Abstract
Macrophages play pivotal roles in both the induction and resolution phases of inflammatory processes. Macrophages have been shown to synthesize anti-inflammatory fatty acids in an LXR-dependent manner, but whether the production of these species contributes to the resolution phase of inflammatory responses has not been established. Here, we identify a biphasic program of gene expression that drives production of anti-inflammatory fatty acids 12–24h following TLR4 activation and contributes to down-regulation of mRNAs encoding pro-inflammatory mediators. Unexpectedly, rather than requiring LXRs, this late program of anti-inflammatory fatty acid biosynthesis is dependent on SREBP1 and results in the uncoupling of NFκB binding from gene activation. In contrast to previously identified roles of SREBP1 in promoting production of IL1β during the induction phase of inflammation, these studies provide evidence that SREBP1 also contributes to the resolution phase of TLR4-induced gene activation by reprogramming macrophage lipid metabolism.
Introduction
Failure to resolve endogenous or extrinsic inflammatory stimuli can lead to a chronic state of low-grade inflammation that results in cellular dysfunction and tissue damage (Tabas and Glass, 2013). Recent studies have shown that the immune and metabolic systems are highly integrated with one another (Cildir et al., 2013). For instance, increased infiltration of pro-inflammatory macrophages in adipose tissue, liver and skeletal muscle and their release of cytokines that impair local insulin signaling contributes to insulin resistance (Lumeng et al., 2008; Osborn and Olefsky, 2012; Tencerova et al., 2015; Varma et al., 2009; Wynn et al., 2013; Xu et al., 2003). In addition, immune cell function itself is coordinately regulated with cellular metabolism (Spann and Glass, 2013). For example, upon inflammatory activation, macrophages rapidly induce glycolysis through HIF-1α and NF-κB, enabling them to trigger microbicidal activity even in a hypoxic inflammatory tissue environment (Huang et al., 2014a; Rodriguez-Prados et al., 2010; Tannahill et al., 2013). In contrast, macrophages display a shift to oxidative metabolism of glucose and fatty acids and acquire an anti-inflammatory phenotype in the context of tissue repair and remodeling (Mantovani et al., 2013; Rodriguez-Prados et al., 2010).
Macrophage activation in response to ligation of TLR4 provides a paradigm for investigation of molecular mechanisms that positively and negatively regulate inflammatory responses (Iyer et al., 2010; Medzhitov and Horng, 2009). TLR4 signaling induces immediate/early gene expression through activation of latent transcription factors that include members of the NFκB, IRF and AP-1 families (Glass and Natoli, 2015; Medzhitov and Horng, 2009). These factors in turn induce secondary response genes via the production of type I interferons, TNFα and other signaling molecules. Collectively, the immediate/early and secondary responses drive expression of inflammatory response genes that support innate immunity and set the stage for adaptive immunity. TLR4 signaling also results in down-regulation of a broad program of gene expression, although molecular mechanisms are less well characterized.
Recent lipidomic analysis in macrophages revealed an immediate reduction of fatty acid synthesis in response to TLR4 activation, followed by an increase in eicosanoid synthesis that was linked to the arachidonic acid pathway and delayed responses characterized by sphingolipid and sterol biosynthesis (Dennis et al., 2010). Lipid uptake is activated by chronic (~24 hours) LPS treatment, leading triglycerides to accumulate in lipid droplets within macrophages (Feingold et al., 2012; Huang et al., 2014b). These changes in lipid metabolism may be linked to changes in macrophage activity over the time-course of the response to LPS.
Macrophages can also synthesize anti-inflammatory fatty acids under the control of liver X receptors (LXRs) α and β (Li et al., 2013; Spann et al., 2012). The LXR pathway is de-repressed following genetic deletion of the nuclear receptor co-repressor NCoR, leading to increased production of 9Z palmitoleic acid and polyunsaturated omega-3 and −9 fatty acids (Li et al., 2013). These fatty acids exert anti-inflammatory functions in macrophages in part by binding to G protein coupled receptors (Oh da et al., 2010). NCoR deletion in macrophages conferred protection of mice from high fat diet-induced inflammation and insulin resistance (Li et al., 2013). Therefore, it is possible that anti-inflammatory fatty acids produced by the macrophage act in an autocrine/paracrine manner to regulate its function autonomously as well as the functions of surrounding parenchymal cells.
Cholesterol and fatty acid homeostasis are regulated at the level of transcription by LXRs and SREBPs 1 and 2 (Goldstein et al., 2006; Hong and Tontonoz, 2014; Horton et al., 2002). Their roles in cholesterol homeostasis are largely antagonistic. SREBPs (primarily SREBP2) drive transcriptional programs that increase cellular cholesterol synthesis and import (Horton et al., 2002), while LXRs induce expression of genes that mediate cholesterol efflux and inhibit import (Hong and Tontonoz, 2014). In contrast, LXRs and SREBPs (primarily SREBP1) function in a coordinate manner to positively regulate fatty acid biosynthesis. LXRs directly activate the expression of SREBP1c, and both LXRs and SREBP1 bind to and activate numerous genes involved in fatty acid biosynthesis (Repa et al., 2000a; Schultz et al., 2000). Further, at co-bound genomic loci, SREBP functions in a permissive manner, allowing signal-specific tailoring of LXR-mediated activation of lipid metabolic gene expression profiles (Spann et al, Cell 2012), resulting in context dependent synthesis and output of select lipid species.
LXRs and SREBPs also play important roles in regulating macrophage activation. LXRs primarily function to inhibit inflammatory responses by antagonizing pro-inflammatory transcription factors such as NFκB (Ghisletti et al., 2009; Hong and Tontonoz, 2014) and by activating genes with anti-inflammatory activities, such as Mer and Abca1 (Gonzalez et al., 2009; Ito et al., 2015). In contrast, SREBP1 has been found to promote the acute inflammatory response by regulating genes involve in the production of active Il1β (Im et al., 2011; Reboldi et al., 2014). Further, the LXR pathway is subject to negative regulation by TLR4 (Castrillo et al., 2003). This suggests that macrophage fatty acid synthesis is influenced by TLR signaling via temporal modulation of LXR activities.
To address the question of whether TLR4 signaling regulates the production of anti-inflammatory fatty acids, we analyzed lipidomic data generated by the LIPD MAPs consortium evaluating the temporal response of primary mouse macrophages to the specific TLR4 agonist Kdo2 LIPID A (KLA) (http://www.lipidmaps.org/)(Dennis et al., 2010). This analyses revealed that the intracellular content of anti-inflammatory mono- and poly- (ω−3,−9) unsaturated fatty acids was rapidly decreased at early time points of TLR4-mediated inflammation; while the resolution phase was characterized by increased intracellular unsaturated fatty acid levels. This temporal pattern of changes in specific lipid species was correlated with changes in mRNAs encoding corresponding biosynthetic enzymes. Unexpectedly, we found that the late up-regulation of unsaturated fatty acid synthesis was independent of LXR, but was instead driven by SREBP1. Anti-inflammatory fatty acid synthesis was compromised in Srebf1−/− macrophages at late time points compared to wild-type macrophages, concomitant with a hyper-inflammatory state due to impaired resolution of NF-κB associated activity and gene expression. Supplementation with exogenous mono- and polyunsaturated fatty acids rescues the late hyper-inflammatory response in both Srebf1−/− macrophages and Srebf1−/− mice. Collectively, these findings provide evidence that SREBP1 contributes to resolution of pro-inflammatory TLR4 signaling by reprogramming fatty acid metabolism.
Results
TLR4 signaling reprograms macrophage fatty acid metabolism
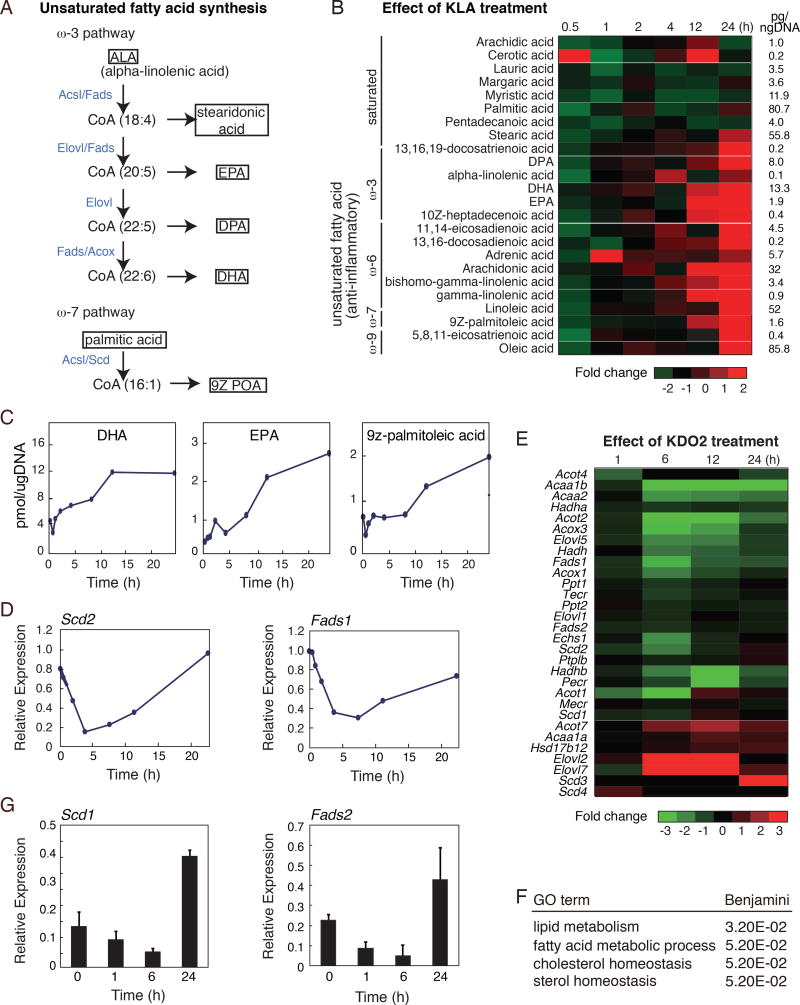
To investigate changes in macrophage fatty acid levels throughout the course of an inflammatory response, we utilized lipidomic data generated by the LIPID MAPS Consortium (http://www.lipidmaps.org/)(Dennis et al., 2010). Metabolic pathways responsible for generation of long chain omega-3 fatty acids and 9Z palmitoleic acid (9Z–POA) are shown in Fig. 1A. Activation of TLR4 by KLA, a chemically defined substructure of bacterial lipopolysaccharide (LPS) that is specifically recognized by Toll-like receptor 4 (Raetz et al., 2006), rapidly and transiently decreased the cellular content of most fatty acids analyzed (Fig.1B). Unexpectedly, in addition to the known up-regulation of omega-6 fatty acids, such as arachidonic acid, the cellular content of anti-inflammatory omega-3 and omega-9 fatty acids was also significantly increased during the late inflammatory response (12–24 hours after KLA treatment) (Fig. 1B and 1C). Based on estimates of cell volume, maximum intracellular concentrations of DHA are on the order of 10 µM and EPA and 9Z-POA are on the order of 2 µM.
Figure 1. Activation of TLR4 reprograms macrophage fatty acid metabolism.
A. Pathway maps illustrating omega-3 and −9 pathways. Enzymes catalyzing each step are highlighted in blue.
B. Lipidomic analysis of saturated and unsaturated fatty acids (omega-3, −6, −7 and −9) in thioglycollate-elicited macrophages treated with KLA for 0, 1, 6, 12, 24 hours.
C. Cellular content of omega-3 (DHA and EPA) and omega-7 (9Z-POA) fatty acids in thioglycollate-elicited macrophages treated with KLA for 0, 1, 6, 12, 24 hours.
D. Relative mRNA expression levels for Scd2 and Fads1 determined by microarray analysis of RNA from thioglycollate-elicited macrophages treated with KLA for 0, 1, 6, 24 hours.
E. Heat map of mRNA expression levels determined by RNA-Seq analysis of bone marrow-derived macrophages with KLA for 0, 1, 6, 24 hours (FDR < 0.01, RPKM > 0.5).
F. Functional annotations associated with genes exhibiting KLA repressed-induced temporal expression patterns.
G. Relative mRNA expression of Scd2 and Fads2 in human monocyte-derived macrophages treated with KLA for 0, 1, 6, and 24 hours.
Values are expressed as mean ± SEM. *p<0.05, **p<0.01.
See also Figure S1.
Analysis of microarray data from the same KLA-treated macrophages, generated by the LIPID MAPS Consortium (http://www.lipidmaps.org/)(Dennis et al., 2010), demonstrated biphasic expression of genes encoding enzymes involved in mono-unsaturated and omega-3 polyunsaturated fatty acid biosynthesis, exemplified by Scd2, Fads1, Acox3, and Elovl5 (Fig. 1D, Suppl. Fig. 1). To independently confirm these findings, we performed RNA-sequencing (RNA-seq), throughout a time course of KLA treatment. These experiments also revealed a common biphasic expression pattern for many of the genes involved in synthesis of unsaturated fatty acids, exemplified by Scd1/2, Elovl5, Fads1, Acsl3, and Acox3 (Fig. 1E). This temporal pattern is characterized by an initial transient reduction within 1–6 hours of TLR4 activation, and subsequent activation in the late phase of the TLR4 response (Fig. 1D, E). The rapid decrease in lipid species observed at 30 minutes (Figure 1B) precedes the decrease in mRNA levels of biosynthetic genes (Fig. 1D, E), indicating that initial phase of reduced fatty acid levels is determined by post-transcriptional mechanisms. However, the increase in levels of mono and polyunsaturated fatty acids between 12 and 24 hours is correlated with increases in mRNA levels of corresponding biosynthetic genes. Whole transcriptome analysis revealed that among the 22,455 measurable transcripts, 2993 genes with RefSeq annotations were reduced >2.0-fold at 6 hours, and subsequently increased 2.0-fold at 24 hours after KLA treatment. We define this subset of KLA-regulated genes as KLA repressed-induced genes. Consistent with temporal changes in macrophage fatty acid content, the entire set of KLA repressed-induced genes was significantly enriched for functional annotations linked to lipid metabolism (Fig. 1F). Experiments in KLA-treated human monocyte-derived macrophages revealed a similar biphasic expression pattern for genes involved in synthesis of unsaturated fatty acids, exemplified by Scd1 and Fads2 (Fig. 1G). The temporal pattern characterized by induction in the late phase of the TLR4 response, suggests that the observed temporal dynamics of specific fatty acid metabolic reprogramming is conserved in humans. Collectively, these findings indicate that TLR4 signaling induces a biphasic reprogramming of fatty acid metabolism in macrophages through transcriptional and post-transcriptional mechanisms.
Biphasic expression of fatty acid biosynthetic genes is independent of LXRs
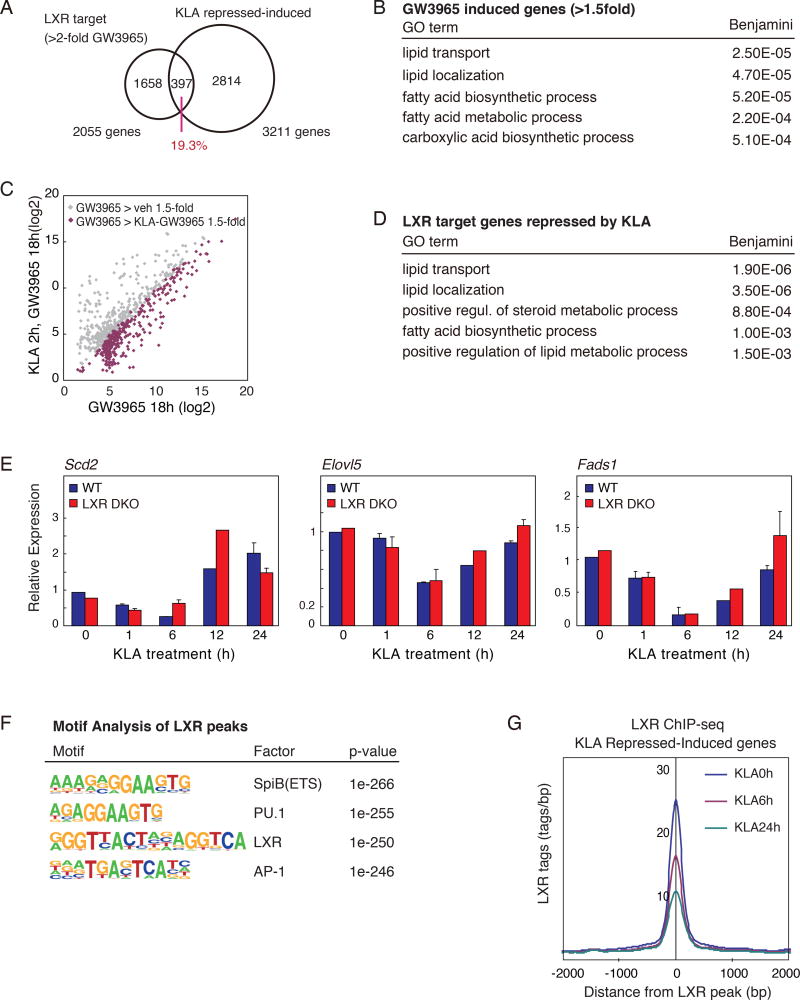
Many enzymes involved in unsaturated fatty acid synthesis are products of LXR-regulated target genes (Calkin and Tontonoz, 2012; Hong and Tontonoz, 2014). Because TLR4 activation can repress LXR induction of gene expression (Castrillo et al., 2003), it is possible that altered LXR activity could account for the biphasic pattern of expression observed for genes involved in mono and polyunsaturated fatty acid biosynthesis. To address this possibility, we performed RNA-seq analysis of RNA recovered from macrophages treated with either vehicle or the synthetic LXR agonist GW3965. Approximately one fifth (19.3%) of LXR target genes (GW3965>vehicle 2-fold), are represented as KLA repressed-induced genes (Fig. 2A). In addition, gene ontology analysis revealed that both LXR-induced and KLA repressed-induced genes are enriched for similar functional annotations, including lipid metabolism and fatty acid metabolic process (Fig. 2B). To further examine the extent to which TLR4-mediated inflammation repressed LXR-dependent gene expression, macrophages were pretreated with vehicle or KLA, followed by treatment with either vehicle or the LXR agonist GW3965. RNA-seq revealed that LXR target gene activation was markedly attenuated by KLA pretreatment (~42% GW3965-induced genes, Fig. 2C), consistent with previous findings (Joseph et al., 2003). These TLR4-compromised, LXR target genes were significantly enriched for functional annotations linked to lipid transport, lipid localization, and fatty acid biosynthetic process (Fig. 2D). These data suggest that the macrophage LXR-regulatory program involved in synthesis of unsaturated fatty acid is repressed in the early phase of TLR4 activation, which could be important for allowing appropriate induction of the inflammatory response.
Figure 2. Genes required for anti-inflammatory fatty acid synthesis demonstrate biphasic temporal expression patterns following TLR4 ligation.
A. Venn diagram of overlap between LXR target genes (GW3965 induced genes >2-fold vs untreated) and KLA repressed-induced genes.
B. Functional annotations associated with LXR target genes induced by GW3965 treatment.
C. Scatter plot depicting the relationship between fold change of LXR target genes (GW3965 > 1.5-fold vs. untreated) comparing RNA-Seq data from thioglycollate-elicited macrophages treated with GW3965 (18 hours), with or without KLA pretreatment (100ng/ml for 2 hours).
D. Functional annotations associated with LXR target genes repressed by KLA treatment.
E. Scd2, Elovl5 and Fads1 mRNA expression in LXRα/β−/− and WT thioglycollate-elicited macrophages treated with KLA for 0, 1, 6, 12, and 24 hours.
F. De novo motif analysis of LXR peaks in WT thioglycollate-elicited macrophages.
G. Normalized distribution LXR ChIP-Seq tag density, at enhancers vicinal to KLA repressed-induced genes, in thioglycollate-elicited macrophages treated with KLA for 0, 1, 6, 24 hours.
See also Figure S2.
To assess whether the repression of LXR activity is required for the early reduction of unsaturated fatty acid related gene expression and production following KLA treatment, we took advantage of LXR-deficient macrophages. The temporal dynamics of TLR4 activation was assessed by expression profiling of KLA treated macrophages prepared from wild type and LXRα/β−/− mice (Repa et al., 2000b). Unexpectedly, quantitative PCR analysis of the temporal mRNA expression patterns of genes involved in unsaturated fatty acid synthesis, exemplified by Scd2, Elovl5 and Fads1, revealed similar patterns in LXRα/β−/− and wild type macrophages (Fig. 2E). We further evaluated effects of TLR4 activation on the genome-wide location of endogenous LXRs by chromatin immunoprecipitation sequencing (ChIP-Seq). These studies revealed co-localization of LXRs with macrophage lineage-determining factors PU.1 and AP-1 based on motif co-enrichment (Fig. 2F), consistent with previous studies using tagged LXRs in RAW264.7 macrophages (Heinz et al., 2010). Further, these studies revealed an unexpected finding that LXR binding at KLA repressed-induced loci significantly decreases in the late phase of inflammation (Fig. 2G, Suppl. Fig. 2). Thus temporal changes in LXR binding are disassociated from late phase induction of KLA repressed-induced genes.
TLR4 signaling reprograms enhancer activities near repressed-induced genes
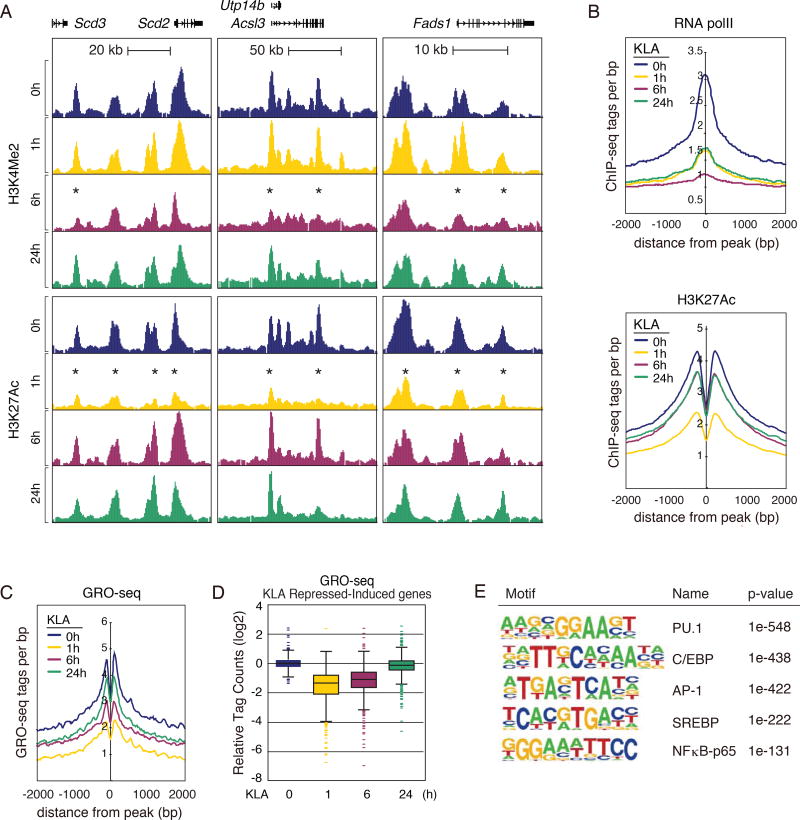
The unexpected finding that LXR is dispensable for late activation of genes directing fatty acid metabolism prompted us to analyze the local enhancer landscapes of these genes for candidate regulators associated with the temporal profile of repressed-induced genes. To identify enhancers exhibiting temporal activities associated with KLA repressed-induced genes, we performed ChIP-seq to analyze the dimethylation status of lysine 4 of histone H3 (H3K4me2), acetylation status of lysine 27 of histone H3 (H3K27ac), and RNA polymerase II (RNA polII) in naive and KLA-stimulated macrophages. Whereas H3K27ac and RNA polII correlate positively with active transcriptional activity (Creyghton et al., 2010; Kaikkonen et al., 2013), deposition of H3K4me2 has been demonstrated as an indicator of both previous and current local transcription (He et al., 2010; Kaikkonen et al., 2013; Ostuni et al., 2013).
Consistent with their reduced mRNA levels (Fig. 1D), genomic loci of representative KLA repressed-induced genes, exemplified by Scd2/3, Acsl3, and Fads1, are associated with decreased H3K27ac and H3K4me2 during the early phase inflammatory response at KLA 1 and 6 hours respectively (asterisks, Fig. 3A). Prior studies revealed that the lineage determining transcription factor (LDTF) PU.1 is necessary for establishing macrophage-specific cistromes for signal responsive transcription factors (Heinz et al., 2010). Centering our analysis on PU.1-bound regions, we analyzed the temporal pattern of relevant features at enhancers associated with repressed-induced genes. Chromatin features of active transcription, defined by RNA polII and H3K27ac, were decreased during the early inflammation phase (at 1 h post KLA), then subsequently increased at 6 and 24 hours post KLA stimulation (Fig. 3B), preceding increased levels of nearby mRNA.
Figure 3. Temporal dynamics of cis-reglatory elements associated with KLA repressed-induced genes.
A. UCSC genome browser images illustrating normalized tag counts for H3K4Me2 and H3K27Ac, at the LXR target genes in bone marrow-derived macrophages treated with KLA for 0, 1, 6 and 24 hours.
B. Distribution of RNA Pol II and H3K27Ac tag densities in vicinity of KLA repressed-induced enhancers in bone marrow-derived macrophages treated with KLA for 0, 1, 6, 24 hours.
C. Distribution of GRO-Seq tags at KLA repressed-induced enhancers in thioglycollate-elicited macrophages treated with KLA for 0, 1, 6, 24 hours.
D. Relative distribution of GRO-Seq tags at gene bodies of KLA repressed-induced genes in thioglycollate-elicited macrophages treated with KLA for 0, 1, 6, 24 hours.
E. Sequence motifs enriched at enhancers associated with KLA repressed-induced genes.
As a more direct analysis of active transcription, we analyzed global run-on sequencing (GRO-seq) data (Kaikkonen et al., 2013), to measure nascent transcript levels at KLA repressed-induced loci. Consistent with enhancer ChIP-seq data, GRO-seq revealed that nascent RNA transcription at KLA repressed-induced enhancers follows a similar temporal profile, exhibited by early transcriptional repression and late response induction (Fig. 3C). Further, GRO-seq analysis revealed a conserved temporal pattern of transcription at associated KLA repressed-induced gene bodies (Fig. 3D). Collectively, these results suggest that KLA repressed-induced enhancer activity and gene transcription is transiently inhibited following inflammatory activation, ensuring decreased unsaturated fatty acid synthesis in macrophages. This is followed by subsequent late phase induction of relevant macrophage transcription and gene expression, culminating in increased unsaturated fatty acid synthesis and output by macrophages.
To define transcription factors potentially determining this temporal regulation of KLA repressed-induced genes, we performed motif analysis on enhancers exhibiting the repressed/induced pattern of chromatin features. As expected, de novo motif analysis identified motifs for the macrophage LDTFs, PU.1, C/EBP and AP-1, as the most highly enriched sequences. Unexpectedly, an SREBP response element (SRE) was also highly enriched in repressed-induced associated enhancers (Fig. 3E). Given its role in regulation of fatty acid metabolism in various cell types, these findings suggested that SREBP1 might be a determinant of late inflammatory phase regulatory dynamics leading to induction of genes necessary for unsaturated fatty acid biosynthesis.
SREBP1 activity is induced during the resolution phase of the inflammatory response
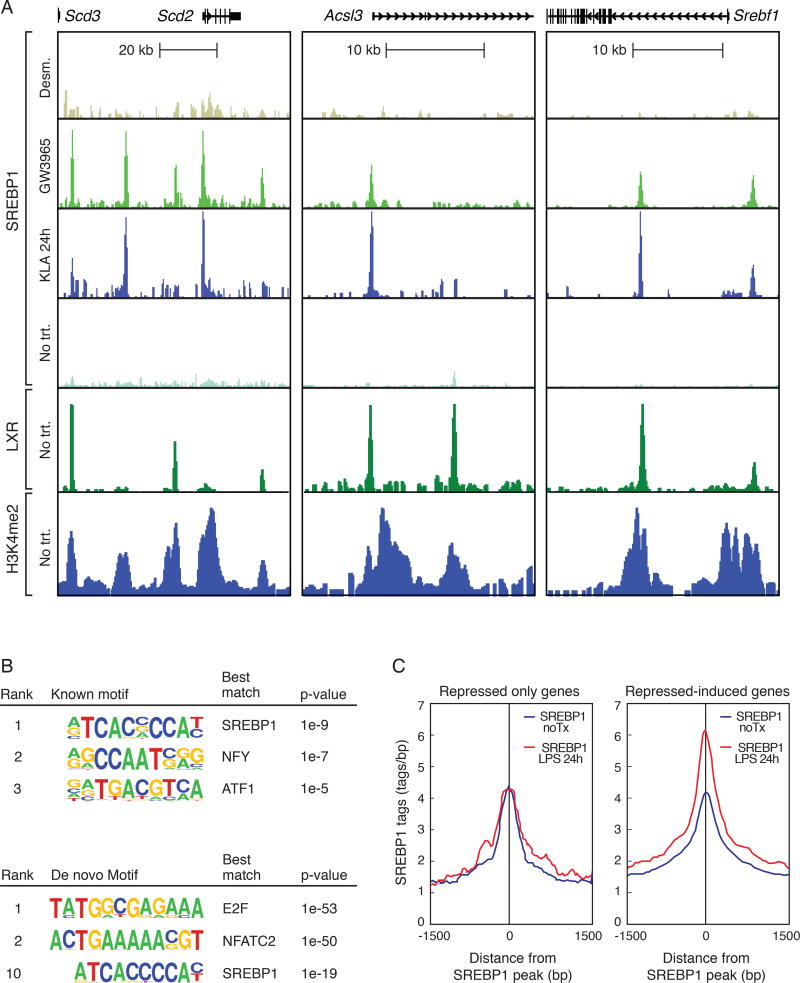
We previously demonstrated that LXR and SREBP1 not only co-localize to representative genes involved in maintaining cholesterol and fatty acid homeostasis, but their coordinate regulatory actions can control context-specific expression profiles (Spann et al., 2012). To investigate the potential relationships of LXR and SREBP1 in controlling macrophage lipid metabolism following TLR4 activation, we performed ChIP-seq of SREBP1 and LXR in mouse primary macrophages stimulated with ligands for LXR and TLR4 for 24hours. As expected, SREBP1 recruitment was observed in the enhancers of lipid synthesis-related genes, as exemplified by Scd2, Acsl3 and Srebf1 following GW3965, but not desmosterol treatment, which is a potent suppressor of SREBP processing (Fig. 4A). Further, LXR and SREBP1 cistromes exhibited significant overlap when comparing genome wide binding profiles (Fig. 4A, Suppl. Fig. 3A). The genes associated with LXR-SREBP1 co-bound sites were enriched for functional annotations for fatty acid metabolism, and fatty acid biosynthesis and elongation (Suppl. Fig. 3B). Further, temporal patterns for direct measurement of enhancer activity levels, demonstrated by H4K5ac and GRO-seq, revealed these LXR-SREBP1 co-bound regions exhibited a coordinate KLA repressed-induced profile (Suppl. Fig. 3C).
Figure 4. SREBP1 is recruited to repressed-induced enhancers during the resolution phase of the inflammatory response.
A. UCSC genome browser images illustrating normalized tag counts for SREBP1, LXR and H3K4Me2, at indicated loci, in thioglycollate-elicited macrophages treated with vehicle, KLA, GW3965, or desmosterol for 24 hours.
B. Known and de novo motifs identified in regions bound by SREBP1 in the late inflammatory response. For ChIP-seq peaks used in motif analysis, peaks for each SREBP ChIP were identified using Homer and calculated the Irreproducible Discovery Rate (IDR) to measure the consistency between replicate experiments for the strength of binding at each loci and retained SREBP1 peaks with IDR < 0.05.
C. Distribution of SREBP1 tag densities, at enhancers associated with genes exhibiting either repressed-repressed or repressed-induced temporal expression patterns, in thioglycollate-elicited macrophages treated with KLA for 24 hours.
See also Figure S3.
Remarkably, KLA treatment also dramatically increased the binding of SREBP1 at enhancer-like regions associated with genes required for mono- and polyunsaturated fatty acid biosynthesis (Fig. 4A), consistent with the enrichment of the SREBP recognition element in repressed-induced enhancers (Fig. 3E). Furthermore, rigorous peak analysis using HOMER defined peaks along with irreproducible discovery rate (IDR) analysis, identified the top known motif in the KLA-induced SREBP1 cistrome as matching the consensus sterol response element (Fig. 4B, top). The sterol response element was independently identified by de novo motif analysis of IDR-defined SREBP1 binding sites (Fig. 4B bottom). Multiple independent experiments indicated that the late phase KLA induction of SREBP1 binding activity was associated with parallel increased nuclear levels of mature SREBP1 protein, as determined by western blotting (Suppl. Fig. 3D). Intriguingly, the late phase increase in SREBP1 recruitment is specific to KLA repressed-induced associated promoters and enhancers, as binding is not changed at regions of solely KLA-repressed genes (Fig. 4C).
SREBP1 drives TLR-responsive late activation of repressed-induced genes
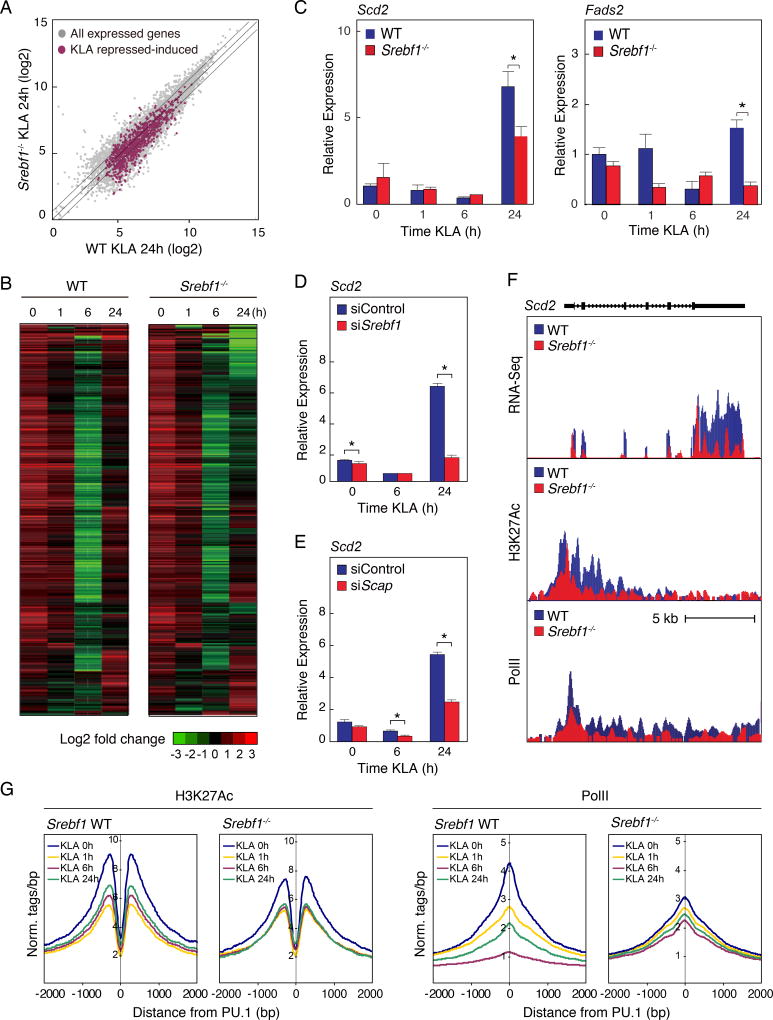
The observation that SREBP1 was recruited to the genes involved in unsaturated fatty acid synthesis in the late inflammatory response led us to examine the consequences of Srebf1 deletion in the inflammatory response of macrophages on a genome-wide scale. We performed RNA-Seq analysis of KLA treated bone marrow-derived macrophages (BMDM) prepared from wild type (WT) and Srebf1−/− mice (Shimano et al., 1997) (Fig. 5A and B). We identified 2995 significantly expressed transcripts with RefSeq annotations exhibiting the KLA repressed-induced phenotype; characterized by reduced levels of >2-fold at 6 hours, and subsequently increased >2-fold at 24 hours after KLA treatment. The expression of 1047 these genes (~35%), in the KLA repressed-induced group, were significantly reduced at 24 hours post KLA treatment in Srebf1−/− macrophages compared to levels in WT cells (Fig. 5A and B). Both RNA-seq and quantitative PCR analysis confirmed that Srebf1−/− macrophages demonstrated significant reduction in the expression of genes mediating mono- and polyunsaturated fatty acid biosynthesis, exemplified by Scd1/2, Acsl3, Fads1/2, and Acot2, during the resolution phase of inflammation at 24 hours post KLA treatment (Fig. 5C and Suppl. Fig. 4A).
Figure 5. Srebf1−/− macrophages exhibit reduced fatty acid biosynthetic gene expression during the resolution phase of the TLR4 response.
A. Scatter plot depicting the relationship between fold change of KLA repressed-induced genes, comparing RNA-seq from wild-type (WT) versus Srebf1−/− bone marrow-derived macrophages treated with KLA for 24 hours. Gray dots show all expressed genes. Red dots represent all KLA repressed-induced genes.
B. Hierarchical clustering and heatmap of the fold change in expression levels of KLA repressed-induced genes in WT and Srebf1−/− bone marrow-derived macrophages treated with KLA for the indicated times (FDR < 0.01, RPKM > 0.5).
C. Relative mRNA expression of Scd2 and Fads2 in WT and Srebf1−/− bone marrow-derived macrophages treated with KLA for the indicated times.
D. Relative mRNA expression of Scd2 mRNA KLA-treated thioglycollate-elicited macrophages, transfected with siRNA control or targeting Srebf1.
E. Relative mRNA expression of Scd2 mRNA KLA-treated thioglycollate-elicited macrophages, transfected with siRNA control or targeting Scap.
F. Distribution of RNA-Seq, H3K27ac and RNA Pol II tag densities at the Scd2 locus in WT and Srebf1−/− bone marrow-derived macrophages treated with KLA for 24 hours.
G. Distribution of H3K27Ac and RNA Pol II tag densities in the vicinity of enhancers associated with KLA repressed-induced genes in WT and Srebf1−/− bone marrow-derived macrophages treated with KLA for the indicated times.
Values are expressed as mean ± SEM. *p<0.05, **p<0.01.
See also Figure S4.
We independently confirmed the requirement of SREBP1 in the regulation of these genes by using small interfering RNAs (siRNAs) specifically targeting Srebf1 or Scap. Quantitative PCR analysis indicated that the Srebf1 knockdown was sufficient to inhibit the late phase inflammation induction of unsaturated fatty acid related gene expressions, exemplified by Scd2 (Fig. 5D). RNA-Seq analysis further confirmed that siRNA-mediated Srebf1 knockdown, led to significantly compromised late phase induction of many genes controlling synthesis of mono- and polyunsaturated fatty acids, including Acsl3 and Fads1/2, in macrophages at 24 hours post KLA treatment (Suppl. Fig. 4B–D). Further, knockdown of Scap, which is required for SREBP processing and activation (Horton et al., 2002), resulted in a similar compromise in late phase induction of gene expression (Fig. 5E).
To gain further insight into the mechanism by which the repressed-induced gene expression is compromised in Srebf1−/− macrophages, we performed ChIP-seq of H3K27ac and RNA pol II to evaluate local enhancer activity at KLA repressed-induced loci. Both H3K27ac and RNA pol II levels were markedly decreased at KLA 24 hours, in Srebf1−/− macrophages, at KLA repressed-induced gene bodies, as exemplified by Scd2 (Fig. 5F). Further, the normal temporal dynamics, characterized by late phase increase in both H3K27ac and RNA pol II levels, was globally compromised in Srebf1−/− macrophages when looking at profiles for all KLA repressed-induced associated enhancers (Fig. 5G), consistent with both the deficient late phase expression recovery, as measured by RNA-seq, and increased late recruitment of SREBP1 to these enhancers. These results suggest a significant role for SREBP1 in late phase induction of repressed-induced genes that control fatty acid biosynthesis.
To investigate whether signaling through other TLRs would exert similar effects on SREBP1 target genes, we assessed relevant gene expression levels throughout a time course of PAM3CSK4 (TLR2 agonist) and Poly(I:C) (TLR3 agonist) treatment, comparing temporal responses in Srebf1−/− and WT macrophages. Similar to the temporal dynamics of TLR4 activation, TLR2 and TLR3 responses of unsaturated fatty acid biosynthetic genes demonstrated a repressed-induced expression profile (Suppl. Fig. 4E–F) dependent on SREBP1. However, the induced phase was much more pronounced in the case of TLR2 activation. This result suggests a predominant role of the MyD88 pathway, which is used by both TLR4 and TLR2, but not TLR3, which instead signals primarily through TRIF.
SREBP1 is necessary for resolution of the TLR-mediated inflammation
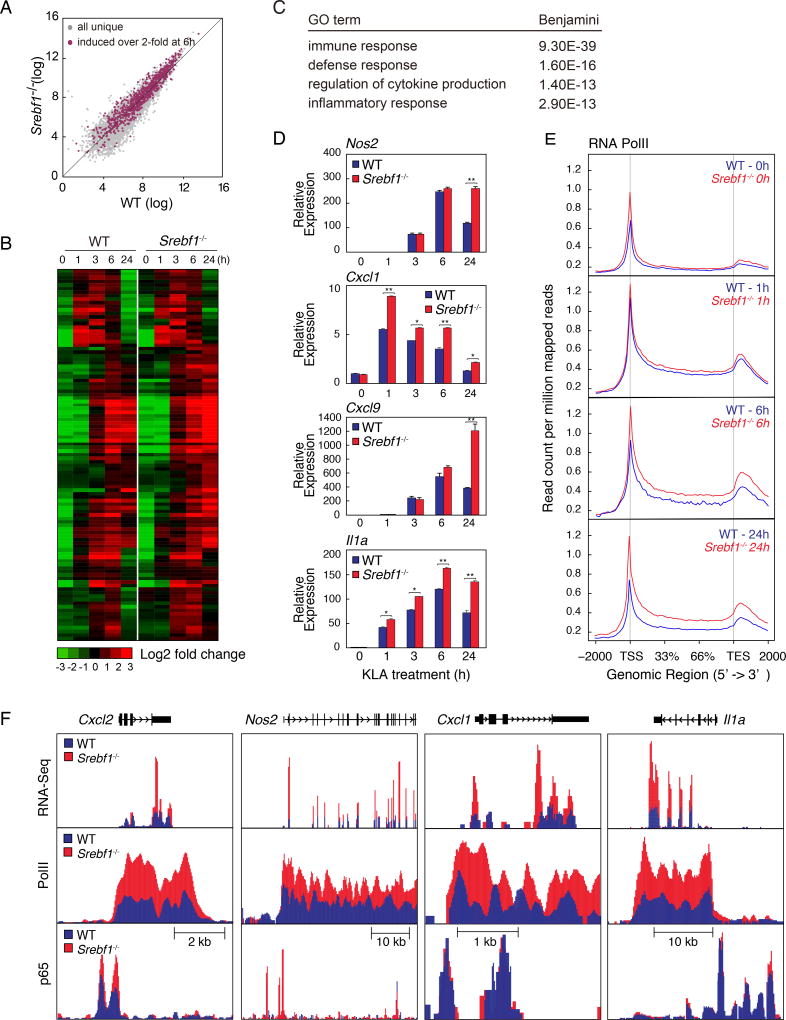
Previous studies demonstrated that unsaturated fatty acids such as EPA, DHA 9Z-POA have potent anti-inflammatory effects in macrophages by antagonizing inflammatory signaling through GPCRs, nuclear receptors and other mechanisms (Cao et al., 2008; Li et al., 2013; Oh et al., 2010). To investigate whether the late phase of expression of genes involved in mono- and polyunsaturated fatty acid biosynthesis contributes to the resolution phase of TLR4 signaling, we evaluated the temporal expression profiles of genes that are induced following KLA treatment in wild type and Srebf1−/− macrophages. Indeed, RNA-seq analysis revealed that Srebf1−/− macrophages demonstrated delayed resolution and often exaggerated gene expression upon TLR4 activation, relative to their WT counterparts (Fig. 6A and B). In WT macrophages, 964 significantly expressed transcripts (with RefSeq annotations) were detected that were increased > 2.0-fold at 6 hours after KLA treatment, and subsequently decreased > 2.0-fold at 24 hours post KLA treatment (defined herein as KLA induced-repressed genes). 247 of these induced-repressed genes demonstrated significantly increased expression, at 24 hours KLA treatment, in Srebf1−/− macrophages compared to WT (Fig. 6A and B). This set of KLA induced-repressed genes had significant enrichment of functional annotations for immune response, regulation of cytokine production and inflammatory response (Fig. 6C). Quantitative PCR analysis confirmed that inflammatory gene expressions, as exemplified by Nos2, Cxcl1, Cxcl9 and Il1a, are significantly increased at 24 hours KLA treatment in Srebf1−/− macrophage (Fig. 6D). Further, siRNA-mediated knockdown experiments confirmed the requirement of SREBP1 for appropriate resolution of inflammatory gene expression, as exemplified by Cxcl2, Nos2, Cxcl1, Il1a, Il12b, and Il6 (Suppl. Fig. 5A).
Figure 6. Srebf1−/− macrophages exhibit a hyper-inflammatory phenotype.
A. Scatter plot depicting the relationship between fold change of KLA induced-repressed genes, comparing RNA-seq from wild-type versus Srebf1−/− bone marrow-derived macrophages treated with KLA for 24 hours. Gray dots show all uniquely expressed genes. Red dots represent all KLA induced-repressed genes.
B. Hierarchical clustering and heatmap of the fold change in expression levels of KLA induced-repressed genes, comparing RNA-seq data from WT and Srebf1−/− bone marrow-derived macrophages treated with KLA for 24 hours (FDR < 0.01, RPKM > 0.5).
C. Functional annotations associated with KLA induced-repressed genes.
D. Relative mRNA expression of inflammatory genes in WT and Srebf1−/− bone marrow-derived macrophages treated with KLA for the indicated times.
E. Distribution of RNA pol II tag densities at loci of KLA induced-repressed genes WT and Srebf1−/− bone marrow-derived macrophages treated with KLA for indicated times.
F. UCSC genome browser image illustrating normalized tag counts for RNA-seq, RNA Pol II and p65 ChIP-seq at loci of inflammatory genes in WT and Srebf1−/− bone marrow-derived macrophages treated with KLA for the indicated times.
Values are expressed as mean ± SEM. *p<0.05, **p<0.01.
See also Figure S5.
We further assessed relevant pro-inflammatory gene expression levels throughout a time course of PAM3CSK4 (TLR2 agonist) and Poly(I:C) (TLR3 agonist) treatment, comparing temporal responses in Srebf1−/− and WT macrophages. Similar to the temporal dynamics of TLR4 activation, TLR2 and TLR3 responses of pro-inflammatory genes demonstrated an induced-repressed expression profile (Suppl. Fig. 5E–F). Further, resolution of TLR2- and TLR3-mediated inflammatory gene expression was drastically compromised in Srebf1−/− macrophages, relative to their WT counterparts (Suppl. Fig. 5E–F). Interestingly, Pam3 induced genes showed delayed resolution, similar to TLR4 response, whereas PolyI:C induced genes were hyper-responsive throughout the time course. These results are consistent with an MyD88-dependent induction of SREBP1 mediating late resolution of TLR2 and TLR4 responses. The hyper-activation of Ifnb1 and Ifna4 in response to TLR3 agonist in Srebf1−/− macrophages may reflect a different mechanism.
To further define the SREBP-dependent temporal regulatory pattern, we analyzed ChIP-seq data for RNA polymerase II (RNA PolII) in KLA-treated Srebf1−/− and WT macrophages. Normalized tag density plots at induced-repressed genes, revealed increased levels of RNA PolII in Srebf1−/− versus WT macrophages (Fig. 6E). The average tag density levels, between Srebf1−/− and WT, demonstrated most significant differentials at 24 hours post KLA (Fig. 6E). These distinct patterns are exemplified for Cxcl2, Nos2, Cxcl1 and Il1a in Fig. 6F. Consistent with ChIP-Seq and mRNA expression data, GRO-seq analysis revealed a similarly conserved temporal pattern of transcription at associated KLA induced-repressed gene bodies (Suppl. Fig. 5B). These results suggest that the temporal dynamics of induced-repressed inflammatory genes are regulated via local enhancer activities driven by KLA responsive transcription factor complexes.
Given the role of NFκB as a primary driver of TLR4-mediated responses, we further performed ChIP-seq of the p65 component of NFκB to determine whether the increased inflammatory gene expression, exhibited by Srebf1−/− macrophages, was due to increased p65 recruitment to the induced-repressed loci. Unexpectedly, the ChIP-seq analysis revealed a strikingly similar pattern of p65 binding in KLA treated Srebf1−/− and WT macrophages (Fig. 6F and Suppl. Fig. 5C, D). Further, the similarity of p65 binding, comparing Srebf1−/− and WT profiles, remains consistent whether looking at all repressed-induced loci (correlation coefficient = 0.949817), or the subset of induced-repressed loci demonstrating the most significant alterations upon loss of Srebp1 (correlation coefficient = 0.9438753) (Suppl. Fig. 5C–D). This finding is consistent with previous studies suggesting that the repressive actions of unsaturated fatty acids on NFκB activity are independent of changes in factor binding (Li et al., 2013).
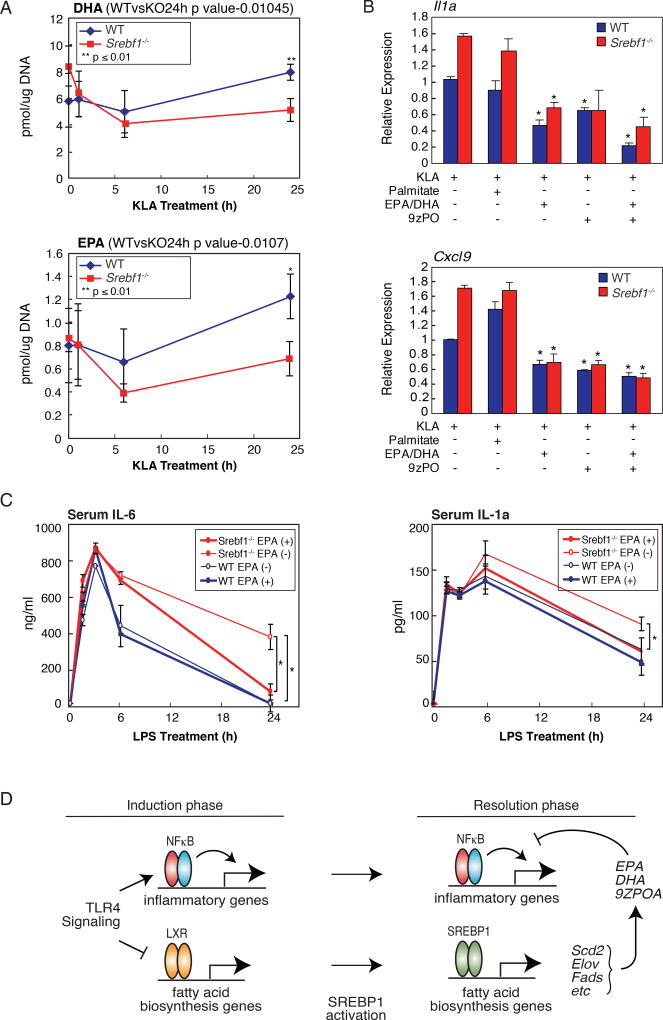
The gene expression pattern observed in Srebf1−/− macrophages predicts that the late phase of mono- and poly-unsaturated fatty acid production would be compromised in these cells. We therefore performed lipidomic analysis of KLA-treated Srebf1−/− and wild type macrophages to assess changes in fatty acid levels. Consistent with the altered gene expression patterns, Srebf1−/− macrophages demonstrated marked decreases in unsaturated fatty acid production, as exemplified by DHA, EPA, and 9Z–POA; with most dramatic differentials, between Srebf1−/− and WT macrophages, occurring at 24 hours post KLA treatment (Fig. 7A, Suppl. Fig. 6A). This late phase-specific decrease is consistent with the possibility that these anti-inflammatory fatty acids contribute to the resolution phase of the TLR4 response.
Figure 7. SREBP1 is necessary for resolution of inflammation by driving appropriate macrophage production of anti-inflammatory unsaturated fatty acids in late inflammatory response.
A. Lipidomics analysis of unsaturated fatty acid (EPA and DHA 9Z–PO) levels in KLA treated WT and Srebf1−/− bone marrow-derived macrophages.
B. Relative mRNA expression of inflammatory genes in WT and Srebf1−/− bone marrow-derived macrophages treated with KLA for 24 hours, with or without supplementation with the indicated exogenous fatty acids (20µM) at 12h post KLA treatment.
C. Serum levels of cytokines IL-6 and IL-1a, as quantified by ELISA, in WT and Srebf1−/− mice treated with 5mg/kg LPS for 0, 1, 3, 6, 24 hours, with or without EPA supplementation as indicated.
D. Model for integrated actions of NFκB, LXRs and SREBP1 during the induction and resolution phases of the TLR4 response.
Values are expressed as mean ± SEM. *p<0.05, **p<0.01.
See also Figure S6.
To further test the link between late phase resolution of inflammation and unsaturated fatty acid output, fatty acid rescue experiments were performed, in which exogenous unsaturated fatty acids were added to KLA-treated Srebf1−/− and WT macrophages. In these experiments, mono- (9Z-POA) and poly-unsaturated (EPA/DHA) fatty acids were supplemented, either alone or simultaneously, at 12 hours post KLA treatment to mimic late phase accumulation. Cells were then harvested at 24 hours after KLA treatment. Addition of exogenous unsaturated fatty acids led to significant reduction of inflammatory gene expression in macrophages (Fig. 7B). Further, this unsaturated fatty acid-specific repressive effect was more pronounced in Srebf1−/− cells, relative to their WT counterparts (Fig. 7B), consistent with intact production of these fatty acid species in WT macrophages. Similar results were observed in siRNA-mediated knockdown cells (Suppl. Fig. 6B).
To investigate the role of SREBP1-mediated unsaturated fatty acid output in modulating the inflammatory response in vivo, Srebf1−/− mice were challenged with a sub-lethal dose of LPS. Consistent with the increased late phase inflammatory gene expression patterns observed in the Srebf1−/− macrophages (Fig. 6B–F), circulating cytokine levels of IL-6 and IL-1α remained significantly higher in Srebf1−/− mice, at 24 hours post LPS injection (Fig. 7C); thus suggesting a compromised resolution of inflammation relative to their WT counterparts. In addition, supplementation of exogenous EPA, prior to the LPS challenge, protected Srebf1−/− mice from an exaggerated inflammatory response, restoring circulating cytokine levels to those seen in WT mice (Fig. 7C).
Discussion
Emerging evidence suggests that the immune system and lipid metabolism are coordinately regulated at multiple levels within the body. Here, we demonstrate a reciprocal relationship between cellular levels of anti-inflammatory fatty acids and the temporal induction and resolution of pro-inflammatory gene expression following TLR4 activation (Fig. 7D). Anti-inflammatory fatty acid levels rapidly fall following KLA treatment, in advance of down regulation of mRNAs encoding corresponding biosynthetic enzymes. Given the ability of these fatty acid species to suppress NF-κB-dependent gene expression, their down regulation is likely to be necessary for a full TLR4 response. At 12-24h following TLR4 ligation, anti-inflammatory fatty acid levels rise, concurrent with increased expression of mRNAs encoding biosynthetic enzymes and decreased expression of mRNAs encoding pro-inflammatory mediators.
Mechanisms responsible for down-regulation of lipid biosynthetic genes remain to be established. Repression of basal LXR-dependent gene expression does not account for this effect because a similar pattern of gene expression was observed in LXR double knockout macrophages. Thus, while TLR signaling blunts the ability of LXR agonists to induce target gene expression, alternative mechanisms must account for the observed down-regulation. During the initial phase of the TLR4 response, the p65 component of NF- κB is recruited to many of the enhancer elements associated with the set of repressed-induced genes. This TLR-induced p65 binding is associated with loss of both coactivator recruitment and active chromatin features at these loci, correlating with their loss of transcriptional activity and expression. Our unpublished results using an NF- κB inhibitor, suggest a requirement for NF- κB activity in mediating the early phase repression of these genes (data not shown). However, a direct role of p65-containing NF-κB complexes in down regulation of these genes remains to be established.
Unexpectedly, the late up-regulation of mRNAs encoding enzymes required for synthesis of anti-inflammatory fatty acids was independent of LXRs and instead required SREBP1. Consistent with these findings, ChIP-Seq experiments indicated a reduction of LXR binding to enhancers associated with repressed-induced genes, but a marked increase in the binding of SREBP1. This KLA-induced binding of SREBP1 to cis regulatory elements at late time points was associated with increases in the total nuclear content of processed SREBP1. Further, the late increase in SREBP1 binding was associated with increases in chromatin features associated with active enhancers. SREBP1 KO macrophages, or macrophages in which siRNAs were used to knock down SREBP1, displayed compromised late induction of repressed-induced genes and reduced production of anti-inflammatory fatty acids. While our data clearly provides evidence for a novel role of SREBP1 in transcriptionally tailoring specific macrophage lipid metabolic output, driving late phase synthesis of anti-inflammatory unsaturated fatty acids, the mechanisms controlling both the late phase induction of SREBP1 recruitment and the SREBP1-target activation specificity are not entirely clear. An understanding of these mechanisms could be of importance in identifying novel targets for development of SREBP1-centric interventions of various inflammatory disease states.
Recent studies provided evidence that SREBP-1a is required for the formation of the inflammasome and secretion of IL-1β in response to systemic inflammation (such as endotoxic shock) (Im et al., 2011). Consistent with these findings, we observed increased secretion of IL-1β protein following KLA treatment of Srebf1−/− macrophages as compared to WT controls (data not shown). However, our studies also demonstrated that a subset of TLR4-responsive, pro-inflammatory genes was hyper-activated in Srebf1−/− macrophages at 12–24 hours following KLA treatment. Similar late hyper-inflammatory trends were observed in Srebf1−/− macrophages stimulated with ligands for TLR2 and TLR3; thus, indicating that SREBP1 is genetically required for the normal resolution phase of varied TLR responses in macrophages. Our findings further suggest that SREBP1-driven synthesis of anti-inflammatory fatty acids contributes to this resolution phase. The late TLR4-mediated increase of these fatty acid species is compromised in Srebf1−/− macrophages, and supplementation of exogenous anti-inflammatory, both in cultured macrophages and in vivo, reverses hyper-induction of pro-inflammatory gene expression caused by loss of SREBP1. This is consistent with the presence of higher concentrations of these species in WT macrophages. Although our studies focused on 9Z-POA, DHA and EPA, it is possible that additional anti-inflammatory metabolites of polyunsaturated fatty acids, such as resolvins protectins and fatty acid hydroxyl fatty acids, are also generated by the late SREBP1-dependent program of gene expression. Interestingly, loss of SREBP1 results in increased recruitment of RNA-Pol II to a subset of inflammatory response genes independent of changes in p65 binding activity. These results are consistent with prior studies suggesting that DHA, EPA and 9Z-POA uncouple NF-κB binding from its transcriptional output (Li et al., 2013). Because p65 binding itself is unchanged, the mechanism of inhibition is unlikely to be through alterations in the IκB kinase cascade required for NF-κB activation.
In concert, our findings provide evidence for a role of SREBP1 in promoting resolution of the transcriptional response of macrophages to TLR signaling by driving the synthesis of anti-inflammatory fatty acids. While we have shown that the SREBP1 pathway also influences resolution following activation of TLR2 and TLR3, the extent to which it is involved in resolution of responses to other pattern recognition receptors or cytokine-dependent inflammatory responses remains to be determined. It will therefore be of interest to investigate this pathway further with respect to control of the resolution phase of inflammation in response to infection and injury, as well as in disease contexts in which inflammation plays a pathogenic role.
Experimental procedures
Cell culture
LXRα/β−/− and Srebf1−/− were generated as described previously (Repa et al., 2000b; Shimano et al., 1997). These mice were backcrossed to the C57BL/6J strain for more than ten generations. Mouse thioglycollate-elicited macrophages and were isolated from male 6- to 9- weeks old C57BL/6J (Charles River laboratories), LXRα/β−/− and Srebf1−/− mice and cultured as previously described (Spann et al., 2012). Peritoneal macrophages were harvested by lavage 3 days after intraperitoneal injection of 3ml of 3% thioglycollate medium (http://www.lipidmaps.org/protocols/), overnight culture and adherence selection. Bone marrow from mice were isolated by perfusion of the medullary cavity of femurs, tibias and iliac bones and cultured in medium containing RPMI-1640, 10% FCS, and 20ug/ml M-CSF (R&D) for 6 days. RAW264.7 cells are maintained in the RPMI 1640 media supplemented with 10% FCS (Hyclone) and used between passage 5–10. For the fatty acid rescue experiments, cells were treated with fatty acids complexed with FA-free low-endotoxin BSA (Sigma, final FA:BSA molar ratio was 5:1).
Animal Study
All mice used in this study has C57BL/6 background. Male, 8–11 weeks old Srebf1−/− mice and age-matched littermate control were individually housed in cages in a 12hr/12hr light/dark cycle with free access to food and water. For supplemental EPA administration study, mice were fed with fish meal-free diet (fish meal-free F1: 4.4% fat; Funabashi Farm, Funabashi, Japan) or fish meal-free diet supplemented with 5% EPA ethyl ester (v/v) for 7 days before single intraperitoneal injection of 5mg/kg LPS (n=5, each group). All animal procedures were in accordance with research guidelines for care and use of laboratory animals of Tokyo Medial and Dental University. Temporal changes of serum IL-6 and IL-1a were quantified by ELISA (R&D).
ChIP-Seq
ChIP from thioglycollate-elicited peritoneal macrophages or bone marrow-derived macrophages were performed as described previously (Spann et al., 2012), with modifications as described in Supplemental Experimental Procedures. ChIP-Seq libraries were prepared from ChIP DNA by blunting, A-tailing, adapter ligation as previously described (Heinz, et al. 2010) using barcoded adapters (NextFlex, Bioo Scientific). Libraries were PCR-amplified for 12–15 cycles, size selected by gel extraction and sequenced on either Illumina Genome Analyzer II Hi-Seq 2000 for 51 cycles.
RNA-Seq
Total RNA was isolated from cells and purified using RNeasy columns and RNase free DNase digestion according to the manufacturer's instructions (QIAGEN). RNA-Seq libraries were prepared from polyA enriched mRNA, either as previously described (Kaikkonen, et al. 2013), or as detailed in Supplemental Experimental Procedures.
High-throughput sequencing and data analysis
All sequencing was conducted using either Illumina Genome Analyzer II or Hi-Seq 2000 sequencers using single-end 50bp reads. All data were aligned to the mm9 assembly of the mouse genome, and all subsequent data analysis was performed using HOMER, and detailed instructions for analysis can be found at http://homer.salk.edu/homer/ (Heinz et al., 2010). Each sequencing experiment was normalized to total of 107 uniquely mapped tags by adjusting the number of tags at each position in the genome to the correct fractional amount given the total tags mapped. Sequence experiments were visualized by preparing custom tracks for the UCSC genome browser. Differentially expressed genes were identified using HOMER as described previously (Li et al., 2013). For SREBP1 ChIP-Seq analysis, ChIP-seq peaks for each SREBP ChIP replicate were identified using Homer and then the strength of binding at each loci was quantified as the position adjusted reads from the start of the peak region (Homer peak score). We calculated the Irreproducible Discovery Rate (IDR) to measure the consistency between replicate experiments for the strength of binding at each loci and retained SREBP peaks with IDR < 0.05. For various ontology analyses, either HOMER or DAVID Bioinformatics Resources 6.7 was used.
Statistical analyses
Statistical analyses were performed using Graph Pad Prism 5 software. The images were prepared using Adobe Illustrator CS5 or Photoshop CS5.1. Data are presented as the mean ± SEM. For experiments involving two factors, data were analyzed by two-way ANOVA followed by Bonferroni post tests. Individual pair-wise comparisons were performed using Student’s t test. p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Timothy Osborne for making anti-SREBP1 antibodies available for these studies. We thank Dr. Kristen Jepsen, Mahdieh Khosroheidari, and Hiroko Matsui for assistance with Illumina sequencing; Jana Collier for technical assistance; Miguel Mooney for technical assistance; and Leslie Van Ael for assistance with manuscript preparation. Y.O. was supported by MEXT KAKENHI Grant Numbers 24641142 and 25H10, grants from Naito Foundation, Japan Heart Foundation, Japan Foundation for applied enzymology, Ono medical foundation and Takeda Science Foundation. M.U.K. was supported by Fondation Leducq Career Development award, Academy of Finland Research Fellowship and grants from Sigrid Jusélius, ASLA-Fulbright, Finnish Foundation for Cardiovascular Research and Finnish Cultural Foundation. T.S. was supported by funding from the Swedish Society for Medical Research. Studies were primarily supported by NIH grants DK091183, CA17390, and DK063491.
Footnotes
Author Contributions
Y.O., N.J.S. and C.K.G. conceived project and designed experiments. Y.O., N.J.S., V.M.L. and J.T. analyzed data. Y.O., N.J.S., T.S., C.E., E.D.M., T.M., S.H., M.U.K., A.F.C., M.J.K. and M.T.L. performed experiments. H.S. provided mice. M.J.K., I.M., and A.S., provided technical expertise. Y.O., N.J.S. and C.K.G. interpreted data and wrote the manuscript.
References
- Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13:213–224. doi: 10.1038/nrm3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, Hotamisligil GS. Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell. 2008;134:933–944. doi: 10.1016/j.cell.2008.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrillo A, Joseph SB, Vaidya SA, Haberland M, Fogelman AM, Cheng G, Tontonoz P. Crosstalk between LXR and toll-like receptor signaling mediates bacterial and viral antagonism of cholesterol metabolism. Mol Cell. 2003;12:805–816. doi: 10.1016/s1097-2765(03)00384-8. [DOI] [PubMed] [Google Scholar]
- Cildir G, Akincilar SC, Tergaonkar V. Chronic adipose tissue inflammation: all immune cells on the stage. Trends Mol Med. 2013;19:487–500. doi: 10.1016/j.molmed.2013.05.001. [DOI] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis EA, Deems RA, Harkewicz R, Quehenberger O, Brown HA, Milne SB, Myers DS, Glass CK, Hardiman G, Reichart D, et al. A mouse macrophage lipidome. J Biol Chem. 2010;285:39976–39985. doi: 10.1074/jbc.M110.182915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold KR, Shigenaga JK, Kazemi MR, McDonald CM, Patzek SM, Cross AS, Moser A, Grunfeld C. Mechanisms of triglyceride accumulation in activated macrophages. J Leukoc Biol. 2012;92:829–839. doi: 10.1189/jlb.1111537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol. 2015;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, DeBose-Boyd RA, Brown MS. Protein sensors for membrane sterols. Cell. 2006;124:35–46. doi: 10.1016/j.cell.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Bensinger SJ, Hong C, Beceiro S, Bradley MN, Zelcer N, Deniz J, Ramirez C, Diaz M, Gallardo G, et al. Apoptotic cells promote their own clearance and immune tolerance through activation of the nuclear receptor LXR. Immunity. 2009;31:245–258. doi: 10.1016/j.immuni.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong C, Tontonoz P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat Rev Drug Discov. 2014;13:433–444. doi: 10.1038/nrd4280. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Everts B, Ivanova Y, O'Sullivan D, Nascimento M, Smith AM, Beatty W, Love-Gregory L, Lam WY, O'Neill CM, et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat Immunol. 2014a;15:846–855. doi: 10.1038/ni.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YL, Morales-Rosado J, Ray J, Myers TG, Kho T, Lu M, Munford RS. Toll-like receptor agonists promote prolonged triglyceride storage in macrophages. J Biol Chem. 2014b;289:3001–3012. doi: 10.1074/jbc.M113.524587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im SS, Yousef L, Blaschitz C, Liu JZ, Edwards RA, Young SG, Raffatellu M, Osborne TF. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 2011;13:540–549. doi: 10.1016/j.cmet.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito A, Hong C, Rong X, Zhu X, Tarling EJ, Hedde PN, Gratton E, Parks J, Tontonoz P. LXRs link metabolism to inflammation through Abca1-dependent regulation of membrane composition and TLR signaling. Elife. 2015;4:e08009. doi: 10.7554/eLife.08009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SS, Ghaffari AA, Cheng G. Lipopolysaccharide-mediated IL-10 transcriptional regulation requires sequential induction of type I IFNs and IL-27 in macrophages. J Immunol. 2010;185:6599–6607. doi: 10.4049/jimmunol.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SB, Castrillo A, Laffitte BA, Mangelsdorf DJ, Tontonoz P. Reciprocal regulation of inflammation and lipid metabolism by liver X receptors. Nat Med. 2003;9:213–219. doi: 10.1038/nm820. [DOI] [PubMed] [Google Scholar]
- Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–325. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Spann NJ, Kaikkonen MU, Lu M, Oh da Y, Fox JN, Bandyopadhyay G, Talukdar S, Xu J, Lagakos WS, et al. NCoR repression of LXRs restricts macrophage biosynthesis of insulin-sensitizing omega 3 fatty acids. Cell. 2013;155:200–214. doi: 10.1016/j.cell.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumeng CN, DelProposto JB, Westcott DJ, Saltiel AR. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–3246. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. J Pathol. 2013;229:176–185. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Oh da Y, Talukdar S, Bae EJ, Imamura T, Morinaga H, Fan W, Li P, Lu WJ, Watkins SM, Olefsky JM. GPR120 is an omega-3 fatty acid receptor mediating potent anti-inflammatory and insulin-sensitizing effects. Cell. 2010;142:687–698. doi: 10.1016/j.cell.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn O, Olefsky JM. The cellular and signaling networks linking the immune system and metabolism in disease. Nat Med. 2012;18:363–374. doi: 10.1038/nm.2627. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Raetz CR, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Jr, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, et al. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. J Lipid Res. 2006;47:1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- Reboldi A, Dang EV, McDonald JG, Liang G, Russell DW, Cyster JG. Inflammation. 25-Hydroxycholesterol suppresses interleukin-1-driven inflammation downstream of type I interferon. Science. 2014;345:679–684. doi: 10.1126/science.1254790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000a;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Turley SD, Lobaccaro JA, Medina J, Li L, Lustig K, Shan B, Heyman RA, Dietschy JM, Mangelsdorf DJ. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science. 2000b;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Prados JC, Traves PG, Cuenca J, Rico D, Aragones J, Martin-Sanz P, Cascante M, Bosca L. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Shimomura I, Hammer RE, Herz J, Goldstein JL, Brown MS, Horton JD. Elevated levels of SREBP-2 and cholesterol synthesis in livers of mice homozygous for a targeted disruption of the SREBP-1 gene. J Clin Invest. 1997;100:2115–2124. doi: 10.1172/JCI119746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spann NJ, Glass CK. Sterols and oxysterols in immune cell function. Nat Immunol. 2013;14:893–900. doi: 10.1038/ni.2681. [DOI] [PubMed] [Google Scholar]
- Tabas I, Glass CK. Anti-inflammatory therapy in chronic disease: challenges and opportunities. Science. 2013;339:166–172. doi: 10.1126/science.1230720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tencerova M, Aouadi M, Vangala P, Nicoloro SM, Yawe JC, Cohen JL, Shen Y, Garcia-Menendez L, Pedersen DJ, Gallagher-Dorval K, et al. Activated Kupffer cells inhibit insulin sensitivity in obese mice. FASEB J. 2015;29:2959–2969. doi: 10.1096/fj.15-270496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma V, Yao-Borengasser A, Rasouli N, Nolen GT, Phanavanh B, Starks T, Gurley C, Simpson P, McGehee RE, Jr, Kern PA, et al. Muscle inflammatory response and insulin resistance: synergistic interaction between macrophages and fatty acids leads to impaired insulin action. Am J Physiol Endocrinol Metab. 2009;296:E1300–1310. doi: 10.1152/ajpendo.90885.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Barnes GT, Yang Q, Tan G, Yang D, Chou CJ, Sole J, Nichols A, Ross JS, Tartaglia LA, et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J Clin Invest. 2003;112:1821–1830. doi: 10.1172/JCI19451. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.