm-PDMS and APTES-PNIPAAm-PDMS substrates confirmed that APTES treatment had no effect on the wetting behavior of the surfaces. Statistical analysis of PNIPAAm-PDMS substrates before and after surface treatment with APTES shows that there are no significant differences in water contact angle at 42 °C and 22 °C (p > 0.05; ANOVA). Water contact angle for PNIPAAm-PDMS substrates have a mean (± s.e.m) water contact angle of 106.5° ± 0.5 at 42 °C that drops to 92.5° ± 1.4 at 22 °C (N=8; p < 0.01; t-test) and APTES-PNIPAAm-PDMS substrates exhibit a drop in mean water contact angle from 102.4° ± 1.1 at 42 °C to 92.5° ± 3.3 at 22 °C (N=8; p < 0.05; t-test).
While APTES modification of PNIPAAm-PDMS surfaces does not alter the rate of VSMC attachment, it does modulate the resulting morphology of attached VSMCs. When seeded onto PNIPAAm-PDMS substrates, cells appear to have low adherence to the surface, while VSMCs on APTES-PNIPAAm-PDMS appear to attach with a more natural morphology. However, more importantly, APTES modification improves the rate of cell sheet formation. While PNIPAAm-PDMS substrates were capable of culturing VSMC cell sheets, APTES treatment greatly increased the success rate of cell sheet formation and successful long-term culture. Unlike PNIPAAm-PDMS surfaces, VSMCs adhere to APTES-PNIPAAm-PDMS surfaces without having to adsorb serum proteins onto the substrate surface before seeding cells, thus simplifying the culture process and removing a potential source for variability.
Beyond the functional improvements towards cell sheet formation, APTES treatment of PNIPAAm-PDMS substrates causes significant changes to the FT-IR absorption spectra that reveal how APTES incorporates into the PNIPAAm-PDMS surface. Interestingly, there is a drop in the 1650 cm−1 amide peak after APTES treatment. Because NIPAAm covalently binds to PDMS, heating PNIPAAm-PDMS substrates in APTES will not reduce the PNIPAAm surface density.[27] Therefore, decreased FT-IR peak absorbance within the amide groups of PNIPAAm-PDMS substrates after APTES treatment indicates structural changes such as the formation of hydrogen bonds between the C=O (1650 cm−1) of the PNIPAAm and N-H of the APTES.[42] This result is consistent with another PNIPAAm based cell culture platform where a mixture of polymerized PNIPAAm and APTES was spin-coated to form a thermo-responsive, glass based APTES-PNIPAAm cell culture platform.[42] In this system, the APTES forms an inner-penetrating network with the PNIPAAm chains via hydrogen bonding.[42]
2.3 Vascular Smooth Muscle Cell Sheet Structure and Mechanics
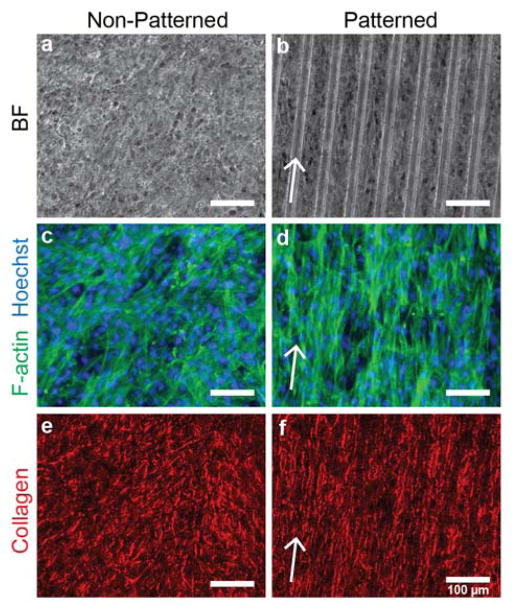
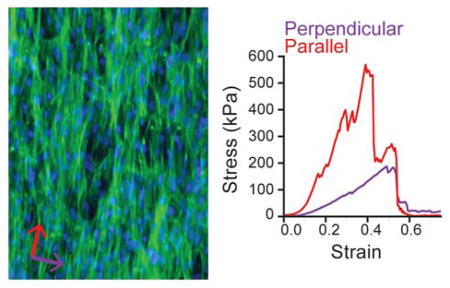
Patterned and non-patterned vascular smooth muscle cell sheets were cultured using the thermo-responsive APTES-PNIPAAm-PDMS cell culture platform. VSMC sheets grown on patterned substrates exhibit clear structural differences compared to sheets grown on non-patterned substrates (Figure 5). Phase contrast microscopy images of patterned cell sheets show alignment in the direction of the micro-pattern (Figure 5a,b). Fluorescent images of the F-actin structure of both patterned and non-patterned substrates show preferential alignment compared to the random organization of the F-actin structure of non-patterned cell sheets (Figure 5c,d). Furthermore, the cell-derived collagenous extracellular matrix shows similar alignment in the cell sheets grown on patterned surfaces, while the collagen structure shows no alignment in the non-patterned cell sheets (Figure 5e,f). Interestingly, the underlying micro-pattern does not influence the concentration of collagen in the extracellular matrix. After 12 days in culture, VSMC sheets cultured on non-patterned substrates have a mean (± s.d.) pepsin-soluble collagen content per wet mass of 9.3 ± 1.7 μg mg−1 compared to 9.4 ± 1.1 μg mg−1 for sheets grown on patterned substrates, indicating that the substrate topography does not impact VSMC collagen synthesis (p = 0.86; t-test). Vascular smooth muscle cell sheets grown on micro-patterned, thermo-responsive substrates have clear cellular and extra-cellular alignment when compared to cells grown on non-patterned VSMC sheets, achieving results similar to other studies using VSMCs.[22,30,32] Interestingly, substrate topography does not modulate collagen synthesis, which is consistent with our previous findings.[32] The combination of these two results is significant: any differences in mechanical properties between patterned and non-patterned VSMC sheets can be attributed to cell sheet structure and not composition.
Figure 5.
Vascular smooth muscle cell sheet structure on non-patterned and patterned substrates. (a,b) Phase contrast images of vascular smooth muscle cell sheets grown on non-patterned and patterned substrates for 17 days. (c,d) F-actin structure for non-patterned and patterned VSMC sheets (green). Cell nuclei are labeled with Hoechst (blue). (e,f) Collagen structure (red) of VSMC sheets after picrosirius red stain. Arrows correspond to direction of micro-pattern. Scale bar indicates 100 μm.
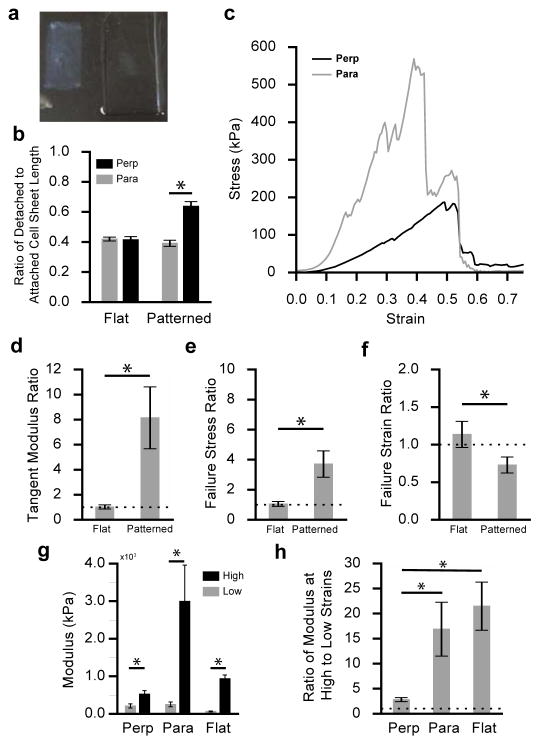
To determine the relationship between VSMC sheet structure and mechanical properties, vascular smooth muscle cell sheets were cultured for 17–21 days on patterned and non-patterned substrates, detached, and then mechanically characterized. Upon detachment from the thermo-responsive substrates into solution, vascular smooth muscle cell sheets contract (Figure 6a). The ratio of detached VSMC sheet geometry (length & width) to attached VSMC geometry quantifies cell sheet shrinkage after detachment. A small ratio indicates increased contraction and a ratio of 1 would mean the VSMC sheet did not contract. Patterned and non-patterned cell sheets contract differently when detached (Figure 6b). As expected, the non-patterned substrates contract equally in both directions (p > 0.05; t-test), while patterned cell sheets contract more in the direction of alignment compared to the transverse direction (perpendicular to alignment). Interestingly, there is no difference in the amount of contraction between aligned VSMC sheets in the direction of alignment compared to non-patterned cell sheets (p=0.3; t-test) whereas patterned VSMC sheets contract less in the perpendicular direction compared to non-patterned cell sheet contraction (p < 0.001; t-test).
Figure 6.
Vascular smooth muscle cell sheet mechanical characterization. (a) Detached non-patterned vascular smooth muscle cell sheet (left) next to APTES-PNIPAAm-PDMS substrate (right). (b) Ratio of detached VSMC sheet geometry (length and width) to attached VSMC sheet geometry in the parallel (gray) and perpendicular (black) direction for flat and patterned sheets (N=9). (c) Engineering stress-strain curve for a patterned VSMC sheet stretched in the aligned (gray) and perpendicular (black) direction. (d) Mean ratio of failure stress (± s.e.m) in the aligned direction compared to the perpendicular direction for patterned and flat VSMC sheets (N=8; p < 0.05; t-test). (e) Mean ratio of failure strain (± s.e.m; N=8) in the aligned direction to the perpendicular direction for patterned and flat VSMC sheets (p < 0.05; t-test). (f) Mean ratio of tangent modulus (± s.e.m) in the aligned direction compared to the perpendicular direction for patterned and flat VSMC sheets (N=8; p < 0.05; t-test). (g) Mean VSMC sheet modulus (± s.e.m) at low strains (0–15% strain) and high strains (maximum tangent modulus before failure) in the perpendicular (N = 8) and parallel directions (N= 8) of aligned sheets and flat, cell sheets (N=8) (p < 0.05; t-test). (h) Mean ratio (± s.e.m) of the modulus measured at high strains to low strains for VSMC sheets in the parallel (N=8) and perpendicular (N=8) direction for patterned sheets and non-patterned sheets (N=8); (p < 0.05; t-test).
To quantify VSMC sheet mechanical anisotropy, patterned and non-patterned sheets were mechanically characterized using a custom uniaxial tensile tester.[43] A stress-strain curve for a patterned VSMC sheet tested in both directions – parallel and perpendicular to alignment – shows increased stiffness in the aligned direction (Figure 6c). The results from the mechanical characterization are summarized in Table 1. Aligned VSMC sheets have a mean stiffness of approximately 3000 kPa and 500 kPa in the aligned and perpendicular directions, respectively, compared to non-patterned sheets that have a mean modulus of approximately 1000 kPa. Correspondingly, patterned sheets failed at higher stresses in the direction of alignment than both the perpendicular direction and the non-patterned sheets. Patterned VSMC sheets had a lower mean failure strain of 0.43 ± 0.1 in the direction of alignment compared to 0.50 ± 0.1 in the perpendicular direction. Interestingly, non-patterned sheets failed at higher strains (0.75 ± 0.05) than patterned sheets in either direction.
Table 1.
Mechanical properties of patterned and non-patterned VSMC sheets (mean ± s.e.m)
| Low Strain Modulus (kPa) | Maximum Modulus (kPa) | Failure Stress (kPa) | Failure Strain | |
|---|---|---|---|---|
| Parallel | 250 ± 65 | 2995 ± 961 | 410 ± 140 | 0.43 ± 0.1 |
| Perpendicular | 208 ± 56 | 522 ± 93 | 155 ± 40 | 0.50 ± 0.06 |
| Non-Patterned | 250 ± 65 | 932 ± 102 | 225 ± 28.5 | 0.75 ± 0.05 |
The ratio of the mechanical properties in the aligned direction relative to the perpendicular direction was used to quantify the degree of mechanical anisotropy in both patterned and non-patterned cell sheets (Figure 6d,e,f). Patterned sheets exhibited an 8.1x increase in stiffness and a 3.7x increase in failure stress in the aligned direction compared to the perpendicular direction (Figure 6d,e). Interestingly, patterned sheets failed at lower strains (0.73x decrease) in the aligned direction compared to the perpendicular direction (Figure 6f). Non-patterned VSMC sheet mechanical properties showed no dependence on direction, as expected given their structural isotropy.
To quantify the nonlinearity of VSMC sheets, the modulus was calculated at low strains (0–15%) and compared to the maximum tangent modulus at high strains (Figure 6g). Interestingly, VSMC sheet modulus increased significantly at higher strains for both non-patterned and patterned sheets (both directions). Patterned sheets stiffen significantly in the aligned direction, increasing from 250 kPa to 3000 kPa, while patterned sheets stiffen modestly in the perpendicular direction, increasing from 208 kPa to 522 kPa. Interestingly, the modulus at low strains for patterned sheets in the aligned and perpendicular directions are not significantly different (p = 0.62; t-test), while the initial modulus for non-patterned sheets is significantly lower than patterned sheets in either direction (p < 0.05; t-test). The ratio of the modulus at high strains to modulus at low strains quantifies the stiffening the sheets experience when stretched to high strains (Figure 6h). For aligned sheets, the stiffness exhibited a 3x increase the perpendicular direction, while exhibiting a much larger 17x increase in the parallel direction (p < 0.05; t-test). Non-patterned sheets exhibited a 21x increase in modulus, which was similar to the increase exhibited by patterned sheets in the aligned direction (p>0.05; t-test). Patterned and non-patterned VSMC sheets both exhibit non-linear mechanical behavior, an essential component of the mechanical response of native vascular tissue. Moreover, the nonlinear mechanical response of aligned VSMC sheets is more pronounced in the aligned direction compared to the perpendicular direction. The nonlinear, anisotropic mechanical response makes aligned VSMC sheets an attractive material for engineering multi-layered vascular tissue that mimics the complex mechanical behavior of native arterial tissue.
The APTES-PNIPAAm-PDMS cell sheet culture platform enables controlled detachment of patterned and non-patterned VSMC sheets at short culture durations to overcome the limitations of earlier research efforts. Previously, patterned and non-patterned VSMC sheets were cultured on PDMS substrates, where controlled detachment was not possible and the sheets were cultured until spontaneous detachment (8–10 wks).[32] As a result of long culture durations, the cell sheets were multiple cells thick; patterned cell sheets didn’t maintain alignment when the sheets grew thicker and in turn only exhibited a modest 1.5x increase in stiffness in the direction of alignment. Using this novel, thermo-responsive cell sheet culture platform to detach aligned VSMC sheets at earlier culture durations, cell sheets were detached as single-layers and retain preferential alignment. Moreover, their uniform alignment is reflected in the mechanical measurements: aligned VSMC sheets exhibit an 8x increase in stiffness in the aligned direction compared to the perpendicular direction. Furthermore, aligned cell sheets grown for shorter time durations are more compliant than sheets grown for longer durations.
Single-layer VSMC sheets exhibit a stress-strain response that shares phenomenological similarities to native vascular tissue where the material is compliant at low strains and then rapidly stiffens at high strains.[4] Having this ideal, non-linear stress-strain response is an essential feature for the underlying, micro-structural unit of a tissue-engineered vascular graft that mimics native mechanical properties. This non-linear stress-strain response is often reduced to one number, the tangent modulus. While this greatly oversimplifies the complex stress-strain behavior, it enables one to compare the properties of tissue-engineered biomaterials to native tissue. Importantly we observe that the tangent moduli of our patterned VSMC sheets are comparable to native arterial tissue. For example, native human coronary arteries have an elastic modulus in the range of 1–4 MPa whereas our patterned VSMC sheets have a moduli of approximately 3 MPa.[44] The observation that we observe physiologically relevant tangent moduli in our single-layer VSMC sheets supports our proposed approach for constructing thicker arterial tissue by stacking multiple patterned VSMC sheets. It is interesting to note that compared to other tissue-engineered vascular tissue, our VSMC sheets have tangent moduli smaller than aligned VSMC sheets cultured for longer durations (8–10 weeks) as well as anisotropic, self-assembled engineered tissue constructs grown using fibroblasts.[32,45]
While VSMC sheets have structural and mechanical features that make them an attractive material for engineering a vascular graft, VSMC sheets exhibit morphological differences from native vasculature. Although individual layers within the tunica media feature aligned collagen, native collagen fibers found in the tunica media are crimped and straighten when exposed to physiological loads.[4] In contrast, collagen produced by VSMC sheets is not crimped, nor a mature collagen fiber (Figure 5e,f). Additionally, fibrillar elastin found within the tunica media give the arterial tissue their compliance and elasticity.[46] However, mature VSMCs produce globular elastin, not fibrillar elastin, and as a result the sheets possess minimal elasticity and plastically deform at high strains.[32] Despite the differences between VSMC sheets and native tissue, a multi-layered vascular graft could be constructed by stacking aligned VSMC sheets with an additional elastic layer to engineer a graft that matches the nonlinear, elastic stress-strain response of native vascular tissue. Additionally, to prevent thrombosis and allow the transmission of signal cues to VSMCs, any vascular graft would need to have a blood contacting endothelial layer that mimics the tunica intima. [8,9] Lastly, a vascular graft would also need the support of an adventitial layer for additional mechanical support as well as providing nutrients to the medial layer via the vasa vasorum.[8]
Although VSMC sheets exhibit structural and mechanical properties suited for vascular tissue, further work is needed to successfully build engineered vascular tissue. Because the mechanical response is driven by cell-derived extracellular matrix, VSMC sheets grown under different conditions (e.g. shorter culture duration, no ascorbic acid treatment, etc.) will secrete ECM with different compositions and potentially have more physiologically tuned stiffness.[47] For example, as previously stated, mature VSMC sheets do not produce mature, fibrillar elastin, the sheets deform inelastically at high strain.[46] Therefore, more work is needed to understand the relationship between cell sheet culture and their micro-structural properties, including identifying and controlling sources for variations in mechanical and structural properties. Although aligned, single-layer, VSMC sheets possess desirable micro-structural mechanical properties, the link between micro and macro-structural properties is complicated and further fundamental work is needed to predict tissue level properties from micro-structural properties.[48]
3. Conclusions
In this study, we developed a thermo-responsive cell culture platform to produce structurally and mechanically anisotropic vascular smooth muscle cell sheets. To accomplish this, we computationally and experimentally optimized a method for grafting NIPAAm onto micro-patterned PDMS substrates to achieve optimal VSMC attachment. Next, the PNIPAAm-PDMS surfaces were adapted by incorporating a secondary network of polymerized APTES to improve VSMC adhesion and cell sheet formation rate. Structural and mechanical characterization of patterned and non-patterned VSMC sheets revealed significant mechanical and structural anisotropy, and patterned sheets were approximately 8x stiffer in the aligned direction compared to the perpendicular direction. These properties make this tissue an attractive micro-structural building block for engineering multi-layered vascular tissue. Moreover, within the context of functional tissue engineering, the results represent initial steps towards designing tissue-engineered materials with well-defined micro- and macro-structural properties.
4. Experimental Section
PDMS Substrate Preparation
Sylgard 184 poly-dimethylsiloxane (PDMS) substrates were prepared by mixing base and curing agent in a 10:1 ratio (Dow Corning, Midland, MI). The PDMS mixture was degassed and cured at 80 °C for 12 hours in either a petri dish or on a micro-patterned silicon wafer to produce 1 mm thick flat and patterned PDMS, respectively. Micro-patterned silicon wafers were prepared to produce PDMS substrates with 5 μm deep grooves that are 50 μm wide with 20 μm wide ridges as previously described.[49]
Thermo-responsive Substrate Preparation
One side of the PDMS substrates was soaked in a solution of 20% (w/v) benzophenone in acetone for 3 min. PDMS substrates were then rinsed with excess deionized (DI) water and dried for 60 min at room temperature. N-isopropylacrylamide (NIPAAm) solution composed of 20% (w/v) N-isopropylacrylamide (Acros Organics, Geel, Belgium), 0.5% benzyl alcohol (Sigma), and 5 mM sodium periodate (Sigma) was degassed under house vacuum for 2 hours. Benzyl alcohol allows the aqueous monomer solution to come into contact with the hydrophobic PDMS surface and also functions as a chain transfer agent.[50] Sodium periodate acts as an oxygen scavenger to prevent dissolved oxygen from inhibiting benzophenone excitation and NIPAAm polymerization.[51] Substrates were then coated with the NIPAAm solution, placed in an air-tight chamber, and purged of oxygen by flowing nitrogen for 1 min. Benzophenone soaked PDMS substrates were exposed to 55 mW cm−2 (measured at 365 nm) UV irradiation for 3 min. PNIPAAm-coated PDMS (PNIPAAm-PDMS) substrates were then rinsed in DI water to remove excess PNIPAAm monomer and soaked in baths of acetone and ethanol for 12 hours each to extract absorbed benzophenone.
APTES Modification of Thermo-responsive PDMS
Thermo-responsive PNIPAAm-PDMS substrates were modified with 3-aminopropyltriethoxysilane (APTES; Sigma). APTES solution was composed of 10% APTES (v/v) dissolved in 200-proof ethanol and activated with 0.5% (v/v) DI water. Substrates were coated with 10% APTES solution and baked at 80 °C for 2 hours under vacuum to incorporate APTES into the PNIPAAm-PDMS surface. After APTES surface modification, the substrates were soaked in DI water for 48 hours to remove unreacted APTES monomer.
Benzophenone Diffusion Model
The mass of benzophenone soaked into the PDMS substrates was estimated by measuring UV absorbance at 345 nm – peak UV absorbance for benzophenone – with a SpectraMax M5 plate reader (Molecular Devices, Sunnyvale, CA). To convert absorbance to benzophenone mass, a calibration curve was experimentally measured. PDMS substrates were soaked with known volumes of benzophenone solution of predetermined concentrations (N=5) to relate benzophenone mass to UV absorbance (345 nm). The relationship between UV absorbance and benzophenone mass was modeled using linear model using R (R Foundation for Statistical Computing, Vienna, Austria). PDMS substrates were soaked in 20% (w/v) benzophenone dissolved in either ethanol or acetone for 1–9 min by exposing only one side of the substrate to solution (N=4). Using the standard curve, UV absorbance measurements estimated benzophenone mass.
The experimental data was fit using a 1-d analytical solution to Fick’s law to determine the diffusion coefficient. Using the analytical benzophenone concentration profile, the drying process was modeled using a finite difference approach and 3.0 × 10−11 m2 s−1 as the diffusion coefficient.[34] During drying, all PDMS surfaces were modeled as a zero-flux boundary condition (dC/dx = 0).
Substrate Characterization
Attenuated total reflection, Fourier-transform infrared (FT-IR) absorbance spectra were acquired using a Nicolet (Thermo-Fisher, Waltham, MA) spectrometer with a resolution of 2.0 cm−1 over the range of 1000 – 4000 cm−1 collected using OMNIC (Version 7.2a). Sessile drop water contact angle measurements were performed on a Krüss DSA100 Goniometer (Krüss GmbH, Hamburg, Germany) within a temperature controlled chamber controlled by a Polyscience Digital Temperature Controller (Polyscience, Niles, IL) using Drop Shape Analysis (DSA) for Windows (Version 1.90.0.14). Substrates were placed directly on the surface of the heating block and allowed to reach a constant temperature before water contact angle measurements were taken (N=8). Sessile drop water contact angle images were taken using Drop Shape Analysis and analyzed using the Tangent-2 method to extract water contact angle.
Cell Sheet Culture
Bovine aortic vascular smooth muscle cells (VSMCs) (Coriell Cell Repositories, Camden, NJ) tested negative for mycoplasma and were cultured in low glucose Dulbecco’s Modified Eagle Medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 100 units mL−1 of penicillin, 100 μg mL−1 streptomycin (Invitrogen), and 2 mM L-Glutamine (Invitrogen). VSMCs (Passage 8–12) were seeded at a density of 45,000 cells cm−2 and 60,000 cells cm−2 onto flat and patterned thermo-responsive substrates, respectively. Different seeding densities ensured that flat and patterned cell sheets would concurrently reach confluence. After 4 days, culture medium was supplemented daily with 50 μg mL−1 L-ascorbic acid (Sigma, St. Louis, MO) for the culture duration.
For the cell attachment studies, PNIPAAm-PDMS substrates were prepared by soaking the substrates in VSMC medium for 2 hours prior to cell seeding to allow serum protein adsorption, while cells were seeded directly onto APTES-PNIPAAm-PDMS substrates without adsorbing serum proteins. VSMCs were seeded onto both PNIPAAm-PDMS and APTES-PNIPAAm-PDMS substrates with a density of 15,000 cells cm−2. Phase contrast microscopy images of cell attachment were taken 18 hours after seeding on an Axiovert S100 microscope (Zeiss) at random locations for each graft density (N=3) at random locations (n=5). Cell attachment efficiency was found by calculating the ratio of mean number of adherent cells per cm−2 to cell seeding density (15,000 cells cm−2).
Cell Sheet Imaging and Collagen Quantification
VSMC sheets were rinsed with PBS, fixed in 4% paraformaldehyde and then permeabilized with 0.1% Triton X-100 (Sigma) in PBS. Cell sheets were then blocked with 1% BSA in PBS for 1 hour and then stained with FITC-phalloidin (Life Technologies, Grand Island, NY) and Hoechst (Life Technologies) to visualize F-actin and cell nuclei, respectively. For collagen staining, VSMC sheets were fixed with 4% paraformaldehyde, rinsed with deionized water, and then incubated in 0.1% picrosirius red dye (Electron Microscopy Sciences, Hatfield, PA). VSMC sheets were then rinsed in 0.5% acetic acid and then dehydrated in baths of ethanol and xylene. Stained VSMC sheets were imaged using an Axiovert S100 microscope. For collagen quantification, VSMC sheets were incubated at 4 °C in 0.1 mg mL−1 of porcine pepsin (Sigma) in 0.5 M acetic acid for 24 hours before measuring collagen content using the Sircol Collagen Assay (Biocolor, UK).
Cell Sheet Mechanical Characterization
Cell sheets were cultured for 17–21 days before mechanical characterization. Cells were transferred onto a gelatin mold composed of 7.5% (w/v) Type A bovine gelatin (Sigma) dissolved into low glucose DMEM by incubating the cell sheet for 20 min at 4 °C. Cell sheets were cut into strips in both the direction of alignment and the direction perpendicular to alignment, while non-patterned cell sheet strips were cut in arbitrary directions. Length and width of cell sheet strips were measured with calipers, and thickness was measured using an Axiovert S100 microscope (Karl Zeiss, Germany). Cell sheet strips were glued to mounts using cyanoacrylate adhesive (Loctite Instant-Bonding Adhesive, #414; Henkel Corporation, Westlake, OH) and then mounted in the tensile tester. Cell sheets were stretched to the zero-strain state by manually stretching the sheets until they began to bear load. Cell sheets were mechanically characterized by subjecting strips to 3 pre-stress load cycles to an engineering strain of 0.2 before stretching the cell sheet to failure at a strain rate of 0.05 s−1. Failure stress and strain were measured, and Young’s modulus was determined over the linear regime of the stress-strain curve.
Statistical Analysis
Statistical analysis was performed using R. T-tests were performed using Welsh’s t-test. Statistical analysis of samples with multiple conditions was performed using 1-way analysis of variance (ANOVA) and a post-hoc Tukey test.
Supplementary Material
Acknowledgments
This work was supported by the Hartwell Foundation (to J.Y.W), NIH pre-doctoral training grant NIGMS 5T32 GM008764 (to D.E.B), NIH training grant T32 HL0007969 (to D.E.B), BU UROP (to B.L.L & S.B.S), and Lutchen Fellowship (to B.L.L. & S.B.S).
Contributor Information
Dr. Daniel E. Backman, Department of Biomedical Engineering, Boston University, 44 Cummington Mall, Boston, MA, 02215, USA
Bauer L. LeSavage, Department of Biomedical Engineering, Boston University, 44 Cummington Mall, Boston, MA, 02215, USA
Shivem B. Shah, Department of Biomedical Engineering, Boston University, 44 Cummington Mall, Boston, MA, 02215, USA
Prof. Joyce Y. Wong, Department of Biomedical Engineering, Boston University, 44 Cummington Mall, Boston, MA, 02215, USA. Division of Materials Science and Engineering, Boston University, 15 Saint Mary’s Street, Boston, MA, 02215, USA
References
- 1.Langer R, Vacanti J. Science. 1993;260:920. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 2.Kumar Va, Brewster LP, Caves JM, Chaikof EL. Cardiovasc Eng Technol. 2011;2:137. doi: 10.1007/s13239-011-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell SL, Niklason LE. Cardiovasc Pathol. 2003;12:59. doi: 10.1016/s1054-8807(02)00183-7. [DOI] [PubMed] [Google Scholar]
- 4.Holzapfel GA. In: Collagen Structure and Mechanics. Fratzl P, editor. Springer; US: 2008. pp. 285–324. [Google Scholar]
- 5.Han HC, Ku DN, Vito RP. Ann Biomed Eng. 2003;31:403. doi: 10.1114/1.1561291. [DOI] [PubMed] [Google Scholar]
- 6.Ballyk PD, Walsh C, Butany J, Ojha M. J Biomech. 1997;31:229. doi: 10.1016/s0197-3975(97)00111-5. [DOI] [PubMed] [Google Scholar]
- 7.Davies AH, Magee TR, Baird RN, Sheffield E, Horrocks M. Br J Surg. 1992;79:1019. doi: 10.1002/bjs.1800791011. [DOI] [PubMed] [Google Scholar]
- 8.Rhodin JAG. Compr Physiol. 2011:1. [Google Scholar]
- 9.Davis EC. Lab Invest. 68:89. [PubMed] [Google Scholar]
- 10.Wagenseil JE, Mecham RP. Physiol Rev. 2009;89:957. doi: 10.1152/physrev.00041.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark JM, Glagov S. Arteriosclerosis. 1985;5:19. doi: 10.1161/01.atv.5.1.19. [DOI] [PubMed] [Google Scholar]
- 12.Sarkar S, Sales KM, Hamilton G, Seifalian AM. J Biomed Mater Res B Appl Biomater. 2007;82:100. doi: 10.1002/jbm.b.30710. [DOI] [PubMed] [Google Scholar]
- 13.Elbert DL. Curr Opin Biotechnol. 2011;22:674. doi: 10.1016/j.copbio.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nichol JW, Khademhosseini A. Soft Matter. 2009;5:1312. doi: 10.1039/b814285h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams C, Xie AW, Yamato M, Okano T, Wong JY. Biomaterials. 2011;32:5625. doi: 10.1016/j.biomaterials.2011.04.050. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Yamato M, Kohno C, Nishimoto A, Sekine H, Fukai F, Okano T. Biomaterials. 2005;26:6415. doi: 10.1016/j.biomaterials.2005.04.061. [DOI] [PubMed] [Google Scholar]
- 17.Okano T, Yamada N, Sakai H, Sakurai Y. J Biomed Mater Res. 1993;27:1243. doi: 10.1002/jbm.820271005. [DOI] [PubMed] [Google Scholar]
- 18.Altomare L, Cochis A, Carletta A, Rimondini L, Farè S. J Mater Sci Mater Med. 2016;27:1. doi: 10.1007/s10856-016-5703-8. [DOI] [PubMed] [Google Scholar]
- 19.Jaeyun J. Enzymatically degradable versatile hydrogel platform for cell sheet engineering. Boston University; 2015. [Google Scholar]
- 20.Wu C, Zhou S. Macromolecules. 1995;28:8381. [Google Scholar]
- 21.Akiyama Y, Kikuchi A, Yamato M, Okano T. Langmuir. 2004;20:5506. doi: 10.1021/la036139f. [DOI] [PubMed] [Google Scholar]
- 22.Sarkar S, Dadhania M, Rourke P, Desai Ta, Wong JY. Acta Biomater. 2005;1:93. doi: 10.1016/j.actbio.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 23.Palchesko RN, Zhang L, Sun Y, Feinberg AW. PLoS One. 2012;7:e51499. doi: 10.1371/journal.pone.0051499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown XQ, Ookawa K, Wong JY. Biomaterials. 2005;26:3123. doi: 10.1016/j.biomaterials.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Ebara M, Hoffman JM, Stayton PS, Hoffman AS. Radiat Phys Chem. 2007;76:1409. [Google Scholar]
- 26.Ma D, Chen H, Shi D, Li Z, Wang J. J Colloid Interface Sci. 2009;332:85. doi: 10.1016/j.jcis.2008.12.046. [DOI] [PubMed] [Google Scholar]
- 27.Lin JB, Isenberg BC, Shen Y, Schorsch K, Sazonova OV, Wong JY. Colloids Surf B Biointerfaces. 2012;99:108. doi: 10.1016/j.colsurfb.2011.10.040. [DOI] [PubMed] [Google Scholar]
- 28.Rayatpisheh S, Li P, Chan-park MB. Macromol Biosci. 2012;12:937. doi: 10.1002/mabi.201100477. [DOI] [PubMed] [Google Scholar]
- 29.Akiyama Y, Yamato M, Okano T. J Robot Mechatronics. 2013;25:631. [Google Scholar]
- 30.Williams C, Tsuda Y, Isenberg BC, Yamato M, Shimizu T, Okano T, Wong JY, Williams BC. Adv Mater. 2009;21:2161. [Google Scholar]
- 31.Isenberg BC, Tsuda Y, Williams C, Shimizu T, Yamato M, Okano T, Wong JY. Biomaterials. 2008;29:2565. doi: 10.1016/j.biomaterials.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Isenberg BC, Backman DE, Kinahan ME, Jesudason R, Suki B, Stone PJ, Davis EC, Wong JY. J Biomech. 2012;45:756. doi: 10.1016/j.jbiomech.2011.11.015. [DOI] [PubMed] [Google Scholar]
- 33.Lee EL, von Recum Ha, Von Recum HA, von Recum Ha. J Biomed Mater Res Part A. 2010;9999A doi: 10.1002/jbm.a.32754. NA. [DOI] [PubMed] [Google Scholar]
- 34.Schneider MH, Tran Y, Tabeling P. Langmuir. 2011;27:1232. doi: 10.1021/la103345k. [DOI] [PubMed] [Google Scholar]
- 35.Dendukuri D, Panda P, Haghgooie R, Kim JM, Hatton TA, Doyle PS. Macromolecules. 2008;41:8547. [Google Scholar]
- 36.Choi BC, Choi S, Leckband DE. Langmuir. 2013;29:5841. doi: 10.1021/la400066d. [DOI] [PubMed] [Google Scholar]
- 37.Takahashi H, Nakayama M, Yamato M, Okano T. Biomacromolecules. 2010;11:1991. doi: 10.1021/bm100342e. [DOI] [PubMed] [Google Scholar]
- 38.Choi S, Choi BC, Xue C, Leckband D. Biomacromolecules. 2013;14:92. doi: 10.1021/bm301390q. [DOI] [PubMed] [Google Scholar]
- 39.Bhattacharya a, Misra BN. Prog Polym Sci. 2004;29:767. [Google Scholar]
- 40.Patel NG, Zhang G. Organogenesis. 2013;9:93. doi: 10.4161/org.25149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuddannaya S, Chuah YJ, Hui M, Lee A, Menon NV, Kang Y, Zhang Y. 2013 doi: 10.1021/am402903e. [DOI] [PubMed] [Google Scholar]
- 42.Patel NG, Cavicchia JP, Zhang G, Zhang Newby B. Acta Biomater. 2012;8:2559. doi: 10.1016/j.actbio.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 43.Backman DE, LeSavage BL, Wong JY. J Biomech. 2016 doi: 10.1016/j.jbiomech.2016.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Claes E, Atienza JM, Guinea GV, Rojo FJ, Bernal JM, Revuelta JM, Elices M. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:3792. doi: 10.1109/IEMBS.2010.5627560. [DOI] [PubMed] [Google Scholar]
- 45.Zaucha MT, Gauvin R, Auger Fa, Germain L, Gleason RL. J R Soc Interface. 2011;8:244. doi: 10.1098/rsif.2010.0228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel A, Fine B, Sandig M, Mequanint K. Cardiovasc Res. 2006;71:40. doi: 10.1016/j.cardiores.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 47.Barone LM, Faris B, Chipman SD, Toselli P, Oakes BW, Franzblau C. Biochim Biophys Acta. 1985;840:245. doi: 10.1016/0304-4165(85)90125-4. [DOI] [PubMed] [Google Scholar]
- 48.Roberts EG, Lee EL, Backman D, Buczek-Thomas JA, Emani S, Wong JY. Ann Biomed Eng. 2014 doi: 10.1007/s10439-014-1210-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sarkar S, Lee GY, Wong JY, Desai Ta. Biomaterials. 2006;27:4775. doi: 10.1016/j.biomaterials.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 50.Richey T, Iwata H, Oowaki H, Uchida E, Matsuda S, Ikada Y. Biomaterials. 2000;21:1057. doi: 10.1016/s0142-9612(99)00281-1. [DOI] [PubMed] [Google Scholar]
- 51.Uchida E, Uyama Y, Ikada Y. J Appl Polym Sci. 1990;41:677. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.