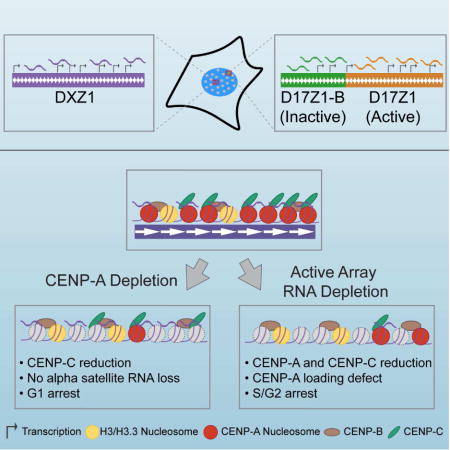
SUMMARY
Human centromeres are defined by alpha satellite DNA arrays that are distinct and chromosome-specific. Most human chromosomes contain multiple alpha satellite arrays that are competent for centromere assembly. Here, we show that human centromeres are defined by chromosome-specific RNAs linked to underlying organization of distinct alpha satellite arrays. Active and inactive arrays on the same chromosome produce discrete sets of transcripts in cis. Non-coding RNAs produced from active arrays are complexed with CENP-A and CENP-C, while inactive array transcripts associate with CENP-B and are generally less stable. Loss of CENP-A does not affect transcript abundance or stability. However, depletion of array-specific RNAs reduces CENP-A and CENP-C at the targeted centromere via faulty CENP-A loading, arresting cells before mitosis. This work shows that each human alpha satellite array produces a unique set of non-coding transcripts, and RNAs present at active centromeres are necessary for kinetochore assembly and cell cycle progression.
Keywords: centromere, kinetochore, RNA, polymerase, repetitive DNA, cell cycle
eTOC Blurb
Non-coding RNAs are required for centromere function in model systems, but the identity and function of human centromeric transcripts are less clear. McNulty et al. show that human centromeres produce array-specific, non-coding alpha satellite RNAs that differentially complex with centromere proteins for centromere assembly and cell cycle progression.
INTRODUCTION
The centromere is the chromosomal locus that orchestrates assembly of the kinetochore and attachment of spindle microtubules during meiosis and mitosis to ensure faithful chromosome segregation and genomic stability. The centromere region in humans is organized into specific chromatin domains, the core centromere comprised of centromeric chromatin and the heterochromatic pericentromere. Each of these domains is defined and maintained by a complex combination of sequence-dependent (Aldrup-MacDonald et al., 2016; Catania et al., 2015; Grimes et al., 2002; Ikeno et al., 1998) and epigenetic factors (Earnshaw and Migeon, 1985; Earnshaw et al., 1989; Warburton et al., 1997). Centromeric (CEN) chromatin is defined by the incorporation of the histone H3 variant CENP-A into a subset of nucleosomes. CENP-A nucleosomes are interspersed with canonical H3 nucleosomes that contain euchromatic histone modifications (H3K4me2, H3K36me2) to create a domain of CEN chromatin on each chromosome (Bergmann et al., 2011; Sullivan and Karpen, 2004). Loading of CENP-A into CEN chromatin occurs each cell cycle, and requires licensing of the chromatin by the Mis18 complex in order to recruit the CENP-A chaperone HJURP (Foltz et al., 2009; Nardi et al., 2016). The Mis18 complex further interacts with the histone acetyltransferase KAT7 to maintain an acetylated chromatin state that promotes CENP-A deposition (Ohzeki et al., 2016).
Studies in various organisms have demonstrated that transcription occurs within centromeres and is involved in CENP-A loading and kinetochore assembly (Blower, 2016; Catania et al., 2015; Grenfell et al., 2016; Liu et al., 2015; Quenet and Dalal, 2014; Rosic et al., 2014; Topp et al., 2004; Wong et al., 2007). In humans, RNA polymerase II (RNAP II) has been observed at centromeres in G1 and at mitosis (Quenet and Dalal, 2014; Wong et al., 2007), implicating transcription in centromere function. Recently, a 1.3kb long non-coding RNA present at ~50% of human centromeres was described (Quenet and Dalal, 2014). The chromosomal origin of the transcript was not identified, nor was it shown if this transcript acted in cis or instead in trans, as has been observed in Drosophila (Rosic et al., 2014). Although it is not impossible that human centromeres might share a homologous RNA component, it is important to consider that alpha satellite DNA sequences at human centromeres differ by both molecular and sequence organization.
Human centromeres are defined by alpha satellite DNA that is based on a 171bp monomeric subunit. Individual monomers that are only 50–70% identical are tandemly organized to form a higher order repeat (HOR) (Willard, 1985) (Figure 1A). A HOR is reiterated hundreds to thousands of times to yield long arrays that stretch for 2–5Mb and contain HORs that are 97–100% identical (Aldrup-Macdonald and Sullivan, 2014). Importantly, the number and order of specific monomers in a HOR confers chromosome-specificity (Willard, 1985). For instance, the DXZ1 array of human chromosome X (HSAX) is defined by a 12-monomer (12-mer) HOR (Figure 1B) (Waye and Willard, 1985), while the DYZ3 array of chromosome Y (HSAY) is defined by a 34-mer HOR (Figure 1C). Therefore, arrays on different chromosomes are molecularly distinguished from one another by variation in the sequence and order of individual monomers and by the overall size of the HOR. It was originally thought that each chromosome had a single large alpha satellite array at which the centromere forms. However, it has been demonstrated that 70% of human chromosomes contain more than one distinct alpha satellite array within their centromere region (Choo et al., 1990; Miga et al., 2014; Slee et al., 2011; Wevrick and Willard, 1991). Human chromosome 17 (HSA17), for example, contains three alpha satellite arrays, D17Z1, D17Z1-B, and D17Z1-C, that are each distinguished by sequence variation and differences in HOR size (Rudd et al., 2003; Rudd and Willard, 2004; Shepelev et al., 2009; Waye and Willard, 1986). Either D17Z1 or D17Z1-B can form an active centromere, depending on the chromosome and the individual (Aldrup-MacDonald et al., 2016; Maloney et al., 2012). Meanwhile, the adjacent alpha satellite arrays on HSA17 do not incorporate CENP-A or recruit other centromere proteins and are assembled into heterochromatin (Hayden et al., 2013; Maloney et al., 2012). The molecular basis for establishment and maintenance of centromeric epialleles is not well understood, but could feasibly include an RNA component.
Figure 1. Human Centromeres Produce Array-Specific Non-Coding RNAs in cis.
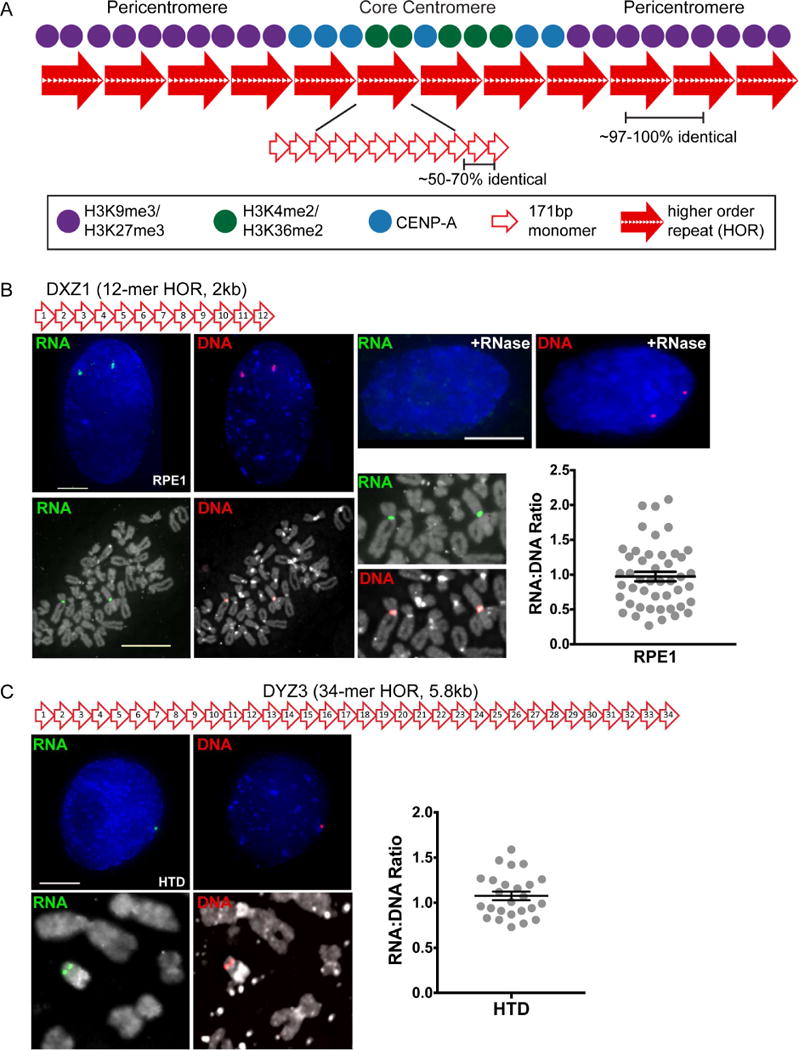
(A) Schematic of the human centromere, including core centromeric chromatin containing CENP-A and euchromatic H3 nucleosomes and flanking heterochromatin enriched for H3K9 and K27 methylation. These domains assemble on alpha satellite DNA, a 171bp monomer (open red arrows) tandemly arranged into higher order repeats (HORs, large red-filled arrows) that are extensively reiterated. A set number of monomers confers chromosome specificity so that alpha satellite DNA at each centromere is distinct.
(B) The centromere of Homo sapiens chromosome X (HSAX) is defined by DXZ1, a single array based on a 12-mer HOR. Fluorescence images were obtained from diploid RPE1 interphase cells (bar, 5μm) and metaphase chromosomes (bar, 15μm) after RNA FISH with a probe spanning the entire DXZ1 HOR (green). Sequential DNA FISH with the same probe labeled in a different fluorophore (red) verified chromosomal location of RNA signal and permitted quantification of alpha satellite RNA:DNA ratio. In each experiment, control slides were treated with RNase to verify that FISH signals detected RNA.
(C) Sequential RNA (green) – DNA (red) FISH on interphase and metaphase cells from male line HTD with a probe spanning DYZ3, the 34-mer alpha satellite HOR on HSAY and quantification of RNA:DNA ratio. In (B) and (C), each data point represents a single interphase centromere and mean ± SEM are shown. See also Figure S1.
Previous studies of human centromeric transcription took a general approach to identify transcripts without accounting for chromosome-specific alpha satellite arrays or the presence of active and inactive alpha satellite arrays on the same chromosome. Moreover, it is not known if active and inactive arrays on normal, endogenous chromosomes exhibit different transcriptional activities. Here, we address the identity of alpha satellite transcripts in the context of chromosome-specific alpha satellite arrays and centromeric epialleles. We found that transcripts are produced from individual alpha satellite arrays, even when they lack kinetochore proteins. Array-specific transcripts localize in cis, occupying distinct spatial domains throughout the cell cycle. Targeted depletion of distinct alpha satellite RNAs reduced centromere proteins in an array-specific manner. Our data indicate that each human centromere produces a chromosome-specific set of non-coding transcripts corresponding to underlying alpha satellite sequence and organization. The transcripts that are associated with centromere proteins contribute to CENP-A recruitment and appear to be stabilized by their incorporation within the centromere-kinetochore complex.
RESULTS
Each alpha satellite array produces array-specific transcripts that localize in cis
Alpha satellite RNAs have been described at the interphase nucleolus (Wong et al., 2007), although other studies did not observe this nucleolar localization (Ideue et al., 2014). Recently, a single long non-coding human centromeric transcript was reported to localize to many human centromeres, although none of the centromeres were chromosomally identified to pinpoint the chromosome of origin (Quenet and Dalal, 2014). We wanted to understand centromere transcription in the context of alpha satellite organization and chromosome-specific transcription. We first searched for RNAs homologous to the alpha satellite arrays on HSAX (DXZ1) and HSAY (DYZ3). The RNA FISH probes used spanned the entire HOR on each chromosome, thereby ensuring array-specificity (Figure 1B, 1C). DXZ1 and DYZ3 alpha satellite transcripts were present as discrete foci in every cell, including interphase cells from different asynchronous cell lines (G1/S/G2) and on metaphase chromosomes (Figure 1, Figure S1A, S1B). FACS analysis verified that multiple cell cycle stages were represented in cell populations (Figure S2A). We used immunostaining for Ki-67, a nuclear antigen that displays cell cycle-specific morphology (Bridger et al., 1998; Kill, 1996) to cytologically stage individual cells in IF-RNA FISH experiments (Figure S2). It was important to verify that RNA FISH signals corresponded to specific alpha satellite arrays, so DXZ1 or DYZ3 DNA were detected in the same cells that the RNA had been visualized using sequential DNA FISH with the identical HOR probe labeled in a different fluorophore. In all instances, DNA signals co-localized with RNA foci in every cell (Figure 1B, 1C, Figure S1B). RNA signals were determined to represent only single-stranded RNA by treating cells with RNase A and RNase T1 before fixation and hybridization, thereby abolishing RNA FISH signal (Figure 1B). These results suggest that alpha satellite transcripts are present at multiple stages of the cell cycle, are chromosome-specific, and localize in cis to the site of transcription.
HSAX and HSAY each have only one alpha satellite array, but we were also interested in measuring transcription on chromosomes like HSA17 that have multiple alpha satellite arrays that are functionally distinct (i.e. epialleles) (Maloney et al., 2012) (Figure 2A). We hypothesized that the inactive array might produce little or no transcripts due to its inactive centromeric state. RNA FISH was done in multiple cell lines under stringency conditions that distinguish D17Z1 or D17Z1-B transcripts (see STAR methods) (Aldrup-MacDonald et al., 2016; Maloney et al., 2012). In RPE1 cells that assemble the centromere at the D17Z1 array on both homologs (Aldrup-MacDonald et al., 2016), transcripts were observed at both the active D17Z1 array and the inactive D17Z1-B array in metaphase and interphase (Figure 2B). In the HSA17 epiallele cell line HTD (Maloney et al., 2012), transcripts from D17Z1 and D17Z1-B in each functional context (active and inactive) were also observed (Figure S1D). These findings indicate that transcription of alpha satellite DNA is not unique to the active centromere. Detection of transcripts at the genomic location of each array demonstrates that both D17Z1 and D17Z1-B transcripts are produced in cis and remain in distinct spatial domains throughout the cell cycle.
Figure 2. Active and Inactive Alpha Satellite Arrays Are Transcribed.
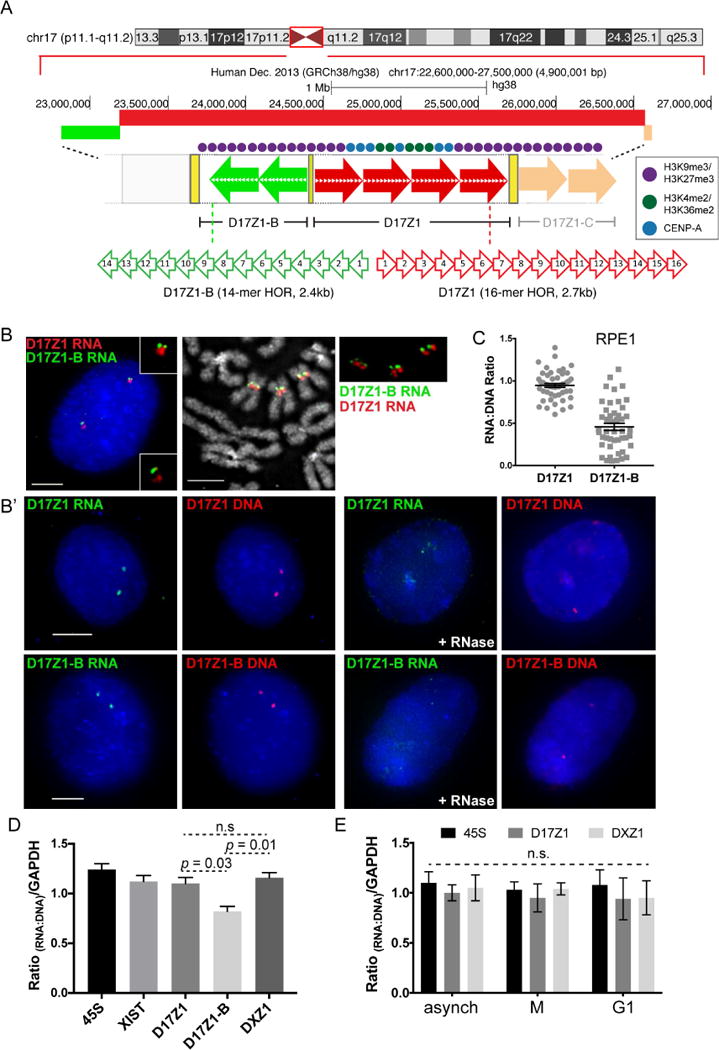
(A) HSA17 has three distinct alpha satellite arrays; either D17Z1 (red arrows) or D17Z1-B (green arrows) can be the active centromere.
(B, B′) In RPE1, D17Z1 is active on both HSA17s. RNA FISH with D17Z1 (red) and D17Z1-B (green) HOR probes on interphase cells and metaphase chromosomes (B) and RNA-DNA FISH in interphase cells (B′). RNase treatment verified detection of RNA. Bars, 5μm.
(C) Dot plots of RNA:DNA ratios of D17Z1 and D17Z1-B from RNA-DNA FISH in (B′) (mean ± SEM).
(D) RT-qPCR of D17Z1, D17Z1-B, and DXZ1 RNA in RPE1 cells relative to qPCR of gDNA of same array. 45S rRNA and XIST lncRNA relative to gDNA are shown for comparison (mean ± SEM). Data represent two biological replicates that each contained three technical replicates.
(E) RT-qPCR of D17Z1 and DXZ1 transcripts from synchronized RPE1 cells (mean ± SEM). No significant differences in RNA:DNA ratios were observed at D17Z1 or DXZ1 across the cell cycle. Data in this figure were statistically analyzed using a t-test. See also Figures S1–S3.
Alpha satellite transcripts are present at the centromere of every chromosome
To confirm that dual transcription at active and inactive arrays is not HSA17-specific, we measured transcription on HSA7, another centromere epiallele chromosome (i.e. centromere activity at either D7Z1 or D7Z2) (Figure S1E). Both arrays were actively transcribed, regardless of the location of the centromere, and array-specific transcripts remained in cis (Figure S1F). These findings show that individual alpha satellite arrays produce unique non-coding RNAs that are concentrated at and maintained at the site of transcription.
Although we did not carry out RNA FISH for every single alpha satellite array in the human genome, we asked if all human centromeres are transcribed. We carried out RNA FISH using a peptide nucleic acid (PNA) probe complementary to the CENP-B box, a 17bp consensus sequence present within a subset of monomers within all alpha satellite arrays, except for the Y chromosome (Masumoto et al., 1989). Using this probe, we observed RNA signals at the centromere of each autosome and HSAX on metaphase chromosomes and in punctate foci in interphase RPE1 cells (Figure 3A, 3A′). We did not observe aggregation of transcripts at nucleoli in interphase cells. These results indicate that transcripts are present at the centromeres of all autosomes and HSAX throughout the cell cycle. We conclude that alpha satellite transcription is a general feature of this class of highly repetitive DNA and is not restricted to centromere-forming arrays.
Figure 3. Chromosome-Specific Alpha Satellite RNAs Have Array-Specific Size Ranges.
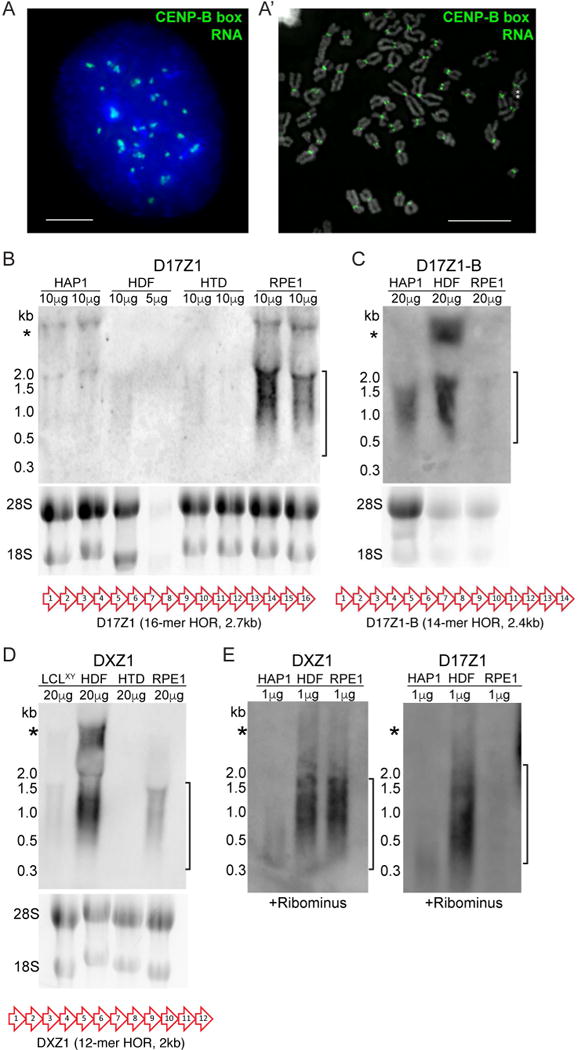
(A, A′) RNA FISH in RPE1 cells with PNA probe (green) recognizing the CENP-B box found in a subset of monomers within all alpha satellite arrays except for the Y. Multiple transcripts were detected in interphase (bar, 5μm), and at every centromere at metaphase (bar, 15μm). Asterisks denote HSAX.
(B) Northern blot detection of D17Z1 RNA in total RNA isolated from multiple cells lines, with each cell line in duplicate. Ethidium bromide stained gel is shown under each blot. Bracket indicates size range for chromosome-specific, array-specific transcripts. A schematic of the entire D17Z1 higher order repeat (HOR) that was used as a probe is shown.
(C) Northern blot of D17Z1-B RNA in multiple cell lines. A schematic of the entire D17Z1-B HOR that was used as a probe is shown.
(D) Northern blot of DXZ1 RNA. A schematic of the entire DXZ1 HOR that was used as a probe is shown.
(E) Northern blot of DXZ1 and D17Z1 RNA after rRNA depletion eliminated contaminating rRNA bands (*) observed in panels (B)–(D).
Alpha satellite transcript abundance is roughly equal to that of the underlying DNA
We next asked if active versus inactive alpha satellite arrays might be distinguished by the quantity of alpha satellite transcripts. To accurately quantitate the abundance of alpha satellite transcripts, we measured individual RNA and DNA foci in interphase cells using sequential RNA-DNA FISH. This approach is essential because alpha satellite array size varies between non-homologous chromosomes, as well as between homologs. By normalizing RNA to DNA, we can directly and more accurately compare alpha satellite transcript abundance among different arrays and different cell lines. RNA levels generally matched the amount of DNA at an active array (Figure 1B, 1C, Figure S1A, S1B). We also measured levels of alpha satellite RNAs on multi-array chromosomes in interphase cells and found that an active D17Z1 or active D17Z1-B array produced RNA at levels roughly equivalent to that of the underlying DNA. However, at inactive arrays, RNA levels varied widely, ranging from lower to equivalent amounts of RNA at the inactive array compared to the active array (Figure 2C, Figure S1C, S1D). Relative RNA:DNA ratios were confirmed by RT-qPCR of total RNA and data normalization to DNA abundance of the same array estimated by qPCR using the Pfaffl method (Pfaffl, 2001) (see STAR Methods). RT-qPCR revealed lower amounts of RNA at inactive arrays compared to active arrays, confirming RNA-DNA FISH (Figure 2D). Our data suggest that inactive alpha satellite arrays produce transcripts that are more variable in abundance.
It has been reported that human centromeric transcripts are produced primarily in early G1 (Quenet and Dalal, 2014). We performed RT-qPCR with array-specific primers on synchronized cell populations using the same comparisons to the underlying genomic DNA as for asynchronous cells (Figure 2E; Figure S3). G1 cells were collected 3 hours after release from nocodazole arrest (Figure S2A′); M phase cells were isolated by mitotic shake-off after treatment with nocodazole. Transcripts were detected in comparable amounts in both G1 and M, confirming RNA FISH results (Figure 1B, 1C, Figure 2B). Based on the RT-qPCR results, combined with RNA FISH on cells immunostained with Ki-67 (Figure S2B), we conclude that alpha satellite RNA is abundant throughout the cell cycle and is related to centromere location and the underlying alpha satellite array.
Array-specific alpha satellite transcripts vary in size
Alpha satellite HOR units vary from 1–6kb depending on the chromosome, raising the possibility that each alpha satellite array might produce differently sized transcripts. We defined the size of chromosome-specific, alpha satellite array-specific transcripts using Northern blotting of total RNA from different human cell lines. Transcripts specific to D17Z1-B and DXZ1 arrays ranged from 300 to 1800 nucleotides, while D17Z1 transcripts ranged from 500 to 2000 nucleotides (Figure 3B–D), suggesting that the size range reflects chromosome-specific, array-specific HOR sizes. Centromeric RNAs over 2000 nucleotides in size have been reported (Bouzinba-Segard et al., 2006). We also observed a large band on all Northerns, but the size suggested it was contaminating rRNA. Indeed, it was eliminated after treatment of samples with RiboMinus (Figure 3E). We noted that the abundance of transcripts produced from the same array differed between cell lines. These results suggest that individual differences in centromeric transcription are related to total array size, genomic variation, and/or cell type.
Alpha satellite transcripts from active arrays are more stable than those from inactive arrays
The identity of the RNA polymerase responsible for centromeric transcription has been an open area of debate, because evidence exists for the involvement of RNA polymerases (RNAP) I, II, and III (Blower, 2016; Chan et al., 2012; Grenfell et al., 2016; Liu et al., 2015; Longo et al., 2015; Quenet and Dalal, 2014; Wong et al., 2007). To identify the polymerase that transcribes the chromosome- and array-specific alpha satellite transcripts, we pharmacologically inhibited RNAPs. Cells were treated with Actinomycin D at low (50 ng/mL) and high (5 μg/mL) concentrations to inhibit RNAP I or RNAP I/II, respectively and with ML-60218 to inhibit RNAP III. RNA-DNA FISH was performed at time points to measure RNA:DNA ratios. Incubation of RPE1 cells with low concentrations of Actinomycin D for up to 16 hours had no effect on the abundance of active array D17Z1 or inactive array D17Z1-B transcripts (Figure 4A). However, treatment with a high concentration of Actinomycin D decreased inactive array D17Z1-B transcript abundance, with no change in RNA at active arrays DXZ1 or D17Z1 (Figure 4B–D′). XIST RNA was detected as a control and was no longer visible after 7 hours of RNAP I and II inhibition (Figure 4C′). Similarly, after 24 hours of treatment with the RNAP III inhibitor ML-60218 or inhibition of all polymerases simultaneously with Actinomycin D and ML-60218, there was no change in the abundance of D17Z1 transcripts (Figure S4A). Transcripts from control genes 45S rRNA and 5S rRNA were reduced by RNAP I or RNAP III inhibition, respectively (Figure S4B). We conclude that alpha satellite transcripts are transcribed by RNAP II and have long half-lives that are reduced if transcripts are produced from inactive arrays.
Figure 4. Active and Inactive Alpha Satellite RNAs Exhibit Different Stabilities.
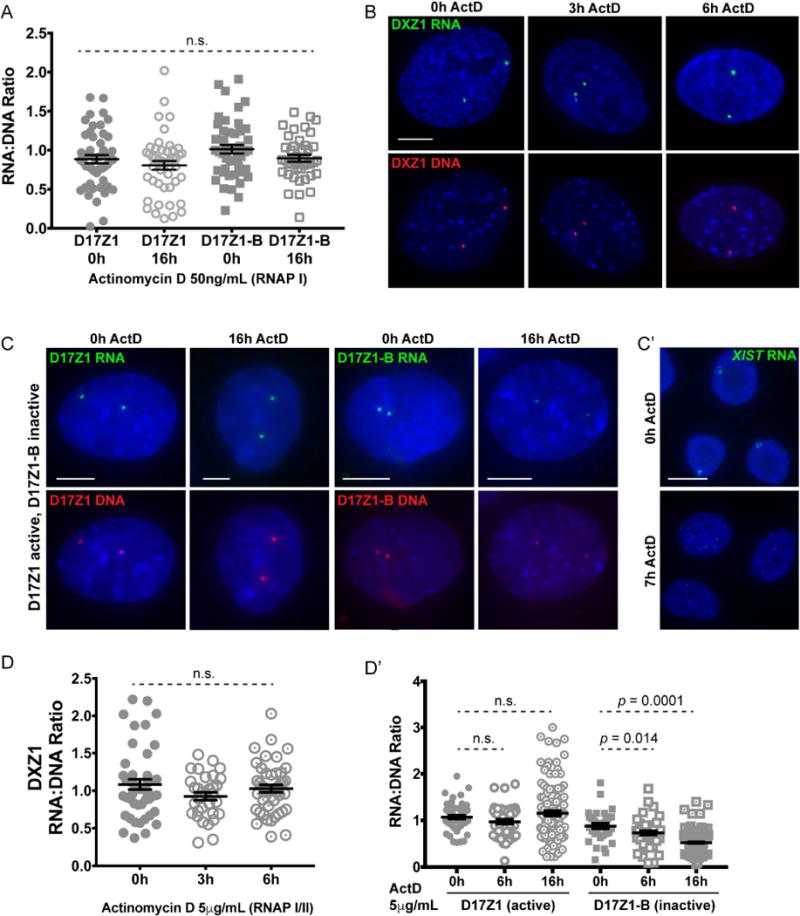
(A) Quantification of alpha satellite RNA:DNA ratios in RPE1 cells after RNA Polymerase I (RNAP I) inhibition with low concentrations of Actinomycin D (ActD) (mean ± SEM).
(B, C) RNA (green) – DNA (red) FISH for DXZ1, D17Z1, and D17Z1-B after RNAP I/II inhibition with high concentrations of ActD. DXZ1 and D17Z1 are active centromere arrays; D17Z1-B is inactive. Bars, 5μm
(C′) XIST RNA FISH (green) served as a control for efficacy of RNAP II inhibition. Bar, 15μm. (D, D′) Quantification of DXZ1 RNA:DNA ratios over 6 hours of ActD treatment, and D17Z1 and D17Z1-B over 16 hours of ActD treatment (mean ± SEM). Data were statistically analyzed using a t-test. In (A) and (D), each data point represents a single interphase centromere. See also Figure S4.
Alpha satellite transcripts from active arrays are complexed with chromatin-bound centromere proteins
The observed stability of active alpha satellite RNAs could reflect association with centromere proteins. We tested if active array transcripts co-localized with CENP-A using sequential immunofluorescence-RNA FISH (IF-RNA FISH). Using a CENP-B box PNA probe that detects both active and inactive arrays, we observed that many, but not all, RNA FISH signals co-localized with CENP-A (Figure 5A). This result was not surprising, since many chromosomes carry both active and inactive arrays, and reinforced our finding that inactive HSA7 and HSA17 arrays are transcribed. When we focused on a specific chromosome like HSAX, we observed that DXZ1 transcripts overlapped with CENP-A (Figure 5A). Only a portion of the DXZ1 transcripts (and other active arrays) co-localized with CENP-A. These results agree with our previous observation that CENP-A is only located on a portion of alpha satellite DNA (Sullivan et al., 2011). Our IF-RNA FISH results suggest that only a fraction of alpha satellite transcripts co-localizes with centromeric chromatin. These findings agree with previous studies reporting that non-coding centromeric RNAs in various organisms are present at the inner centromere (Blower, 2016; Ideue et al., 2014; Liu et al., 2015; Wong et al., 2007).
Figure 5. Alpha Satellite Transcripts Are Complexed with Centromere Proteins and are Unaffected by CENP-A Depletion.
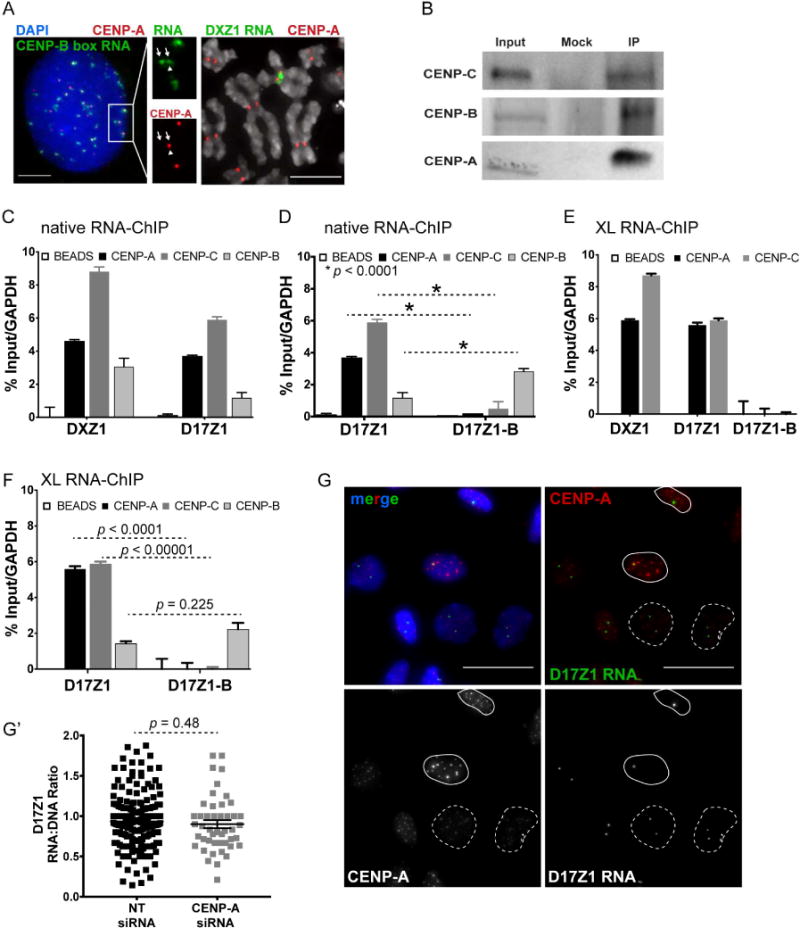
(A) Left, CENP-A immunostaining (red) and RNA FISH on RPE1 interphase cell using a CENP-B box PNA probe (green). CENP-A and ~50% of CENP-B box RNA FISH signals overlap. Inset emphasizes that chromosomes with multiple arrays produce transcripts from all arrays (arrows) yet only one array is active/associated with CENPs (arrowhead). Bar, 5μm.
Right, CENP-A immunostaining (red) and RNA FISH with DXZ1 probe (green) on cell line LCLXiX showing that CENP-A is associated with a portion of DXZ1 transcripts. Bar, 15μm.
(B) Western blots of input, mock (no antibody), and native chromatin fractions from HAP1 cells immunoprecipitated (IP) with antibodies specific for CENP-A, CENP-B, CENP-C.
(C) Native RNA-ChIP and RT-qPCR showing association of DXZ1 and D17Z1 transcripts with CENP-A and CENP-C (mean ± SEM; n=4).
(D) Native RNA-ChIP and RT-qPCR showing that transcripts from the centromere inactive array D17Z1-B are associated with CENP-B that does not discriminate between active and inactive centromeres (mean ± SEM, n = 4).
(E, F) Crosslinked RNA-ChIP and RT-qPCR showing that transcripts from active arrays DXZ1 and D17Z1, but not inactive D17Z1-B, are associated with CENP-A and CENP-C (mean ± SEM, n = 4). Active and inactive array transcripts are associated with CENP-B.
(G, G′) CENP-A immunostaining (red) and RNA FISH for D17Z1 transcripts (green) after CENP-A depletion by siRNA in HT1080 cells. Nuclei with >50% CENP-A knockdown are denoted by dashed outlines; those with little CENP-A depletion have solid outlines. Quantitation of D17Z1 RNA:DNA ratio after CENP-A depletion (mean ± SEM). Each data point represents a single interphase HSA17 centromere. Data were statistically analyzed using a t-test. See also Figures S3 and S4.
To determine if alpha satellite transcripts are physically associated with centromere proteins, we performed RNA immunoprecipitation (RIP) on native and UV crosslinked chromatin using antibodies to CENP-A, CENP-B, and CENP-C, followed by RT-qPCR to amplify chromosome-specific alpha satellite arrays (Figure 5B, Figure S4C). Both D17Z1 and DXZ1 transcripts were enriched in CENP-A and CENP-C pulldown fractions (Figure 5C, 5E). Transcripts from the inactive array D17Z1-B were not immunoprecipitated with CENP-A and CENP-C (Figure 5D, 5F). All alpha satellite transcripts were physically associated with CENP-B, a DNA binding protein that recognizes the CENP-B box sequence within active and inactive arrays (Earnshaw et al., 1989) (Figure 5C, 5D, 5F). We conclude that alpha satellite transcripts originating from the active array are physically associated with chromatin-bound centromeric proteins that define active kinetochores. Transcripts produced from inactive arrays appear to be present in a different protein complex that includes CENP-B.
Alpha satellite transcription and RNA localization is independent of CENP-A
Association of active array transcripts with CENP-A and CENP-C raised the question of when transcripts appear at the centromere, that is, before or after centromere assembly. We depleted CENP-A using siRNA transfection and measured active array alpha satellite RNA levels in individual cells. Due to the extreme stability of CENP-A protein and transfection efficiency, we observed a range in CENP-A knockdown, with a substantial (>50%) reduction in CENP-A levels observed in a subset of cells (Figure 5G, Figure S4D). As expected, CENP-C was also reduced upon loss of CENP-A (Figure S4E, S4E′) (Fachinetti et al., 2013). Even in cells most significantly depleted for CENP-A, there was no change in the abundance or localization of D17Z1 transcripts (Figure 5G′). We conclude that alpha satellite RNAs produced from active arrays are physically associated with CENP-A, but the interaction with CENP-A or CENP-C is not required for the production or retention of the transcripts at the centromere. These results suggest that alpha satellite transcription is upstream of CENP-A and CENP-C recruitment and kinetochore assembly.
Targeted depletion of alpha satellite transcripts leads to loss of key centromere proteins
To further explore the relationship between centromere assembly and alpha satellite RNAs, we depleted chromosome-specific transcripts at active arrays using double-stranded RNAs (dsRNAs) generated from in vitro transcribed alpha satellite HORs and reduced to siRNA-sized fragments. Array-specific small dsRNAs were introduced into human cells by transfection and “boosted” twice over a period of six days to deplete the long-lived alpha satellite transcripts at a single centromere. Cells were then analyzed by CENP-A and CENP-C immunostaining and RNA-DNA FISH. Depletion of DXZ1 transcripts decreased the amount of DXZ1 RNA and was accompanied by 30% (average) reduction in CENP-A and 44% (average) reduction in CENP-C (Figure 6A–6D, Figure S5). Centromere proteins were only lost at targeted DXZ1; RNAs and CENPs at a non-targeted array (D7Z1 on HSA7) were not significantly decreased (Figure 6A–6D; replicate data in Figure S5), further emphasizing the array- and chromosome-specific nature of alpha satellite transcripts.
Figure 6. Targeted Depletion of Alpha Satellite Transcripts Disrupts Centromere Assembly.
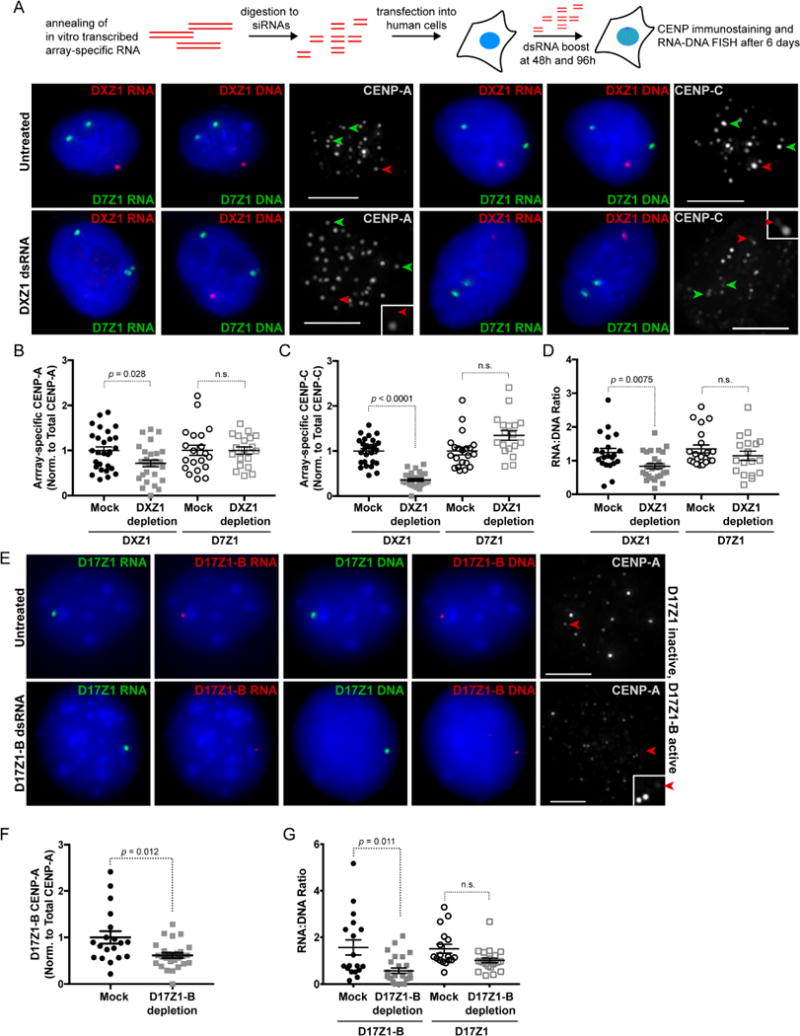
(A) HT1080 cells were transfected with array-specific dsRNAs and assayed by immunostaining and RNA-DNA FISH after 6 days. Depletion of DXZ1 transcripts using array-specific dsRNAs decreases DXZ1 RNA and reduces CENP-A only at DXZ1 (red arrowhead). Transcripts and CENP-A (green arrowheads) at control array D7Z1 (HSA7) are not decreased. Bars, 5μm.
(B, C) Quantitation of CENP-A and CENP-C at targeted DXZ1 array and control array D7Z1 (mean ± SEM).
(D) RNA:DNA ratios of targeted DXZ1 and control array D7Z1 measured from RNA-DNA FISH experiments (mean ± SEM).
(E) Depletion of active D17Z1-B transcripts in a somatic cell hybrid line Z12.3B containing a single epiallele HSA17 chromosome led to reduction of CENP-A (red arrowhead) and RNA at D17Z1-B (red) but not at inactive D17Z1 (green).
(F) Quantitation of CENP-A at D17Z1-B after dsRNA depletion (mean ± SEM). Each data point represents a single interphase HSA17.
(G) Quantitation of RNA:DNA ratios at targeted (D17Z1-B) and control (D17Z1) arrays after D17Z1-B dsRNA depletion (mean ± SEM). For (B), (D), (F), and (G) each data point represents a single centromere at interphase. Data were statistically analyzed using a t-test. See also Figure S5 for replicate data.
We also depleted transcripts on an epiallele chromosome, specifically a version of HSA17 in which the D17Z1-B is the active centromere and D17Z1 is “inactive” (Figure 6E–G). For these experiments, a single HSA17 had been placed within a somatic cell hybrid background (Maloney et al., 2012). The specificity of alpha satellite RNAs for their array of origin was especially notable in that reduction of active D17Z1-B transcripts did not change levels or localization of transcripts produced from the nearby inactive D17Z1 array, although CENP-A was decreased by an average of 36% at targeted D17Z1-B (Figure 6E–6G; replicate data in Figure S5). The effect of alpha satellite RNA depletion on an epiallele chromosome raised the possibility that centromere assembly might shift to the neighboring array. However, we did not observe movement of centromere proteins to D17Z1 when D17Z1-B transcripts were depleted (Figure 6E). These results indicate that the centromere null phenotype is not rescued via de novo centromere assembly at the neighboring array, or at least does not occur immediately after disruption of the active centromere on a normal, endogenous chromosome.
Loss of alpha satellite RNAs at a single chromosome prevents new CENP-A loading and leads to cell cycle arrest
Global loss of CENP-A leads to mitotic defects and cell cycle arrest in G1 (Fachinetti et al., 2013; Li et al., 2011; Maehara et al., 2010). In Drosophila, depletion of centromere transcripts present at all centromeres also triggers mitotic defects (Rosic et al., 2014). However, when we depleted array-specific alpha satellite transcripts, mitotic defects and, in fact, mitotic cells were not observed, even though CENP-A and CENP-C were clearly reduced at the targeted centromere (Figure 6, Figure S5). We used Ki-67 immunostaining to identify the cell cycle stage of individual cells within the population of cells depleted for DXZ1 RNA. When compared to mock transfected cells, cells lacking DXZ1 RNA were arrested in S and G2 phases of the cell cycle (Figure 7A–7C, Figure S6D, S6E). In contrast and as expected, CENP-A depleted cells were arrested in G1 (Figure S5F–S5H). Our results suggest that loss of non-coding alpha satellite transcripts at a single centromere impairs kinetochore assembly and arrests cells just prior to mitosis.
Figure 7. Cell Cycle Arrest and Impaired CENP-A Loading in the Absence of Alpha Satellite Transcripts.
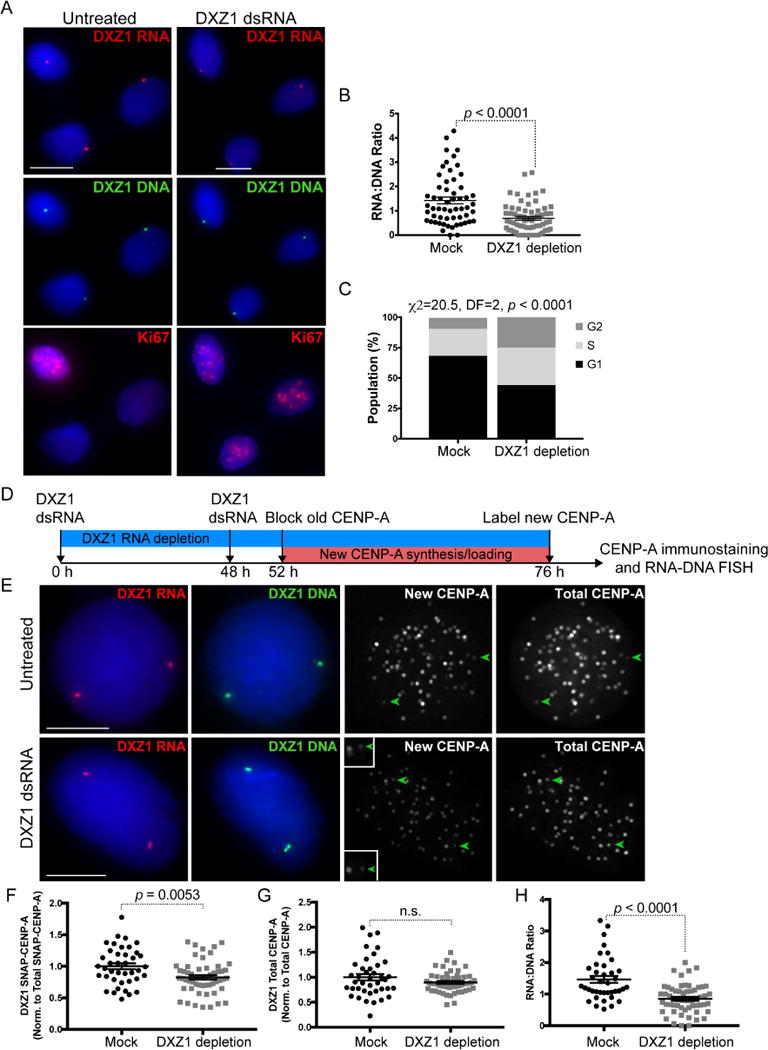
(A) Ki-67 immunostaining (red) determined cell cycle phase of individual nuclei from HT1080 cells mock-treated or depleted for DXZ1 transcripts. DXZ1 RNA (red) and DXZ1 DNA (green) were detected by sequential RNA-DNA FISH.
(B) Quantitation of DXZ1 RNA:DNA ratios at targeted DXZ1 arrays (mean ± SEM). Each data point is a single centromere at interphase.
(C) Quantitation of Ki-67-staged (G1/S/G2) interphase cells from mock-treated and DXZ1 RNA depleted cells. Bars, 10μm. N ≥ 60 cells analyzed for each treatment.
(D) Schematic of experimental approach to measure nascent SNAP-tagged CENP-A incorporation on HSAX after DXZ1 dsRNA depletion in HT1080 cells.
(E) Depletion of DXZ1 transcripts using array-specific dsRNAs decreases DXZ1 RNA (red) and reduces SNAP-CENP-A only at DXZ1 (green arrowhead). Total CENP-A is shown in far-right panel. Because DXZ1 dsRNA knockdown arrests cells [see (A)], dsRNA treatment was done for 52 hours to deplete transcripts by 50% while allowing cells to continue cycling to measure new CENP-A loading. Bars, 5μm.
(F, G) Quantitation of new (SNAP) CENP-A and total CENP-A at mock-treated and targeted DXZ1 arrays (mean ± SEM). Each data point in (C) through (E) represents a single centromere at interphase. Replicate data is presented in Figure S6.
(H) RNA:DNA ratios of mock treated and dsRNA-targeted DXZ1 (mean ± SEM). Each data point in (F) through (H) represents a single centromere at interphase. Data were statistically analyzed using a t-test, except for (C), in which the Chi-square test was used. See also Figure S6 for replicate data.
To gain additional insight into the effect of alpha satellite RNA depletion on centromere proteins, we measured loading of nascent CENP-A using the established SNAP labeling system (Jansen et al., 2007; Ross et al., 2016). In our experiments, significant depletion of alpha satellite RNA occurred by 76–144 hours after introduction of dsRNAs, resulting in notable reduction in CENP-A (Figure 6B, 6F) and cell cycle arrest. Therefore, we shortened the timeframe of array-specific alpha satellite transcript targeting experiments to deplete transcripts by 50% while still allowing cells to cycle in order to capture new CENP-A loading (Figure 7D). After 52 hours of depletion of DXZ1 transcripts, new CENP-A was detected by blocking old CENP-A and staining for nascent CENP-A using TMR-Star (Figure 7E–7H, Figure S6A–S6C). Compared to mock transfected cells, newly incorporated CENP-A at DXZ1 was reduced by ~20% (average), indicating that even partial destruction of alpha satellite RNA affects centromere assembly (Figure 7F, 7G, Figure S6). Total CENP-A at the targeted array did not differ between mock and dsRNA treated cells after 48 hours, suggesting that, in this shortened depletion window, maintenance of CENP-A was not significantly affected by reduction in DXZ1 transcripts, despite the observed defect in new CENP-A loading (Figure 7G, Figures S6B).
DISCUSSION
In this study, we identified non-coding transcripts produced in cis and in a chromosome-specific manner from distinct alpha satellite arrays. The transcripts associate with centromere proteins, and those incorporated into the functional kinetochore are more stable than those produced from inactive arrays. At alpha satellite arrays that build a kinetochore, centromere protein assembly and cell cycle progression is dependent on the presence of the transcripts.
Transcription is an important requirement of centromere assembly in various eukaryotes (Blower, 2016; Carone et al., 2009; Chan et al., 2012; Chen et al., 2015; Grenfell et al., 2016; Lu and Gilbert, 2007; Molina et al., 2016; Rosic et al., 2014; Wong et al., 2007). Our study also emphasizes that alpha satellite transcription is a key feature of eukaryotic centromeres. However, our results argue against a single, defining transcript at all human centromeres or even at a single centromere (Quenet and Dalal, 2014). Instead, we provide new evidence that alpha satellite transcripts are array-specific and are also produced from arrays that are not the site of kinetochore assembly. These findings emphasize the importance of the unique, chromosome-specific organization of alpha satellite arrays and the genomic characteristics that differentiate between arrays located on the same chromosome. Human centromeres represent gaps in the genome assembly that are currently best represented by graphical, but not contiguous or linear, maps (Miga, 2015; Miga et al., 2014). Sequence and size variation within alpha satellite DNA is wide-spread between chromosomes and individuals (Miga et al., 2014; Warburton and Willard, 1992; Wevrick and Willard, 1989; Willard and Waye, 1987). These factors could affect the relationship of the transcripts and the protein complexes with which they are associated, as well as the efficiency of centromere assembly and long-term chromosome stability.
Centromeric chromatin and heterochromatin assembly are in constant competition at human centromeres (Hall et al., 2012; Molina et al., 2016). Too much heterochromatin inactivates centromeres, preventing new CENP-A loading (Nakano et al., 2008; Ohzeki et al., 2016). Centromeres also require H3K4me2 for centromeric transcription (Molina et al., 2016), and replenishment of CENP-A within centromeric chromatin is ensured by the antagonistic activity of the KAT7 acetyltransferase that counteracts SUV39H1-mediated pericentric heterochromatin assembly (Ohzeki et al., 2016). We showed that transcription also occurs at inactive alpha satellite arrays that are enriched for H3K9 and H3K27 methylation (Maloney et al., 2012). This raises the possibility that alpha satellite RNAs partner with distinct protein complexes that have different chromosomal functions, and that “inactive” arrays are merely devoid of centromere activity but are not functionally inert. Our RNA-IP experiments showed that inactive array transcripts were only associated with CENP-B, the alpha satellite DNA binding protein. CENP-B can efficiently create and maintain heterochromatin on ectopic alpha satellite (Okada et al., 2007), thus, CENP-B-bound inactive array transcripts could be part of establishing and maintaining pericentric heterochromatin. However, CENP-B can also promote CENP-A assembly within an acetylated chromatin environment (Ohzeki et al., 2012; Okada et al., 2007), stabilizing CENP-A nucleosomes and recruiting CENP-C (Fachinetti et al., 2015). This dichotomic behavior might be important in the context of human centromeric epialleles, in which two juxtaposed arrays each produce transcripts. The chromatin context (KAT7-mediated acetylation versus SUV39H1-mediated methylation) and differential role of CENP-B may be key factors that maintain chromatin domains and functional states through distinct interactions with alpha satellite transcripts, with levels of array-specific transcripts fine-tuning the balance between efficient centromere and heterochromatin assembly (Carone et al., 2013). Future characterization of proteins associated with RNAs produced from arrays that lack centromeric proteins will extend our understanding of the functional roles of human alpha satellite DNA.
Like proteins in the constitutive centromere-associated network (CCAN), alpha satellite RNAs are present throughout the cell cycle, and not only in G1. Alpha satellite transcripts may interact distinctly with proteins that are constitutively and transiently associated with the centromere. Our data suggest that alpha satellite transcripts at active centromeres are components of centromeric chromatin complexes. Their role in kinetochore assembly appear to be high in the assembly hierarchy. Nascent protein labeling indicated that loading of new CENP-A is impaired at alpha satellite arrays that are devoid of transcripts, suggesting that the non-coding RNAs themselves act on a chromosome-specific level to recruit CENP-A, its chaperone HJURP, or licensing factors such as MIS18BP1. RNA has been implicated in CENP-A recruitment by binding prenucleosomal CENP-A/HJURP (Quenet and Dalal, 2014), however, our findings support physical association of alpha satellite transcripts with chromatin-bound centromere proteins.
In our experiments, CENP-C was particularly sensitive to alpha satellite RNA depletion. Targeted depletion of RNAs produced from one chromosome-specific array led to a reduction in CENP-A but an even more remarkable reduction in CENP-C. Chromatin-bound RNAs may recruit or stabilize specific centromere proteins. CENP-C is a particularly interesting candidate because it is a known RNA-binding protein (Du et al., 2010; Topp et al., 2004; Wong et al., 2007) that interacts with MIS18BP1 for new CENP-A loading and stabilizes other centromeric proteins by linking inner and outer kinetochore protein complexes (Falk et al., 2015; Klare et al., 2015; Moree et al., 2011). CENP-A itself is required to mediate CENP-C assembly at centromeres, and a distinctive CENP-B/CENP-C interaction further enhances centromere assembly (Fachinetti et al., 2013; Fachinetti et al., 2015). Based on our results, we envision a model in which the absence of alpha satellite transcripts that are normally part of the CENP-A/-B/-C complex results in impaired centromere assembly and altered kinetochore architecture. These defects are highlighted by the inability to load new CENP-A and recruit and/or stabilize CENP-C, as well as additional kinetochore proteins.
Depletion of non-coding alpha satellite transcripts at a single chromosome (DXZ1 on HSAX) reduced essential centromere proteins at that centromere and arrested cells prior to mitosis. Previous studies have shown that a single structurally or functionally defective centromere is sufficient to trigger the spindle assembly checkpoint (SAC) (Kerscher et al., 2003; Rieder et al., 1994; Spencer and Hieter, 1992). Disruption of CENP-ACID function in Drosophila delays cells in interphase at a ‘kinetochore assembly’ checkpoint (Blower and Karpen, 2001), presumably to protect against an error-prone mitosis. Human cells that lack CENP-A but have functional p53 arrest also arrest in interphase and become senescent (Maehara et al., 2010). Our results, combined with these other studies, suggest that in normal cells, mechanisms exist to avoid mitotic defects in the absence of centromeric RNAs or important centromere proteins, both globally and at a single centromere.
To fully understand centromere assembly, it is important to distinguish between the act of transcription, such as chromatin remodeling, nucleosome eviction, and polymerase recruitment, and the non-coding products themselves. In Drosophila, the act of transcription and associated transcription factors are required for de novo centromere assembly (Chen et al., 2015). Our approach of depleting array-specific RNAs after their production separates the action of transcription through the centromere from the physical transcripts and suggests that the transcripts themselves are important for maintenance of full centromere function and kinetochore assembly. Additionally, the observation that dsRNA depletion did not alter centromere proteins at untargeted chromosomes further highlights the cis-acting nature of array-specific transcripts. We did not observe trans effects of alpha satellite RNAs, as described in Drosophila and Xenopus (Blower, 2016; Rosic et al., 2014). This was most notable on multi-array, epiallele chromosomes (HSA7 and HSA17) exhibiting transcripts from both the active and inactive array. When active centromere D17Z1-B transcripts were depleted, CENP-A did not move to the neighboring D17Z1 array and D17Z1 transcripts did not relocate to the crippled D17Z1-B centromere. How centromeric epialleles are established and why they exist is not clear. Comparable levels of transcription from each array may promote epiallele activity, such that if one centromere becomes compromised or mutated, a back-up array exhibiting appropriate transcription is available. However, the relationship between alpha satellite organization and transcription may also contribute to centromeric epiallele. Genomic variation within D17Z1 is unfavorable for centromere assembly and function (Aldrup-MacDonald et al., 2016). Variation may alter the structure or function of alpha satellite RNAs so that variant transcripts cannot interact properly with centromere proteins. Dissecting differences in transcription of wild-type and variant alpha satellite arrays could be important for determining mechanisms by which alpha satellite transcripts confer centromere assembly and chromosome inheritance.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Additional information and requests for reagents and protocols should be directed to and will be fulfilled by the Lead Contact, Beth Sullivan (beth.sullivan@duke.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines and Culture Conditions
Human dermal fibroblast HDF cells (male), human fibrosarcoma HT1080 (HTD; male) cells, and human retinal pigment epithelial RPE1 cells (female) were maintained in Minimum Essential Medium (MEM) Alpha. HCT116 cells (male) were grown in McCoy’s medium. HAP1 (haploid; single X) cells were grown in IMDM. B-lymphocyte GM07357 (LCLXY; male) and GM03543 (LCLXiX; female) cells were maintained in RPMI 1640 medium supplemented with 15% fetal bovine serum. The human-mouse somatic cell hybrid Z12.3B line containing a single HSA17 with an active D17Z1-B array (Aldrup-MacDonald et al., 2016) was maintained in MEM alpha supplemented with hypoxanthine-aminopterin-thymidine (HAT) and ouabain. Unless otherwise indicated, all cell culture media was supplemented with 10% FBS and 1X antibiotic-antimycotic. All cell lines were grown at 37°C with 5% CO2.
METHOD DETAILS
Cell Cycle Assessment
HT1080 cells were treated with 50ng/mL nocodazole overnight. Mitotic cells were harvested by shake off, re-plated, and grown in complete medium for 3 hours. Cells were harvested by trypsinization and 1×106 cells were fixed in 100% EtOH, then stained with staining solution (1X PBS, 50 g/mL propidium iodide, 0.1mg/mL RNase A, 0.05% Triton X-100) or a control solution omitting propidium iodide. Propidium iodide fluorescence was analyzed using the PE-Cy5 channel on the FACSCanto System (BD Biosciences) and FlowJo 10 analysis software.
RPE1 cells were transfected with 10nM of each CENP-A Trilencer-27 siRNA knockdown duplex using Oligofectamine with two additional boosts at 48 hours and 96 hours after initial transfection. Cells were harvested for Ki-67 immunostaining and CENP-A IF 144 hours (6 days) after the first transfection. HT1080 cells were treated with 50nM DXZ1 dsRNA using Oligofectamine and boosted 48 hours after the initial transfection. Cells were harvested for Ki-67 immunostaining and RNA-DNA FISH 96 hours (4 days) after the first transfection. Ki-67 staining patterns were used to classify cell cycle phase of individual cells.
Probe Preparation for FISH and Northern Blotting
Plasmid probes were labeled by nick translation with biotin, digoxygenin, or AlexaFluor dUTP. Hybridization was performed in 50–68% formamide buffer. One microliter of 200mM ribonucleoside vanadyl complex (RVC) was added for RNA FISH experiments. For RNA and DNA FISH experiments, biotin labeled probes were detected with AlexaFluor 488 or 594 Streptavidin; digoxygenin labeled probes were detected with anti-digoxigenin conjugated to FITC or Cy3.
Northern Blotting
Total RNA was isolated from RPE1, HAP1, HDF, HTD, and GM7357 (LCLXY) cells using TRIzol Reagent and, in some cases, ribosomal RNA was removed from 10μg total RNA with the RiboMinus Eukaryote Kit. Total RNA (10–20μg) or 1μg rRNA-depleted RNA was diluted in 2X RNA Loading Dye and denatured at 65°C. Samples containing ethidium bromide were separated on a 1.2% agarose, 6.6% formamide, 1X MOPS gel. After imaging the rRNA subunits, RNA was transferred to a Hybond N+ nylon membrane. RNA was UV-crosslinked to the membrane using the autocrosslink setting on the Stratalinker (Stratagene). The membrane was prehybridized in ExpressHyb at 68°C, hybridized with 150ng digoxigenin-labeled probe overnight at 68°C, then washed twice with 2X SSC/0.1% SDS and once with 0.2X SSC/0.1% SDS, both at 68°C. Digoxigenin-probe hybridization was detected using DIG-High Prime DNA Labeling and Detection Protocol. After addition of Tropix CDP-Star chemiluminescent substrate, probe signal was visualized using the G:Box CHEMI XT4 imaging system and GeneSys software (Syngene).
RNA-DNA FISH
Cells were harvested by trypsinization, diluted in hypotonic buffer, and centrifuged onto slides treated with RNase AWAY. Control slides were treated with 7.5μL/mL RNase Cocktail diluted in Cytoskeletal (CSK) buffer for 2 hours at 37°C. Slides were permeabilized on ice in CSK buffer (100mM NaCl, 300mM sucrose, 3mM MgCl2, 10mM PIPES, pH 6.8, containing 2mM RVC), then CSK buffer/RVC/0.5% Triton X-100, and finally CSK buffer/RVC. Cells were fixed in 4% paraformaldehyde (PFA) in PBS at room temperature and washed twice with 70% ethanol/RVC before storing overnight in 70% ethanol/RVC. Cells were dehydrated in ice-cold ethanol (70%/80%/95%/100%) and hybridized with the appropriate denatured probe at high stringency conditions (65–70% hybridization mix) to specifically distinguish alpha satellite arrays. Stringency conditions below 65% can result in cross-hybridization of array-specific probes with other centromeric arrays due to the presence of similar monomer sequence on multiple chromosomes. Slides were hybridized overnight with denatured probe at 37°C. Slides were washed in 2X SSC/0.05% Tween-20 containing 65–70% formamide, 2X SSC/0.05% Tween-20, 1X SSC/0.05% Tween-20, and finally 4X SSC/0.1% Tween-20. Slides were mounted in VECTASHIELD containing 1μg/mL DAPI.
For hybridization with PNA CENP-B box probe, cells were prepared as described above. After dehydration in the ethanol series, cells were hybridized for 1 hour with 2ng biotin-labeled CENP-B box PNA probe in 70% formamide, 10mM Tris HCl, pH 7.5, and 10X Blocking Reagent. Slides were washed twice in 10mM Tris HCl pH 7.5, 70% formamide, 0.1% BSA and once in 4X SSC/0.01% Tween-20.
After imaging RNA signals, DNA detection was done. Coverslips were removed, and cells were fixed in 10% formalin, then permeabilized in KCM. Slides were denatured in 70% formamide/2X SSC, pH7 for up to 5 minutes at 70°C and hybridized overnight with denatured probe under a sealed coverslip at 37°C. Slides were washed and mounted as described above.
IF-RNA FISH
Cells treated with hypotonic were spun onto RNase AWAY-treated slides at 800 rpm (interphase cells) or 2000 rpm (metaphase chromosomes). Slides were permeabilized in KCM at room temperature, fixed in 4% PFA in PBS, and incubated again in KCM. Slides were blocked in 1X PBS/0.5% Triton X-100/1% BSA before incubation overnight at 4°C with primary antibodies to human CENP-A (Abcam ab13939 or custom polyclonal AP3497) (Maloney et al., 2012), human CENP-C (Abcam ab50974), or mouse Cenp-A (custom polyclonal D601AP) overnight. Slides were washed in PBS/0.1% Triton X-100, incubated with fluorophore-conjugated secondary antibodies for 2 hours at room temperature, and washed as before. Slides were rinsed in 1X PBS before proceeding to RNA FISH, as described above, beginning at the permeabilization step.
RNA Polymerase Inhibition
All polymerase inhibitions were done in RPE1 cells. RNA Polymerase I inhibition was achieved using 50ng/mL Actinomycin D at 60–70% confluency for up to 16 hours. RNA Polymerases I and II were jointly inhibited using 5μg/mL Actinomycin D for up to 16 hours. Inhibition of RNA Polymerase III was performed for up to 24 hours using 25μM ML-60218.
Semi-Quantitative RT-PCR
RPE1 cells were seeded in 6 well plates and treated with 50ng/μL Actinomycin D for 16 hours or 25μM ML-60218 for 24 hours to inhibit RNA Polymerase I or RNA Polymerase III, respectively. Total RNA was isolated from an equal number of cells treated with Actinomycin D or untreated cells using the RNeasy Mini Kit. Genomic DNA was eliminated by on-column DNase I digestion. 45S rRNA and GAPDH were reverse transcribed from 50 ng of total RNA and amplified using the SuperScript One-Step RT-PCR system with Platinum Taq DNA Polymerase. Small RNA-containing total RNA was isolated from an equal number of cells treated with ML-60218 or untreated cells using the mirVana miRNA Isolation Kit and genomic DNA was eliminated by treatment with the TURBO DNA-free Kit. 5S rRNA and GAPDH were reverse transcribed from 100 ng of total RNA and amplified using the SuperScript One-Step RT-PCR system with Platinum Taq DNA Polymerase. All RT-PCR products were run on an ethidium bromide-stained 2% TAE agarose gel alongside a Low Molecular Weight DNA Ladder. Gels were visualized using the G:Box CHEMI XT4 system and GeneSys software. Individual bands were quantified using the Gel Analyzer tool (ImageJ). 45S or 5S amplification products were normalized to GAPDH levels from the same sample. GAPDH, 45S RNA, and DXZ1 primers were described previously (Stimpson et al., 2014; Sullivan et al., 2011)
RT-qPCR
RPE1 and HTD cells were seeded into 6 well plates (75,000 – 125,000 cells/well) and grown for ~24 hours before synchronization. Asynchronous cells were left untreated for the duration. For metaphase synchronization, cells were treated with nocodazole (100ng/mL) for 6 hours, then mitotic cells were collected by shake-off. To acquire G1 cells, cells were incubated in nocodazole (50ng/mL) for 5 hours, then mitotic cells were shaken off, re-plated, and grown for an additional 3 hours. Stabilized RNA lysates were isolated from asynchronous/synchronous cells using reagents from the FastLane Cell cDNA Kit and purified using the RNeasy Mini Kit before continuing with the FastLane procedure. Lysates were purified again using the gDNA wipeout step of the FastLane Cell cDNA Kit, followed by first-strand cDNA synthesis. Each time-course or asynchronous experiment was repeated at least once. Reactions lacking reverse transcriptase (RT-) were included for each sample. Genomic DNA (gDNA) from asynchronous, mitotic, and G1 cells, grown using the manufacturer’s growth and treatment conditions for RNA extraction/cDNA synthesis was isolated using the Quick-gDNA Mini-prep Kit. RT-qPCRs were performed using the QuantiFast SYBR Green PCR kit with primers specific to 45S RNA, DXZ1, D17Z1, D17Z1-B, and XIST. Each primer pair was optimized for efficiency and specificity prior to analyzing experimental samples (Figure S3). RT-qPCR was performed in triplicate for each sample within the respective run. Ct values (triplicates) were averaged and SEM was calculated. Abundance of RNA to DNA relative to GAPDH was determined using Pffafl methodology. Genomic DNA served as a transformer, and RT-minus values served as controls in the calculation: Eˆ(Ct(QueryRT−) − (QueryRT+)/Ct(QuerygDNA))/Eˆ(Ct(RefRT−) − (RefRT+)/Ct(RefgDNA)).
siRNA and dsRNA Depletions
For CENP-A RNAi, 10nM of each CENP-A Trilencer-27 siRNA knockdown duplex was introduced into RPE1 cells using Oligofectamine with additional boosts at 48 hours and 96 hours after initial transfection. Double-stranded RNAs recognizing specific alpha satellite array transcripts were generated by in vitro transcription (IVT) of plasmids containing a single DXZ1 or D17Z1-B HOR insert flanked by T3 and T7 promoters. T3 RNA Polymerase and T7 RNA Polymerase were used to produce sense and anti-sense DXZ1 and D17Z1-B HOR single-stranded RNAs. Complementary IVT products were annealed and treated with TURBO DNase and RNase Cocktail to remove single-stranded RNA. Long dsRNAs were treated with RNase III to generate ~21 base pair dsRNAs that were transfected into HT1080 or Z12.3B cells at a final concentration of 50nM using Oligofectamine. Additional dsRNA boosts were done at 48 hours and 96 hours after initial transfection. For both CENP-A siRNA and dsRNA depletions, cells were harvested for CENP-A or CENP-C IF and RNA-DNA FISH 144 hours (6 days) after the first transfection.
dsRNA Depletion and SNAP-CENP-A Detection
HT1080 cells stably expressing SNAP-CENP-A-HA (Ross et al., 2016) were treated with 50nM DXZ1 dsRNA using Oligofectamine and boosted with additional DXZ1 dsRNA 48 hours after initial transfection. Existing SNAP-CENP-A was blocked for 4 hours after the second dsRNA transfection by the addition of bromothenylpteridine (BTP) (SNAP-Cell Block) in complete growth medium for 30 minutes at 37°C. BTP was removed by washing twice with 1X PBS and once with complete growth medium, and incubating cells in complete growth medium for 30 minutes to allow excess compound to diffuse from cells (Ross et al., 2016). Cells were again washed twice with 1X PBS and once with complete growth medium, then grown in complete growth media for 24 hours to allow for synthesis and incorporation of new CENP-A. New CENP-A was pulse labeled by incubation with 2μM SNAP-Cell TMR-Star for 30 minutes at 37°C. Cells were washed and complete media was added. After an additional 30 minutes, washes were performed again to remove any excess TMR-Star that had diffused. Cells were then harvested for CENP-A IF and RNA-DNA FISH.
RIP/RNA-ChIP
Irreversibly crosslinked samples
Cells were UV crosslinked in cold 1X PBS, pelleted, and re-suspended in lysis buffer for 10 minutes at room temperature with mechanical shearing. CENP-A (custom polyclonal AP3497; 2μg/IP), CENP-B (Abcam, ab25734, 5μg/IP), or CENP-C (Abcam, ab30334, 8μg/IP) antibodies were pre-incubated with Dynabeads prior to overnight incubation with chromatin. Chromatin complexes were treated with DNase I and RNase Inhibitors, washed, and then RNA was heated to 55°C with 1.2mg/mL RNase-free Proteinase K for 30 minutes. Proteinase K was inactivated at 65°C prior to RNA elution. RNA-Bee was used to purify RNA. After genomic DNA wipeout, RNA was reverse transcribed using Quantitect Reverse Transcription kit. RT− samples were matched to each RT+ sample.
Native chromatin samples
Native chromatin was isolated from nuclei using micrococcal nuclease in the presence of RNase-free reagents (Mravinac et al., 2009). Ten micrograms of chromatin were used in each immunoprecipitation with antibodies to CENP-A (0.5μg), CENP-B (8μg), and CENP-C (5–8μg) (described above). RNA-ChIP products were isolated using a modified MAGnify ChIP protocol (Aldrup-MacDonald et al., 2016) that included the addition of DNase I and RNase Inhibitors during the chromatin-antibody binding step. RNA-protein complexes were immunoprecipitated in replicate within the same experiment and/or by repeating the experiment at least once. RNA was extracted using either RNeasy or RNA-Bee. Two consecutive rounds of gDNA removal were performed prior to RT (first round: RNeasy cleanup; second round: Qiagen’s gDNA wipeout reagent for 5m at 42°C). RT was performed using Qiagen’s Quantiscript RT. RT-minus samples were included. RT-qPCR was performed on all samples using primers to 28S rRNA and/or 45S rRNA (Stimpson et al., 2014)to confirm that RT samples produced Ct values and that RT-minus samples yielded a value of “No Ct” before proceeding with query regions of interest. RNA quality was verified on a 1.1% agarose gel containing 1% household bleach. For all RNA-ChIP, RT-qPCR was performed using Qiagen’s QuantiFast SYBR Green kit and 2μL of cDNA per reaction. RT-qPCR for each sample replicate was performed in duplicate or triplicate, depending on sample availability. For D17Z1 and D17Z1-B runs, 500pg-1ng of p17H8 (D17Z1) and p2.5-3 (D17Z1-B) plasmids were included as specificity controls. Percent Input was calculated as follows: Eˆ(Adjusted inputquery - Ct Mean (IP)query)/Eˆ(Adjusted inputGAPDH - Ct (IP)GAPDH).
Western Blot Confirmation of IP Pull-down
Western Blot (WB) confirmation was performed for MAGnify ChIP Input, Mock, CENP-A, CENP-B, and CENP-C. For Input, 6μg (for CENP-A) and 10μg (for CENP-B and CENP-C) of chromatin were combined with equal volumes of Laemmli Sample Buffer (LSB) supplemented with 2-mercaptoethanol (CENP-A) or dithiothreitol (DTT; CENP-B, CENP-C) and boiled for 10 minutes at 95°C. IP magnetic beads (chromatin-antibody bound or mock chromatin bound) were re-suspended in 10μl–50μl of LSB after the last IP wash step and boiled for 10 minutes at 95°C. Following denaturation, the magnetic beads were pelleted and the supernatant was collected. This step was repeated to ensure complete bead removal from the sample. For CENP-A (custom polyclonal AP3497), a single reaction was sufficient for WB detection. For CENP-B and CENP-C (commercial antibodies), up to five IP reactions were combined. Samples were separated on a BioRad Mini-PROTEAN TGX stain-free precast gel (4–20% for CENP-A; 7.5% for CENP-B and CENP-C) at 70V for 2–2.5hr and visualized on the G:Box. Proteins were then wet-transferred in 8% methanol onto a nitrocellulose membrane at 70V for 3 hours at 4°C and imaged again. The membrane was blocked in either 5% non-fat dry milk in TBST for 1 hour (CENP-A) or overnight at 4°C in StartingBlock T20 (CENP-B, CENP-C). Primary antibodies were applied and incubated overnight at 4°C with rocking in either 5% milk/TBST (CENP-A, AP3497 1:500) or StartingBlock T20 (CENP-B ab25734 at 1:750; CENP-C ab33034 at 1:750). The membrane was then washed 3 × 5 minutes in TBST with rocking at room temperature. For CENP-A, a goat anti-rabbit HRP antibody (1:2000 in 5% milk/TBST) was incubated with the membrane for 2 hours at room temperature with rocking, followed by 3 × 5 minute washes in TBST at room temperature. For CENP-B and CENP-C, Clean-Blot IP Detection Reagent (HRP) was applied at 1:40 in StartingBlock T20 and incubated overnight at 4°C with rocking followed by 3×5m washes in TBST at room temperature with rocking. Bands were detected using WesternBright ECL solution at a 1:1 ratio (CENP-A) for 2 minutes or Pierce ECL Detection Reagents at a 1:1 ratio (CENP-B, CENP-C) for 5 minutes. Imaging was performed using the G:Box’s chemiluminescence settings.
Microscopy and Imaging
All microscopic images were collected using a DeltaVision Elite imaging system (Applied Precision/GE Healthcare) inverted Olympus IX-71 microscope coupled to the Photometrics CoolSNAP HQ2 CCD camera. Images were acquired using 60X (PLAN APO 1.42 NA) or 100X (PLAN APO 1.40 NA) oil objectives (Olympus) with 0.2μm z sections. Images were deconvolved using the conservative algorithm with 10 iterations, and images stacks were viewed as quick projections that were saved as TIFF and PSD formats.
Quantifications and Statistical Analyses
TIFF images of RNA-DNA FISH signals and centromere protein immunofluorescence signals were analyzed in ImageJ using a custom macro. Fluorescent signals in each image channel were segmented after automatic background subtraction and defined by thresholding manually or using the ImageJ Intermodes settings. The integrated density of each segmented region of interest (ROI, i.e. RNA or DNA FISH signal or CENP spot) was measured and values were exported to Excel. For RNA-DNA FISH experiments, the RNA and DNA signals for a specific centromere were identified and matched in the Excel file, and the RNA:DNA ratio was calculated. For CENP-A siRNA experiments, CENP-A signals were segmented by thresholding and the sum total integrated density of all CENP-A signals in the nucleus that was defined by DAPI signal were measured and values were exported to an Excel file. The total CENP-A value for a specific nucleus was matched to the D17Z1 RNA-DNA ratios calculated for that same cell. Nuclei with significant CENP-A depletion were defined as those containing <50% of the average amount of total CENP-A present in NT (control) siRNA-treated cells. This same approach was used to quantify CENP-C signals in CENP-A siRNA-treated cells and NT siRNA-treated cells. For quantification of CENP-A, SNAP-CENP-A and CENP-C in alpha satellite RNA depletion experiments, CENPs associated with specific centromeric arrays were identified by DNA FISH, that is, matching the DNA FISH signal to a specific CENP signal/ROI. The integrated density of the array-specific CENP signal was measured and matched to a column in an Excel file containing the sum total integrated density of all CENP signals for that particular nucleus, defined by DAPI signal. These two values were used to calculate the amount of CENP at an individual array by comparing it to the total CENP signal in the nucleus, presenting it as a percentage of the total amount of CENP in the nucleus. To normalize the CENP measurements in cells treated with dsRNAs, the average ratio (CENPs at targeted centromere to CENPs at all centromeres) that had been calculated in control cells was set to 1, and each dsRNA-treated ratio was divided by this average. At least 20 independent nuclei or chromosomes were measured per experiment, and the standard error of the mean was determined for each set of data points. Data were statistically analyzed in Excel or GraphPad Prism using a Student’s t-test and all data were presented as graphs using GraphPad Prism. Data in Figure 7C, Figure S4H, and Figure S6E were analyzed using the Chi-square test. The p-values are indicated on figure panels.
Supplementary Material
HIGHLIGHTS.
Human centromeres produce array- and chromosome-specific non-coding RNAs in cis
Both active and inactive arrays produce non-coding RNAs; the latter are less stable
Alpha satellite RNAs are physically associated with centromere proteins
Array-specific RNAs are necessary for new CENP-A loading and localization of CENP-C
Acknowledgments
We thank Hunt Willard for alpha satellite plasmids and So Young Kim (Duke Functional Genomics Core) for HAP1 cells. We are grateful to Jack Keene and Jeff Blackinton (Duke University) for helpful advice during the early stages of this project. We acknowledge Stefanie Sarantopoulos and Jonathan Poe (Duke University) for assistance with FACS analysis and Rachel O’Neill (University of Connecticut) for the crosslinked RNA-IP protocol. We sincerely thank Jeffrey Fergus and Loni Pickle at ThermoFisher for advice on optimizing Western Blot conditions following MAGnify ChIP. This work was supported by National Science Foundation Graduate Research Fellowship DGE-1644868 (S.M.M), National Institutes of Health grant R01 GM098500 (B.A.S.), and March of Dimes Foundation grant 1-FY13-517 (B.A.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
S.M.M, L.L.S., B.A.S conceived and designed the experiments; S.M.M and L.L.S. performed the experiments; S.M.M., L.L.S., B.A.S. analyzed the data; B.A.S directed the research; S.M.M. and B.A.S. wrote the manuscript.
References
- Aldrup-MacDonald ME, Kuo ME, Sullivan LL, Chew K, Sullivan BA. Genomic variation within alpha satellite DNA influences centromere location on human chromosomes with metastable epialleles. 2016 doi: 10.1101/gr.206706.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldrup-Macdonald ME, Sullivan BA. The past, present, and future of human centromere genomics. Genes (Basel) 2014;5:33–50. doi: 10.3390/genes5010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann JH, Rodriguez MG, Martins NM, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LE, Earnshaw WC. Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. EMBO J. 2011;30:328–340. doi: 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD. Centromeric Transcription Regulates Aurora-B Localization and Activation. Cell Rep. 2016;15:1624–1633. doi: 10.1016/j.celrep.2016.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Karpen GH. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat Cell Biol. 2001;3:730–739. doi: 10.1038/35087045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouzinba-Segard H, Guais A, Francastel C. Accumulation of small murine minor satellite transcripts leads to impaired centromeric architecture and function. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8709–8714. doi: 10.1073/pnas.0508006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridger JM, Kill IR, Lichter P. Association of pKi-67 with satellite DNA of the human genome in early G1 cells. Chromosome Res. 1998;6:13–24. doi: 10.1023/a:1009210206855. [DOI] [PubMed] [Google Scholar]
- Carone DM, Longo MS, Ferreri GC, Hall L, Harris M, Shook N, Bulazel KV, Carone BR, Obergfell C, O’Neill MJ, et al. A new class of retroviral and satellite encoded small RNAs emanates from mammalian centromeres. Chromosoma. 2009;118:113–125. doi: 10.1007/s00412-008-0181-5. [DOI] [PubMed] [Google Scholar]
- Carone DM, Zhang C, Hall LE, Obergfell C, Carone BR, O’Neill MJ, O’Neill RJ. Hypermorphic expression of centromeric retroelement-encoded small RNAs impairs CENP-A loading. Chromosome Res. 2013;21:49–62. doi: 10.1007/s10577-013-9337-0. [DOI] [PubMed] [Google Scholar]
- Catania S, Pidoux AL, Allshire RC. Sequence Features and Transcriptional Stalling within Centromere DNA Promote Establishment of CENP-A Chromatin. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan FL, Marshall OJ, Saffery R, Kim BW, Earle E, Choo KH, Wong LH. Active transcription and essential role of RNA polymerase II at the centromere during mitosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1979–1984. doi: 10.1073/pnas.1108705109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Bowers S, Lipinszki Z, Palladino J, Trusiak S, Bettini E, Rosin L, Przewloka MR, Glover DM, O’Neill RJ, et al. Establishment of Centromeric Chromatin by the CENP-A Assembly Factor CAL1 Requires FACT-Mediated Transcription. Dev Cell. 2015;34:73–84. doi: 10.1016/j.devcel.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo KH, Earle E, Vissel B, Filby RG. Identification of two distinct subfamilies of alpha satellite DNA that are highly specific for human chromosome 15. Genomics. 1990;7:143–151. doi: 10.1016/0888-7543(90)90534-2. [DOI] [PubMed] [Google Scholar]
- Du Y, Topp CN, Dawe RK. DNA binding of centromere protein C (CENPC) is stabilized by single-stranded RNA. PLoS Genet. 2010;6:e1000835. doi: 10.1371/journal.pgen.1000835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw WC, Migeon BR. Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma. 1985;92:290–296. doi: 10.1007/BF00329812. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Ratrie H, 3rd, Stetten G. Visualization of centromere proteins CENP-B and CENP-C on a stable dicentric chromosome in cytological spreads. Chromosoma. 1989;98:1–12. doi: 10.1007/BF00293329. [DOI] [PubMed] [Google Scholar]
- Fachinetti D, Folco HD, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol. 2013;15:1056–1066. doi: 10.1038/ncb2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Han JS, McMahon MA, Ly P, Abdullah A, Wong AJ, Cleveland DW. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Dev Cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk SJ, Guo LY, Sekulic N, Smoak EM, Mani T, Logsdon GA, Gupta K, Jansen LE, Van Duyne GD, Vinogradov SA, et al. Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science (New York, NY) 2015;348:699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Bailey AO, Yates JR, 3rd, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenfell AW, Heald R, Strzelecka M. Mitotic noncoding RNA processing promotes kinetochore and spindle assembly in Xenopus. The Journal of cell biology. 2016;214:133–141. doi: 10.1083/jcb.201604029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimes BR, Rhoades AA, Willard HF. Alpha-satellite DNA and vector composition influence rates of human artificial chromosome formation. Molecular therapy: the journal of the American Society of Gene Therapy. 2002;5:798–805. doi: 10.1006/mthe.2002.0612. [DOI] [PubMed] [Google Scholar]
- Hall LE, Mitchell SE, O’Neill RJ. Pericentric and centromeric transcription: a perfect balance required. Chromosome Res. 2012;20:535–546. doi: 10.1007/s10577-012-9297-9. [DOI] [PubMed] [Google Scholar]
- Hayden KE, Strome ED, Merrett SL, Lee HR, Rudd MK, Willard HF. Sequences associated with centromere competency in the human genome. Mol Cell Biol. 2013;33:763–772. doi: 10.1128/MCB.01198-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ideue T, Cho Y, Nishimura K, Tani T. Involvement of satellite I noncoding RNA in regulation of chromosome segregation. Genes to cells: devoted to molecular & cellular mechanisms. 2014;19:528–538. doi: 10.1111/gtc.12149. [DOI] [PubMed] [Google Scholar]
- Ikeno M, Grimes B, Okazaki T, Nakano M, Saitoh K, Hoshino H, McGill NI, Cooke H, Masumoto H. Construction of YAC-based mammalian artificial chromosomes. Nature biotechnology. 1998;16:431–439. doi: 10.1038/nbt0598-431. [DOI] [PubMed] [Google Scholar]
- Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. The Journal of cell biology. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Crotti LB, Basrai MA. Recognizing chromosomes in trouble: association of the spindle checkpoint protein Bub3p with altered kinetochores and a unique defective centromere. Mol Cell Biol. 2003;23:6406–6418. doi: 10.1128/MCB.23.18.6406-6418.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kill IR. Localisation of the Ki-67 antigen within the nucleolus. Evidence for a fibrillarin-deficient region of the dense fibrillar component. J Cell Sci. 1996;109(Pt 6):1253–1263. doi: 10.1242/jcs.109.6.1253. [DOI] [PubMed] [Google Scholar]
- Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A. CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. The Journal of cell biology. 2015;210:11–22. doi: 10.1083/jcb.201412028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhu Z, Zhang S, Yu D, Yu H, Liu L, Cao X, Wang L, Gao H, Zhu M. ShRNA-targeted centromere protein A inhibits hepatocellular carcinoma growth. PLoS One. 2011;6:e17794. doi: 10.1371/journal.pone.0017794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Qu Q, Warrington R, Rice A, Cheng N, Yu H. Mitotic Transcription Installs Sgo1 at Centromeres to Coordinate Chromosome Segregation. Molecular cell. 2015;59:426–436. doi: 10.1016/j.molcel.2015.06.018. [DOI] [PubMed] [Google Scholar]
- Longo MS, Brown JD, Zhang C, O’Neill MJ, O’Neill RJ. Identification of a recently active mammalian SINE derived from ribosomal RNA. Genome Biol Evol. 2015;7:775–788. doi: 10.1093/gbe/evv015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Gilbert DM. Proliferation-dependent and cell cycle regulated transcription of mouse pericentric heterochromatin. The Journal of cell biology. 2007;179:411–421. doi: 10.1083/jcb.200706176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehara K, Takahashi K, Saitoh S. CENP-A reduction induces a p53-dependent cellular senescence response to protect cells from executing defective mitoses. Mol Cell Biol. 2010;30:2090–2104. doi: 10.1128/MCB.01318-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney KA, Sullivan LL, Matheny JE, Strome ED, Merrett SL, Ferris A, Sullivan BA. Functional epialleles at an endogenous human centromere. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13704–13709. doi: 10.1073/pnas.1203126109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto H, Masukata H, Muro Y, Nozaki N, Okazaki T. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. The Journal of cell biology. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miga KH. Completing the human genome: the progress and challenge of satellite DNA assembly. Chromosome Res. 2015;23:421–426. doi: 10.1007/s10577-015-9488-2. [DOI] [PubMed] [Google Scholar]
- Miga KH, Newton Y, Jain M, Altemose N, Willard HF, Kent WJ. Centromere reference models for human chromosomes X and Y satellite arrays. Genome research. 2014;24:697–707. doi: 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina O, Vargiu G, Abad MA, Zhiteneva A, Jeyaprakash AA, Masumoto H, Kouprina N, Larionov V, Earnshaw WC. Epigenetic engineering reveals a balance between histone modifications and transcription in kinetochore maintenance. Nat Commun. 2016;7:13334. doi: 10.1038/ncomms13334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. The Journal of cell biology. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mravinac B, Sullivan LL, Reeves JW, Yan CM, Kopf KS, Farr CJ, Schueler MG, Sullivan BA. Histone modifications within the human X centromere region. PLoS One. 2009;4:e6602. doi: 10.1371/journal.pone.0006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, Kandels-Lewis S, Larionov V, Earnshaw WC, Masumoto H. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardi IK, Zasadzinska E, Stellfox ME, Knippler CM, Foltz DR. Licensing of Centromeric Chromatin Assembly through the Mis18alpha-Mis18beta Heterotetramer. Molecular cell. 2016;61:774–787. doi: 10.1016/j.molcel.2016.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Bergmann JH, Kouprina N, Noskov VN, Nakano M, Kimura H, Earnshaw WC, Larionov V, Masumoto H. Breaking the HAC Barrier: histone H3K9 acetyl/methyl balance regulates CENP-A assembly. EMBO J. 2012;31:2391–2402. doi: 10.1038/emboj.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohzeki J, Shono N, Otake K, Martins NM, Kugou K, Kimura H, Nagase T, Larionov V, Earnshaw WC, Masumoto H. KAT7/HBO1/MYST2 Regulates CENP-A Chromatin Assembly by Antagonizing Suv39h1-Mediated Centromere Inactivation. Dev Cell. 2016;37:413–427. doi: 10.1016/j.devcel.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Ohzeki J, Nakano M, Yoda K, Brinkley WR, Larionov V, Masumoto H. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenet D, Dalal Y. A long non-coding RNA is required for targeting centromeric protein A to the human centromere. eLife. 2014:e03254. doi: 10.7554/eLife.03254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. The Journal of cell biology. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosic S, Kohler F, Erhardt S. Repetitive centromeric satellite RNA is essential for kinetochore formation and cell division. The Journal of cell biology. 2014;207:335–349. doi: 10.1083/jcb.201404097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JE, Woodlief KS, Sullivan BA. Inheritance of the CENP-A chromatin domain is spatially and temporally constrained at human centromeres. Epigenetics Chromatin. 2016;9:20. doi: 10.1186/s13072-016-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd MK, Schueler MG, Willard HF. Sequence organization and functional annotation of human centromeres. Cold Spring Harb Symp Quant Biol. 2003;68:141–149. doi: 10.1101/sqb.2003.68.141. [DOI] [PubMed] [Google Scholar]
- Rudd MK, Willard HF. Analysis of the centromeric regions of the human genome assembly. Trends in genetics: TIG. 2004;20:529–533. doi: 10.1016/j.tig.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Shepelev VA, Alexandrov AA, Yurov YB, Alexandrov IA. The evolutionary origin of man can be traced in the layers of defunct ancestral alpha satellites flanking the active centromeres of human chromosomes. PLoS Genet. 2009;5:e1000641. doi: 10.1371/journal.pgen.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slee RB, Steiner CM, Herbert BS, Vance GH, Hickey RJ, Schwarz T, Christan S, Radovich M, Schneider BP, Schindelhauer D, et al. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene. 2011;31:3244–3253. doi: 10.1038/onc.2011.502. [DOI] [PubMed] [Google Scholar]
- Spencer F, Hieter P. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:8908–8912. doi: 10.1073/pnas.89.19.8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stimpson KM, Sullivan LL, Kuo ME, Sullivan BA. Nucleolar organization, ribosomal DNA array stability, and acrocentric chromosome integrity are linked to telomere function. PLoS One. 2014;9:e92432. doi: 10.1371/journal.pone.0092432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH. Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nature structural & molecular biology. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LL, Boivin CD, Mravinac B, Song IY, Sullivan BA. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 2011;19:457–470. doi: 10.1007/s10577-011-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN, Zhong CX, Dawe RK. Centromere-encoded RNAs are integral components of the maize kinetochore. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15986–15991. doi: 10.1073/pnas.0407154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, Stetten G, Gimelli G, Warburton D, Tyler-Smith C, Sullivan KF, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Current biology: CB. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Willard HF. PCR amplification of tandemly repeated DNA: analysis of intra- and interchromosomal sequence variation and homologous unequal crossing-over in human alpha satellite DNA. Nucleic Acids Res. 1992;20:6033–6042. doi: 10.1093/nar/20.22.6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Chromosome-specific alpha satellite DNA: nucleotide sequence analysis of the 2.0 kilobasepair repeat from the human X chromosome. Nucleic Acids Res. 1985;13:2731–2743. doi: 10.1093/nar/13.8.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye JS, Willard HF. Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol. 1986;6:3156–3165. doi: 10.1128/mcb.6.9.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick R, Willard HF. Long-range organization of tandem arrays of alpha satellite DNA at the centromeres of human chromosomes: high-frequency array-length polymorphism and meiotic stability. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:9394–9398. doi: 10.1073/pnas.86.23.9394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wevrick R, Willard HF. Physical map of the centromeric region of human chromosome 7: relationship between two distinct alpha satellite arrays. Nucleic Acids Res. 1991;19:2295–2301. doi: 10.1093/nar/19.9.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willard HF. Chromosome-specific organization of human alpha satellite DNA. Am J Hum Genet. 1985;37:524–532. [PMC free article] [PubMed] [Google Scholar]
- Willard HF, Waye JS. Chromosome-specific subsets of human alpha satellite DNA: analysis of sequence divergence within and between chromosomal subsets and evidence for an ancestral pentameric repeat. J Mol Evol. 1987;25:207–214. doi: 10.1007/BF02100014. [DOI] [PubMed] [Google Scholar]
- Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome research. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


