Abstract
Oncogenic gene fusions drive many human cancers, but tools to more quickly unravel their functional contributions are needed. Here we describe methodology permitting fusion gene construction for functional evaluation. Using this strategy, we engineered the known fusion oncogenes, BCR-ABL1, EML4-ALK, and ETV6-NTRK3, as well as 20 previously uncharacterized fusion genes identified in TCGA datasets. In addition to confirming oncogenic activity of the known fusion oncogenes engineered by our construction strategy, we validated five novel fusion genes involving MET, NTRK2, and BRAF kinases that exhibited potent transforming activity and conferred sensitivity to FDA-approved kinase inhibitors. Our fusion construction strategy also enabled domain-function studies of BRAF fusion genes. Our results confirmed other reports that the transforming activity of BRAF fusions results from truncation-mediated loss of inhibitory domains within the N-terminus of the BRAF protein. BRAF mutations residing within this inhibitory region may provide a means for BRAF activation in cancer, therefore we leveraged the modular design of our fusion gene construction methodology to screen N-terminal domain mutations discovered in tumors that are wild-type at the BRAF mutation hotspot, V600. We identified an oncogenic mutation, F247L, whose expression robustly activated the MAPK pathway and sensitized cells to BRAF and MEK inhibitors. When applied broadly, these tools will facilitate rapid fusion gene construction for subsequent functional characterization and translation into personalized treatment strategies.
Keywords: Cancer genetics, Fusion genes, Oncogenomics, Cloning strategies, Chromosomal translocations: genomic aspects, Functional genomics
INTRODUCTION
Cancer genome profiling efforts by large consortia such as The Cancer Genome Atlas (TCGA) are cataloging the complex genomic landscape of diverse tumors, which are comprised of numerous somatically-acquired genetic alterations ranging from point mutations to ploidy change. Similar to activating mutations in oncogenes, chromosomal rearrangements also lead to oncogene activation through formation of gene fusion transcripts by, for example, increasing oncogene expression via a fused hyper-active transcription promoter or creation of chimeric hypermorphic or neomorphic alleles (1). Indeed, oncogenic fusion genes represent an important class of cancer driver aberrations, some of which have been exploited clinically for cancer therapy. For example, the functional role of BCR-ABL1 in promoting chronic myeloid leukemia led to successful therapies incorporating ABL inhibitors such as imatinib and dasatinib (2,3). Similarly, use of ALK inhibitors crizotinib and ceritinib has significantly improved clinical outcome in non-small cell lung cancers driven by ALK fusions (4,5).
The discovery of gene fusions has been accelerated by advances in next generation sequencing (NGS) technologies (6). While the overall frequency of recurrent fusion transcripts is lower than activating mutations in oncogenes, the oncogenic role of individual fusion genes is suggested by their presence in multiple tumor types as well as the anti-correlation between their presence and that of cancer driver mutations in known oncogenes (7). More importantly, several recent reports describing the oncogenic behavior and therapeutic response of tumors driven by extremely rare fusions highlight their clinical impact. For example, individual cases of myeloid neoplasms driven by fusions involving JAK2 and FLT3 are sensitive to JAK inhibitor (ruxolitinib) (8) and tyrosine kinase inhibitor (sorafenib) (9), respectively. Likewise, we recently reported an oncogenic fusion involving the RET kinase in a single medullary thyroid carcinoma patient whose activity is highly sensitive to multiple tyrosine kinase inhibitors (10). Together, these examples highlight the importance of identifying the subset of rare, oncogenic gene fusions and assessing their sensitivity to therapeutics.
The functional interrogation of fusion genes is complicated given their large number, inability to accurately predict those with driver activity and technical roadblocks preventing efficient fusion gene construction for biological assays. To address these challenges, we report here a method enabling rapid and accurate fusion gene construction using a multi-fragment, recombineering-based strategy. We used this approach to construct known oncogenic fusion genes BCR-ABL1, EML4-ALK and ETV6-NTRK3, all of which exhibited strong driver activity consistent with their role in cancer. We next scaled our fusion gene construction strategy to successfully build a pilot set of 20 fusion genes identified from TCGA datasets. Functional validation assays revealed that five of these fusion genes, which contained portions of MET, NTRK2, and BRAF kinases, exhibited robust transforming activity and marked responsiveness to inhibitors targeting their activated pathways. To illustrate another use of our fusion gene cloning strategy that leverages its versatility and modular design, we performed domain-function studies of BRAF fusion genes by differentially recombining N-terminal segments/domains of BRAF onto BRAF’s C-terminal kinase domain. Data resulting from this work support previous reports indicating that the transforming activity by BRAF fusion genes results from truncation-mediated loss of inhibitory domains located within the N-terminus of BRAF (11–14). Because gene mutations residing within this inhibitory domain might serve as a means to activate BRAF in cancer, we leveraged the modular design of our construction methodology to fuse onto BRAF’s kinase domain a set of inhibitory domains, each containing individual patient mutations, to screen for those capable of attenuating kinase inhibition. Using this approach, we identified an oncogenic mutation, F247L, whose expression robustly activates the MAPK pathway and sensitizes cells to inhibitors of BRAF and MEK.
MATERIALS AND METHODS
Fusion gene construction
The DNA sequences of positive control fusion genes (BCR-ABL1, EML4-ALK, and ETV6-NTRK3) and the 20 uncharacterized fusion genes (Supplementary Table 1–2) were obtained from The Cancer Genome Atlas (TCGA) and other published sources (7,15–17). PCR templates were from sequence-verified ORF collections by the ORFeome collaboration (18–20), Mammalian Gene Collection (21), and commercial ORF sources (Life Technologies). Based on the sequence of each fusion arm, primers were designed to contain recombination sequences (B1/B2/B2r/B4) followed by 18-nucleotide ORF-specific sequence (Supplementary Table 3). As illustrated in Figure 1A, the forward primer for ORF 1 carried a B1 site (5′-GGGGACAACTTTGTACAAAAAAGTTGGC) while the reverse primer carried a B2 site (5′-GGGGACCACTTTGTACAAGAAAGCTGGGT). As to ORF 2, the forward primer carried a B2r site (5′-GGGGACCCAGCTTTCTTGTACAAAGTGGTTA) while the reverse primer carried a B4 site (5′-GGGGACAACTTTGTATAGAAAAGTTGGGTG). Depending on the given ORF sequence combined with each recombination site sequence, it is possible that the fusion primer would introduce an unwanted, in frame stop codon that would ultimately reside between the left and right fusion gene arms. One prevents this by manually checking the fusion primer sequence for stop codons, which would be removed by altering the wobble position, or simply substituting the following B2 site and B2r site sequences that eliminates the possibility of stop codons: B2 site (5′-GGGGACCACTTTGTACAAGAAAGCTGGGA); B2r site (5′-GGGGACCCAGCTTTCTTGTACAAAGTGGTTC). The resulting PCR products were incorporated into compatible pDONR vectors (P1/P2 for Fusion fragment 1, P2r/P4 for Fusion fragment 2) through BP recombination (Life Technologies) following the manufacturer’s recommendations. The resulting Fusion fragment 1 will be flanked with L1/L2 recombination sites, while Fusion fragment 2 will be flanked with R2/L4 recombination sites. These products were subsequently transferred into pLenti-EF1α-DEST and pHAGE-EF1α-DEST vectors containing R1/R4 sites through multi-site LR recombination reaction (Life Technologies). The reaction mixtures were transformed into STBL3 (Life Technologies) competent bacteria. The 5′ and 3′ pDONR vectors as well as pLenti-EF1α-DEST and pHAGE-EF1α-DEST vectors required for fusion gene construction/expression are available from Addgene (www.addgene.org).
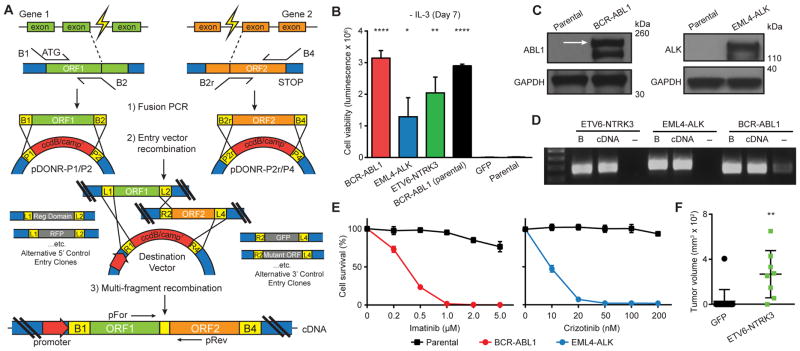
Figure 1. Multi-fragment recombineering of fusion genes.
(A) Schematic illustration of fusion gene construction. ATG = translation start site; B1/P1, B2/P2, B2r/P2r, B4/P4 = recombination sites for BP recombination; L1/R1, L2/R2, L4/R4 = recombination sites for LR recombination; pFor and pRev = PCR detection primers. (B) Ba/F3 cell survival assay for BCR-ABL1, EML4-ALK, and ETV6-NTRK3 seven days following IL3 depletion (mean luminescence, error bars denote standard deviation, N=3). (C) Immunoblots of BCR-ABL1 and EML4-ALK expression in Ba/F3. Arrow denotes the correct size of BCR-ABL1. (D) PCR detection of the indicated fusion transcripts from Ba/F3 RNA/cDNA extracts. B = fusion DNA backbone (positive control); - = cDNA from GFP-expressing cells as negative control. (E) Dose-dependent survival assays of Ba/F3 cells expressing BCR-ABL1 and EML4-ALK treated with imatinib and crizotinib, respectively, for 72 hours (mean percentage of cell survival, error bars denote standard deviation, N=4). (F) Endpoint volumes (Day 59 post-injection) of xenograft tumors by HMLER cells expressing ETV6-NTRK3 (N=8) and GFP control (N=15). Horizontal bars denote mean volumes; error bars denote standard deviation. All p-values calculated by t-test; *, p<0.05; **, p<0.01; ****, p<0.0001.
BRAF mutagenesis
Site-directed mutagenesis was performed based on the HiTMMoB platform (22,23). Mutagenesis primers were designed as listed in Supplementary Table 4. Mutant fragments and C-terminal BRAF kinase domain (BRAF-ex9) in pDONR vectors were transferred into pHAGE-EF1α-DEST vectors containing R1/R4 sites through multi-site LR recombination reaction (Life Technologies).
Cell culture and transduction
All cell lines were propagated at 37°C and 5% CO2 in humidified atmosphere. MCF10A cells were obtained from the ATCC and cultured as described previously (24). HMLER cells were provided by S. Mani (MD Anderson Cancer Center, Houston, TX) and maintained in MEGM medium (Lonza) without serum and antibiotics. Parental Ba/F3 cells were cultured in RPMI1640 medium supplemented with final concentration of 5% fetal bovine serum and 2.5 ng/ml recombinant mouse IL3 (R&D Systems). Lentivirus production and cell transduction were described earlier (25,26). Cells lines were fingerprinted prior to use on April 21, 2015 by the MD Anderson Cancer Center Characterized Cell Line Core using STR testing platform. Ba/F3 is a mouse-originated cell line, thus STR testing could not be performed.
Ba/F3 cell viability and inhibitor assays
As described previously (25,27), Ba/F3 cell viability was determined at 7 days after IL3 depletion. Cells were treated with DMSO or respective inhibitors at the indicated concentrations for 72 hours, and cell viability was determined using CellTiter-Glo (Promega). All inhibitor compounds were purchased from Selleck Chemicals.
MCF-10A anchorage independent growth assay
Soft agar assays were performed in 6-well plates in triplicate. First, bottom layers were prepared at 0.8% Noble agar (Affymetrix, Inc.) with complete MCF-10A growth medium. After solidification, 10,000 cells were mixed with 0.45% agar in complete growth medium and laid on top of the bottom layer. Two milliliters medium was added in each well after 3 days and medium was refreshed every 3 days. Colonies were counted 2 weeks after seeding.
Immunoblotting
The following antibodies were used to detect protein expression: c-Abl (Cell signaling), ALK (Cell signaling), Raf-B (Santa Cruz), MET (Cell signaling), TrkB (abcam), Phospho-Stat1 (Y701; Cell signaling), Stat1 (Cell signaling), Phospho-Stat3 (Y705; Cell signaling), Phospho-AKT (S473 & T308; Cell signaling), Phospho-ERK1/2 (T202/Y204; Cell signaling), ERK1/2 (Cell signaling), Vinculin (Cell signaling), and GAPDH (Santa Cruz).
Immunofluorescence
MCF-10A cells and those expressing MET fusions and wild-type MET were seeded at 20,000 cells/well on Millicell EZ Slide (EMD Millipore). Cells are fixed in 2% formalin and permeabilized by 0.5% Triton X-100/PBS. The following antibodies were used to detect protein expression and localization: MET (Cell signaling) and GM130 (Golgi marker; abcam). Slides were mounted by SlowFade® Gold Antifade Mountant with DAPI (Thermo Fisher Scientific) to label DNA.
Quantitative PCR and fusion detection PCR
Total RNA was isolated from transduced cells for cDNA synthesis using SuperScript IV First-Strand Synthesis System (Life Technologies) as described previously (26,28). For qPCR, coding regions within wild type gene transcript were amplified using gene specific primers (Supplementary Table 5) and SYBR Green PCR Master Mix (Life Technologies). Expression levels were normalized to mouse β-actin and comparative cycle threshold method was used to quantify mRNA copy number. For fusion detection PCR, coding regions in fusion transcripts were amplified using primers (Supplementary Table 5) annealing to either fusion fragment by PCR and the products were run on 1.5% agarose gel.
Animal studies
All studies using mice were performed in accordance with our IACUC-approved animal protocol (AN-5428) at Baylor College of Medicine. For xenograft tumor assays, primary, non-transformed cells of the defined lineages (106 cells per injection site) were suspended in a 1:1 Hank’s balanced salt solution (Life Technologies) and Matrigel (BD Biosciences) and injected into female athymic mice (CrTac:NCr-Foxn1nu; 4–6 weeks in age) subcutaneously at bilateral flanks (28). Mice were monitored twice a week and tumors were measured and calculated by length × width2/2. The assays were terminated (assay endpoint) once the first animal presents a tumor at maximal burden as allowed in our animal protocol.
RESULTS
Fusion gene construction
To address technical challenges related to fusion gene cloning, we developed an approach incorporating a Gateway™-based (Life Technologies) phage recombination strategy permitting highly efficient and accurate fusion gene construction (Fig. 1A). This strategy incorporates use of two PCR primer sets, each containing requisite recombination sites (attB1/B2/B2r/B4) adjacent to nucleotides complementary to open reading frame (ORF) sequences comprising the target fusion fragment. These primers are entered into a high-fidelity PCR reaction using total cellular cDNA or ORF gene clones as template, and the resulting two PCR amplicons are subsequently recombined into separate entry clones by recombination. The preparation of separate entry clones permits simultaneous assembly of control fusion clones containing optional fluorescent proteins, regulatory elements, etc. (see GFP fusion studies below). Each set of two entry clones are compatible with secondary multi-fragment recombination into a variety of compatible destination vectors, permitting fusion gene expression in desired cell models. Recombination provides fusion fragment assembly in the appropriate order to model gene breakpoints while maintaining fragment reading frames. The strategy results in a fixed 27-nucleotide linker between the two fusion fragments, which when translated leads to an in-frame, 9-amino acid segment (TQLSCTKWL). Inclusion of the short linker did not impact functional studies using positive control fusion constructs (see positive control studies below). Moreover, the modularity of fusion genes whose activity are commonly based on gain/loss of protein functional domains, as well as the observation that breakpoint locations vary widely among recurrent fusion oncoproteins, further supports the limited impact of the linker on the ability to assess the activity of fusion genes.
Functional validation of known fusion oncogenes
Use of sequence-validated ORF collections made available by the ORFeome collaboration (20) and others (21) as fragment PCR template permits the scaling of fusion gene construction. As proof-of-concept, we leveraged the ORFeome collection as well as commercial ORF sources to build three known fusion oncogenes based on published sequences: BCR-ABL1, EML4-ALK, and ETV6-NTRK3. To validate activity of these fusion genes, we first employed murine pro-B Ba/F3 cells, which die in the absence of exogenous interleukin 3 (IL3) (29), to quantify the ability of driver fusion genes to rescue cell survival and proliferation in the absence of IL3. The ability to assay transfer of Ba/F3 addiction from IL3 to oncogenes has been used by us (23,25,27) and others (30) to investigate activity and therapeutic sensitivity by kinase proteins. Viral delivery of BCR-ABL1 promoted robust 245-fold (p<0.0001) Ba/F3 cell proliferation in the absence of IL3 compared to undetectable growth by parental cells and those expressing green fluorescent protein (GFP; negative control), and this activity was at a level comparable to parental BCR-ABL1 cloned directly from a patient sample (Fig. 1B). Similarly, expression of EML4-ALK and ETV6-NTRK3 led to a 100- (p=0.0217) and 159- (p=0.0022) fold increase in Ba/F3 cell growth, respectively, compared to GFP-expressing cells (Fig. 1B). We next confirmed BCR-ABL1 and EML4-ALK fusion gene expression in Ba/F3 cells by immunoblot analysis (Fig. 1C). Given our difficulty identifying a suitable antibody for detection of ETV6-NTRK3, we confirmed mRNA transcription of the chimeric gene by PCR-amplifying over the 27-nucleotide linker from cDNA preparations (Fig. 1D) using DNA primers annealing to either arm of the fusion transcript (pFor and pRev, Fig. 1A). This simple transcript detection strategy can be used to confirm fusion gene construct expression across transduced cell lines.
Ba/F3 has been widely used to evaluate kinase inhibitors (30). Therefore, we next used BCR-ABL1- and EML4-ALK-expressing Ba/F3 cells to measure response to clinically-approved inhibitors of ABL1 and ALK. Treatment of Ba/F3 cells addicted to BCR-ABL1 and EML4-ALK exhibited robust sensitivity to ABL inhibitor imatinib (IC50 = 0.3012μM) and ALK inhibitor crizotinib (IC50 = 0.0097μM), respectively, compared to parental Ba/F3 cells (Fig. 1E).
ETV6-NTRK3 is a recurrent event in secretory breast carcinomas (31). We chose to evaluate ETV6-NTRK3 activity in primary, non-tumorigenic human mammary epithelial cells (HMECs) modified to express the telomerase catalytic subunit, SV40 large T and small t antigens and H-RasV12 (HMLER) (32). Expression of ETV6-NTRK3 in HMLER cells led to robust tumor formation (N=7/8; p=0.0013) when implanted into athymic mice, whereas control cells stably expressing GFP failed to form significant numbers of tumors (N=1/15; Fig. 1F).
Construction of previously uncharacterized fusion genes
We resourced recent published literature (7,15–17) on TCGA datasets to select 20 functionally-uncharacterized fusion genes based on mRNA expression profiles and frame status (Supplementary Table 1–2). We scaled fusion PCR reactions to a 96-well format to amplify the unique individual 5′ and 3′ DNA fragments corresponding to each of the 20 fusion genes using the required fusion primers (Fig. 1A and Supplementary Table 3). Each PCR reaction was used for BP recombination into corresponding fusion donor clones (5′ and 3′ fragments recombined into pDONR-P1/P2 and pDONR-P2r/P4, respectively; Fig. 1A) by direct transfer in a 96-well format. We selected, on average, 3 bacterial colonies following each donor recombineering reaction for DNA sequencing, which revealed a 94.7% success rate (both fusion arms) for proper PCR synthesis and recombination (Supplementary Table 6). A sequence-verified plasmid isolate representing each 5′ and 3′ fusion arm was combined with destination vector for multi-fragment LR recombination (Fig. 1A). Sequencing an average of 3.9 colonies per LR reaction indicated an 86.7% success rate for proper recombination across all fusion clones (Supplementary Table 6). In total, we achieved 100% of fusion clones attempted in a single fusion construction run with an overall efficiency of 91.5% considering correctly-sequenced clones analyzed throughout each step of the construction strategy
We next sought to examine each of the 20 fusion genes for driver activity; however, systematic testing for driver activity across groups of genes is cofounded by differences in their encoded protein function and differing roles in the recognized hallmarks of cancer (33). Therefore, there are no true “generalizable” driver testing systems applicable to detecting oncogenic or oncogene effector activity across all gene and cancer lineage types. Considering this caveat, we again employed the Ba/F3 cell model acknowledging a potential high false negative rate for detecting activity for bona fide fusion gene drivers of cancer. Nevertheless, viral delivery of the 20 fusion genes revealed five (BAIAP2L1-MET, TFG-MET, AFAP1-NTRK2, SQSTM1-NTRK2 and FAM114A2-BRAF) that promoted robust cell proliferation in the absence of IL3 compared to GFP-expressing control cells (described below). Immunoblotting and fusion transcript PCR using Ba/F3 extracts confirmed expression of all 20 fusion genes (Supplementary Fig. 1–3 and below).
MET fusion genes
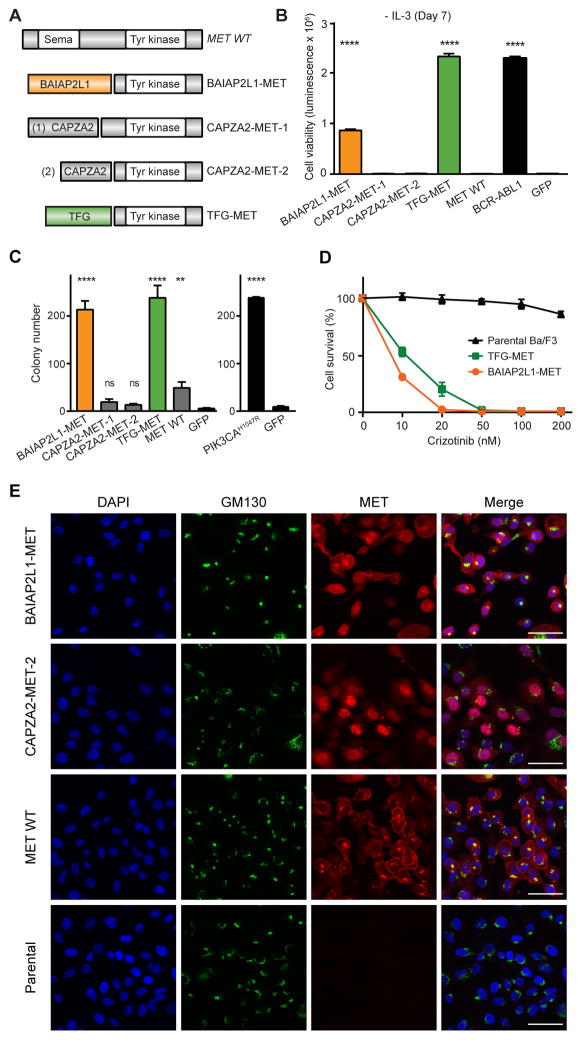
Among the 20 uncharacterized fusion genes selected for construction were four that involved the MET receptor tyrosine kinase (34) (Fig. 2A). BAIAP2L1-MET and TFG-MET, which were identified to be expressed in kidney papillary cell carcinoma (TCGA-BQ-7049) and thyroid carcinoma (TCGA-FK-A3S3), respectively, strongly enhanced Ba/F3 cell survival and proliferation (79- and 213-fold compared to GFP-expressing and wild-type MET expressing cells (p<0.0001; Fig. 2B). In contrast, both variants of CAPZA2-MET failed to stimulate Ba/F3 growth (Fig. 2B) despite encoding the complete tyrosine kinase domain of MET and being expressed in Ba/F3 (Supplementary Fig. 2A). Control assays revealed that expression of the individual gene fragments encoding N-terminal gene partners (BAIAP2L1 and TFG) of the active MET fusion genes failed to promote Ba/F3 growth (Supplementary Fig. 2B) despite their verified expression (Supplementary Fig. 2C). We next assayed MET fusion driver activity using normal MCF-10A breast epithelial cells (35), which are widely used for anchorage-independent growth assays to assess oncogene activity. Expression of BAIAP2L1-MET and TFG-MET significantly increased MCF-10A colony formation (39- and 43-fold compared to GFP-expressing cells, respectively; p<0.0001) similar to PIK3CAH1047R oncogenic control (23) (Fig. 2C). Both variants of CAPZA2-MET failed to stimulate transformation and colony growth in MCF-10A (Fig. 2C) similar to our findings with Ba/F3. Expression of wild-type MET moderately increased MCF-10A cell colony formation by 9-fold (p=0.005; Fig. 2C) while it failed to relieve IL3 dependency of Ba/F3 cells (Fig. 2B). Immunoblotting of MCF-10A extracts confirmed expression of each fusion gene and revealed heightened phosphorylation of ERK1/2 (T202/Y204), STAT1 (Y701), and STAT3 (Y705) in cells expressing BAIAP2L1-MET and TFG-MET compared to control and both CAPZA2-MET variants (Supplementary Fig. 2D–E). Finally, we performed Ba/F3 dose-response assays to examine the response of BAIAP2L1-MET and TFG-MET to crizotinib, which is known to inhibit the MET kinase (36,37). Ba/F3 cells expressing BAIAP2L1-MET or TFG-MET exhibited marked sensitivity to crizotinib (IC50, BAIAP2L1-MET = 8.42nM; TFG-MET = 10.53nM) compared to control cells (IC50, = 675nM; Fig. 2D).
Figure 2. Oncogenic validation of MET fusions.
(A) Schematic illustration of MET fusion genes. (B) Ba/F3 cell survival assay for MET fusions (mean luminescence, error bars denote standard deviation, N=3) compared to positive control, BCR-ABL1, GFP = negative control. (C) MCF-10A anchorage-independent colony formation assays for all MET fusions (mean colony count from 10 random areas, error bars denote standard deviation, N=3). PIK3CAH1047R = positive control; GFP = negative control. (D) Dose-dependent survival assays of Ba/F3 cells expressing BAIAP2L1-MET and TFG-MET treated with crizotinib for 72 hours (mean percentage of cell survival, error bars denote standard deviation, N=4). (E) MCF-10A cells expressing BAIAP2L1-MET, CAPZA2-MET-2, wild-type MET, and parental were immunostained for MET (red) and Golgi body marker GM130 (green). DNA was labeled with DAPI. Scale bar: 50μM. All p-values calculated by t-test; ns, not significant; **, p<0.01; ****, p<0.0001.
We next sought to investigate why CAPZA2-MET fusion genes failed to promote Ba/F3 growth and MCF-10A colony formation. The MET protein is normally processed by the Golgi apparatus and related protein processing machinery for deposition at the plasma membrane where it interacts with its ligand, hepatocyte growth factor (HGF) (34). One explanation for CAPZA2-MET inactivity might relate to its inability to reach the cell surface due to altered localization by CAPZA2. To test this possibility, we performed immunofluorescence assays using an antibody specific to the C-terminus of MET. Staining of MCF-10A cells expressing active BAIAP2L1-MET and wild-type MET revealed MET localization at the Golgi apparatus and plasma membrane, whereas signal was undetectable in vector control (parental) cells suggesting low endogenous MET expression (Fig. 2E). In contrast, cells expressing CAPZA2-MET exhibited MET localization at the nucleus (Fig. 2E), an observation consistent with our hypothesis. However, we cannot eliminate a possible oncogenic role of CAPZA2-MET protein in the nucleus that might be discernable with use of other models to assess oncogenic activities.
NTRK2 fusion genes
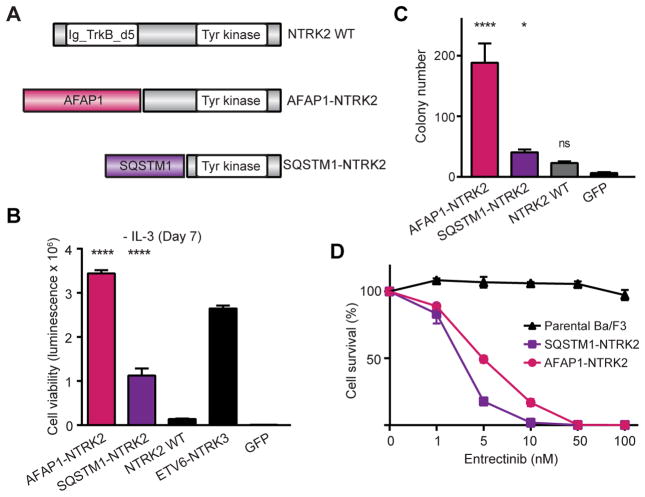
Our series of 20 fusion genes also included AFAP1-NTRK2 and SQSTM1-NTRK2, which were identified in low grade glioma specimens (TCGA-HT-7680 and TCGA-DU-A76L, respectively; Fig. 3A). AFAP1-NTRK2 and SQSTM1-NTRK2 strongly enhanced Ba/F3 cell survival and proliferation (316- and 103-fold, respectively, compared to GFP-expressing cells; p<0.0001; Fig. 3B) in the absence of IL3 at a level comparable to positive control, ETV6-NTRK3. In contrast, wild-type NTRK2 exhibited much weaker activity in the Ba/F3 model (7-fold, p=0.306; Fig. 3B). We also observed no activity for N-terminal partners of the two NTRK2 fusions (AFAP1 and SQSTM1; Supplementary Figure 3A) compared to GFP and BRAFV600E positive control cells despite verified expression by qPCR (Supplementary Figure 3B). We observed a similar trend using the MCF-10A colony formation assay, as expression of AFAP1-NTRK2 exhibited greater transforming activity (34-fold, p<0.0001) than SQSTM1-NTRK2 (7-fold, p=0.0216; Fig. 3C). Wild-type NTRK2 did not significantly promote colony formation (Fig. 3C) despite verified expression by qPCR (Supplementary Fig. 3C). Immunoblot analysis confirmed fusion gene expression and activation of MAPK signaling through elevated phosphorylation of ERK1/2 (T202/Y204) (Supplementary Fig. 3D–E). Interestingly, the same anti-NTRK2 (Trk-B) antibody used to detect AFAP1-NTRK2 and SQSTM1-NTRK2 protein expression did not detect wild-type NTRK2 (Supplementary Fig. 3D–E) despite the fact all were expressed from the same vector type and qPCR analysis of RNA extracts from the same cells indicated similar transcript levels for all three constructs (Supplementary Fig. 3C). Finally, dose-response assays using Ba/F3 revealed both NTRK2 fusions sensitized cells to a pan-NTRK inhibitor, entrectinib (38) (IC50, AFAP1-NTRK2 = 4.57nM; SQSTM1-NTRK2 = 2.26nM; parental > 1μM; Fig. 3D).
Figure 3. Oncogenic validation of NTRK2 fusions.
(A) Schematic illustration of NTRK2 fusion genes. (B) Ba/F3 cell survival assay for NTRK2 fusions (mean luminescence, error bars denote standard deviation, N=3) compared to positive control, ETV6-NTRK3, GFP = negative control. (C) MCF-10A anchorage-independent colony formation assays for NTRK2 fusions (mean colony count from 10 random areas, error bars denote standard deviation, N=3). GFP = negative control. (D) Dose-dependent survival assays of Ba/F3 cells expressing AFAP1-NTRK2 and SQSTM1-NTRK2 treated with entrectinib for 72 hours (mean percentage of cell survival, error bars denote standard deviation, N=4). All p-values calculated by t-test; ns, not significant; *, p<0.05; ****, p<0.0001.
BRAF fusion genes
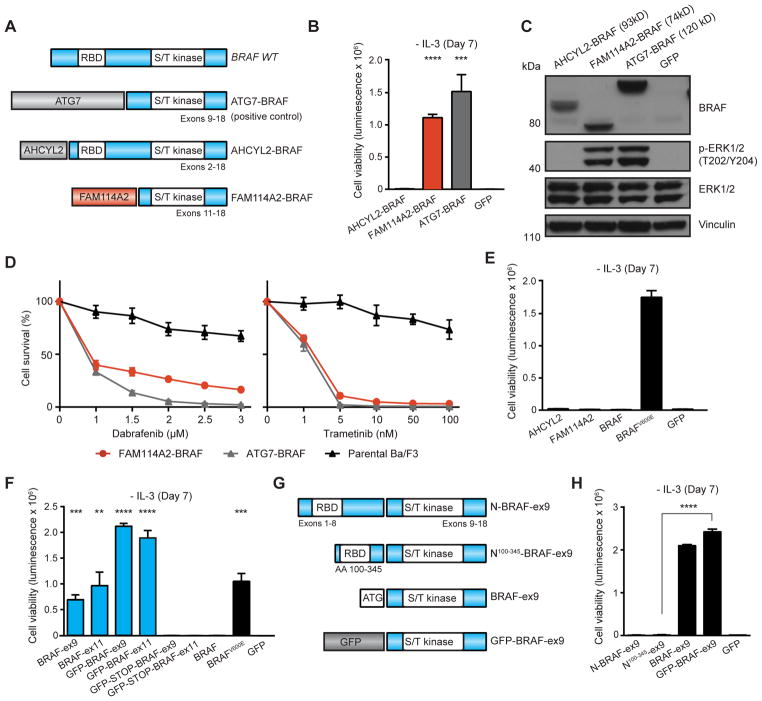
We also constructed two BRAF fusion genes discovered in cutaneous melanoma and thyroid carcinoma specimens, AHCYL2-BRAF (TCGA-D3-A3C3) and FAM114A2-BRAF (TCGA-ET-A3BN), both of which contain an intact serine/threonine kinase domain encoded by BRAF (Fig. 4A). Examination in Ba/F3 revealed potent growth-promoting activity for FAM114A2-BRAF (336-fold increase compared to GFP control cells in the absence of IL3; p<0.0001) similar to ATG7-BRAF previously reported by TCGA and other studies (7,15) (Fig. 4B). Unlike FAM114A2-BRAF, AHCYL2-BRAF provided no growth advantage in Ba/F3 at the same time point following IL3 removal (Fig. 4B). Consistent with this observation, immunoblot analysis of cell lysates from Ba/F3 cells expressing FAM114A2-BRAF and ATG7-BRAF revealed activation of MAPK signaling evidenced by phosphorylation of ERK1/2 (T202/Y204), and this response was absent in cells expressing AHCYL2-BRAF and GFP (Fig. 4C). Previous studies reported sensitivity of melanocytic tumors harboring BRAF fusion genes to targeted therapies (39,40). Indeed, dose-response assays using Ba/F3 revealed marked sensitivity to clinically-approved BRAF and MEK inhibitors dabrafenib and trametinib, respectively, by Ba/F3 cells expressing FAM114A2-BRAF (dabrafenib, IC50 = 0.714μM; trametinib, 1.458nM) and ATG7-BRAF (dabrafenib, IC50 = 0.794μM; trametinib, 1.170nM) as predicted from the fusion’s strong activation of MAPK signaling (Fig. 4D).
Figure 4. Oncogenic validation and domain-function studies of BRAF fusion genes.
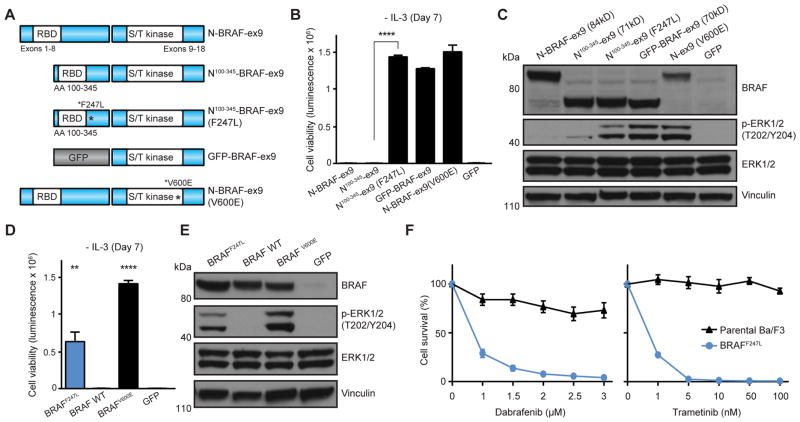
(A) Schematic illustration of BRAF fusion genes. (B) Ba/F3 cell survival assay for BRAF fusions (mean luminescence, error bars denote standard deviation, N=3 respectively). ATG7-BRAF = positive control; GFP = negative control. (C) Immunoblots of BRAF fusions expression and MAPK signaling activation in Ba/F3. (D) Dose-dependent survival assays of Ba/F3 cells expressing FAM114A2-BRAF fusions treated with dabrafenib and trametinib for 72 hours (mean percentage of cell survival, error bars denote standard deviation, N=4 respectively). ATG7-BRAF = positive control. (E) Ba/F3 cell survival assay for the indicated full-length, wild-type genes (mean luminescence, error bars denote standard deviation, N=3 respectively) compared to BRAFV600E (positive control). GFP = negative control. (F) Ba/F3 cell survival assay for BRAF kinase domain (BRAF-ex9: Exons 9–18; BRAF-ex11: Exons 11–18) and corresponding GFP-BRAF-ex9/11 fusions with and without STOP codon following GFP (mean luminescence, error bars denote standard deviation, N=3) compared to full-length, wild-type BRAF. BRAFV600E = positive control; GFP = negative control. (G) Schematic illustration of construct structures and (H) activities in Ba/F3 cell survival assay: BRAF kinase domain (Exons 9–18) fused to i) full-length BRAF N-terminus = N-BRAF-ex9; ii) fragment corresponding to BRAF AA100-345 = N100-345-BRAF-ex9; iii) kinase domain only = BRAF-ex9; iv) GFP = GFP-BRAF-ex9. Shown mean luminescence, error bars denote standard deviation, N=3. All p-values calculated by t-test; **, p<0.01; ***, p<0.001; ****, p<0.0001.
We next investigated why the AHCYL2-BRAF fusion gene failed to promote Ba/F3 growth despite its inclusion of an intact kinase domain. Previous studies suggest that the oncogenic potential of BRAF kinase fusion proteins can be attributed to the kinase domains but not to its N-terminal partner (40–42). Indeed, stable expression of individual gene fragments encoding the N-terminal gene partners of both BRAF fusion genes (AHCYL2 and FAM114A2) failed to promote Ba/F3 growth (Fig. 4E) despite verified expression (Supplementary Fig. 4A). Expression of full-length BRAF, which contains the wild-type kinase domain, similarly failed to influence Ba/F3 growth in contrast to expression of oncogenic BRAFV600E (Fig. 4E). We complemented these studies by expressing BRAF exons included in active BRAF fusion genes ATG7-BRAF and FAM114A2-BRAF: exons 9–18 (BRAF-ex9; as encoded by ATG7-BRAF) and exons 11–18 (BRAF-ex11; as encoded by FAM114A2-BRAF). Expression of these BRAF fragments led to a significant Ba/F3 growth (average 145-fold for BRAF-ex9/11) compared to control cells (BRAF-ex9, p=0.0002; BRAF-ex11, p=0.0032; Fig. 4F).
We next leveraged the versatility afforded by the modular design of our fusion cloning strategy (Fig. 1A) to examine the consequence of fusing active BRAF-ex9/11 to N-terminal sequence other than FAM114A2, AHCYL2, or ATG7. Expression of each kinase fragment as a fusion to monomeric GFP (GFP-BRAF-ex9/ex11) robustly stimulated Ba/F3 growth (average 350-fold, p<0.0001), whereas expressing similar constructs engineered with a stop codon following GFP coding sequence (GFP-STOP-BRAF-ex9/11) did not enhance Ba/F3 growth as expected (Fig. 4F). Interestingly, immunoblot analysis and qPCR of outgrowth Ba/F3 indicated lower expression for GFP-BRAF proteins but higher activation of MAPK signaling compared to truncated BRAF-ex9/11 proteins (Supplementary Fig. 4B–C) consistent with their greater activity in Ba/F3 (Fig. 4F), suggesting that addition of protein sequence N-terminal to BRAF kinase domain may perturb and, in fact, increase kinase activity.
Our data along with the structure of the inactive AHCYL2-BRAF fusion gene, which preserves not only the kinase domain but also a significant portion of the N-terminus of BRAF, also suggests that BRAF fusion genes like ATG7-BRAF and FAM114A2-BRAF are activated through removal of an inhibitory component within the N-terminus of BRAF. This notion is supported by previous findings (11–13) indicating that RAF kinases are regulated by the N-terminal autoinhibitory component and truncation of the N terminus can lead to activation of RAF kinases. To examine this further, we again resourced the modular design of our fusion construction strategy to fuse segments of BRAF N-terminus onto the active C-terminal (exons 9–18) fragment of BRAF (Fig. 4G). Fusion of full-length N-terminus back to the kinase domain (N-BRAF-ex9) completely abolished BRAF-ex9 activity in Ba/F3 (Fig. 4H) compared to the truncated kinase (BRAF-ex9) and GFP fusion (GFP-BRAF-ex9). Fusion of an N-terminal BRAF fragment (AA100-345), which is present in the inactive fusion gene AHCYL2-BRAF, back to the kinase domain (N100-345-BRAF-ex9) similarly attenuated BRAF-ex9 activity in Ba/F3 (Fig. 4H; versus GFP-BRAF-ex9, p<0.0001) and showed no evidence of MAPK activation in early passage Ba/F3 cell lysates (Supplementary Fig. 4D) thus supporting earlier findings (11–13) that a portion of BRAF’s N-terminus serves to inactivate kinase activity.
BRAF mutation studies
The presence of an inhibitory domain located within BRAF’s N-terminus raises the possibility that mutation of this region might serve as another mechanism for BRAF activation during tumorigenesis. To investigate this possibility, we mined cBioPortal for Cancer Genomics (43,44) for tumors and cell lines that contained a missense mutation within the coding sequence corresponding to AA100-345 but devoid of V600 hotspot mutations (Supplementary Table 4). Among all 1438 missense mutations identified across 146 tumors, our analysis revealed 11 cases corresponding to 12 individual mutations, which were engineered into the N-terminal AA100-345 inhibitory fragment used for the studies in Figure 4G–H.
We once again leveraged the modular design of our fusion gene construction strategy to fuse each mutated N-terminal fragment to the C-terminal BRAF kinase domain (BRAF-ex9), followed by testing all mutated fragments for ability to attenuate kinase suppression mediated by A100-345 in Ba/F3. Of the 12 mutations examined, only one (F247L; identified in TCGA case TCGA-AG-A002) promoted Ba/F3 growth in the absence of IL3 (184-fold compared to N100-345-BRAF-ex9, p<0.0001) to a level comparable with GFP (GFP-BRAF-ex9) and V600E fusion genes [N-BRAF-ex9 (V600E); Fig. 5A–B]. Immunoblot analysis indicated that the F247L mutation in the fusion construct [N-BRAF-ex9 (F247L)] restored activation of MAPK signaling through phosphorylation of ERK1/2 (T202/Y204; Fig. 5C). To confirm these findings, we engineered the F247L mutation into full-length BRAF. Expression of BRAFF247L in Ba/F3 led to a significant IL3-dependent growth (116-fold compared to GFP-expressing cells, p=0.0011), albeit to a lesser extent compared to BRAFV600E (Fig. 5D), and immunoblot analysis confirmed MAPK activation by both BRAFF247L and BRAFV600E (Fig. 5E). Importantly, BRAFF247L sensitized Ba/F3 cells to both dabrafenib (IC50 = 0.6646μM) and trametinib (IC50 = 0.5412nM; Fig. 5F) suggesting potential use of these therapeutics in patients whose tumor harbor the BRAFF247L mutation.
Figure 5. BRAF fusion modularity and mutation studies.
(A) Schematic illustration and (B) activities in Ba/F3 of BRAF kinase domain (Exons 9–18) fused to BRAF AA100-345 with or without F247L mutation (mean luminescence, error bars denote standard deviation, N=3 respectively). GFP-BRAF-ex9 and N-BRAF-ex9 (V600E) = positive control; N-BRAF-ex9 and GFP = negative control. (C) Immunoblots of BRAF structural constructs expression and MAPK signaling activation in Ba/F3. (D) Ba/F3 cell survival assay of full-length BRAFF247L; shown mean luminescence, error bars denote standard deviation, N=3; BRAFV600E = positive control; GFP = negative control. (E) Immunoblots of expression of full-length BRAFF247L, wild-type BRAF, and BRAFV600E in Ba/F3. (F) Dose-dependent survival assays of Ba/F3 cells expressing full-length BRAFF247L treated with dabrafenib and trametinib for 72 hours (mean percentage of cell survival, error bars denote standard deviation, N=4 respectively). All p-values calculated by t-test; **, p<0.01; ****, p<0.0001.
DISCUSSION
Over the past few years, cooperative NGS studies have identified more than 9,000 previously uncharacterized gene fusions (http://cgap.nci.nih.gov/Chromosomes/Mitelman); however, the functional consequence of the majority of these events remains to be determined. The functional interrogation of fusion genes is hampered by difficulties cloning such events using traditional molecular biology techniques, which involves obtaining tissues harboring the desired fusion gene or complex multi-step PCR as well as restriction digestion and ligation reactions that are inefficient.
Recent development of CRISPR/Cas9 genome editing technologies have enabled engineering of chromosomal rearrangements within endogenous loci (45–49). While these strategies offer great promise for characterizing fusion genes, inability to cover all types of chromosomal rearrangements (e.g. tandem duplication) and potential off-target complications render it less efficient for functional screening of the thousands of fusion genes identified from NGS studies. Publications (45–49) on modeling chromosomal rearrangements using CRISPR/Cas9 system reported a very low efficiency (1–8%) for engineering events including EML4-ALK (8% and 4%), KIF5B-RET (1.6%), CD74-ROS1 (1.04%) and BCR-ABL (0.78%) into standard cell lines such as HEK293T and murine embryonic fibroblasts. The potential insertions and deletions resulted from non-homologous end joining (NHEJ) could easily cause mutations and/or frameshift in cases involving exon-exon fusions. Moreover, the incompatibility of mouse chromosomal orientations with human orthologs renders some fusion genes extremely difficult to engineer in animal models. On the other hand, we successfully made all 23 fusion genes in our study and achieved a >91% average success rate across all clones resulting from each recombineering step of our strategy. Resulting fusion genes are present in expression-ready vectors that can be immediately delivered to mouse and human cells for use in diverse downstream applications. DNA synthesis offers another route to fusion gene design, the cost of commercial gene synthesis ranges from $0.20 to 0.50/base depending on gene sequence length plus additional service fees due to structural complexity thus eliminating this approach for most investigators wanting to build multiple constructs. For example, we were quoted $18,400 by a leading gene synthesis service company to provide the 23 fusion genes highlighted in our study, and this cost would not have covered additional work required for sub-cloning the synthesized DNA into expression vectors.
We applied our fusion gene construction strategy to a pilot study for construction of 3 bona fide fusion gene drivers and 20 uncharacterized fusion genes identified by TCGA. Our functional investigation revealed 5 fusion genes containing kinase domains encoded by MET, NTRK2, and BRAF. While the majority of the previously-uncharacterized fusion genes appeared inactive in our test assays, it is likely that some of the remaining 15 fusion genes represent false negatives given that they were simply inactive in the Ba/F3 platform that is most sensitive to kinases (30). Future use of the described construction technology would perhaps be best applied for scaled production of cancer lineage-specific fusion genes, followed by functional screening in cancer context-specific model systems.
Our mechanistic studies on BRAF fusion genes corroborates other studies suggesting that loss of an N-terminal regulatory domain of wild-type BRAF contributes to the oncogenic activity of BRAF fusion events. Our fusion construction strategy can therefore be used to map inhibitory or activating protein sequences whose removal or ectopic insertion can activate oncogenes through gene fusion. Importantly, we also leveraged the versatility and modular design of our fusion gene construction strategy to discover a single missense mutation (F247L) within the N-terminus of BRAF that could potentially abolish its inhibitory function, leading to activation of the C-terminal kinase domain.
Another interesting observation made during this study relates to our finding that, despite being expressed from the same vector as AFAP1-NTRK2 and SQSTM1-NTRK2, we did not detect wild-type NTRK2 protein expression in our cell models even though its RNA transcript was expressed at levels similar to the NTRK2 fusion constructs. Based on this observation, we hypothesize that truncating NTRK2 through the process of fusion genesis results in increasing the overall stability of the fusion gene-encoded protein (and associated NTRK2 kinase domain) compared to wild-type protein, whose normal levels may be tightly regulated by proteasome or related machinery.
Our hypothesized means of NTRK2 activation, if proven, along with our confirmation that BRAF fusion activation occurs through loss of an N-terminal kinase inhibitory domain illustrates the notion that fusion genes can function through diverse mechanisms. Moreover, our observations related to differential subcellular localization and activity of MET fusion gene products further highlight the importance of understanding the biological functions of fusion gene proteins beyond their activity as a driver. While there have been numerous attempts to build computational algorithms to predict the functional impact of mutations on protein function, to our knowledge similar algorithms currently do not exist for predicating fusion proteins that are active in cancer. Such functional prediction algorithms for fusion proteins would greatly facilitate filtering of candidate fusion transcripts from the many identified from NGS datasets. Given the diverse mechanisms by which fusion genes activate, the systematic functional testing of numerous fusion gene events using construction pipelines such as the one presented here would ultimately inform development and enable refinement of functional prediction algorithms.
Supplementary Material
Acknowledgments
Financial support: This work was supported by the Cancer Prevention and Research Institute of Texas (CPRIT) RP120046 (K.L. Scott), R21CA198320 from NIH (K.L. Scott), U01CA168394 from NIH (G.B. Mills and K.L. Scott), P30CA016672 from NIH (UT MD Anderson), and P30CA125123 from NIH (Baylor College of Medicine). H. Lu and N. Villafane were supported by the CPRIT Pre-Doctoral Fellowship (RP140102) and Medical Resident Training Grant (RP14010), respectively.
We thank Zayed Institute for Personalized Cancer Therapy at UT MD Anderson Cancer Center for reagents and technical support.
References
- 1.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nature reviews Cancer. 2007;7:233–45. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2010;362:2260–70. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- 3.Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–7. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 4.Shaw AT, Kim DW, Nakagawa K, Seto T, Crino L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–94. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–97. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nature reviews Genetics. 2010;11:685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 7.Yoshihara K, Wang Q, Torres-Garcia W, Zheng S, Vegesna R, Kim H, et al. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2014 doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwaab J, Knut M, Haferlach C, Metzgeroth G, Horny HP, Chase A, et al. Limited duration of complete remission on ruxolitinib in myeloid neoplasms with PCM1-JAK2 and BCR-JAK2 fusion genes. Ann Hematol. 2015;94:233–8. doi: 10.1007/s00277-014-2221-y. [DOI] [PubMed] [Google Scholar]
- 9.Falchi L, Mehrotra M, Newberry KJ, Lyle LM, Lu G, Patel KP, et al. ETV6-FLT3 fusion gene-positive, eosinophilia-associated myeloproliferative neoplasm successfully treated with sorafenib and allogeneic stem cell transplant. Leukemia. 2014;28:2090–2. doi: 10.1038/leu.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grubbs EG, Ng PK, Bui J, Busaidy NL, Chen K, Lee JE, et al. RET fusion as a novel driver of medullary thyroid carcinoma. J Clin Endocrinol Metab. 2015;100:788–93. doi: 10.1210/jc.2014-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanton VP, Jr, Cooper GM. Activation of human raf transforming genes by deletion of normal amino-terminal coding sequences. Mol Cell Biol. 1987;7:1171–9. doi: 10.1128/mcb.7.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stanton VP, Jr, Nichols DW, Laudano AP, Cooper GM. Definition of the human raf amino-terminal regulatory region by deletion mutagenesis. Mol Cell Biol. 1989;9:639–47. doi: 10.1128/mcb.9.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran NH, Wu X, Frost JA. B-Raf and Raf-1 are regulated by distinct autoregulatory mechanisms. J Biol Chem. 2005;280:16244–53. doi: 10.1074/jbc.M501185200. [DOI] [PubMed] [Google Scholar]
- 14.Shin CH, Grossmann AH, Holmen SL, Robinson JP. The BRAF kinase domain promotes the development of gliomas in vivo. Genes & cancer. 2015;6:9–18. doi: 10.18632/genesandcancer.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Network. Electronic address imo, Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–96. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stransky N, Cerami E, Schalm S, Kim JL, Lengauer C. The landscape of kinase fusions in cancer. Nature communications. 2014;5:4846. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang L, Lee MS, Lu H, Oh DY, Kim YJ, Park D, et al. Analyzing Somatic Genome Rearrangements in Human Cancers by Using Whole-Exome Sequencing. Am J Hum Genet. 2016;98:843–56. doi: 10.1016/j.ajhg.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome research. 2004;14:2128–35. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamesch P, Li N, Milstein S, Fan C, Hao T, Szabo G, et al. hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics. 2007;89:307–15. doi: 10.1016/j.ygeno.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, et al. A public genome-scale lentiviral expression library of human ORFs. Nature methods. 2011;8:659–61. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Temple G, Gerhard DS, Rasooly R, Feingold EA, Good PJ, Robinson C, et al. The completion of the Mammalian Gene Collection (MGC) Genome research. 2009;19:2324–33. doi: 10.1101/gr.095976.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsang YH, Dogruluk T, Tedeschi PM, Wardwell-Ozgo J, Lu H, Espitia M, et al. Functional annotation of rare gene aberration drivers of pancreatic cancer. Nature communications. 2016;7:10500. doi: 10.1038/ncomms10500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dogruluk T, Tsang YH, Espitia M, Chen F, Chen T, Chong Z, et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer research. 2015;75:5341–54. doi: 10.1158/0008-5472.CAN-15-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–68. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 25.Liang H, Cheung LW, Li J, Ju Z, Yu S, Stemke-Hale K, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome research. 2012;22:2120–9. doi: 10.1101/gr.137596.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott KL, Nogueira C, Heffernan TP, van Doorn R, Dhakal S, Hanna JA, et al. Proinvasion metastasis drivers in early-stage melanoma are oncogenes. Cancer cell. 2011;20:92–103. doi: 10.1016/j.ccr.2011.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung LW, Yu S, Zhang D, Li J, Ng PK, Panupinthu N, et al. Naturally occurring neomorphic PIK3R1 mutations activate the MAPK pathway, dictating therapeutic response to MAPK pathway inhibitors. Cancer cell. 2014;26:479–94. doi: 10.1016/j.ccell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wardwell-Ozgo J, Dogruluk T, Gifford A, Zhang Y, Heffernan TP, van Doorn R, et al. HOXA1 drives melanoma tumor growth and metastasis and elicits an invasion gene expression signature that prognosticates clinical outcome. Oncogene. 2014;33:1017–26. doi: 10.1038/onc.2013.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Palacios R, Steinmetz M. Il-3-dependent mouse clones that express B-220 surface antigen, contain Ig genes in germ-line configuration, and generate B lymphocytes in vivo. Cell. 1985;41:727–34. doi: 10.1016/s0092-8674(85)80053-2. [DOI] [PubMed] [Google Scholar]
- 30.Warmuth M, Kim S, Gu XJ, Xia G, Adrian F. Ba/F3 cells and their use in kinase drug discovery. Curr Opin Oncol. 2007;19:55–60. doi: 10.1097/CCO.0b013e328011a25f. [DOI] [PubMed] [Google Scholar]
- 31.Tognon C, Knezevich SR, Huntsman D, Roskelley CD, Melnyk N, Mathers JA, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer cell. 2002;2:367–76. doi: 10.1016/s1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 32.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes & development. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 34.Furlan A, Kherrouche Z, Montagne R, Copin MC, Tulasne D. Thirty years of research on met receptor to move a biomarker from bench to bedside. Cancer research. 2014;74:6737–44. doi: 10.1158/0008-5472.CAN-14-1932. [DOI] [PubMed] [Google Scholar]
- 35.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer research. 1990;50:6075–86. [PubMed] [Google Scholar]
- 36.Christensen JG, Zou HY, Arango ME, Li Q, Lee JH, McDonnell SR, et al. Cytoreductive antitumor activity of PF-2341066, a novel inhibitor of anaplastic lymphoma kinase and c-Met, in experimental models of anaplastic large-cell lymphoma. Mol Cancer Ther. 2007;6:3314–22. doi: 10.1158/1535-7163.MCT-07-0365. [DOI] [PubMed] [Google Scholar]
- 37.Zou HY, Li Q, Lee JH, Arango ME, McDonnell SR, Yamazaki S, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer research. 2007;67:4408–17. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 38.Rolfo C, Ruiz R, Giovannetti E, Gil-Bazo I, Russo A, Passiglia F, et al. Entrectinib: a potent new TRK, ROS1, and ALK inhibitor. Expert Opin Investig Drugs. 2015;24:1493–500. doi: 10.1517/13543784.2015.1096344. [DOI] [PubMed] [Google Scholar]
- 39.Botton T, Yeh I, Nelson T, Vemula SS, Sparatta A, Garrido MC, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res. 2013;26:845–51. doi: 10.1111/pcmr.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hutchinson KE, Lipson D, Stephens PJ, Otto G, Lehmann BD, Lyle PL, et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin Cancer Res. 2013;19:6696–702. doi: 10.1158/1078-0432.CCR-13-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer research. 2008;68:8673–7. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cin H, Meyer C, Herr R, Janzarik WG, Lambert S, Jones DT, et al. Oncogenic FAM131B-BRAF fusion resulting from 7q34 deletion comprises an alternative mechanism of MAPK pathway activation in pilocytic astrocytoma. Acta Neuropathol. 2011;121:763–74. doi: 10.1007/s00401-011-0817-z. [DOI] [PubMed] [Google Scholar]
- 43.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discovery. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nature communications. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nature communications. 2014;5:3964. doi: 10.1038/ncomms4964. [DOI] [PubMed] [Google Scholar]
- 47.Blasco RB, Karaca E, Ambrogio C, Cheong TC, Karayol E, Minero VG, et al. Simple and rapid in vivo generation of chromosomal rearrangements using CRISPR/Cas9 technology. Cell reports. 2014;9:1219–27. doi: 10.1016/j.celrep.2014.10.051. [DOI] [PubMed] [Google Scholar]
- 48.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–7. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lekomtsev S, Aligianni S, Lapao A, Burckstummer T. Efficient generation and reversion of chromosomal translocations using CRISPR/Cas technology. BMC genomics. 2016;17:739. doi: 10.1186/s12864-016-3084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.