Abstract
TDP-43 can form pathological proteinaceous aggregates linked to ALS and FTLD. Within the putative aggregation domain, engineered repeats of residues 341–366 can recruit endogenous TDP-43 into aggregates inside cells; however, the nature of these aggregates is a debatable issue. Recently, we showed that a coil to β-hairpin transition in a short peptide corresponding to TDP-43 residues 341–357 enables oligomerization. Here we provide definitive structural evidence for amyloid formation upon extensive characterization of TDP-43(341–357) via chromophore and antibody binding, electron microscopy (EM), solid-state NMR, and X-ray diffraction. On the basis of these findings, structural models for TDP-43(341–357) oligomers were constructed, refined, verified, and analyzed using docking, molecular dynamics, and semiempirical quantum mechanics methods. Interestingly, TDP-43(341–357) β-hairpins assemble into a novel parallel β-turn configuration showing cross-β spine, cooperative H-bonding, and tight side-chain packing. These results expand the amyloid foldome and could guide the development of future therapeutics to prevent this structural conversion.
Graphical Abstract
TDP-43 (transactive response DNA binding protein 43 kDa) is a protein implicated in RNA regulation.1 Its binding to nucleic acids takes place through two RNA recognition motifs (RRM) located at the N-terminus, where sequences for nuclear localization and export are also present. TDP-43 is also associated with neurodegenerative diseases, as aggregates of this protein have been found in patients with amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration2 (FTLD). TDP-43 may also play role(s) in the neurotoxicity of Aβ oligomers.3–5 The nature of the TDP-43 aggregates and their pathological role(s) is still unclear. Although most TDP-43 aggregates do not resemble amyloid,2,6 some do under certain conditions.7,8 For certain, short peptides corresponding to segments from other regions of TDP-43 can form amyloid-like fibrils in vitro9–13 (Table S1 in the Supporting Information, SI). Moreover, the toxicity of the full-length protein has recently been attributed to the formation of noxious oligomers.14 On the contrary, sequestering of all endogenous TDP-43 molecules into aggregates has also been suggested to cause a harmful loss of the native protein’s function in its control of RNA metabolism.15 From a pathway point of view, aggregate formation could be driven by several factors that range from simple overexpression of this protein or the elimination of neighboring “self chaperoning” elements, as has been observed in RNase A.16 For a review on the subject, interested readers are referred to Buratti and Baralle.1
In summary, TDP-43 aggregates might: (i) be due to the elimination of neighboring “self chaperoning” elements, as has been observed in RNase A,16 (ii) play physiological roles in RNA and stress granules but eventually act as “TDP-43 sinks” to induce loss-of-function damage,15 or (iii) be neurotoxic through a gain-of-function mechanism. Most importantly, these scenarios do not necessarily exclude each other and may all be pathologically relevant. Therefore, understanding the dynamics of TDP-43 aggregation may represent a key step in the search and design of natural, synthetic, or semisynthetic compounds able to correct this process in patients’ cells.
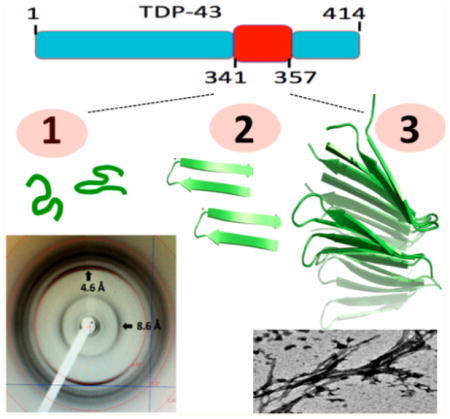
The localization of several pathological point mutations to the 341–366 segment suggests that it is relevant for disease processes.10 The finding that a TDP-43 construct expressing multiple repeats of the C-terminal Q/N rich region spanning residues 341–366 is able to reproduce most of the pathological effects of the full-length protein aggregates in cells17,15 constitutes strong evidence that this segment is relevant for formation of these aggregates. Recently, on the basis of experimental results from circular dichroism (CD) and nuclear magnetic resonance (NMR), we have characterized the structural transformation of a peptide corresponding to this segment in TDP-43, from a disordered monomer to an aggregating conformer rich in β-structure.18 Using molecular dynamics (MD) simulations to test the stability of different plausible structural models, we showed that residues 341–357 formed a β-hairpin and residues 358–367 adopted a turn, followed by a well-defined yet noncanonical structure; however, how these monomers assemble and how the resulting oligomer’s structure is formed remains unknown. Regarding this issue, the preliminary observation that a short peptide corresponding to residues 341–357 aggregates more rapidly than 341–366 suggests that these residues are the most crucial for self-association.
The main objective of this work is to characterize the structure of the oligomer formed by a peptide corresponding to residues 341–357 in TDP-43 using a variety of biophysical, biochemical, and computational methods. We also aim to rigorously test whether or not this aggregate has amyloid-like properties.
The short segment 341–357 of TDP-43 has been recently proposed to adopt a β-hairpin conformation.18 Its sequence, ASQQNQSGPSGNNQNQG, consists of two Gln/Asn rich regions (bold) separated by a turn (underlined). Because the peptide corresponding to this segment of TDP-43 protein aggregates quickly at concentrations appropriate for solution NMR, we studied a diluted sample by CD spectroscopy. The CD spectra indicated that the β-structure content increases over time (Figure 1A and Table S2 in the SI).
Figure 1.
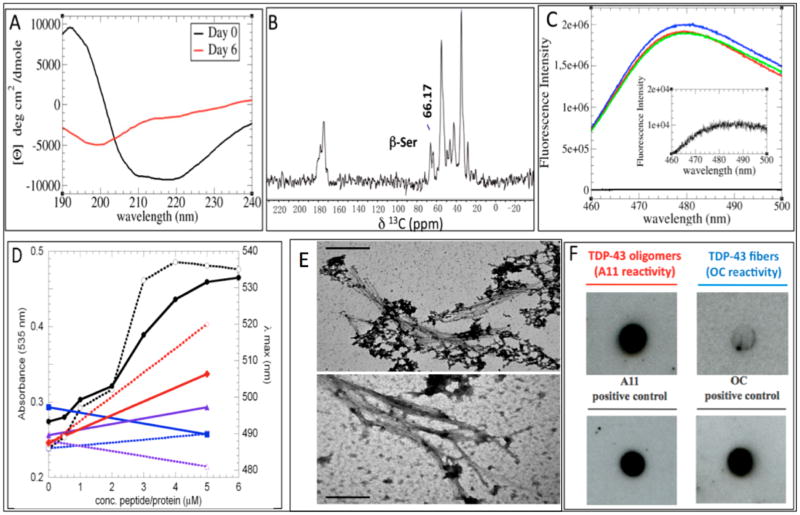
(A) CD spectra of a TDP-43(341–357), contrasting freshly dissolved samples (black line) with samples aged for 6 days (red line); an enrichment of β-secondary structure content occurs. (B) 13C ssNMR spectrum; a peak arising from a Ser 13Cβ atom in a β-conformation is indicated. (C) ThT fluorescence assay of three different samples (blue, red, and green curves) with a blank (black line) magnified, indicating the presence of amyloid structures. (D) Congo Red absorbance at 535 nm (solid symbols, solid lines, left y axis) and absorbance maxima (open symbols, dotted lines, right y axis) in the presence of TDP-43(341–357) (black circles), RNase A (blue squares), KIAβW (purple triangles), and Aβ1–40 (red diamonds). (E) Representative TEM micrograph shows lateral association of TDP-43(341–357) aggregates (top) that resemble those formed by expanded PolyQ (Q62) segments (bottom). The bar scale in both cases is 0.2 μm. (F) Antibody A11 recognizes TDP-43(341–357), which is consistent with the formation of smaller amyloid-like oligomers; in contrast, the OC antibody (specific for amyloid-like fibrils) shows weak/negligible binding. Prefibrillar oligomers and fibrillar species of Aβ1–42 were used as positive controls for A11 and OC reactivity, respectively.
To gain additional structural information on the conformation of this peptide after aggregation, we used solid-state NMR. The 1D 13C ssNMR spectrum of aggregated forms of TDP-43(341–357) shows significant resonance overlap, but a peak at 66.17 ppm can be unambiguously assigned to a Ser 13Cβ (Figure 1B). The low-field shifted value of this nucleus is consistent with the presence of β-sheet structure. To determine whether these β-rich aggregates are amyloid-like, we first performed Thioflavin T (ThT) and Congo Red (CR) binding assays, as amyloid-like fibers are well-known to bind these two dyes. ThT fluorescence is strongly enhanced upon binding to amyloid fibrils, whereas CR increases its absorbance of visible light and experiences a red shift. As shown in Figure 1C,D, residues 341–357 of TDP-43 induce these spectral changes, which suggests that the aggregate behaves as an amyloid-like protein.
On the basis of these findings, we decided to determine the morphology of these aggregates with transmission electron microscopy (TEM). The fibrils were several hundred nanometers long and approximately 15–25 nm wide (Figure 1E). We observed flat, wide fibrils with a marked tendency to associate laterally. It is interesting that the morphology of these fibrils is very similar to that formed by expanded PolyQ (Q62) segments19 (Figure 1E) and distinct from the thinner (9 nm wide) nonbranching fibrils formed by the Aβ peptide.20
The amyloid-like nature of these aggregates was further probed with two different conformational antibodies A11 and OC, which recognize amyloid-like oligomers and fibrils, respectively (Figure 1F). The A11 assay was positive, which strongly suggests the presence of TDP-43(341–357) oligomers similar to the small, diffusible multimeric species formed by several neurotoxic peptides,21 including polyQ22 and Aβ.23 In contrast, the binding of TDP-43(341–357) to the OC antibody was negligible, in agreement with Wang et al.,24 who showed that the C-terminal motif of TDP-43 aggregates is not recognized by the OC antibody. This result is intriguing because the TEM images showed strong similarity with PolyQ fibers, which are recognized by the OC antibody25 and thus may suggest a subtle structural difference between these fibrils.
Considering our previous observations, therefore, these novel analyses have confirmed that the TDP-43(341–357) peptide evolves structurally, gaining β-structure over time and assembling into A11-reactive oligomers. Moreover, to test possible contributions from neighboring segments, we studied the conformation of TDP-43(322–366) using NMR and found evidence of a random coil ensemble that aggregates more slowly that TDP-43(341–357) (Figure S1 in the SI); these results strongly suggest that flanking segments are not key for amyloid formation by TDP-43(341–357).
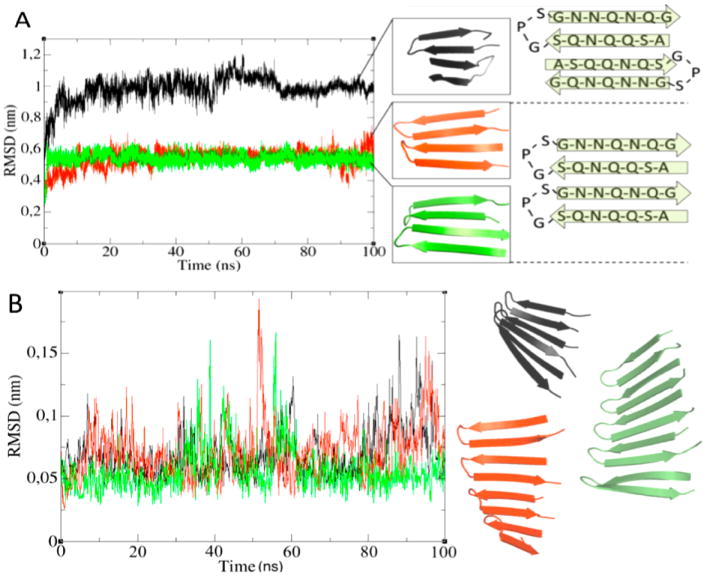
To gain further insight into this conversion and make an atomistic model of the process, we carried out computational studies. Two TDP-43(341–357) monomers were placed in a simulation box and studied using three independent 100 ns MD runs. The three simulations started with different initial velocities on each atom. During all three runs, β-sheet formation occurs through the lateral association of the β-hairpins. Two different types of interactions were found, which we denoted as in-register (observed in two out of three runs) and out-of-register interactions (observed in one of three runs). The in-register β-strands permit each residue to interact with its counterpart in the strands above and below it, which we consider to be an optimal arrangement (Figure 2A). The out-of-register disposition involves an asymmetric packing of the residues within the strands and did not seem to be a plausible intermediate toward the formation of a mature fibril due to these less optimal interactions. Nevertheless, to corroborate the plausibility of both binding modes, we utilized replica exchange molecular dynamics (REMD) to improve sampling around these configurations. The results obtained provide additional evidence in favor of the in-register mode of β-hairpin association (Figure S2 in the SI).
Figure 2.
(A) RMSD corresponding to three independent MD simulations where lateral association occurs (left). Note that the term lateral refers to the side by side packing of peptide backbones. Snapshots from the last frame of each of the structures are shown together with graphical representations of the in-register and out-of-register binding modes (right). The distance root-mean-square deviation (dRMSD) shows the same trend (Figures S3–S5 in the SI). (B) dRMSD of the trimer (black), tetramer (red) and pentamer (green) fitted to their respective time-averaged structures over all of the frames. Snapshots at t = 100 ns are represented for each system. All RMSD calculations were performed with respect to the corresponding time-zero structures.
The in-register binding mode involves an extraordinary inter-residue complementarity because one strand of each β-hairpin is paired with the same residue above and below, which allows for optimal van der Waals interactions and the formation of an extensive network of hydrogen bonds throughout the Asn and Gln side chains. The stability of the interactions between these monomers is such that even upon heating in silico the system remained stable at temperatures up to 350 K. Interestingly, interactions of the strands within the β-hairpins were the first to dissociate, while the interstrand interactions between different TDP-43(341–357) units were maintained. That is, the intermolecular interactions seem to be stronger than the intramolecular ones. Heating at 400 K corroborated this, where prior to dissociation the H-bonds were observed to break in the same order (Figure S6 in the SI).
The quality of this structure was analyzed with PROCHECK, 26 which showed that 91.7% of the residues are in the most favored regions, whereas the remaining 8.3% were in additional allowed regions of the Ramachandran map (Figure S7 in the SI). This ensures the stereochemical quality of the system. On the basis of these results, we decided to dock additional monomers onto this pair of β-hairpins. Up to five monomers laterally associated were built this way, and their structural stability was checked by MD simulations. After each 100 ns run, all three systems (trimer, tetramer, and pentamer) were found to be twisted and showed a remarkable stability (Figure 2B), as gauged by low dRMSD values. Supplementary Figure S8 in the SI provides further evidence of the structural stability of these systems, where the structures corresponding at times 0, 50, and 100 ns are superimposed to highlight the persistence of both twisting and the H-bond networks.
Amyloid-like oligomers typically display a remarkable resistance to chemical denaturants27 and mechanical unfolding. 28 This exceptional conformational stability has been attributed to hyperpolarization of amide groups forming the H-bond network.29,30 The elevated stability of the TDP-43(341–357) observed in the simulation runs previously mentioned prompted us to test whether these structures are also able to exhibit the typical H-bonding cooperativity (HBC) present in amyloid-like fibrils. Semiempirical quantum mechanics methods have been proposed as an appropriate choice to study such interactions as they provide a good balance between computational cost and accuracy.31 We utilized the AM1 method to compute the energetics upon oligomerization through lateral association because this is the most rigorous function applicable to a system of this size. Because this method is intended to reproduce heats of formation, we analyzed the HBC effect in terms of average formation energies. The results of these calculations show that it is indeed more favorable to form systems of increasing size, which means that as more and more β-hairpins dock onto the growing aggregate, their H-bonds become stronger and stronger (Figure S9 and Table S3 in the SI).
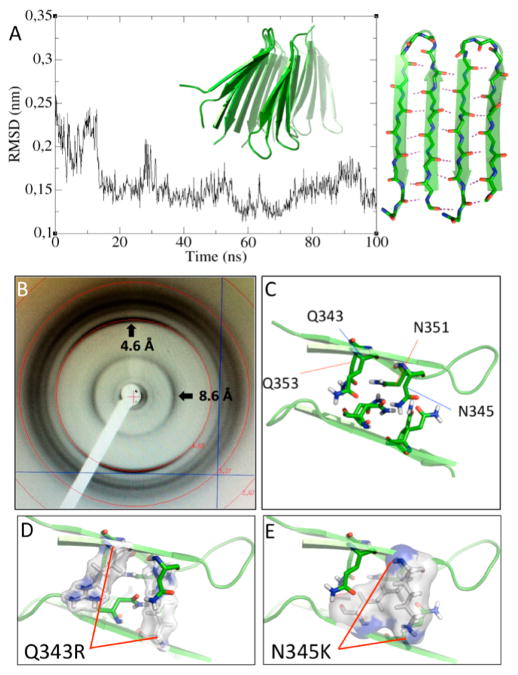
These results are able to reproduce both the H-bonding cooperativity and twisting that are characteristic of amyloid-like fibrils. Moreover, these findings are consistent with an aggregation pathway wherein the oligomer grows through lateral association of monomers. Considering the spectroscopy-based evidence for β-conformation derived from CD and ssNMR experiments and given the positive results of the ThT and CR binding assays, the A11 immunoreactivity, and the morphology of the aggregates as revealed by TEM, which are in-line with amyloid-like aggregates, we tested whether these oligomeric β-sheets can pack to form a dry interface. To this end, we used HADDOCK32 to perform fully flexible, solvated docking on two of TDP-43(341–357) pentamers. We obtained a cross-β spine structure, where the two β-sheets form a dry interface through precise side-chain intermeshing with a high shape complementarity value33 of 0.84. This value approaches that found (0.86) for the GNNQQNY heptapeptide of Sup3534 and is significantly higher than typical Sc values for protease/protease inhibitors (0.71 to 0.76), subunit interfaces (0.70 to 0.74), or antibody/antigen contacts (0.64 to 0.68).33 A 100 ns MD simulation showed not only that this structure is stable but also that the twist is preserved (Figure 3A).
Figure 3.
(A) RMSD of two TDP-43(341–357) pentamers packed forming a cross β-spine from a 100 ns MD simulation. Each pentamer is a ten-stranded β-sheet formed by five antiparallel β-hairpins, whose stability is evidenced by low RMSD values during MD simulations, calculated with respect to the time-zero structure. (B) X-ray diffraction pattern collected from fibers confirming the presence of a cross β-spine. The reflection at 4.6 Å corresponds to the interstrand distance, whereas that at 8.6 Å is the intersheet distance. (C) Gln and Asn side chains contact and form a dry interface between the two β-sheets, with a high shape complementarity value of 0.84. Q343R (D) and N345 K (E) substitutions are incompatible with this supramolecular organization, and, in consequence, peptides carrying these substitutions do not oligomerize.18
Finally, the fiber diffraction of the peptide confirmed the cross-β association of the TDP-43(341–357) monomers, which shows intra-β-sheet and inter-β-sheet reflections at 4.6 and 8.6 Å, respectively (Figure 3B). The latter value is smaller than the same reflection observed in most amyloid fibrils formed by other polypeptides, except for polyQ aggregates. (See Table S4 in the SI.) These data establish the amyloid-like nature of the TDP-43(341–357) fibrils. X-ray powder diffraction of the fibers that were used for the ssNMR experiments showed the same two major reflections (Figure S10 in the SI), with d-spacing corresponding to the above distances, which makes this motif unique with respect to previously reported amyloid structures. This result can reconcile all of our experimental findings with the predicted aggregative pathway based on β-hairpin formation and lateral association.
Furthermore, inspecting the dry interface (Figure 3C) of this aggregative motif clarifies why the pathological substitutions Q343R and N345 K do not aggregate in vitro.18 In both cases, the length of the mutated side chains will not allow for the proper side-chain intermeshing, as can be inferred from Figure 3D,E because they are too long for maintain the two mating β-sheets at the distance of 8.6 Å observed here and in polyQ fibrils35 or even the longer distance of 10 Å seen in most amyloid fibrils (Table S4 in the SI).
On the basis of all of the experimental and computational results, we conclude that TDP-43(341–357) forms an amyloid-like fibril with a novel β-turn topology in which each β-hairpin associates in parallel to contribute two β-strands to the same β-sheet. It is interesting that in this structural model TDP-43(341–357) shows a “β-turn” configuration, which has been recently reported for polyQs36 and in contrast with the “β-arc” topology observed for Aβ, in which β-hairpins contribute one β-strand to two different β-sheets. This β-arc fold is not likely to occur in TDP-43(341–357), as it would imply breakage of the interstrand H-bonds favored by the Gly-Pro turn. In fact, it was not observed in our REMD and MD simulations. (See Figure S11 in the SI). Nonetheless, TDP-43 and polyQ aggregates share a similar morphology, as seen by EM and β-turn topology; however, the configuration of consecutive hairpins is parallel in the case of TDP-43(341–357) and antiparallel for polyQ aggregates.
In conclusion, we have shown that the TDP-43(341–357) peptide can form oligomeric aggregates that are recognized by the conformation-specific A11 antibody and fibrils with amyloid-like dye-binding characteristics. In contrast with most amyloid-like fibrils, but akin to those formed by polyglutamine, TDP-43(341–357) fibrils show a rather unusual flat morphology, as seen by EM, and short inter β-sheet separation, as measured by X-ray diffraction. Finally, we advance a mechanism for oligomerization and an atomic resolution structural model for the TDP-43(341–357) amyloid-like fibril in which β-hairpins combine in a novel parallel β-turn configuration. These results advance our understanding of aggregates in TDP-43 pathologies and could aid the development of therapeutics specifically aimed at slowing down or inhibiting this process.
EXPERIMENTAL AND COMPUTATIONAL METHODS
A peptide whose sequence ASQQNQSGPSGNNQNQG corresponds to TDP-43(341–357) was purchased from Genescript (Piscataway Township, NJ). Its purity was over 95% as assessed by HPLC. NMR spectroscopy and mass spectrometry confirmed the peptide’s composition. The peptide’s concentration in solution was determined by weight and by far-UV absorbance, using ε205nm = 48 080 cm−1·M−1 at 205 nm calculated using the sequence and the parameters of Anthis and Clore.37
PolyQ-containing protein was cloned by PCR using a plasmid containing 62 Qs as template, kindly provided by Dr. Yoshitaka Nagai.38 The PCR insert was cloned into the pET28a vector (Novagen) using the NheI and XhoI restriction sites. With this cloning strategy, the MGSSHHHHHHSSGLVPRGSHMAS amino acid sequence remained at the N-terminus of the protein. Expression and purification of the protein is fully described elsewhere.28
Circular Dichroism Spectroscopy
Far UV-CD spectra were recorded using a JASCO J-710 spectropolarimeter. The instrument bandwidth was 1.2 nm and the scan speed was 20 nm·min−1. Spectra were recorded over the wavelength range of 190–260 nm at 25.0 °C using a 0.1 cm path length, with a peptide concentration of 105 μM in 3 mM KH2PO4/K2HPO4 buffer (pH 6.8) on freshly dissolved and aged samples. Eight scans were recorded and averaged for each spectrum, and the buffer reference spectrum was subtracted. For a detailed description of the CD spectral analysis, we refer the reader to the SI Methods.
Solid-State NMR Spectroscopy
The TDP-43(341–357) peptide (25 mg) was dissolved in 10 mM KH2PO4/K2HPO4 (pH 6.3) and allowed to form fibrils during 7 days. Following solvent removal, the fibrils were packed in a 4 mm Varian rotor and measured on a Chemagnetics Infinity 300 MHz instrument in H/C double resonance mode with an APEX double-resonance magic-angle spinning (MAS) probe. Cross-polarization (CP) experiments were performed with a Hahn-Echo to suppress broad background signals. TPPM decoupling (80 kHz) was applied during acquisition.
Thioflavin T Fluorescence
Following the procedure of LeVine,39 a 1.0 mM stock solution of ThT (Sigma, St. Louis, MO) was prepared in 3 mM KH2PO4/K2HPO4 buffer, pH 6.8. Fluorescence samples contained 5 μL of ThT stock solution, whose final concentration was 50 μM, with 25 μM of TDP-43 peptides. Amyloid Aβ1–40 (rPeptide) and ribonuclease A (Sigma type XII-A) were used as positive and negative controls, respectively. Spectra were recorded at 25 °C on a Jobin-Yvon Fluoromax-4 instrument using 3 nm excitation and emission slit widths. The excitation wavelength was 440 nm and emission was recorded over 460–500 nm at a scan speed of 2 nm·s−1.
Congo Red Binding
Congo Red spectroscopic assays were performed as described by Klunk et al.;40 5 μM of CR stock solution (Sigma) was prepared in 5 mM KH2PO4 and 150 mM NaCl and used to test binding to TDP-43(341–357) at concentrations ranging from 1 to 5 μM. Amyloid Aβ1–40 (rPeptide) was used as a positive control, whereas ribonuclease A (Sigma type XII-A) and KIAβ, a nonaggregating β-hairpin peptide,41 were used as negative controls. Congo Red absorption spectra were recorded from 440 to 600 nm in a dual beam Cary 210 UV–vis spectrometer at room temperature at a 1/2 nm·s−1 scan speed. The samples were aged for 6 days to ensure aggregate formation and study their possible amyloid-like nature.
Transmission Electron Microscopy
TDP-43 peptide samples were incubated for ~14 days at 37 °C at a concentration of 50 μM in PBS [pH 7.0] in the presence of 5% DMSO and with no stirring. PolyQ samples were incubated for 20–30 days at 37 °C at a concentration of 10–40 μM in PBS [pH 7.4] in the presence of 0.02% NaN3 and with no stirring. For fibrillo-genesis assay, samples diluted to concentrations of 5–50 μM (10 μL) were adsorbed onto carbon-coated 300-mesh copper grids (Ted Pella) and negatively stained for 60 s using 1 to 2% uranyl acetate. Immediately before use, the carbon-coated grids were glow-discharged to enhance their hydrophilicity using an Emitech K100X apparatus (Quorum Technologies). The formation of amyloid fibrils was analyzed on a JEOL 1200EX II (Jeol Limited) electron microscope equipped with a CCD Megaview III camera (Olympus Soft Imaging). The images were acquired at a magnification of 120.000× and a voltage of 80 kV.
Dot Blot Analysis
TDP-43 samples were incubated for approximately 2 weeks at 37 °C at a concentration of 50 μM in PBS with 5% DMSO, pH 7.0 and without stirring. For Dot Blot analysis, 2 μL of each TDP-43 peptide sample were spotted onto a nitrocellulose membrane. After blocking for 1 h at RT with 10% nonfat milk in Tris-buffered saline (TBS) containing 0.01% Tween 20 (TBS-T), the membrane was incubated with the polyclonal specific antioligomer A11 antibody specific for toxic oligomers21 (Life Technologies) or the monoclonal antibody OC (Millipore), which recognizes mature amyloid-like fibrils,25 diluted 1:1000 in 3% BSA TBS-T, at RT for 1 h. The membranes were washed three times for 5 min each with TBS-T before incubating with antirabbit HRP conjugated antirabbit IgG (GE Healthcare) diluted 1:5000 in 3% BSA/TBS-T at room temperature for 1 h. After washing the membranes thrice in TBS-T buffer, the blots were developed with ECL Plus chemiluminescence kit from Amersham-Pharmacia (GE Healthcare). Prefibrillar oligomers and fibrillar species of Aβ1–42, served as positive controls for A11 and OC reactivity, respectively.
X-ray Fiber Diffraction
In an Eppendorf tube, 2 mg of peptide powder were dissolved in 0.8 mL of water. The solution was vortexed at room temperature. The resulting cloudy solution was allowed to settle for 7 days to form fibers, after which the tube was spun down and the aqueous solvent was decanted. Residual water was further removed slowly in a dehydration chamber. After 48 h, the dehydrated fibers were cryo-protected in 30% glycerol and flash-frozen in liquid nitrogen. The fiber X-ray diffraction pattern was collected using an in-house sealed tube microfocus X-ray generator (Micro-Max-003, Rigaku Corporation) with Saturn944 CCD and Cu Kα (1.54 Å) radiation at 100 K. This result is shown in Figure 3B. In another experiment, TDP-43(341–357) fibers were prepared as previously described, except that they were left in the dehydration chamber for 24 h. The resulting fibers were analyzed with an X-ray powder diffractometer (PANalytical X’Pert 3) on a spinning sample holder. The data were collected over a 2θ range from 1 to 50°. This is result is shown in Figure S10 in the SI.
MD and REMD Simulations
All simulations were performed using amber99sb-ildn force-field parameters42 as implemented in the GROMACS package,43 version 4.5.5. The systems were placed in TIP3P water44 cubic boxes and simulated under NpT conditions. All individual runs ranged from 100 to 110 ns. The peptide model included N-terminal acetyl and C-terminal amide groups to avoid artificially favoring β-hairpin formation through attractive electrostatic interactions. For specific details we refer the reader to the SI Methods.
Additionally, implicit-solvent REMD45 simulations were carried out to enhance sampling. Temperatures were exponentially separated ranging from 280 to 450 K, using six replicas (50 ns each replica for a total of 300 ns), and with exchange probabilities ranging from 0.3 to 0.5. The REMD simulation was initiated using 2 TDP-43(341–357) in the β-hairpin conformation placed at random starting positions. Molecular dynamics (with 50 ns per replica) from NVT pre-equilibrated systems was performed, and all of the interactions were calculated by setting a cutoff distance of 0, which for GROMACS is equivalent to computing all interactions using an infinite cutoff. Additional computational details and a figure (Figure S12) illustrating the starting peptide configuration are given in the SI.
Protein–Protein Docking
Docking of two ten-stranded systems was performed with the HADDOCK32 program, using data from solvent-accessible surface (SAS) calculations. The HADDOCK program was used to generate the 20-stranded oligomer with side-chain intermeshing (dry interface), made up by a pair of TDP-43 pentamers (each sheet contains 5 β-hairpins and thus 10 β-strands). For specific details we refer the reader to the SI Methods.
Semiempirical QM Calculations
To study the energetics of oligomer formation, we performed AM146 calculations in gas phase using the Gaussian 0947 suite package (revision D.01). The choice of a semiempirical method was based on the differences in size from the smallest to the biggest system, which span up to 10-stranded β sheets (corresponding to five TDP-43(341–357) β-hairpins). Figure S9 and Table S4 in the SI contain a complete description of the procedures used.
Supplementary Material
Acknowledgments
We thank Dr. Margarita Menéndez for use of the Cary 210 spectrophotometer. This work was supported by Grants CTQ2010-21567-C02-02 (FPI Fellowship to M.M.), SAF2013-49179-C2-2-R (D.V.L.), and SAF2013-49179-C2-1-R (M.C.-V.) and an EU JPND AC14/00037 (D.V.L. and M.C.-V.), AriSLA TARMA (F.B.), Thierry Latran Foundation REHNPALS (E.B.), the EU Joint Programme-Neurodegenerative Diseases JPND RiMod-FTD, Italy, Ministero della Sanita’ (E.B.). The in-house X-ray diffraction instrument at Columbia University was purchased with an NIH grant to L.T. (S10OD012018). M.M. is thankful for the kind hospitality of the members of the McDermott and Tong laboratories during his stay in NY during the summer of 2014 (funded by FPI fellowship CTQ2010-21567-C02-02).
Footnotes
Notes
The authors declare no competing financial interest.
Detailed experimental and computational procedures, NOESY and 1H-13C HSQC NMR spectra of TDP-43(322–366), free-energy landscapes in the phi and psi dihedral angle space, dRMSD for lateral associations I–III, in silico thermal heating, PROCHECK structure validation, structural stability of the TDP-43(341–357) oligomers, cooperative hydrogen-bonding formation, TDP-43(341–357) X-ray fiber diffraction, β-arc and β-turn topologies, initial configuration used in the simulations, TDP-43 peptides that form amyloid-like aggregates, CD analysis, interaction energy values and X-ray diffraction patterns in known amyloid and amyloid-like fibrils. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpclett.5b00918.
References
- 1.Buratti E, Baralle FE. TDP-43: Gumming up Neurons through Protein-Protein and Protein-RNA Interactions. Trends Biochem Sci. 2012;37:237–247. doi: 10.1016/j.tibs.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 3.Josephs KA, Whitwell JL, Weigand SD, Murray ME, Tosakulwong N, Liesinger AM, Petrucelli L, Senjem ML, Knopman DS, Boeve BF, Ivnik RJ, Smith GE, Jack CR, Jr, Parisi JE, Petersen RC, Dickson DW. TDP-43 is a Key Player in the Clinical Features Associated with Alzheimer’S Disease. Acta Neuropathol. 2014;127:811–824. doi: 10.1007/s00401-014-1269-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu L, Xu M, Yang M, Yang Y, Li Y, Deng J, Ruan L, Liu J, Du S, Liu X, Feng W, Fushimi K, Bigio EH, Mesulam M, Wang C, Wu JY. An ALS-Mutant TDP-43 Neurotoxic Peptide Adopts an Anti-Parallel β-Structure and Induces TDP-43 Redistribution. Hum Mol Genet. 2014;23:6863–6877. doi: 10.1093/hmg/ddu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, Blanchard J, Grundke-Igbal I, Wegiel J, Deng HX, Siddique T, Igbal K. Alzheimer Disease and Amyotrophic Lateral Sclerosis: An Etiopathogenic Connection. Acta Neuropathol. 2014;127:243–256. doi: 10.1007/s00401-013-1175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T. TDP-43 is a Component of Ubiquitin-Positive Tau-Negative Inclusions in Frontotemporal Lobar Degeneration and Amyotrophic Lateral Sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 7.Robinson JL, Geser F, Stieber A, Umoh M, Kwong LK, Van Deerlin VM, Lee VM, Trojanowski JQ. TDP-43 Skeins Show Properties of Amyloid in a Subset of ALS Cases. Acta Neuropathol. 2013;125:121–131. doi: 10.1007/s00401-012-1055-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bigio EH, Wu JY, Deng HX, Bit-Ivan EN, Mao Q, Ganti R, Peterson M, Siddique N, Geula C, Siddique T, Mesulam M. Inclusions in Frontotemporal Lobar Degeneration with TDP-43 Proteinopathy (FTLD-TDP) and Amyotrophic Lateral Sclerosis (ALS), but not FTLD with FUS Proteinopathy (FTLD-FUS), Have Properties of Amyloid. Acta Neuropathol. 2013;125:463–465. doi: 10.1007/s00401-013-1089-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen AKH, Lin RYY, Hsieh EZJ, Tu PH, Chen RP, Liao TY, Chen W, Wang CH, Huang JJ. Induction of Amyloid Fibrils by the C-Terminal Fragments of TDP-43 in Amyotrophic Lateral Sclerosis. J Am Chem Soc. 2010;132:1186–1187. doi: 10.1021/ja9066207. [DOI] [PubMed] [Google Scholar]
- 10.Saini A, Chauhan VS. Delineation of the Core Aggregation Sequences of TDP-43 C-Terminal Fragment. ChemBioChem. 2011;12:2495–2501. doi: 10.1002/cbic.201100427. [DOI] [PubMed] [Google Scholar]
- 11.Guo W, Chen Y, Zhou X, Kar A, Ray P, Chen X, Rao EJ, Yang M, Ye H, Zhu L, Liu J, Xu M, Yang Y, Wang C, Zhang D, Bigio EH, Mesulam M, Shen Y, Xu Q, Fushimi K, Wu JY. An ALS-Associated Mutation Affecting TDP-43 Enhances Protein Aggregation, Fibril Formation and Neurotoxicity. Nat Struct Mol Biol. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun CS, Wang CY, Chen BP, He RY, Liu GC, Wang CH, Chen W, Chern Y, Huang JJ. The Influence of Pathological Mutations and Proline Substitutions in TDP-43 Glycine-Rich Peptides on Its Amyloid Properties and Celular Toxicity. PLoS One. 2014;9(8):e103644. doi: 10.1371/journal.pone.0103644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saini A, Chauhan VS. Self-Assembling Properties of Peptides Derived from TDP-43 C-Terminal Fragment. Langmuir. 2014;30:3845–3856. doi: 10.1021/la404710w. [DOI] [PubMed] [Google Scholar]
- 14.Fang YS, Tsai KJ, Chang YJ, Kao P, Woods R, Kuo PH, Wu CC, Liao JY, Chou SC, Lin V, Jin LW, Yuan HS, Cheng IH, Tu PH, Chen YR. Full-Length TDP-43 Forms Toxic Amyloid Oligomers That Are Present in Frontotemporal Lobar Dementia-TDP Patients. Nat Commun. 2014;5:4824. doi: 10.1038/ncomms5824. [DOI] [PubMed] [Google Scholar]
- 15.Budini M, Romano V, Quadri Z, Buratti E, Baralle FE. TDP-43 Loss of Cellular Function through Aggregation Requires Additional Structural Determinants Beyond its C-Terminal Q/N Prion-Like Domain. Hum Mol Genet. 2015;24:9–20. doi: 10.1093/hmg/ddu415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teng PK, Anderson NJ, Goldschmidt L, Sawaya MR, Sambashivan S, Eisenberg D. Ribonuclease a Suggests How Proteins Self-Chaperone against Amyloid Fiber formation. Protein Sci. 2012;21:26–37. doi: 10.1002/pro.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Budini M, Buratti E, Stuani C, Guarnaccia C, Romano V, De Conti L, Baralle FE. Cellular Model of TAR DNA-Binding Protein 43(TDP-43) Aggregation Based on Its C-Terminal Gln/Asn-Rich Region. J Biol Chem. 2012;287:7512–7525. doi: 10.1074/jbc.M111.288720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mompeán M, Buratti E, Guarnaccia C, Brito RMM, Chakrabartty A, Baralle FE, Laurents DV. Structural Characterization of the Minimal Segment of TDP-43 Competent for Aggregation. Arch Biochem Biophys. 2014;545:53–62. doi: 10.1016/j.abb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Kar K, Hoop CL, Drombosky KW, Baker MA, Kodali R, Arduini I, van der Wel PC, Horne WS, Wertzel R. β-Hairpin Mediated Nucleation of Polyglutamine Amyloid formation. J Mol Biol. 2013;425:1183–1197. doi: 10.1016/j.jmb.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirschner DA, Inouye H, Duffy LK, Sinclair A, Lind M, Selkoe DJ. Synthetic Peptide Homologous to Beta Protein from Alzheimer Disease Forms Amyloid-Like Fibrils. in vitro Proc Natl Acad Sci USA. 1987;84:6953–6957. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common Structure of Soluble Amyloid Oligomers Implies Common Mechanism of Pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 22.Kar K, Jayaraman M, Sahoo B, Kodali R, Wetzel R. Critical Nucleus Size for Disease-Related Polyglutamine Aggregation Is Repeat Length Dependent. Nat Struct Mol Biol. 2011;18:328–336. doi: 10.1038/nsmb.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lashuel HA, Hartley DM, Petre BM, Wall JS, Simon MN, Walz T, Lansbury PT., Jr Mixtures of Wild-Type and a Pathogenic (E22G) Form of Abeta40 in Vitro Accumulate Protofibrils, Including Amyloid Pores. J Mol Biol. 2003;332:795–805. doi: 10.1016/s0022-2836(03)00927-6. [DOI] [PubMed] [Google Scholar]
- 24.Wang YT, Kuo PH, Chiang CH, Liang JR, Chen YR, Wang S, Shen JC, Yuan HS. The Truncated C-Terminal RNA Recognition Motif of TDP-43 Protein Plays a Key Role in Forming Proteinaceous Aggregates. J Biol Chem. 2013;288:9049–9057. doi: 10.1074/jbc.M112.438564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kayed R, Head E, Sarsoza F, Saing T, Cotman CW, Necula M, Margol L, Wu J, Breydo L, Thompson JL, Rasool S, Gurlo T, Butler P, Glabe CG. Fibril Specific, Conformation Dependent Antibodies Recognize a Generic Epitope Common to Amyloid Fibrils and Fibrillar Oligomers That Is Absent in Prefibrillar Oligomers. Mol Neurodegener. 2007;2:18. doi: 10.1186/1750-1326-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. PROCHECK: a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 27.MacPhee CE, Dobson CM. Chemical Dissection and Reassembly of Amyloid Fibrils Formed by a Peptide Fragment of Transthyretin. J Mol Biol. 2000;297:1203–1215. doi: 10.1006/jmbi.2000.3600. [DOI] [PubMed] [Google Scholar]
- 28.Hervás R, Oroz J, Galera-Prat A, Goñi O, Valbuena A, Vera AM, Gómez-Sicilia A, Losada-Urzáiz F, Uversky VN, Menéndez M, Laurents DV, Bruix M, Carrión-Vázquez M. Common Features at the Start of the Neurodegeneration Cascade. PLoS Biol. 2012;10(5):e1001335. doi: 10.1371/journal.pbio.1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsemekhman K, Goldschmidt L, Eisenberg D, Baker D. Cooperative Hydrogen Bonding in Amyloid Formation. Protein Sci. 2007;16:761–764. doi: 10.1110/ps.062609607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mompeán M, González C, Lomba E, Laurents DV. Combining Classical MD and QM Calculations to Elucidate Complex System Nucleation: A Twisted, Three-Stranded, Parallel β-Sheet Seeds Amyloid Fibril Conception. J Phys Chem B. 2014;118:7312–7316. doi: 10.1021/jp503742p. [DOI] [PubMed] [Google Scholar]
- 31.Rosseti G, Magistrato A, Pastore A, Carloni P. Hydrogen Bonding Cooperativity in PolyQ β-Sheets from First Principle Calculations. J Chem Theory Comput. 2010;6:1777–1782. doi: 10.1021/ct900476e. [DOI] [PubMed] [Google Scholar]
- 32.Dominguez C, Boelens R, Bonvin AMJJ. HADDOCK: a Protein-Protein Docking Approach Based on Biochemical and/or Biophysical Information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence MC, Colman PM. Shape Complementarity at Protein/Protein Interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 34.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Structure of the Cross-Beta Spine of Amyloid-Like Fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma D, Shinchuk LM, Inouye H, Wetzel R, Kirschner DA. Polyglutamine Homopolymers Having 8–45 Residues Form Slablike Beta-Crystallite Assemblies. Proteins. 2005;61:398–411. doi: 10.1002/prot.20602. [DOI] [PubMed] [Google Scholar]
- 36.Buchanan LE, Carr JK, Fluitt AM, Hoganson AJ, Moran SD, de Pablo JJ, Skinner JL, Zanni MT. Structural Motif of Polyglutamine Amyloid Fibrils Discerned with Mixed-Isotope Infrared Spectroscopy. Proc Natl Acad USA. 2014;111:5796–5801. doi: 10.1073/pnas.1401587111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anthis NJ, Clore GM. Sequence-Specific Determination of Protein and Peptide Concentrations by Absorbance at 205 nm. Protein Sci. 2013;22:851–858. doi: 10.1002/pro.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagai Y, Tucker T, Ren H, Kenan DJ, Henderson BS, Keene JD, Strittmatter WJ, Burke JR. Inhibition of Polyglutamine Protein Aggregation and Cell Death by Novel Peptides Identified by Phage Display Screening. J Biol Chem. 2000;275:10437–10442. doi: 10.1074/jbc.275.14.10437. [DOI] [PubMed] [Google Scholar]
- 39.LeVine H., 3rd Thioflavine T Interaction with Synthetic Alzheimer’S Disease Beta-Amyloid Peptides: Detection of Amyloid Aggregation in Solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klunk WE, Pettegrew JW, Abraham DJ. Quantitative Evaluation of Congo Red Binding to Amyloid-Like Proteins with a Beta-Pleated Sheet Conformation. J Histochem Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 41.Diez-García F, Pantoja-Uceda D, Jiménez MÁ, Chakrabartty A, Laurents DV. Structure of a Simplified β-Hairpin and Its ATP Complex. Arch Biochem Biophys. 2013;537:62–71. doi: 10.1016/j.abb.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 42.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Kepleis JL, Dror RO, Shaw DE. Improved Side-Chain Torsion Potentials for the Amber ff99SB Protein Force Field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, And Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 44.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 45.Sugita Y, Okamoto Y. Replica-Exchange Molecular Dynamics Method for Protein Folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 46.Dewar MJS, Zoebisch EG, Healy EF, Stewart JJP. Development and Use of Quantum Mechanical Molecular Models. 76. AM1: A New General Purpose Quantum Mechanical Molecular Model. J Am Chem Soc. 1985;107:3902–3909. [Google Scholar]
- 47.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, et al. Gaussian 09, revision D.01. Gaussian, Inc; Wallingford, CT: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.