Abstract
The bacterial cell wall is a highly conserved essential component of most bacterial groups. It is the target for our most frequently used antibiotics and provides important small molecules that trigger powerful innate immune responses. The wall is composed of glycan strands cross-linked by short peptides. For many years, the penicillin binding proteins were thought to be the key enzymes required for wall synthesis. RodA and possibly other proteins in the wider SEDS family have now emerged as a previously unknown class of essential glycosyltranferase enzymes, which play key morphogenetic roles in bacterial cell wall synthesis. We provide evidence in support of this role and the discovery of small natural product molecules that probably target these enzymes. The SEDS proteins have exceptional potential as targets for new antibacterial therapeutic agents.
The bacterial cell wall is an ancient structure that was probably invented early in the evolution of cells and may have contributed to the explosive bacterial radiation that occurred about 3-4 billion years ago1. The wall is essential for bacterial viability and an important target for antibiotics2. The major conserved component of the wall is peptidoglycan (PG), which comprises long strands of alternating amino sugars cross-linked by peptide bridges. The key enzymes needed for assembly of the wall are glycosyltransferases (GTase), which generate the glycan strands and transpeptidases (TPase), which generate the cross-links3. Penicillin-binding proteins (PBPs), first identified because they bind β-lactam antibiotics that prevent the TPase reaction, were considered to be the key enzymes in PG assembly because one major class (class A) also carry a GTase domain capable of synthesising PG strands. A second major class of PBPs (class B) lack the GTase domain. Curiously, the class B PBPs more frequently display genetic essentiality than the class A PBPs. Furthermore, their mutant phenotypes suggest that they play specialised roles in morphogenesis; thus, mutants of rod-shaped bacteria such as Escherichia coli can be specifically affected in either cell elongation (e.g. pbpA gene) or cell division (ftsI gene)4. Bacillus subtilis has two redundant elongation enzymes, PBP2A and PBPH5, and an essential division enzyme, PBP2B6.
pbp genes show a remarkable level of functional redundancy, possibly reflecting the selective pressure exerted by antibiotic use, either in the natural environment or the human sphere. About 10 years ago, the Popham lab reported the unexpected finding that in B. subtilis, the genes encoding all four class A PBPs could be inactivated without significantly affecting cell growth or division7. This pointed to the existence of a “missing GTase”. Here we identify RodA, a highly conserved member of the SEDS (shape, elongation, division and sporulation8) family of proteins, as a strong candidate for the missing GTase. Our findings are in line with those of other labs published while this work was under review9. We go on to describe the results of a chemical genetics screen for inhibitors of the missing GTase, which has led to the isolation of a novel antibacterial compound that probably targets RodA.
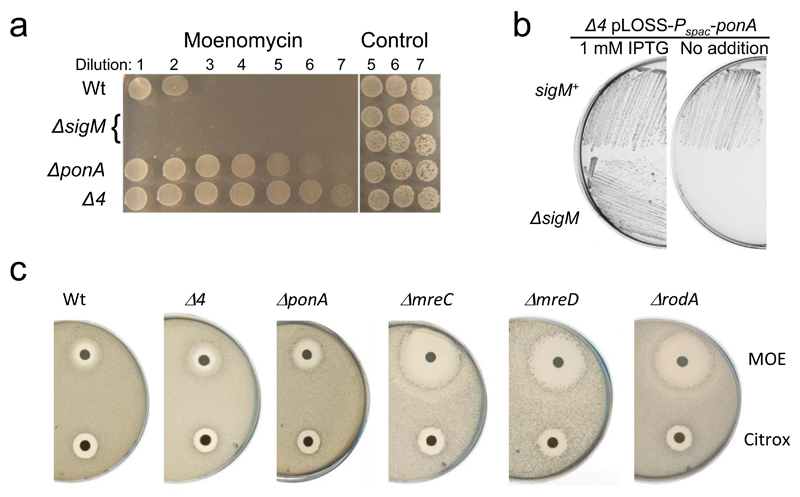
The multiple class A PBP mutant is resistant to moenomycin and hypersensitive to loss of σM
We repeated the Popham lab construction of a B. subtilis strain bearing deletions of the genes encoding all 4 class A PBPs (ponA, pbpD, pbpF and pbpG; strain AG157, hereafter “Δ4”)7 and showed that it is indeed viable, though the cells grow relatively slowly and require elevated levels of Mg++. The strain closely resembles a single ponA mutant, which presumably encodes the “major” class A PBP activity under normal conditions, in having cells that are slightly longer and thinner than wild type. If the missing GTase had a similar GTase active site to that of the class A PBPs it should be sensitive to the known GTase inhibitor, moenomycin (MOE)10. However, Popham’s lab previously reported that the Δ4 mutant is relatively insensitive to MOE and that its growth resembles that of the wild type treated with MOE7, raising the possibility that the missing GTase is MOE resistant. Significantly, we found that the Δ4 mutant was actually more resistant to MOE than the wild type under certain conditions (Fig. 1a), suggesting that an MOE resistant form of GTase is upregulated in the Δ4 mutant, in response to loss of the class A PBP activities. In preliminary RNA-Seq experiments comparing wild type and Δ4 strains many of the higher expressed genes in the Δ4 strain overlapped with genes belonging to the cell wall stress ECF sigma factor σM11 (Supplementary Data Table 1). Other genes belonged to a gene set recognised as being stimulated by treatment of cells with the antibiotic MOE12. Salzberg et al.12 had shown that σM is upregulated in response to MOE stress and that a sigM mutant is hypersensitive to MOE. To test whether σM upregulation was indeed contributing to the increased MOE resistance of the Δ4 mutant, we tried to introduce a sigM mutation into the Δ4 strain but this combination was apparently lethal. Instead, we introduced the sigM mutation into a Δ4 mutant that carried a plasmid containing an inducible copy of ponA, generating a strain in which sigM could be knocked out. As shown in Fig. 1b, growth of this strain was dependent on the expression of ponA (+IPTG), confirming that sigM is essential in the Δ4 mutant (Fig. 1b). Consistent with the notion that PBP1A, the product of the ponA gene, is the major bifunctional PBP in B. subtilis7,13, it was also not possible to introduce the sigM mutation into cells bearing only a ponA mutation.
Figure 1. Use of MOE sensitivity to screen for missing GTase candidate genes.
a, Relative MOE sensitivity of the sigM mutant and resistance of the ponA and Δ4 mutant compared to the wild type. Overnight cultures of each strain were diluted (10-fold series; left to right) and 10 µl spots were placed on NA containing MOE at 10 µg/ml. Right hand spots show the last three dilutions plated on NA with no MOE. sigM spottings were done in duplicate.
b, Synthetic lethality of a Δ4 mutant in a ΔsigM background. Growth on NA plates with or without 1 mM IPTG of strains AG221 (Δ4 pLOSS-Pspac-ponA) and AG831 (Δ4 pLOSS-Pspac-ponA ΔsigM::kan) at 30°C.
c, Hypersensistivity of rodA, mreC and mreD mutants compared to wild type, ponA and Δ4 strains. Disc diffusion assays on MSM plates using 10 µg MOE per disc.
The experiments have been repeated several times during the course of the project, producing consistent results. The figures are representative of at least two independent experiments.
rodA is the σM regulon gene responsible for enhanced resistance to MOE
We took a candidate gene approach to look for genes that might be responsible for the σM-dependence of MOE resistance. Supplementary Data Table 1 summarises the genes identified on the basis of σM-dependence and/or MOE-stimulation, supported by the preliminary RNA-Seq data (further details in Supplementary Data Table 1). Knockouts of all of the non-essential genes with unknown or poorly understood function (51 in total; mainly “y genes”) were examined but, with one exception, none of them showed significantly increased sensitivity to MOE. As previously described14, mutants of yabM did show significant sensitivity. However, mutation of this gene did not affect viability when introduced into the Δ4 background, suggesting that it is not essential for the alternative GTase: indeed, Meeske et al.15 recently showed that YabM belongs to a family of probable lipid II flippases, rather than being a GTase. We therefore considered the essential gene candidates. Of these, most have already had biochemical functions assigned to them, particularly around lipid II (cell wall) synthesis (Supplementary Data Table 1). Five genes of unknown biochemical function remained: divIB, divIC, mreC, mreD, and rodA. The first two of these genes appear to be required specifically for cell division16, and so were excluded from further study. The remaining three genes, mreC, mreD and rodA, are all required for cell elongation and generate a similar bloated cell phenotype when depleted8,17,18. All three genes were initially considered to be essential in B. subtilis but null mutants can be constructed and propagated in media containing osmoprotectants and high levels of Mg++18,19. Mutants in each of the three genes turned out to be hypersensitive to MOE (Fig. 1c), making them contenders for the missing GTase. mreC and mreD are usually located in an operon downstream of the gene encoding the bacterial actin homologue, mreB, and so have been assumed to have some role in coupling MreB function to other proteins involved in cell wall synthesis. Alignments of MreC and MreD sequences from a range of organisms revealed relatively weak sequence conservation, especially for the very hydrophobic MreD protein (not shown). Neither protein seemed a good candidate for an enzyme, since no amino acid group of character typically required for catalysis (e.g. C, D, E, H, K, R, S, T) was conserved in the alignments. In contrast, sequence alignments of RodA and related SEDS proteins revealed several highly conserved, potentially catalytic amino acids, including (B. subtilis RodA numbering): E117 (2/62 exceptions), K120 (1/62 exceptions), R229, D280 and S360 (Supplementary Fig. 1). Until recently, SEDS proteins were thought to encode lipid II flippases, but the MurJ and Amj proteins now appear to fulfil this role in E. coli and B. subtilis15, so we initially focussed on RodA.
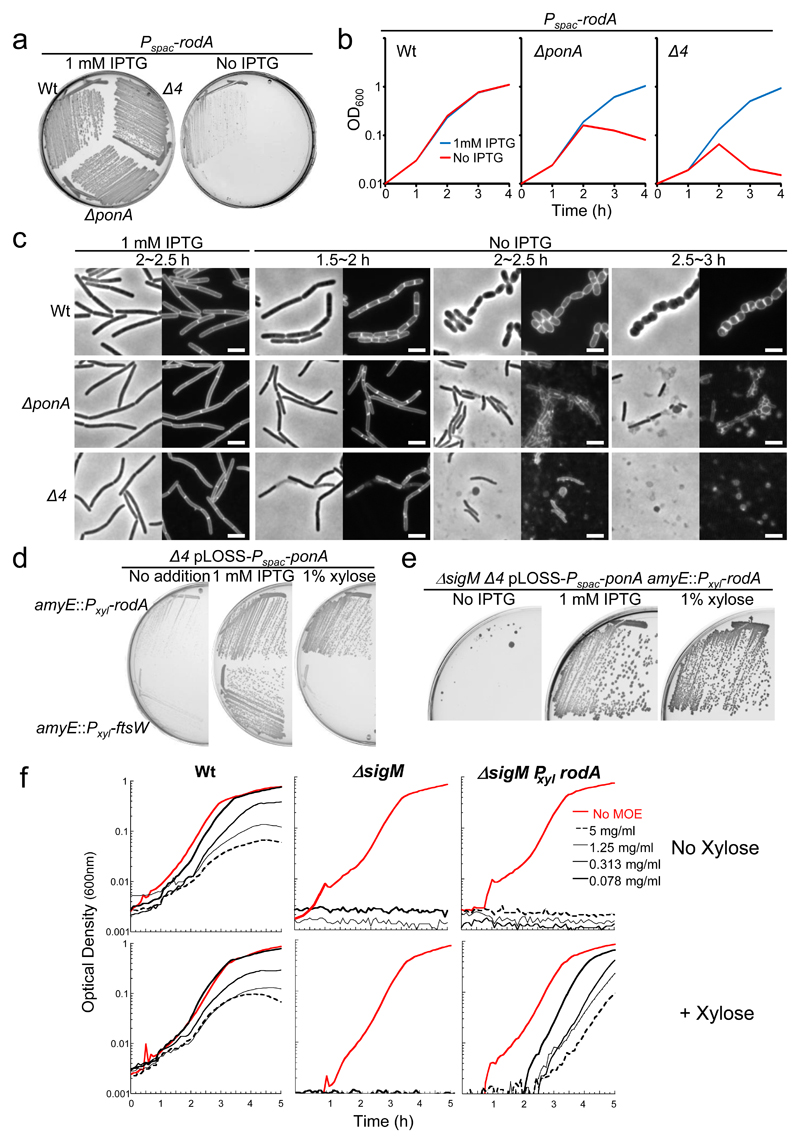
We built conditional and overexpression systems for rodA and used them to test for effects on sensitivity to MOE and ability to impair or enhance the growth of ponA or Δ4 mutants. Previous work showed that depletion of rodA (using an IPTG-dependent promoter, Pspac) can be tolerated in the presence of osmoprotectant (sucrose) and 20 mM Mg++ (19 and Fig. 2a, “Wt”). However, RodA depletions in ΔponA or Δ4 backgrounds were lethal (Fig. 2a). Figure 2b shows an equivalent set of experiments in which sensitivity to depletion of RodA was tested in liquid culture, again in the presence of sucrose and Mg++. Growth of the control strain continued over at least 4 hours of rodA repression, whereas for either the ponA or Δ4 background strains, culture optical densities declined after about 2 hours. Microscopic examination of rodA depletion in an otherwise wild type background revealed a gradual loss of the normal cylindrical phenotype, as expected for a classic rodA mutant (Fig. 2c, upper panels). Surprisingly, when RodA was depleted in either of the class A PBP defective strains the characteristic gradual rounding did not occur: instead the cells stopped growing but remained cylindrical and then underwent extensive lysis (the rounded objects in the figure are phase contrast “pale”, having undergone lysis). Thus, class A PBP activity is required for both continued growth and cell integrity, and for development of the aberrant morphology characteristic of rodA mutant cells.
Figure 2. Evidence for RodA as a candidate for the missing GTase.
a, Synthetic lethality of a rodA mutation in a ΔponA and Δ4 mutants. Growth on NA/MSM plates with or without 1 mM IPTG of strains YK2245 (Pspac-rodA), YK2246 (ΔponA::spc Pspac-rodA) and YK2242 (Δ4 Pspac-rodA) at 30°C.
b, Growth profiles of YK2245 (Pspac-rodA), YK2246 (ΔponA::spc Pspac-rodA) and YK2242 (Δ4 Pspac-rodA) strains in NB/MSM with or without 1 mM IPTG at 37°C.
c, Morphological effects of RodA depletion in ΔponA and Δ4 mutants. Phase-contrast and the corresponding cell membrane-stained images of typical fields of cells of YK2245 (Pspac-rodA), YK2246 (ΔponA::spc Pspac-rodA) and YK2242 (Δ4 Pspac-rodA) cultured in NB/MSM medium with or without 1 mM IPTG at 37°C. Time (h) of cultivation of cells after removal of IPTG is indicated. Scale bar represents 3 μm.
d, Rescue of the growth defect of a Δ4 mutant by overexpression of rodA, but not ftsW. Growth on PAB plates with or without 1 mM IPTG, or 1% xylose, of strains YK2256 (Δ4 pLOSS-Pspac-ponA amyE::Pxyl-rodA) and YK2257 (Δ4 pLOSS-Pspac-ponA amyE::Pxyl-ftsW) at 30°C.
e, Rescue of the synthetic lethality of Δ4 and ΔsigM strains by the overexpression of rodA. Growth on NA plates with or without 1 mM IPTG, or 1% xylose of strain YK2259 (Δ4 pLOSS-Pspac-ponA amyE::Pxyl-rodA ΔsigM::kan) at 30°C.
f, Restoration of resistance to MOE in a sigM mutant overproducing RodA. Wild type (168CA), AG494 (ΔsigM) or YK2268 (ΔsigM amyE::Pxyl-rodA) were grown in the presence (bottom) or absence (top) of xylose, and treated with MOE as indicated.
All the results in the figure are representative of at least two independent experiments.
We previously noted that ponA mutants grow particularly poorly on PAB medium, which supports virtually normal growth of wild type cells20. The Δ4 mutant had a similar growth defect on PAB medium, which could be corrected by induction of ponA expression from a plasmid-borne copy (Fig. 2d, 1 mM IPTG). Importantly, robust growth of the Δ4 mutant on PAB medium could also be achieved when rodA was overexpressed from an extra copy of the gene under xylose-inducible control. Thus, overproduction of RodA seems to ameliorate at least some of the growth deficiencies of the class A PBP defective mutants. B. subtilis (and most bacteria) have a second essential SEDS protein, FtsW, which is required specifically for cell division16. This gene does not seem to be subject to σM control11,12 and indeed, overexpression of ftsW did not rescue growth (Fig. 2d, 1% xylose).
We showed above that it is not possible to combine sigM and ponA mutations under normal conditions. Figure 2e shows that overexpression of rodA was also sufficient to enable robust growth of a sigM Δ4 multiple mutant, consistent with rodA encoding the sole sigM regulated product needed for growth in the absence of class A PBPs.
MOE is a specific inhibitor of the GTase activity of class A PBPs10. If rodA encoded the sigM dependent GTase responsible for growth in the presence of MOE, artificial upregulation of rodA should restore MOE resistance in a sigM mutant. Figure 2f shows that a sigM mutant was at least 10-fold more sensitive to MOE than the isogenic wild type strain. Introduction of a xylose-inducible copy of rodA into the sigM mutant made no difference in the uninduced state but in the presence of xylose, resistance to MOE was boosted almost to the wild type level. In a control experiment, a xylose-inducible copy of the related ftsW gene was induced in the sigM mutant but no such rescue was detected (not shown). Thus, induction of rodA is sufficient to restore growth to a sigM mutant when the GTase activity of class A PBPs is inhibited, highlighting this gene as a strong contender for the missing GTase.
To rule out MreC and/or MreD as the missing GTase, we carried out a series of similar experiments in which we depleted or overproduced the proteins. Neither mreC nor mreD generated a synthetic lethal phenotype when combined with a ponA mutation (Supplementary Fig. 2). Furthermore, overexpression of mreBCD does not rescue ponA lethality on PAB medium, nor provide resistance to MOE in a sigM mutant. Since MreC and MreD are probably members of a large protein complex required for elongation, including not just RodA but also RodZ, class B PBPs and the MreB proteins21, it seems possible that the MOE sensitivity of mreC and mreD mutants is due to indirect effects on function of the elongation complex.
A potential antibiotic inhibitor of the missing GTase
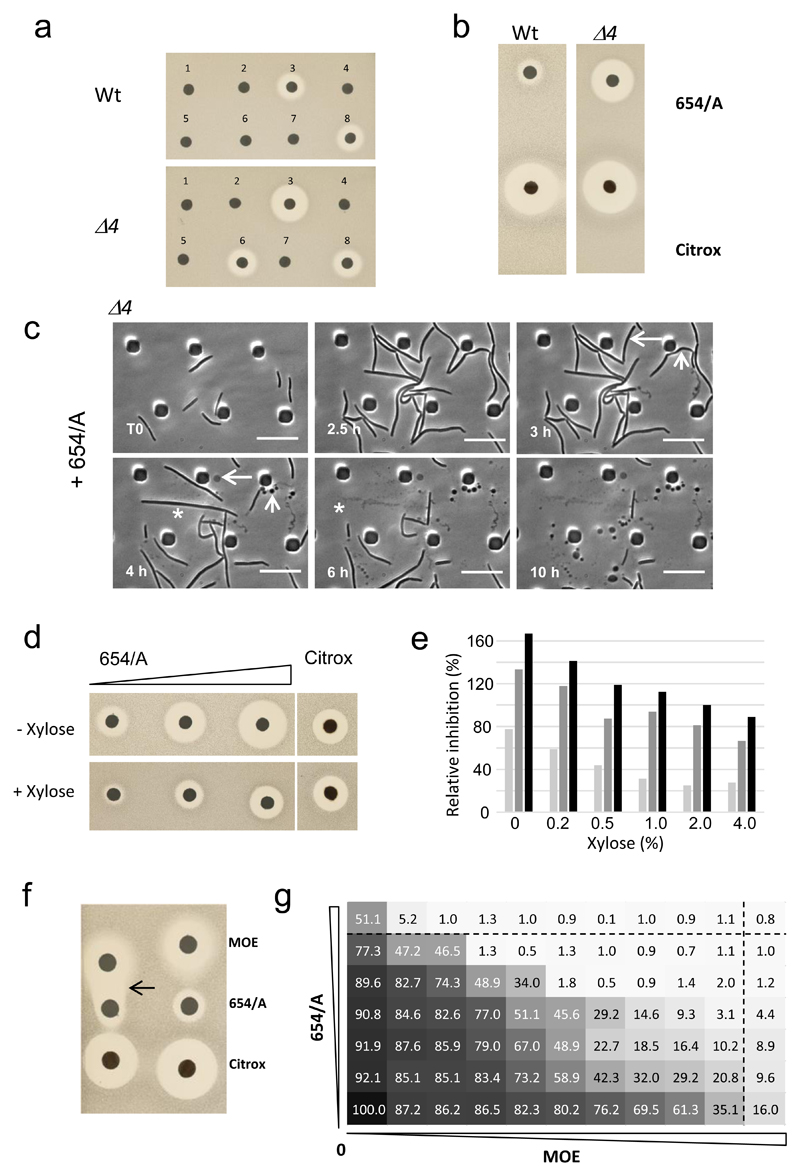
In parallel with the above experiments, we carried out a chemical genetic screen for inhibitors of the missing GTase. Culture extracts from 2,387 different actinomycete strains were screened at Demuris Ltd for differential antibiotic activity against wild type B. subtilis (168CA) and the Δ4 strain. An example of a pair of plates challenged with discs containing extracts from 8 different candidates is shown in Figure 3a. Discs 3 and 6 showed reproducible differential activity (larger zones of inhibition) on the Δ4 vs the Wt. Other extracts showed either no significant activity or large zones on both strains (disc 8). Strain DEM20654 (disc 3) was chosen for further study, based on its robust production of soluble inhibitory activity in liquid medium and reasonably rapid growth properties.
Figure 3. Isolation and characterization of a putative inhibitor of the missing GTase.
a, Validation of a selection of hits from rescreening for antimicrobial activity using disc diffusion test. Fresh culture media extracts of different Actinomycete strains were prepared and tested against B. subtilis Wt and the Δ4 mutant. Discs 3 (Strain DEM20654) and 6 showed reproducible differential activity (larger zones of inhibition) on the Δ4 vs the Wt strain. Other extracts showed either no significant activity or large zones on both strains (disc 8).
b, Confirmation of the differential activity of purified 654/A on Δ4 relative to a control by disc diffusion assay.
c, Effects of purified 654/A on the Δ4 mutant. Individual frames are extracted from the video in Supplementary Video 1. Numbers in the bottom left corner of each frame represent time elapsed in the video. Examples of different lytic events are shown by asterisks (complete cell destruction), arrows (formation of phase pale bulbous cells), and arrowheads (formation of chains of small phase dark “minicells”).. Scale bar represents 10 µm.
d,e, Increased 654/A resistance in ponA mutant cells containing an extra xylose-inducible copy of rodA (Pxyl-rodA). d, Disk diffusion tests were done with three different concentrations of 654/A (corresponding to 2, 5 or 10 µl amounts of purified material, left to right). Citrox was used as a control. Duplicate plates without or with 4% xylose (to induce overexpression of rodA), as indicated, were inoculated with strain YK2251 and incubated overnight. e, Dose response results for three different concentrations of 654/A, and at a range of concentrations of xylose. Average zones of inhibition were measured for duplicate disc diffusion tests and expressed as a percentage of the inhibition zone for the control antibiotic (citrox) measured on the same plate.
f,g, Synergy between 654/A and MOE as demonstrated by disc diffusion (f) or checkerboard analysis (g) with the wild type strain (168CA). f, MOE, 654/A and a control antibiotic (citrox) were placed on a lawn of bacteria on an NA plate. The arrow points to an enhanced zone of clearing at the interface of the MOE and 654/A sub-inhibitory regions. g, Serial dilutions (2-fold) of MOE and 654/A were transferred to a 96 well plate, with MOE along the horizontal axis and 654/A in the vertical as indicated below and to the left. A dilute culture of wild type bacteria was used to inoculate each well and the plate was incubated for 24 h with continuous optical density monitoring. Dotted lines identify the minimum inhibitory concentrations derived by reference to the wells containing only one antibiotic (top row for MOE and far right column for 654/A). OD600 readings at the 18 h time point were used to calculate percentage inhibition values (shown in each rectangular segment) relative to the no antibiotic control.
All the results in the figure are representative of at least two independent experiments.
In preliminary experiments strain DEM20654 appeared to make at least three different bioactive molecules. One was a polyene compound, with strong antifungal properties. A second was antibacterial but triggered the DNA damage response on a B. subtilis reporter strain and so was thought not likely to be the compound of interest. The third activity, designated 654/A, showed a substantially larger zone of inhibition on the Δ4 strain than on the wild type (Fig. 3b). A control antibiotic (citrox) produced equal zones of inhibition on both strains, as did various other antibiotics tested (Supplementary Fig. 3). The activity of interest also inhibited the growth of Staphylococcus aureus but not the eukaryotic fission yeast, Schizosaccharomyces pombe.
Purified 654/A causes cell lysis and has enhanced activity on the Δ4 mutant
The HPLC purified active material contains three UV peaks that run very close to each other (Supplementary Fig. 4). Peak 1 and the minor peak 3 are probably closely related as they have similar UV absorbance profiles. We do not yet know whether any single peak corresponds to the active molecule, or whether a combination is needed for the activity. Nevertheless, availability of this bulk material enabled more extensive analysis of effects of the putative novel antibiotic on cells. Cultures of the wild type and Δ4 mutants were examined by time lapse microscopy in a microfluidic device before and after exposure to a growth inhibitory concentration of 654/A. Representative videos are shown in Supplementary Videos 1-3, and individual frames from the Δ4 strain are shown in Figure 3c. Compound treatment caused an arrest in growth of the Δ4 mutant followed by various lytic events culminating in: complete cell destruction, formation of phase pale bulbous cells, or formation of chains of small phase dark “minicells”. In general, lysis tended to occur at cell poles or mid cell potential division sites. As expected, at this concentration, wild type cells showed only mild morphological defects but at a higher concentration of 654/A, the effects were similar to those shown for the mutant in Figure 3c (Supplementary Video 4). The phenotypic events documented are typical of mutants affected in cell wall synthesis, consistent with the notion that 654/A might work on the missing GTase.
Effects of 654/A activity on the Δ4 mutant are dependent on rodA expression levels
If the 654/A compound acted on the SEDS proteins, overexpression of the rodA and/or ftsW genes should ameliorate the effects of the compound. It emerged that overexpression of rodA did indeed increase resistance of the ponA mutant to 654/A. Figure 3d shows a typical experiment in which a ponA strain carrying an extra copy of rodA under control of Pxyl was exposed to 654/A. As expected, increasing concentrations of 654/A resulted in progressively larger zones of inhibition. However, addition of xylose to induce overproduction of RodA resulted in greatly reduced zones of inhibition at all three concentrations of 654/A. Quantitation of the effects of different levels of RodA overproduction (Fig. 3e) revealed a gradual amelioration of inhibition with increasing xylose concentration. Similar results were obtained in liquid growth experiments (Supplementary Fig. 5), which also showed that the rescue was not due to xylose addition. These results demonstrated that overproduction of RodA counteracts the inhibitory effects of 654/A on B. subtilis.
Moenomycin and 654/A act synergistically
If MOE and 654/A acted on different forms of GTases with overlapping functionality the two antibiotics should behave synergistically. Indeed, in disc diffusion assays, cells exposed to sub-lethal concentrations of the combination of 654/A and MOE were killed in the zone between the two antibiotic discs (arrowed; Fig. 3f). No such synergistic killing effect was evident in the zone between 654/A and the citrox control. We extended this analysis to a checkerboard experiment (Fig. 3g) in which cultures in microwell plates were exposed to the two antibiotics across a range of combinatorial concentrations. Severe inhibition of growth occurred in many of the wells containing sub-MIC concentrations of both antibiotics (Fig. 3g, below and to the left of the MIC lines), indicating a strong synergistic effect.
Discussion
The genes encoding SEDS proteins were identified decades ago through mutations generating a round cell, division or sporulation defective phenotype8,22. The mutant phenotypes are thus very similar to those generated by mutations in genes encoding the class B PBPs. Indeed, in endospore forming bacteria, there is also a sporulation specific class B PBP called SpoVD, and a SEDS protein with indistinguishable mutant phenotype called SpoVE23. In many cases the SEDS gene lies immediately adjacent to a class B PBP gene, for example the mrdA and mrdB genes of E. coli24, suggesting that their functions might be connected. There are even examples of organisms in which the two moieties are naturally fused. We obtained DNA from one such organism, Lachnoclostridium phytofermentans ISDg (accession WP_012199367.1) and confirmed by direct sequencing that the two genes are indeed fused in a single open reading frame (not shown).
The primary sequences of the various classes of SEDS proteins (RodA, FtsW, SpoVE), as well as patterns of transmembrane predictions and conserved residues, are very similar, suggesting that they are functionally equivalent. It has been suggested, based on the extremely hydrophobic nature and the multi-transmembrane character, that SEDS proteins might act as flippases, responsible for delivering the membrane associated PG precursor molecule, lipid II to the class B PBP, ready for transpeptidation25. However, recently, alternative candidates for the lipid II flippases have been identified in the MurJ and Amj proteins, in E. coli and B. subtilis15,26. Our results suggest, instead, that RodA, and possibly the other SEDS proteins8, are GTases that work together with their cognate class B PBP transpeptidases to catalyse key steps in bacterial cell wall morphogenesis. In support of this idea, Ovchinnikov et al.27 reported that the predicted transmembrane structure of FtsW shares similarity with dolichyl-diphosphooligosaccharide-protein glycosyltransferase, which is a GTase that transfers an oligosaccharide from a lipid-linked precursor to nascent glycoproteins. Recently published papers have provided direct biochemical evidence9,28 for GTase activity. It remains possible that the SEDS proteins have both flippase and GTase activities. If so, this would mean that, in principle, the SEDS proteins could take a lipid II molecule from inside the cytoplasmic membrane, flip it and then directly transfer the substrate onto a glycan strand in the wall. This would contrast with the class A PBPs, which, due to their GTase domain lying outside the membrane, presumably rely on a flippase to provide the substrate.
Consistent with the notion of a direct role for the RodA protein in cell wall elongation, depletion of rodA in a mutant lacking class A PBPs results in growth arrest. However, the rounded phenotype characteristic of rodA mutants of both B. subtilis and E. coli, did not emerge. We interpret this to support the idea that RodA is essential for the cell elongation process and that in its absence the class A PBPs manufacture PG in an uncontrolled manner, leading to the rounded phenotype. This is reminiscent of our previous finding that in mreB mutants of B. subtilis delocalization of PBP1A is responsible for the cell wall bulging that leads to lysis20. It seems unlikely that this loss of growth could be attributed to loss of flippase activity, given that B. subtilis has multiple flippases of the MurJ family and an alternative flippase in Amj15. It thus appears that RodA (and possibly FtsW during cell division) are the key GTases responsible for controlled (morphogenic) wall synthesis. The role of the class A PBPs may be to provide bulk PG synthesis when wall synthesis is damaged or perturbed, or to increase the overall growth rate of cells. Altogether, this would be a paradigm shifting change in our thinking about the roles of PBPs in cell wall synthesis.
If RodA and the other SEDS proteins do indeed turn out to be GTases, they may represent one of the most important new antibiotic targets discovered in several decades. The genes are very widely conserved and essential for growth and viability under normal conditions. The crucial advantages of these kinds of proteins as targets for antibiotics lies in their availability on the outer surface of the permeability barrier imposed by the cytoplasmic membrane, and on the absence of equivalent functions in humans. Furthermore, inhibitors with only a single lethal protein target tend to suffer from problems of resistance, which is another reason why dual targeting fluoroquinolones and muti-targeting β-lactams have been so successful. Assuming that RodA and FtsW are both GTases, it may be possible to obtain inhibitors of both, and thus achieve dual lethal targeting. Indeed, we have not excluded the possibility that 654/A has some activity on FtsW. Finally, the existence of a naturally occurring bioactive compound that targets RodA reinforces the notion that these proteins are crucial for cell viability and fitness, and the compound may provide a rich new source of starting points for future antibiotics.
Methods
Bacterial strains, plasmids, primers and growth conditions
The bacterial strains, plasmid constructs and primers for PCR analysis in this study are shown in Supplementary Data Table 2. DNA manipulations were carried out using standard methods. Nutrient agar (Oxoid), LB or Difco antibiotic medium 3 (PAB) plates were used for growth on solid medium supplemented, as required, with MSM (0.5 M sucrose, 20 mM maleic acid and 20 mM MgCl2), IPTG or xylose at 30 or 37°C. Liquid cultures of Bacillus strains were grown at 30 or 37° C in LB medium or nutrient broth (NB, Oxoid) with MSM at 37° C. For genetic selections, antibiotics were added to media at the following concentrations: 100 μg/ml ampicillin, 1 μg/ml erythromycin, 2 or 5 μg/ml kanamycin (or 15 μg/ml with 20 mM Mg++), 50 μg/ml spectinomycin and 5 μg/ml chloramphenicol. Citrox was obtained from Citrox Ltd, Middlesbrough, UK.
pG+host10 and the derivatives used for marker free mutants of Class A PBPs
pG+host10 plasmid was constructed as a derivative of pG+host9 carrying an ampicillin resistance cassette and thermosensitive replication29. The bla gene was amplified by PCR using RE05 / 06 primers and plasmid pLOSS* as substrate. The PCR product and pG+host9 were digested by ClaI and XhoI and cloned into E. coli TG1 to generate plasmid pG+host10. Upstream and downstream DNA regions (500-600 bp) of ponA coding sequence were amplified by PCR using primers RE01 / 02 and RE03 / 04, respectively and double digested by NotI/EcoRI and EcoRI/KpnI, respectively. Digested PCR products were ligated to digested plasmid pG+host9 (NotI/KpnI), cloned into E. coli TG1 to obtain plasmid pG+host9::ΔponA. Similarly, upstream DNA regions of pbpD and pbpF coding sequences were amplified by PCR with primers RE07-08 and RE11 / 12 whereas downstream region of pbpD and pbpF were amplified using primers RE09-10 and RE13 / 14, respectively. Upstream and downstream PCR products were double digested by NotI/EcoRI and EcoRI/HindIII respectively. Each of the pbpD or pbpF flanking DNA-fragments were ligated to double digested NotI/HindIII pG+host10 plasmid, then cloned into E. coli TG1 to generate plasmids pG+host10::ΔpbpD and pG+host10::ΔpbpF. Plasmids obtained were checked by digestion and PCR sequencing with primers AG21 to 23.
The marker-free deletion mutants of the B. subtilis vegetative class A PBPs were obtained by consecutive multi-step reactions using pG+host9 derivatives as described previously (see also above and Supplementary Data Table 2). All class A PBP mutants were selected on NA plates with 10 mM or 20 mM MgSO4. Marker-free deletions were confirmed by PCR using primers listed in Supplementary Data Table 2.
pLOSS-Pspac-ponA
To construct pLOSS-Pspac-ponA plasmid, a 2846 bp fragment of ponA coding sequence and its putative terminator was amplified by PCR using primers AG11 / 12. PCR product and pLOSS* plasmid30 were digested by NotI and SpeI, then ligated and cloned in E. coli DH5α to generate plasmid pLOSS-Pspac-ponA. The presence of the insert was verified by plasmid digestion and plasmid sequencing using primers AG18 / 19.
Pspac-rodA
To generate a strain with conditional expression of rodA the 5’ end of the coding sequence was cloned as an XbaI-EcoRV fragment from pRD163, ligated into pSG441 digested with XbaI and blunted ClaI. The resulting plasmids, pRD159, once integrated into genome of B. subtilis 168CA, gave kanamycin resistant strains that are dependent on IPTG (including strain YK2245).
amyE::Pxyl-rodA and -ftsW
For overexpression of the ftsW and rodA genes each gene was cloned into an amyE integrating construct under the control of the Pxyl promoter. PCR generated DNA fragments (using oligo pairs A328 / A329 for ftsW and A330 / A331 for rodA) containing the entire coding sequences of FtsW and RodA together with their native ribosome binding sites, were cloned into plasmid pJPR1 to link them to the Pxyl promoter, using the restriction sites XbaI and NotI. The resulting derivatives of pJPR1 (pRD164 encoding ftsW and pRD163, for Pxyl-rodA) were then transformed into 168CA selecting for double crossover insertions on the basis of chloramphenicol resistance and loss of amylase activity, to generate strains YK2250 (ftsW) and YK2249 (rodA).
RNA isolation and RNA-seq
Wild-type (168CA) and Δ4 (AG157) strains were cultured in NB at 37°C until mid-exponential phase. Total RNA was isolated using a Total RNA purification Plus Kit (Norgen Bioteck). RNA quality was assessed by Agilent RNA 6000 Nano Kit. The RNA sequencing was performed at PrimBio Research Institute (USA).
Microscopic imaging
For snapshot live cell imaging, cells were mounted on microscope slides covered with a thin film of 1.2% agarose in water, essentially as described previously31. Images were acquired with a Rolera EM-C2 (Q-imaging) camera attached to a Nikon TiE microscope. The images were acquired and analysed with METAMORPH version 6 (Molecular Devices).
For the time lapse images (Fig. 3c) the strains were cultured overnight on LB medium supplemented with MgSO4 to a final concentration of 20 mM, then diluted to an OD600nm of 0.05. Samples of cells were loaded into B04A microfluidic plates (ONIX, CellASIC) at 4 psi for 15 sec and the loading channel was subsequently washed for 30 sec at 3 psi. Throughout imaging, the media flow rate and temperature were maintained at 2 psi and 37°C, respectively. Compound was added to the cells by switching the media flow channel using the CellASIC ONIX FG software (v. 5.0.1.0). Cells were visualised using a Nikon Ti microscope equipped with a Nikon Plan Apo 100x/1.40 oil objective. Images were captured every 5 mins using FRAP-AI v. 7.7.5.0 software (MAG Biosystems, Molecular Devices). Images and videos were processed using ImageJ (NIH).
Moenomycin (MOE) growth assays
Strains were grown in a suitable culture medium overnight at 30°C, then diluted ~1:200 into fresh medium, grown until mid-exponential at 37°C and then diluted again in fresh medium to a suitable OD600nm (usually between 0.03 and 0.1). Using a multichannel pipette, 100 μl of diluted-cells were dispensed into each well of a 96-well plate containing 100 μl volumes of the antibiotic to be tested pre-diluted in the culture medium. The microtiter plate was then incubated with shaking for up to 16 h at 37°C in a microtiter plate reader (FluoStar Galaxy, BMG Lab Technologies). Readings of the OD600nm was taken every 5 to 6 min. For the determination of MOE sensitivity a wide range of MOE concentrations were employed ranging from 4 ng/ml to 20 μg/ml. For moenomycin and 654/A synergy test, MOE was diluted in a 2-fold series along the horizontal axis and 654/A in the vertical.
Inhibitor screening methods
Establishment of optimal growth conditions for screening
Screening was carried out by comparing the effects of actinomycete natural products on growth of wild type (168CA) and Δ4 (AG157) strains. A range of pilot experiments were used to define the optimal conditions for culture growth and extract testing (using a randomly selected 96 well plate containing actinomycete extracts). From these experiments a protocol suitable for high throughput screening was established, as follows.
Actinomycete samples
Actinomycetes were cultured on suitable agar plates, typically GYM medium (glucose 4 gl-1, yeast extract 4 gl-1, malt extract 10 gl-1, pH 7.0) at 30°C for several days until stationary phase was reached. The total agar plus biomass was harvested and crushed by passage through a syringe then frozen at -80°C and thawed. The material was centrifuged at 5000 x g for 10 min. The supernatant was collected and stored at -80°C until use.
Indicator cell preparation
Wild type and Δ4 strains were cultured overnight in LB + 20 mM MgSO4 (LB/Mg) with shaking at 30°C. On the day of screening the overnight culture was diluted to an OD600nm of 0.05 in 10 ml fresh LB/Mg and incubated (160 rpm) at 30°C. When the culture reached an OD600nm of 0.1 ± 0.02, the cells were serially diluted to the required density.
Agar plate screening
For agar plate screening, 50 ml of NA + 20 mM MgSO4 (NA/Mg) was poured into square 100 mm x 100 mm petri dishes. After drying, 3 ml of a 10-3 dilution of test culture was poured onto the plate and swirled to cover the plate. Excess liquid was poured off and the plates were allowed to dry. Extracts from actinomycete plate or liquid cultures (approx. 2 μl) were spotted onto the plates using a pinning device, allowed to dry and then the plates were incubated overnight at 30°C. Zones of inhibition were recorded after 15 h.
Microplate (liquid culture) screening
Thawed Actinomycete extract samples (10 µl) were dispensed into 96-well plates in duplicate and to each well was added 100 µl of the wild type or Δ4 strain (OD600nm of 0.05). Plates were incubated in orbital shakers overnight (18 h). The final OD600nm was measured using a plate reader and wells with significantly reduced density were identified.
Disc diffusion assay
These were carried out as described under agar plate screening except that 10 µl of crude or fractionated extract was spotted onto 5 mm discs cut from Fisher chromatography paper (CHR200), which were then placed on dried, seeded NA plates. Plates were incubated at 30°C for 18 h and the diameter of any zone of inhibition measured. Citrox was used as a control antibiotic for convenience and because it gave very reproducible inhibition under a wide range of conditions and for many different organisms.
The high throughput screen
Based on preliminary results, the microplate (liquid culture) screening method was chosen for the full screen. Extracts from a total of 2387 Actinomycete strains from the Demuris Ltd. collection were screened for activity against the wild type and Δ4 strains. Out of 2387 extracts, 679 showed some B. subtilis killing: a typical hit rate for actinomycete extracts on Firmicutes. A total of 102 candidate hits (reducing OD600nm in the Δ4 strain compared with the wild type) were regrown and crude culture supernatants were screened again for differential activity on the two strains using a disc diffusion assay.
Hit prioritization
Based on comparison of the liquid culture and disc diffusion results three strains, DEM20654, DEM21374 and DEM31701 were prioritised as making compounds with substantially higher activity on the Δ4 strain compared to the wild type. Each of the three strains were cultured in a range of different growth media and retested. DEM20654 was chosen for further work based on its robust production of a specific inhibitor in a range of different liquid media.
Bulk purification of 654/A
DEM20654 was grown in 16 l of GMY medium (glucose, malt & yeast extract), supplemented with 0.5 g l-1 oat flour, and a series of chemical fractionation procedures were used to isolate nearly pure material for further study. After growth to stationary phase, the mycelia were separated from the culture supernatant by filtration. Amberlite XAD-16 resin (20 g l-1) was applied to the supernatant. After overnight incubation under agitation, the beads were filtered off and washed with deionised water. Elution was carried out using methanol (4 l). The solvent was evaporated to an aqueous extract. The pH of the aqueous extract was adjusted to 10 using sodium hydroxide followed by twice extracting with 2 vols of ethyl acetate. The organic layer was then dried under vacuum in 10 g of silica gel. The dried material was subjected to: silica flash chromatography (Biotage® 100 g SNAP Ultra cartridge), using an ethyl acetate / methanol (0-100%) gradient; size exclusion chromatography using methanol and LH20 resin as stationary phase; and high performance liquid column chromatography (HPLC) on an Agilent 1260 Infinity II LC system (Agilent Technologies INC, Santa Clara, CA, USA) equipped with a reverse-phase C18 column (Phenomenex, CA, USA). The HPLC separated fractions were eluted with a gradient of acetonitrile in water from 0 to 100% in 45 min at 1 ml min-1 flow rate. Individual fractions were subjected to activity tests against B. subtilis strains by disc diffusion assays.
Supplementary Material
Acknowledgements
This work was funded by grants from the BBSRC (BB/G015902/1; RAD, JE, AG), ERC (670980; JE, YK, KE) and the Wellcome Trust (WT098374AIA; JE, LJW), and core resources in Demuris Ltd. We thank Robyn Emmins, who constructed some of the mutants, David Roberts for help with microfluidic microscopy, and Ryan Sweet for help with the large scale fermentation run.
Footnotes
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Author contributions
KE, AG, YK, JD, LJW and RAD did the experiments. All authors contributed to experimental design and concepts. JE wrote the main text with contributions from all other authors.
Author conflicts
JE is a Director and shareholder at Demuris Ltd. NA and JD are employees at Demuris Ltd.
References
- 1.Errington J. L-form bacteria, cell walls and the origins of life. Open biology. 2013;3:120143. doi: 10.1098/rsob.120143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lovering AL, Safadi SS, Strynadka NC. Structural perspective of peptidoglycan biosynthesis and assembly. Annu Rev Biochem. 2012;81:451–478. doi: 10.1146/annurev-biochem-061809-112742. [DOI] [PubMed] [Google Scholar]
- 4.Spratt BG. Distinct penicillin-binding proteins involved in the division, elongation and shape of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei Y, Havasy T, McPherson DC, Popham DL. Rod shape determination by the Bacillus subtilis class B penicillin-binding proteins encoded by pbpA and pbpH . J Bacteriol. 2003;185:4717–4726. doi: 10.1128/JB.185.16.4717-4726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daniel RA, Williams AM, Errington J. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis . J Bacteriol. 1996;178:2343–2350. doi: 10.1128/jb.178.8.2343-2350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McPherson DC, Popham DL. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J Bacteriol. 2003;185:1423–1431. doi: 10.1128/jb.185.4.1423-1431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henriques AO, Glaser P, Piggot PJ, Moran CP., Jr Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol Microbiol. 1998;28:235–247. doi: 10.1046/j.1365-2958.1998.00766.x. [DOI] [PubMed] [Google Scholar]
- 9.Meeske AJ, et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature. 2016 doi: 10.1038/nature19331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Heijenoort Y, Leduc M, Singer H, van Heijenoort J. Effects of moenomycin on Escherichia coli. J Gen Microbiol. 1987;133:667–674. doi: 10.1099/00221287-133-3-667. [DOI] [PubMed] [Google Scholar]
- 11.Eiamphungporn W, Helmann JD. The Bacillus subtilis σM regulon and its contribution to cell envelope stress responses. Mol Microbiol. 2008;67:830–848. doi: 10.1111/j.1365-2958.2007.06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salzberg LI, Luo Y, Hachmann AB, Mascher T, Helmann JD. The Bacillus subtilis GntR family repressor YtrA responds to cell wall antibiotics. J Bacteriol. 2011;193:5793–5801. doi: 10.1128/JB.05862-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McPherson DC, Driks A, Popham DL. Two class A high-molecular-weight penicillin-binding proteins of Bacillus subtilis play redundant roles in sporulation. J Bacteriol. 2001;183:6046–6053. doi: 10.1128/JB.183.20.6046-6053.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vasudevan P, McElligott J, Attkisson C, Betteken M, Popham DL. Homologues of the Bacillus subtilis SpoVB protein are involved in cell wall metabolism. J Bacteriol. 2009;191:6012–6019. doi: 10.1128/JB.00604-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meeske AJ, et al. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis . Proc Natl Acad Sci U S A. 2015;112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- 17.Wachi M, Doi M, Okada Y, Matsuhashi M. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J Bacteriol. 1989;171:6511–6516. doi: 10.1128/jb.171.12.6511-6516.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leaver M, Errington J. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol. 2005;57:1196–1209. doi: 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- 19.Kawai Y, et al. A widespread family of bacterial cell wall assembly proteins. EMBO J. 2011;30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawai Y, Daniel RA, Errington J. Regulation of cell wall morphogenesis in Bacillus subtilis by recruitment of PBP1 to the MreB helix. Mol Microbiol. 2009;71:1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x. [DOI] [PubMed] [Google Scholar]
- 21.Errington J. Bacterial morphogenesis and the enigmatic MreB helix. Nat Rev Microbiol. 2105;13:241–248. doi: 10.1038/nrmicro3398. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda M, et al. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation respectively. J Bacteriol. 1989;171:6375–6378. doi: 10.1128/jb.171.11.6375-6378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fay A, Meyer P, Dworkin J. Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J Mol Biol. 2010;399:547–561. doi: 10.1016/j.jmb.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamaki S, Matsuzawa H, Matsuhashi M. Cluster of mrdA and mrdB genes responsible for the rod shape and mecillinam sensitivity of Escherichia coli. J Bacteriol. 1980;141:52–57. doi: 10.1128/jb.141.1.52-57.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Höltje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sham LT, et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ovchinnikov S, et al. Large-scale determination of previously unsolved protein structures using evolutionary information. eLife. 2015;4:e09248. doi: 10.7554/eLife.09248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho H, et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat Microbiol. 2016:16172. doi: 10.1038/nmicrobiol.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maguin E, Prevost H, Ehrlich SD, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claessen D, et al. Control of the cell elongation-division cycle by shuttling of PBP1 protein in Bacillus subtilis. Mol Microbiol. 2008;68:1029–1046. doi: 10.1111/j.1365-2958.2008.06210.x. MMI6210 [pii] [DOI] [PubMed] [Google Scholar]
- 31.Glaser P, et al. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 1997;11:1160–1168. doi: 10.1101/gad.11.9.1160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.