Abstract
Numerous genetic and environmental insults impede the ability of cells to properly fold and posttranslationally modify secretory and transmembrane proteins in the endoplasmic reticulum (ER), leading to a buildup of misfolded proteins in this organelle—a condition called ER stress. ER-stressed cells must rapidly restore protein-folding capacity to match protein-folding demand if they are to survive. In the presence of high levels of misfolded proteins in the ER, an intracellular signaling pathway called the unfolded protein response (UPR) induces a set of transcriptional and translational events that restore ER homeostasis. However, if ER stress persists chronically at high levels, a terminal UPR program ensures that cells commit to self-destruction. Chronic ER stress and defects in UPR signaling are emerging as key contributors to a growing list of human diseases, including diabetes, neurodegeneration, and cancer. Hence, there is much interest in targeting components of the UPR as a therapeutic strategy to combat these ER stress–associated pathologies.
Keywords: protein misfolding, unfolded protein response, apoptosis, neurodegeneration, cancer, diabetes
THE ENDOPLASMIC RETICULUM: A PROTEIN-FOLDING FACTORY
One of the largest organelles in eukaryotic cells, the endoplasmic reticulum (ER) is a network of branching tubules and flattened sacs interconnected through an enclosed space called the ER lumen, which is separated from the surrounding cytosol by a single intracellular lipid bilayer, the ER membrane. Thus, the ER membrane is a boundary between the cytosol and the ER lumen and governs the passage of molecules between these two compartments (1).
The ER plays a major role in the synthesis, folding, and structural maturation of more than a third of all proteins made in the cell (2). In particular, nearly all proteins destined for residence in the ER, plasma membrane, Golgi apparatus, and lysosomes are translated on ER membrane–bound ribosomes and injected into the ER lumen. Likewise, most proteins that are ultimately secreted from the cell begin their journey in the ER. Proteins targeted to the ER have an N-terminal signal sequence that directs them to the ER membrane while they are still being synthesized on ribosomes. These proteins are cotranslationally translocated through the translocon complex, whereupon the signal sequence is removed by a protease as translation of the polypeptide is completed. Once in the ER lumen, proteins must be folded into their unique three-dimensional shapes and undergo various posttranslational modifications, including glycosylation and disulfide bond formation. These processes are catalyzed by abundant ER-resident enzymes such as chaperones, glycosylating enzymes, and oxidoreductases (3, 4). Furthermore, the ionic and electronic milieu of the ER is ideally suited for these protein-folding activities. Compared with the cytosol, the ER maintains a much higher calcium concentration and a more oxidizing redox potential (5, 6). Cells expend large amounts of energy to maintain the unique environment and functions of the ER. Chaperones bind to and help client proteins negotiate and avoid energetically unfavorable twists and turns on the way to their final native conformation (7). In many cases, protein folding also involves the covalent addition and trimming of sugars onto proteins through the process of glycosylation. Together, these enzymatic processes ensure that secretory proteins are properly folded, modified, and assembled into multiprotein complexes in the ER before they traffic further downstream in the secretory pathway.
Despite the effort of these protein-folding machines, the success rate for proper folding is quite low (under 20%) for many proteins translocated into the ER. Because proteins of the secretory pathway often mediate crucial signaling roles (e.g., cell surface receptors, transporters, or polypeptide hormones), incompletely folded forms are not tolerated by the cell and instead are disposed of by stringent quality control systems. Through a process called ER-associated degradation (ERAD), unfolded proteins are removed to the cytosol for subsequent ubiquitylation and degradation by the 26S proteasome (8–10). Ultimately, this may lead to a deficiency of important proteins and a loss of the functions they serve. Moreover, folding efficiency for an individual protein can be further compromised by the presence of an inherited mutation in the gene that encodes it. For example, specific mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) cause problems in its folding within the ER, leading to depletion of this essential ion channel that normally transports chloride across epithelial tissues and to the life-threatening disease cystic fibrosis (11).
ENDOPLASMIC RETICULUM STRESS: WHEN THE CAPACITY TO FOLD PROTEINS WITHIN THE ENDOPLASMIC RETICULUM FAILS TO KEEP UP WITH DEMAND
The capacity for folding proteins within the ER varies greatly among cells types. Cells with the potential to secrete high protein loads are able to do so because they contain a large, well-developed ER. For example, each β-cell of the endocrine pancreas is capable of synthesizing and secreting up one million molecules of insulin per minute; in insulin-resistant states, this enormous protein synthetic load becomes even greater (12). Moreover, plasma cells can secrete their own weight in antibodies every day (13). In contrast, cells that do not routinely secrete large protein loads conserve resources by maintaining a relatively small ER with limited protein-folding capacity.
Regardless of the size of their ER, cells appear to work near the limits of their secretory capacity and often encounter conditions during which the workload imposed on the ER protein-folding machinery exceeds its capability. When the ER protein-folding capacity is overwhelmed, cells are said to be experiencing ER stress (14). A wide range of cellular disturbances can disrupt the efficiency of protein folding in the ER and lead to the accumulation of misfolded proteins within this organelle, including nutrient deprivation, hypoxia, point mutations in secreted proteins that stabilize intermediate folding forms or cause aggregation, and loss of calcium homeostasis with detrimental effects on ER-resident calcium-dependent chaperones (7, 15, 16). In the case of pancreatic β-cells, ER stress can occur as a result of the inability to fold the increased levels of insulin intermediates needed to maintain blood glucose (17). In other cells, such as neurons, the chronic expression of folding-defective secretory proteins can put unsustainable demands on the protein-folding machinery and lead to ER stress (18). Under ER stress, secretory proteins start to accumulate in improperly modified and unfolded forms within the organelle. Therefore, cells have evolved a sophisticated surveillance system to sense and respond to ER stress before it becomes a threat to their survival.
THE UNFOLDED PROTEIN RESPONSE: A SIGNALING PATHWAY THAT DETERMINES CELL FATE UNDER ENDOPLASMIC RETICULUM STRESS
To ensure that protein-folding capacity is in balance with demand, cells constantly monitor the amount of misfolded protein in the ER lumen and initiate corrective responses. When misfolded proteins in the ER accumulate above a critical threshold, this accumulation signals incipient problems in protein folding and sets in motion a signal transduction pathway called the unfolded protein response (UPR) that attempts to remedy the situation. An ancient signaling pathway conserved all the way from yeasts to mammalian cells, the UPR is initiated by three ER transmembrane proteins: IRE1α (inositol-requiring enzyme 1α), PERK (pancreatic endoplasmic reticulum kinase), and ATF6 (activating transcription factor 6) (19, 20). The common theme among these three ER stress sensors is that they all contain an ER-luminal domain believed to be capable of directly or indirectly sensing misfolded proteins when the latter reach critically high concentrations (Figures 1–3) Luminal domain sensing of misfolded proteins leads to changes in the oligomerization state of each sensor and activation of their associated downstream activities, thus transducing a signal from the ER lumen into the cytoplasm. For IRE1α and PERK, luminal domain dimerization is sufficient to initiate their activation, and this event may normally be prevented in unstressed cells by the binding of an ER chaperone called BiP. This inhibition of dimerization may be relieved when BiP is titrated away from the luminal domain by its high affinity for misfolded proteins (21). Furthermore, through direct binding to the luminal domain of IRE1α and PERK, misfolded proteins may act as activating ligands for these ER stress sensors—a mechanism analogous to a wide range of extracellular ligands that activate various receptors present on the plasma membrane (22, 23).
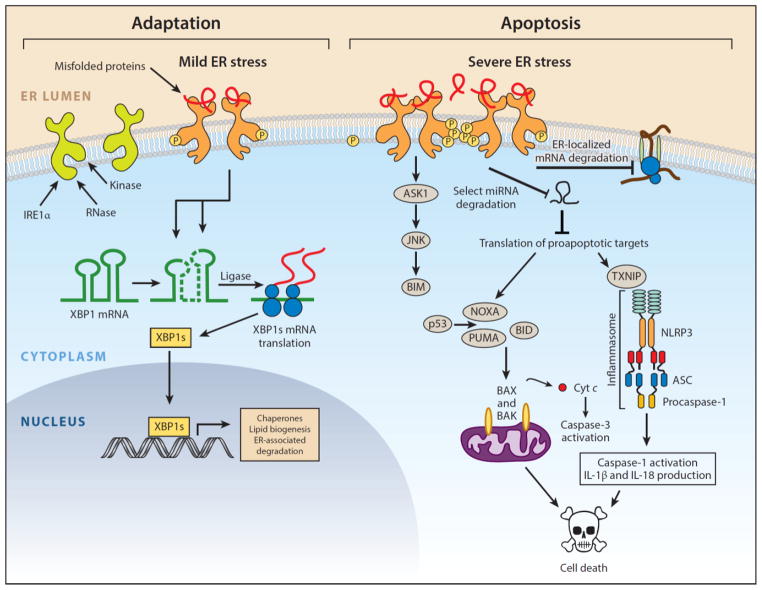
Figure 1.
IRE1α arm of the unfolded protein response. IRE1α is an endoplasmic reticulum (ER) transmembrane protein that becomes activated when misfolded proteins in the ER lumen bind to its luminal domains. The cytoplasmic tail of IRE1α has two enzymatic activities—a serine/threonine kinase domain and an endoribonuclease (RNase) domain. Upon luminal binding of misfolded proteins, IRE1α’s kinase becomes activated and trans-autophosphorylates multiple serine/threonine residues on the cytosolic tail. IRE1α phosphorylation leads to allosteric activation of the adjacent RNase. The consequences of IRE1α activation vary depending on the level of ER stress. In response to low levels of ER stress, IRE1α’s RNase excises a 26-nt intron from the mRNA encoding the XBP1 (X-box protein 1) transcription factor to produce the homeostatic transcription factor XBP1s. XBP1s then translocates to the nucleus and induces transcription of many genes that augment ER size and function in an attempt to restore ER homeostasis. However, if ER stress is irremediable, IRE1α becomes hyperactivated and undergoes homo-oligomerization. Under sustained oligomerization, IRE1α’s RNase endonucleolytically degrades hundreds of ER-localized mRNAs containing an N-terminal signal sequence, which depletes ER cargo and protein-folding components to further worsen ER stress. Moreover, when hyperactivated, IRE1α’s RNase directly cleaves select microRNAs that normally repress proapoptotic targets. In addition to signaling through RNA substrates, IRE1α oligomerization has been shown to induce activation or upregulation of a number of proinflammatory proteins, including the pro-oxidant protein TXNIP (thioredoxin-interacting protein), to activate the inflammasome and its Caspase-1-dependent prodeath pathway. Finally, sustained IRE1α oligomerization serves as an activation platform for ASK1 (apoptosis signal–regulating kinase) and its downstream target JNK (c-Jun NH2-terminal kinase). Phosphorylation by JNK has been reported to both activate proapoptotic BIM and inhibit antiapoptotic BCL-2. Once activated, BH3-only proteins such as BID and BIM disable mitochondrial protecting proteins (e.g., BCL-2, BCL-XL, MCL-1) and in some cases directly trigger the multidomain proapoptotic BAX and BAK proteins to permeabilize the outer mitochondrial membrane and release toxic mitochondrial proteins, such as cytochrome c, into the cytoplasm, where they lead to activation of downstream effector caspases (e.g., Caspase-3) and cell demise. Modified with permission from Reference 149. Copyright © 2014 by Elsevier.
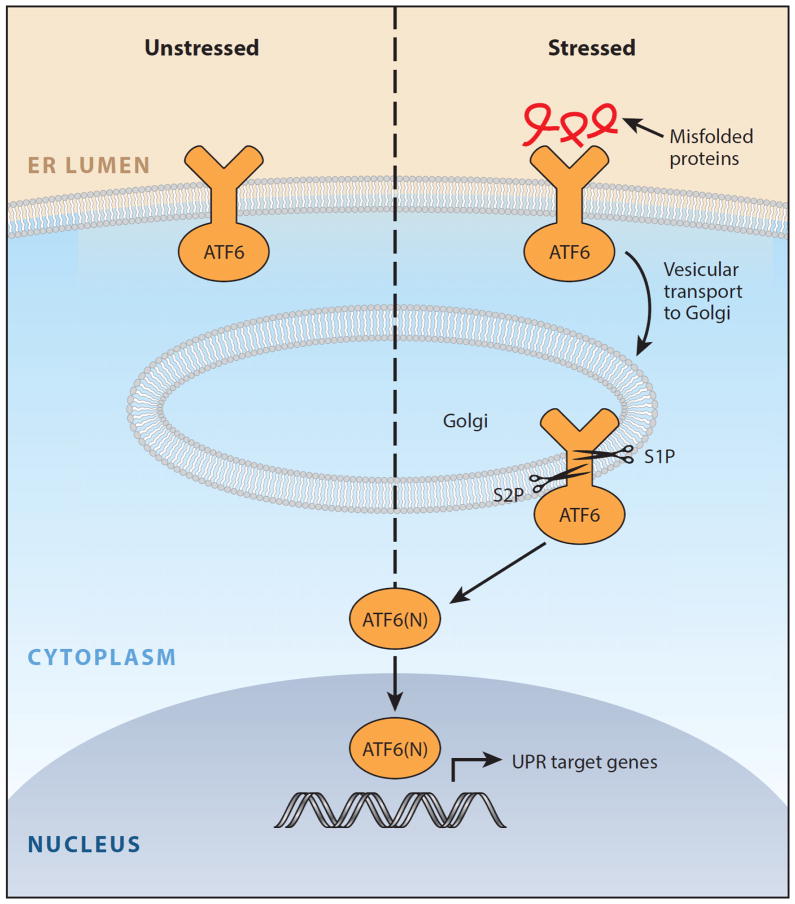
Figure 3.
ATF6 arm of the unfolded protein response. In the presence of misfolded proteins, ATF6 translocates to the Golgi and is cleaved by the Site-1 and Site-2 proteases to release the ATF6(N) transcription factor contained within its cytoplasmic tail. Together with XBP1s, ATF6(N) increases transcription of targets that expand endoplasmic reticulum (ER) size and increase its protein-folding capacity to promote cell survival. Modified with permission from Reference 149. Copyright © 2014 by Elsevier.
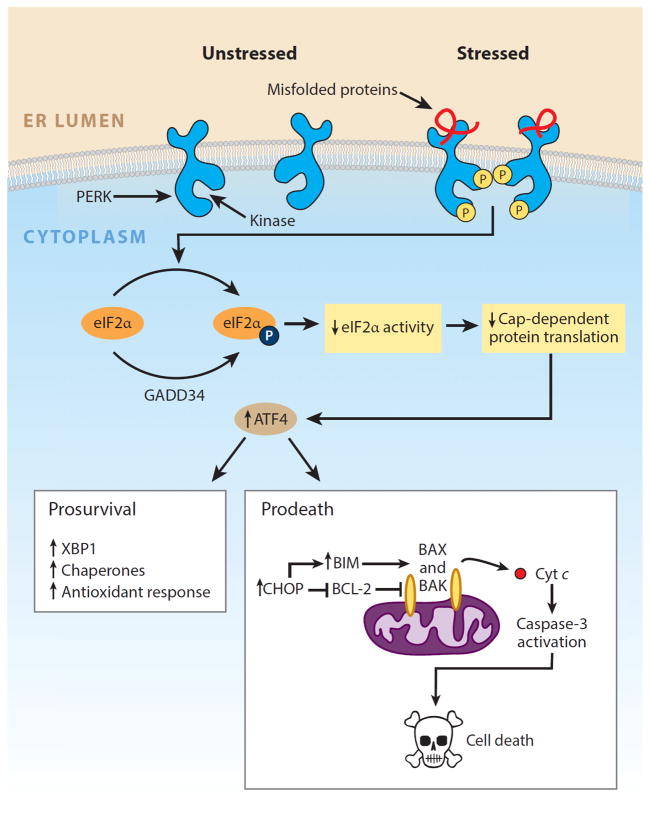
All three UPR sensors have outputs that first attempt to realign protein-folding demand and capacity back into homeostasis so that the cell can continue to survive and function (24). In order to increase protein-folding capacity, the homeostatic UPR expands the size of the ER through increased biogenesis of components (e.g., protein and lipid) and upregulates the transcription of ER chaperones. Concurrently, the combined outputs of the homeostatic UPR increase transcription of ER-resident enzymes and structural components that lead to the removal and degradation of misfolded proteins from the ER lumen. However, despite their common mechanism of luminal activation, each UPR sensor signals in a unique way and has a distinct set of targets. The cytoplasmic tail of IRE1α has two enzymatic activities—a serine/threonine kinase domain and an endoribonuclease (RNase) domain—that work together as a team (25, 26). Upon luminal binding of misfolded proteins, IRE1α’s kinase becomes activated and trans-autophosphorylates; this event leads to allosteric activation of its adjacent RNase domain (27). When active, IRE1α’s RNase excises a 26-nt intron from the mRNA encoding the XBP1 (X-box protein 1) transcription factor. Cytosolic splicing of the two resulting mRNA fragments by a yet-to-be-identified ligase produces the homeostatic transcription factor XBP1s that contains a transactivation domain encoded in the altered reading frame (28, 29). XBP1s then translocates to the nucleus and induces transcription of many genes that augment ER size and function (30). In contrast, PERK contains a single cytosolic kinase that phosphorylates eukaryotic translation initiation factor 2α (eIF2α) (31, 32). Phosphorylation inhibits eIF2α activity and hence slows down global protein translation, giving the cell extra time to attempt to fold the backlog of proteins already present in the ER lumen (33). In the presence of misfolded proteins, ATF6 translocates to the Golgi and is cleaved by the Site-1 and Site-2 proteases to release the ATF6(N) transcription factor contained within its cytoplasmic tail (34). Together with XBP1s, ATF6(N) increases transcription of targets that expand ER size and increase its protein-folding capacity (30). Collectively, these transcriptional events act in a coordinated fashion as homeostatic feedback loops to temper ER stress. If successful in reducing the amount of misfolded protein, UPR signaling is attenuated and the cell survives.
However, if its adaptive responses prove inadequate to restore protein-folding homeostasis, UPR signaling will be sustained, indicating high or chronic ER stress. In the setting of irremediable ER stress, the UPR transforms into an alternate signaling platform called the terminal UPR that actively promotes cell death (35). Although the molecular details are still being worked out, evidence suggests that each of the three UPR sensors has a distinct set of proapoptotic outputs that contribute to cell demise if ER stress cannot be resolved. For example, although a temporary pause in protein translation due to eIF2α phosphorylation can be beneficial for cells under ER stress, a protracted block in translation from sustained PERK signaling is incompatible with survival. Moreover, PERK hyperactivation can upregulate the CHOP (C/EBP-homologous protein; also known as GADD153) transcription factor, which inhibits expression of the gene encoding antiapoptotic BCL-2 to hasten cell death (36, 37). Likewise, when hyperactivated by chronic ER stress, phosphorylated IRE1α transitions from homodimers into high-order oligomers, allowing its RNase to acquire affinity for additional RNA substrates aside from XBP1 mRNA. Under sustained oligomerization, IRE1α’s RNase causes endonucleolytic decay of hundreds of ER-localized mRNAs containing an N-terminal signal sequence, which depletes ER cargo and protein-folding components to further worsen ER stress (38, 39). In addition to reducing key protein-folding components in this way, IRE1α oligomerization has been shown to induce activation or upregulation of a number of proinflammatory and prodeath proteins. For example, when hyperactivated, IRE1α’s RNase reduces the levels of select microRNAs (possibly by directly cleaving their precursors at the ER membrane) that normally repress proapoptotic targets such as the pro-oxidant protein TXNIP (thioredoxin-interacting protein), leading to their rapid upregulation (40, 41). Increased TXNIP expression then activates the inflammasome and its Caspase-1-dependent prodeath pathway (41). Finally, sustained IRE1α oligomerization may serve as an activation platform for ASK1 (apoptosis signal–regulating kinase 1) and its downstream target JNK (c-Jun NH2-terminal kinase) (42, 43). Phosphorylation by JNK has been reported to both activate proapoptotic BIM and inhibit antiapoptotic BCL-2. ATF6α likely has proapoptotic targets as well, but these have not been well defined.
Many, but not all, of the prodeath signals sent from the UPR sensors ultimately converge on the mitochondrial apoptotic pathway. This apoptotic pathway is initiated when toxic mitochondrial proteins, such as cytochrome c and Smac/Diablo, are forcibly released into the cytoplasm, where they lead to activation of downstream effector caspases (e.g., Caspase-3) (44). The BCL-2 family, a large class of both pro- and antideath proteins, governs the intrinsic apoptotic pathway by regulating the integrity of the outer mitochondrial membrane (45). This pathway is engaged when cell injury leads to the expression and/or posttranslational activation of one or more BH3-only proteins, a structurally diverse collection of prodeath proteins that contain a short α-helix known as the BH3 (Bcl-2 homology 3) domain necessary for their killing activity (46). Once activated, BH3-only proteins disable mitochondrial protecting proteins (e.g., BCL-2, BCL-XL, MCL-1) and in some cases directly trigger the multidomain proapoptotic BAX and BAK proteins to permeabilize the outer mitochondrial membrane.
The terminal UPR has been reported to activate at least four distinct BH3-only proteins (BID, BIM, NOXA, PUMA) that then signal the mitochondrial apoptotic machinery (47–49). Each of these BH3-only proteins is activated by ER stress in a unique way. For example, BIM is transcriptionally upregulated by PERK and its protein product stabilized through JNK dephosphorylation in response to ER stress (48). However, it remains unknown whether these BH3-only proteins are simultaneously set in motion by all forms of ER stress or whether only a subset is activated under specific forms of pathological insult that injure this organelle. Researchers are working hard to unravel the molecular details of how ER damage is communicated to the apoptotic machinery, as these signals may represent therapeutic targets in some of the ER stress–related diseases discussed below (50).
THE ROLE OF ENDOPLASMIC RETICULUM STRESS IN DISEASE
Over the past decade, cell injury secondary to chronic ER stress has been increasingly implicated as a central contributor to the pathophysiology of a wide range of prevalent human diseases (20). For example, ER stress and sustained UPR signaling have been well documented in affected tissues in diabetes, neurodegeneration, stroke, pulmonary fibrosis, viral infection, inflammatory disorders, cancer, and heart disease. The common theme among these seemingly disparate diseases is the presence of intracellular and/or extracellular conditions that disrupt protein folding and lead to the accumulation of misfolded proteins in the ER. In strong support of the notion that ER stress can contribute to pathology, inherited mutations in the UPR pathway have been associated with rare forms of diabetes and other diseases in humans (see below). For many of the above-mentioned diseases, genetic manipulation of specific UPR components has been shown to influence disease outcome in rodent models. The existing preclinical data linking ER stress to disease and the emergence of potential targets in the UPR will likely lead to human clinical trials of UPR-directed drugs within the next few years. A few of the diseases most strongly associated with ER stress are discussed below.
Diabetes Mellitus
Pancreatic β-cells synthesize, store and secrete large amounts of the polypeptide hormone insulin; in fact, it is estimated that each human β-cell produces on average about one million molecules of insulin every minute (51). In response to increases in ambient blood glucose levels, prepackaged insulin in secretory granules is released by the β-cell and is replenished by synthesis. A signal transduction cascade ensues when insulin binds its receptor on insulin-responsive cells in peripheral tissues. Upon insulin binding to a target cell, glucose consequently enters, causing energy production. Simultaneously, as blood glucose levels normalize, the stimulus for further insulin release from pancreatic β-cells is removed. This glucostatic cycle is dysregulated in the disease diabetes mellitus, ultimately because of an insufficient mass of functioning β-cells needed to produce the requisite amounts of insulin in the fasted and postprandial states in order to maintain normoglycemia (Figure 4).
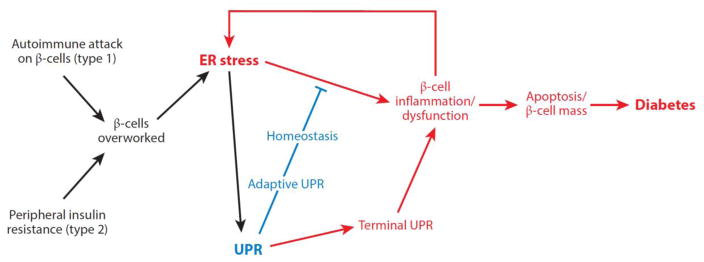
Figure 4.
Model for the role of endoplasmic reticulum (ER) stress in diabetes. ER stress is emerging as an important form of β-cell injury in both type 1 and type 2 diabetes. Overwork of β-cells under conditions of insulin resistance, such as secondary to obesity, may promote attrition of β-cells through unfolded protein response (UPR)-mediated apoptosis in type 2 diabetes. Similarly, as the islet becomes inflamed under autoimmune attack in type 1 diabetes, the per-cell workload of the remaining β-cells to secrete proinsulin increases. Taken together, these combined insults may lead to critically enhanced ER stress in the remaining β-cells, thus hastening their demise through a vicious cycle that ultimately leads to diabetes. Modified with permission from Reference 149. Copyright © 2014 by Elsevier.
To support high-level insulin secretion, β-cells contain highly developed ERs. Insulin biogenesis requires a complex series of molecular biosynthetic events that initiate in the β-cell ER (52). The precursor to insulin, preproinsulin, is cotranslationally translocated into the ER lumen, where its signal sequence is subsequently clipped off, yielding proinsulin. ER-resident oxidoreductases catalyze the formation of three intramolecular disulfides in proinsulin, which allow it to fold to its native shape. This critical oxidative folding step is interrupted in the Akita diabetic mouse mutant, which expresses a proinsulin variant gene, Ins2 (C96Y)—Akita insulin. Ins2 (C96Y) lacks a cysteine needed to form one of the intramolecular disulfide bonds that helps it fold in the ER; its trafficking is therefore impeded, unlike wild-type proinsulin, which is properly trafficked to downstream Golgi and secretory granules, where it is further processed by endoproteases that remove its C-peptide to generate mature insulin (53–55, 56).
Akita vividly links ER stress to the death of β-cells and diabetes. Despite retaining three normal insulin gene copies (mice possess two distinct insulin-encoding genes), Akita mice suffer from insufficient insulin production secondary to β-cell loss. Akita insulin dominantly causes a toxic gain-of-function diabetic syndrome (55). By accumulating in the ER as a conformationally altered immature species, Akita insulin acts as a proteotoxin that exhausts homeostatic UPR outputs and instead triggers a terminal UPR (53). This causes β-cells in the Akita mouse to deterministically enter the apoptotic pathway, leading to frank diabetes within 4 to 5 weeks after birth (15). Rare infantile diabetes–causing Akita-like insulin mutations have been recently described in humans (57). Interestingly, genetic removal of either CHOP, a proapoptotic transcription factor downstream of PERK, or the IRE1α target TXNIP ameliorates β-cell loss and diabetes in the Akita background, emphasizing the central role the terminal UPR plays in β-cell degeneration (15, 41).
A recessive example of diabetogenic UPR dysregulation is evident in Perk-knockout mice. Homozygous deletion in mice of the gene encoding the UPR sensor PERK causes massive and rapid β-cell apoptosis, leading to infantile diabetes (58, 59). Perk-knockout mice further develop pancreatic exocrine insufficiency and exhibit growth defects early in life. These defects are believed to be secondary to the dysfunction and death of several different important professional secretory cell types; intriguingly, diabetes mellitus is one of the earliest and most severe phenotypes in the mutant animals, again highlighting the susceptibility of the β-cell to ER stress. A rare human diabetic syndrome caused by PERK null gene mutations (called Wolcott–Rallison syndrome) phenocopies many of the features of the Perk-knockout animals.
Underlying mechanisms of cell degeneration due to PERK deficiency are understood in considerable detail, although many important questions remain. As mentioned above, PERK is an ER transmembrane kinase that dimerizes under ER stress, which consequently increases phosphorylation activity against its downstream substrate, the eIF2α translation initiation factor. Activated PERK phosphorylates eIF2α on the Ser51 residue, which causes translation to cease globally because it depletes the eIF2·GTP·Met-tRNAi complex needed to initiate cap-dependent mRNA translation. It has been proposed that in the absence of PERK, β-cells cannot properly attenuate translation to match ER protein-folding capacity. As a consequence, they suffer deposition of unfolded proteins in the ER (58). Consistent with this notion, the ERs of β-cells in Perk−/− mice are distended with electron-dense proteinaceous material, and the islets exhibit a high rate of apoptosis. Additionally, there appears to be a marked decline in β-cell proliferation early in neonatal life in the knockout animals. The inability to compensate for increased rates of apoptosis through increasing β-cell proliferation is likely to be an important component in the endocrine pancreatic failure in the Perk−/− animals (60). The homeostatic arm of the IRE1α pathway has also been found to be necessary for β-cell function. For example, mice with a β-cell-specific deletion of the Xbp1 gene show impaired proinsulin processing and decreased insulin secretion (61). Likewise, ATF6 is critical to prevent β-cell loss in a mouse model of diabetes (62).
Translation attenuation through phosphorylation of eIF2α by PERK kinase may be used by the β-cell as a glucose-sensing mechanism to prime the secretory apparatus for upcoming synthesis and structural maturation of proinsulin in response to changes in glucose levels in the blood (63). However, the strategy of limiting translation is fraught with the danger that should upstream stress remain unrelieved, cells may never resume translation at levels needed to recover viability. Thus, there are safety valves that extinguish PERK signaling after a time window has elapsed.
The importance for glycemic control of an escape from this global translational block is seen in two mouse mutant models. The first, a knock-in mouse mutant expressing an unphosphorylatable eIF2α version (S51A), develops a severe wasting syndrome shortly after birth life due to arrested hepatic gluconeogenesis in the homozygote (64, 65), and a milder insulin-resistant hyperglycemic syndrome in the heterozygote (65). The second model involves p58IPK, a cochaperone produced several hours after the UPR has been initiated. p58IPK may help to close a timing loop necessary to turn off the UPR by inhibiting eIF2α kinases, including PERK. Homozygous loss of the gene encoding p58IPK causes diabetes (66–68). By the time p58IPK is produced, if cells have not yet returned to homeostasis, the UPR may switch from promoting homeostasis to an apoptotic mode. In the absence of p58IPK, a continued translational block through PERK may signal a frustrated UPR cycle. Together, the p58IPK, eIF2α (S51A), and PERK genetic models offer fascinating insights into the importance of temporal control of UPR signaling for cell fate.
Another genetic example of ER stress–induced diabetes comes from the Wfs1−/− mouse, and in humans with Wolfram syndrome. WFS1 is an ER transmembrane protein whose loss causes early-onset diabetes, neurodegeneration, and optic and auditory defects (69). The WFS1 protein is widely expressed in diverse tissues and is thought to aid protein assembly and ERAD (70) as well as control processing of the UPR sensor ATF6 (70, 71). As with PERK and p58IPK deficiency, the earliest defects manifest in pancreatic β-cells.
The aforementioned rare disease examples vividly show that accumulation of unfolded protein in the ER and removal of key UPR functions promote apoptosis in β-cells to cause diabetes. Although fascinating from an academic standpoint, the excitement of these experimental rare diseases stems from the question of whether they inform the understanding of common human diabetic syndromes—i.e., type 1, type 2, and gestational diabetes. In all three of these diseases, underlying mechanisms of β-cell functional shutdown and degeneration are evident. As individual units, functioning β-cells in a pancreas may experience increasing and unresolvable ER stress as they compensate for neighboring β-cells that have become dysfunctional through disease and aging. Studies confirm that β-cells of mice may already be functioning (even in healthy states) at levels of UPR activation that are significantly greater than in other professional secretory cells (72). Therefore, without a wide margin for further homeostatic adjustment, β-cells could quickly cross a threshold that puts them at risk for dedifferentiation and apoptosis through a terminal UPR. Thus, as per-cell ER stress levels rise, and terminal UPR outputs stochastically cause the death of individual cells, vicious cycles leading inexorably to whole pancreatic organ failure may set in. In this unifying scheme, the upstream stresses differ for type 1 and type 2 diabetes, but the downstream outcomes will be common: For type 1 diabetes, a disease that results from autoimmune attack by T lymphocytes against β-cells, as β-cell function degenerates, remaining cells in islets would necessarily have to compensate by overworking and may themselves experience critical thresholds of ER stress. Thus, UPR-mediated apoptosis from within the β-cell may synergize with prodeath processes initiated by autoimmune attack from without through cells of the innate and adaptive immune system (73).
For type 2 diabetes, a disease provoked by peripheral insulin resistance, β-cells are forced to compensate by increasing insulin production to abnormally high levels (74). Successful β-cell compensation may prevent progression to frank diabetes, but in some insulin-resistant patients the dysfunction and death of enough β-cells (roughly half of the original β-cell mass) occurring over many years may lead to a tipping point and organ failure beyond which normoglycemia cannot be maintained. The inability to compensate for declining numbers of β-cells through cell proliferation in some populations may contribute to this process. Interestingly, although a prediabetic (compensated) state may exist stably for years, progression to diabetes occurs on a much shorter timescale (perhaps weeks) (75, 76), indicating that an acute-on-chronic organ shutdown may be occurring; the often dramatic initial presentation of type 1 diabetes is also consistent with such dynamics. Intriguingly, both disorders may have a long metastable period of failing compensation, which perhaps could be exploited through intervention in those at risk (as in the Diabetes Prevention Program) (77). Although type 1 diabetes has traditionally been regarded primarily as a discrete and homogeneous disorder (when compared with the greater heterogeneity evident in β-cell function of patients with type 2 diabetes), recent findings of persistent C-peptide production and preserved β-cell function decades after diagnosis in distinct groups of patients may indicate inherent differences in populations with regard to the ability to tolerate stress in the β-cell (78).
Neurodegeneration
A pathologic hallmark of many neurodegenerative diseases is the accumulation of misfolded proteins and protein aggregates within affected neurons and surrounding supporting cells (79). For example, in Alzheimer’s disease (AD), intracellular deposits of tau are observed in neurofibrillary tangles, and extracellular aggregates of amyloid β are seen in senile plaques. The pathology of progressive supranuclear palsy (PSP) shows intracellular tangles of tau protein throughout the neocortex, basal ganglia, and brainstem. In Parkinson’s disease (PD), ubiquitinated protein deposits called Lewy bodies (composed of α-synuclein) are seen in neuronal cytosol. Some cases of inherited amyotrophic lateral sclerosis (ALS) are caused by toxic, gain-of-function point mutations in SOD1 (superoxide dismutase 1) that lead to its aggregation. Other neurodegenerative disorders, such as Huntington’s disease (HD), result from mutant proteins (e.g., huntingtin) containing expanded glutamate repeat sequences. Both mutant SOD1 and mutant huntingtin proteins aggregate, exhaust 26S proteasome activity, and result in secondary accumulations of misfolded proteins in the ER (80, 81). Prions are well known to organize into protein aggregates and are causally involved in the pathogenesis of a group of transmissible spongiform encephalopathies, including Creutzfeldt–Jakob disease and kuru (82). In addition to rare inherited mutations in a single protein that disrupt its proper folding, degenerating neurons are exposed to numerous other insults (e.g., oxidative stress, inflammation, metabolic disturbances) that can compromise protein folding and lead to ER stress.
How protein aggregates contribute to selective neuronal loss in each of these neurodegenerative diseases remains under active investigation. There is evidence that large protein aggregates (e.g., inclusion bodies) in neurodegenerating tissues are protective, whereas smaller misfolded protein species (e.g., fibrils) are toxic (83, 84). Accumulation of toxic protein species can kill neurons (85), and there is growing evidence that ER stress is an important mechanism driving this neurotoxicity (Figure 5) (86–88). IRE1α activation and UPR induction are present in postmortem brain and spinal cord tissues in AD (89–92), PD (93, 94), and ALS (95). Moreover, the accumulation of protein aggregates in cellular and animal models of HD (43), PD (96), and ALS (97–99) strongly correlate with UPR activation. A recent genome-wide association study for PSP found a risk locus that mapped to the gene encoding PERK (100), and an independent study found evidence for PERK hyperactivation in disease-affected brain regions in PSP (101). Brain samples from patients who have succumbed to Creutzfeldt–Jakob disease show activation of a number of ER chaperones and other ER stress markers. Spinal cord segments taken at autopsy of patients with sporadic ALS show that ER stress results in the induction of the UPR, chaperones, and apoptotic markers (95, 102). Using an elegant in vivo reporter system in mutant SOD1 mice, researchers found that the UPR is activated in selectively vulnerable (but not disease-resistant) motor neurons 25–30 days before the earliest signs of neuromuscular denervation (103). Coupled with evidence that mutant SOD1, but not wild-type SOD1, leads to secondary accumulations of misfolded proteins in the ER, these findings strongly suggest that affected neurons attempt to manage the accumulation of misfolded proteins in the ER by activating the UPR (98). Importantly, UPR upregulation is observed prior to the onset of symptoms, suggesting an active role in the disease (97).
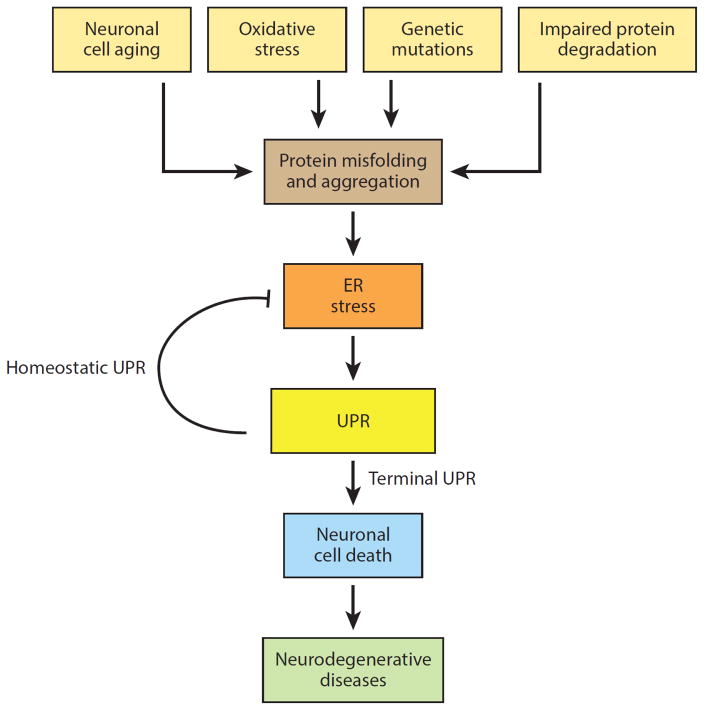
Figure 5.
Model for the role of endoplasmic reticulum (ER) stress in neurodegeneration. In addition to rare inherited mutations in a single protein that disrupt its proper folding, degenerating neurons are exposed to numerous other insults (e.g., oxidative stress, inflammation, metabolic disturbances) that can compromise protein folding and lead to ER stress. Abbreviation: UPR, unfolded protein response. Modified with permission from Reference 149. Copyright © 2014 by Elsevier.
Based on these data, researchers have recently begun to manipulate the UPR in mouse models of neurodegeneration and have uncovered some promising results. For example, conditional deletion of Perk ameliorated synaptic dysfunction in the APP/PSI transgenic model of AD (104). Furthermore, a recent study found that oral administration of a highly selective PERK inhibitor that efficiently crosses the blood-brain barrier significantly reduced neurodegeneration and clinical disease in prion-infected mice (105). In combination with the strong evidence for UPR activation in human patients, studies such as these are bringing great attention to the idea that pharmacologic manipulation of the UPR may have disease-modifying benefits for a variety of neurodegenerative diseases.
Heart Disease, Stroke, and Ischemia-Reperfusion Injury
The link between ER stress and ischemia-reperfusion injury has been implicated on a number of levels (106). Reductions in blood flow as a result of arterial occlusion or hypotension cause tissue hypoxia and hypoglycemia, two conditions that rapidly induce protein misfolding and ER stress. When blood flow is restored, reperfusion of the affected tissues leads to oxidative stress and alterations in the redox status of the ER that disrupt protein disulfide formation and cause ER protein misfolding. UPR activation has been documented in atherosclerotic plaques in both humans and various animal models (107, 108). There is also evidence that high cholesterol, fatty acids, and oxidative stress can trigger ER stress–induced apoptosis of macrophages and endothelial cells associated with atherosclerotic plaques and worsen the progression of atherosclerosis (109). Cardiac myocytes within and adjacent to the field of myocardial infarction activate the UPR. Moreover, genetic deletion of Ask1 in mice partially preserves left ventricular function after coronary artery ligation, implicating the terminal UPR as an important contributor to myocyte loss during myocardial infarction (110). Similarly, brain regions affected by stroke also show evidence of ER stress–induced apoptosis, and mice deficient in Chop show decreased neuronal loss after stroke injury compared with wild-type controls (111).
Cancer
Tumor cells often invade or metastasize into foreign environments where unfavorable conditions, such as hypoxia, glucose deprivation, lactic acidosis, oxidative stress, and inadequate amino acid supplies, compromise protein folding in the ER (112–115). Consequently, many studies have found evidence of sustained and high-level activation of all three branches of the UPR (PERK, ATF6, IRE1α) in a wide range of primary human tumor types, including glioblastoma, multiple myeloma, and carcinomas of the breast, stomach, esophagus, and liver (22, 116–120). Genomic screens have identified rare somatic mutations in IRE1α in a small percentage of human solid tumors (121). Numerous reports have also shown that components of the ER protein-folding machinery, most notably the chaperone Bip/GRP78, are overexpressed in cancers at levels that correlate with disease progression (122, 123).
However, despite the overwhelming evidence of ongoing ER stress and UPR activation in many forms of cancer, whether these processes ultimately inhibit or promote tumor growth in patients remains an area of intense inquiry. Most of the evidence arguing that the UPR supports tumor growth comes from xenograft studies in mice, in which genetically deleting one or more branches of the UPR or altering the expression of the ER chaperone Bip/GRP78 inhibits the in vivo growth of tumor cells (124–127). For example, genetic deletion of IRE1α in a human glioma cell line resulted in reduced angiogenesis and decreased tumor growth when these cells were subsequently injected into mice (128). The IRE1α-XBP1 signaling pathway has been found to induce a number of proangiogenic factors, such as VEGF (vascular endothelial growth factor), which may be one mechanism through which this arm of the UPR can promote the growth of solid tumors (129). These findings suggest that not only is the homeostatic UPR frequently activated in tumors, but it may be necessary for the survival and/or growth of the cancer cells under conditions that stress the ER.
Myeloma, a highly secretory tumor composed of malignant plasma cells, is one cancer for which the UPR is frequently mentioned as a potentially attractive target on the basis strong evidence that this pathway is essential for plasma cell development. In mice, IRE1α and its homeostatic target XBP1 are both required for the differentiation of B lymphocytes into plasma cells (130, 131), illustrating a critical role for the secretory pathway in the health of this cell type. Interestingly, up to 50% of primary myelomas show unusually high levels of XBP1s (116). Moreover, mice expressing a transgene of Xbp1s (that is missing the 26-nt intron and hence requires no further processing by IRE1α) in B lymphocytes develop a plasma cell malignancy closely resembling myeloma (116). There is also evidence to suggest that proteasome inhibition with bortezomib (Velcade), which is approved by the US Food and Drug Administration as first-line therapy for myeloma, leads to myeloma cell death in part by preventing disposal of misfolded proteins through the ERAD pathway and thus triggering ER stress–induced apoptosis (132, 133). On the basis of these findings, several pharmacologic inhibitors of the IRE1α RNase activity have recently been tested on human myeloma xenografts and were found to have antimyeloma activity (134, 135); however, the specificity and off-target effects of these pharmacological agents are not yet well understood.
Although the above findings suggest an oncogenic role for XBP1s in the development of myeloma, recent data have emerged that challenge this notion. First, downregulation of XBP1s expression in myeloma correlates with resistance to bortezomib (136, 137). Second, using a combination of whole-genome and whole-exome sequencing of primary tumors from 38 myeloma patients, researchers discovered XBP1 mutations in two of these patients (138). On further analysis, these mutations were shown to inactivate XBP1s, arguing against an obligate role for this transcription factor in myeloma. Finally, it was recently reported that genetic knockdown of IRE1α or XBP1 in human myeloma cell lines is well tolerated and leads to bortezomib resistance (139), challenging the rationale for using IRE1α inhibitors in this disease. Overall, the lessons from myeloma to date suggest that the effects of the UPR (or at least its IRE1α-XBP1 branch) on tumor development and maintenance are more complicated and nuanced than originally anticipated.
Does the Unfolded Protein Response Present Drug Targets?
Given the evidence of UPR deregulation across a range of human disease, there is great interest in the possibility of pharmacologically modulating its outputs to control cell fate under ER stress. For cell degenerative diseases such as type 2 diabetes and neurodegeneration, the biasing of the UPR’s homeostatic-apoptotic switch to favor cell survival may potentially be disease modifying. However, the parallel and cross-wired networking of the UPR may require multiple nodes to be targeted simultaneously to achieve the desired benefits. One approach may be to prolong the adaptive phases of the UPR to maximize chances of recovery (examples of such targets would include the XBP1 and ATF6 transcription factors); such a regime may involve preconditioning the secretory pathway by preemptive UPR activation to make it more robust. A different approach is to inhibit key mediators of apoptosis (CHOP, TXNIP). Perhaps potential timers of the UPR, such as GADD34 and p58IPK, may also pose attractive targets. In this vein, a small molecule called salubrinal was shown to block phosphatases mediating eIF2α dephosphorylation and hence to enhance cell survival under ER stress (140).
Preemptive preconditioning was demonstrated to be partially cytoprotective in proof-of-concept experiments using synthetic dimerizable modules of PERK in combination with small-molecule dimerizers in cell culture models of ER and oxidative stress (141). It is unclear, however, whether long-term PERK activation, with its attendant consequences of inhibiting translation, would be efficacious in vivo. Related to such an approach, we (142) and others (143) have demonstrated that preempting IRE1α’s homeostatic activation mode can partially prolong cell survival under ER stress. The basis for this particular approach rests on a highly unusual relationship that we discovered between IRE1α’s two catalytic domains (144); the kinase domain of IRE1α can be engaged with a designer kinase inhibitor called 1NM-PP1 to enforce an active ATP-binding conformation while bypassing autophosphorylation (145), which spontaneously triggers RNase activity even without ER stress and forces splicing of XBP1 mRNA, leading to production of the prosurvival Xbp1 transcription factor. Cells subjected to these maneuvers preemptively enjoy a small, but significant, measure of cytoprotection when they are challenged to ER stress (142). It remains to be seen whether these proof-of-concept strategies will be applicable to the wild-type versions of IRE1α and PERK.
Irremediable ER stress hyperactivates both PERK and IRE1α, leading to entry into apoptosis. Thus, a diametrically opposite strategy is to inhibit the hyperactivated state by using inhibitors of PERK (146), RNase inhibitors of IRE1α (134), or allosteric RNase-inhibitory type II kinase inhibitors of IRE1α (145). Early reports suggest that a PERK inhibitor may protect against preclinical models of neurodegeneration (105), but much more work needs to be done to understand the potential benefits and risks of inhibiting ER stress–induced cell degeneration in vivo.
Finally, it has been demonstrated that small chemical chaperones such as tauroursodeoxycholic acid, which act as templates for protein folding and have global effects on stabilizing protein conformations, may also afford significant protection against preclinical models of diabetes (147, 148).
Although they are by no means exhaustive, these examples illustrate emerging concepts about the role of ER stress in disease and the therapeutic potential of targeting components of the UPR to control cell fate under conditions of ER stress.
CONCLUSIONS
The UPR is a highly conserved signal transduction pathway that is activated when cells are unable to keep up with the protein-folding demands on the ER—a form of cell injury called ER stress. In response to ER stress, the UPR initially sends out adaptive outputs that decrease the load and increase the capacity of the ER secretory pathway in an effort to restore ER homeostasis. However, under irremediable ER stress, the UPR assembles into a platform that sends out proinflammatory and prodeath signals to cause cell demise. Cell injury from chronic ER stress is emerging as central to the pathophysiology of a wide range of prevalent human diseases, including diabetes, neurodegeneration, stroke, and cancer. Recent advances in our understanding of how the UPR switches from life to death signaling will hopefully lead to new strategies to combat these ER stress–associated diseases.
Figure 2.
PERK arm of the unfolded protein response. PERK is an endoplasmic reticulum (ER) transmembrane protein that contains a single cytosolic kinase. When its luminal domains are dimerized in the presence of misfolded proteins, PERK phosphorylates eukaryotic translation initiation factor 2α (eIF2α). Phosphorylation inhibits eIF2α activity and hence slows down global protein translation, giving the cell extra time to attempt to fold the backlog of proteins already present in the ER lumen. In contrast, translation of the transcription factor ATF4 (activating transcription factor 4) is selectively upregulated when the amount of active eIF2α is limiting. ATF4 expression transcriptionally upregulates CHOP (C/EBP-homologous protein; also known as GADD153), which tips the ER toward homeostasis through induction of a number of corrective genes, including XBP1 and chaperones. Although a temporary pause in protein translation due to eIF2α phosphorylation can be beneficial for cells under ER stress, a protracted block in translation from sustained PERK signaling is incompatible with survival. Moreover, high levels of CHOP/GADD153 transcription factor can inhibit the expression of antiapoptotic BCL-2 to hasten cell death and upregulate proapoptotic BIM to trigger activation of the mitochondria-dependent apoptotic pathway. Modified with permission from Reference 149. Copyright © 2014 by Elsevier.
TERMS AND DEFINITIONS
- ASK1
apoptosis signal–regulating kinase 1
- ATF6
activating transcription factor 6
- Chaperones
a family of enzymes that bind maturing client proteins, prevent their aggregation, and catalyze their folding to native structures
- eIF2α
eukaryotic translation initiation factor 2
- ER stress
a form of intracellular stress that occurs whenever the protein-folding capacity of the ER is overwhelmed
- ERAD
ER-associated degradation
- Inflammasome
a multiprotein component of the innate immune system that in response to various signals activates procaspase-1 and inflammatory cytokines
- IRE1α
inositol-requiring enzyme 1α
- JNK
c-Jun NH2-terminal kinase
- Mitochondrial apoptotic pathway
genetically controlled cell suicide program initiated when toxic mitochondrial proteins, such as cytochrome c, are released into the cytoplasm
- PERK
pancreatic endoplasmic reticulum kinase
- TXNIP
thioredoxin-interacting protein
- Unfolded protein response (UPR)
intracellular signaling pathway that responds to an accumulation of misfolded proteins in the ER
- XBP1
X-box protein 1
LITERATURE CITED
- 1.Fagone P, Jackowski S. Membrane phospholipid synthesis and endoplasmic reticulum function. J Lipid Res. 2009;50(Suppl):S311–16. doi: 10.1194/jlr.R800049-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anelli T, Sitia R. Protein quality control in the early secretory pathway. EMBO J. 2008;27:315–27. doi: 10.1038/sj.emboj.7601974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tu BP, Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–46. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–47. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 5.van Anken E, Braakman I. Versatility of the endoplasmic reticulum protein folding factory. Crit Rev Biochem Mol Biol. 2005;40:191–228. doi: 10.1080/10409230591008161. [DOI] [PubMed] [Google Scholar]
- 6.Merksamer PI, Trusina A, Papa FR. Real-time redox measurements during endoplasmic reticulum stress reveal interlinked protein folding functions. Cell. 2008;135:933–47. doi: 10.1016/j.cell.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma Y, Hendershot LM. ER chaperone functions during normal and stress conditions. J Chem Neuroanat. 2004;28:51–65. doi: 10.1016/j.jchemneu.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 8.McCracken AA, Brodsky JL. Evolving questions and paradigm shifts in endoplasmic-reticulum-associated degradation (ERAD) Bioessays. 2003;25:868–77. doi: 10.1002/bies.10320. [DOI] [PubMed] [Google Scholar]
- 9.Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–90. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: the long road to destruction. Nat Cell Biol. 2005;7:766–72. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 11.Rowe SM, Miller S, Sorscher EJ. Cystic fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 12.Seino S, Shibasaki T, Minami K. Dynamics of insulin secretion and the clinical implications for obesity and diabetes. J Clin Investig. 2011;121:2118–25. doi: 10.1172/JCI45680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Anken E, Pena F, Hafkemeijer N, Christis C, Romijn EP, et al. Efficient IgM assembly and secretion require the plasma cell induced endoplasmic reticulum protein pERp1. PNAS. 2009;106:17019–24. doi: 10.1073/pnas.0903036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabas I, Ron D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat Cell Biol. 2011;13:184–90. doi: 10.1038/ncb0311-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, et al. Targeted disruption of the Chop gene delays endoplasmic reticulum stress–mediated diabetes. J Clin Investig. 2002;109:525–32. doi: 10.1172/JCI14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang HM, Ye X, Su Y, Yuan J, Liu Z, et al. Coxsackievirus B3 infection activates the unfolded protein response and induces apoptosis through downregulation of p58IPK and activation of CHOP and SREBP1. J Virol. 2010;84:8446–59. doi: 10.1128/JVI.01416-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flamment M, Hajduch E, Ferre P, Foufelle F. New insights into ER stress–induced insulin resistance. Trends Endocrinol Metab. 2012;23:381–90. doi: 10.1016/j.tem.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Gestwicki JE, Garza D. Protein quality control in neurodegenerative disease. Prog Mol Biol Transl Sci. 2012;107:327–53. doi: 10.1016/B978-0-12-385883-2.00003-5. [DOI] [PubMed] [Google Scholar]
- 19.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–29. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol. 2012;197:857–67. doi: 10.1083/jcb.201110131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pincus D, Chevalier MW, Aragon T, van Anken E, Vidal SE, et al. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLOS Biol. 2010;8:e1000415. doi: 10.1371/journal.pbio.1000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner BM, Walter P. Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science. 2011;333:1891–94. doi: 10.1126/science.1209126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Credle JJ, Finer-Moore JS, Papa FR, Stroud RM, Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. PNAS. 2005;102:18773–84. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–86. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 25.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–24. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang XZ, Harding HP, Zhang Y, Jolicoeur EM, Kuroda M, Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–17. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korennykh A, Walter P. Structural basis of the unfolded protein response. Annu Rev Cell Dev Biol. 2012;28:251–77. doi: 10.1146/annurev-cellbio-101011-155826. [DOI] [PubMed] [Google Scholar]
- 28.Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto K, Sato T, Matsui T, Sato M, Okada T, et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6α and XBP1. Dev Cell. 2007;13:365–76. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 31.Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–85. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 32.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–32. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 33.Harding HP, Zhang Y, Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–74. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 34.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–64. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 35.Shore GC, Papa FR, Oakes SA. Signaling cell death from the endoplasmic reticulum stress response. Curr Opin Cell Biol. 2011;23:143–49. doi: 10.1016/j.ceb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCullough KD, Martindale JL, Klotz LO, Aw TY, Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–59. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, et al. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev. 2004;18:3066–77. doi: 10.1101/gad.1250704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han D, Lerner AG, Vande Walle L, Upton JP, Xu W, et al. IRE1α kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–75. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollien J, Weissman JS. Decay of endoplasmic reticulum–localized mRNAs during the unfolded protein response. Science. 2006;313:104–7. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 40.Upton JP, Wang L, Han D, Wang ES, Huskey NE, et al. IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science. 2012;338:818–22. doi: 10.1126/science.1226191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lerner AG, Upton JP, Praveen PV, Ghosh R, Nakagawa Y, et al. IRE1α induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012;16:250–64. doi: 10.1016/j.cmet.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, et al. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–66. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 43.Nishitoh H, Matsuzawa A, Tobiume K, Saegusa K, Takeda K, et al. ASK1 is essential for endoplasmic reticulum stress–induced neuronal cell death triggered by expanded polyglutamine repeats. Genes Dev. 2002;16:1345–55. doi: 10.1101/gad.992302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet. 2009;43:95–118. doi: 10.1146/annurev-genet-102108-134850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giam M, Huang DC, Bouillet P. BH3-only proteins and their roles in programmed cell death. Oncogene. 2008;27(Suppl 1):S128–36. doi: 10.1038/onc.2009.50. [DOI] [PubMed] [Google Scholar]
- 47.Upton JP, Austgen K, Nishino M, Coakley KM, Hagen A, et al. Caspase-2 cleavage of BID is a critical apoptotic signal downstream of endoplasmic reticulum stress. Mol Cell Biol. 2008;28:3943–51. doi: 10.1128/MCB.00013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puthalakath H, O’Reilly LA, Gunn P, Lee L, Kelly PN, et al. ER stress triggers apoptosis by activating BH3-only protein Bim. Cell. 2007;129:1337–49. doi: 10.1016/j.cell.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 49.Li J, Lee B, Lee AS. Endoplasmic reticulum stress–induced apoptosis: multiple pathways and activation of p53-up-regulated modulator of apoptosis (PUMA) and NOXA by p53. J Biol Chem. 2006;281:7260–70. doi: 10.1074/jbc.M509868200. [DOI] [PubMed] [Google Scholar]
- 50.Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–30. doi: 10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 51.Scheuner D, Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with β-cell failure and diabetes. Endocr Rev. 2008;29:317–33. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steiner DF. New aspects of proinsulin physiology and pathophysiology. J Pediatr Endocrinol Metab. 2000;13:229–39. doi: 10.1515/jpem.2000.13.3.229. [DOI] [PubMed] [Google Scholar]
- 53.Ron D. Proteotoxicity in the endoplasmic reticulum: lessons from the Akita diabetic mouse. J Clin Investig. 2002;109:443–45. doi: 10.1172/JCI15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Izumi T, Yokota-Hashimoto H, Zhao S, Wang J, Halban PA, Takeuchi T. Dominant negative pathogenesis by mutant proinsulin in the Akita diabetic mouse. Diabetes. 2003;52:409–16. doi: 10.2337/diabetes.52.2.409. [DOI] [PubMed] [Google Scholar]
- 55.Wang J, Takeuchi T, Tanaka S, Kubo SK, Kayo T, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic β-cell dysfunction in the Mody mouse. J Clin Investig. 1999;103:27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu M, Li Y, Cavener D, Arvan P. Proinsulin disulfide maturation and misfolding in the endoplasmic reticulum. J Biol Chem. 2005;280:13209–12. doi: 10.1074/jbc.C400475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoy J, Edghill EL, Flanagan SE, Ye H, Paz VP, et al. Insulin gene mutations as a cause of permanent neonatal diabetes. PNAS. 2007;104:15040–44. doi: 10.1073/pnas.0707291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 59.Delepine M, Nicolino M, Barrett T, Golamaully M, Lathrop GM, Julier C. EIF2AK3, encoding translation initiation factor 2-α kinase 3, is mutated in patients with Wolcott–Rallison syndrome. Nat Genet. 2000;25:406–9. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 60.Gupta S, McGrath B, Cavener DR. PERK regulates the proliferation and development of insulin-secreting beta-cell tumors in the endocrine pancreas of mice. PLOS ONE. 2009;4:e8008. doi: 10.1371/journal.pone.0008008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee AH, Heidtman K, Hotamisligil GS, Glimcher LH. Dual and opposing roles of the unfolded protein response regulated by IRE1α and XBP1 in proinsulin processing and insulin secretion. PNAS. 2011;108:8885–90. doi: 10.1073/pnas.1105564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Usui M, Yamaguchi S, Tanji Y, Tominaga R, Ishigaki Y, et al. Atf6α-null mice are glucose intolerant due to pancreatic β-cell failure on a high-fat diet but partially resistant to diet-induced insulin resistance. Metabolism. 2012;61:1118–28. doi: 10.1016/j.metabol.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 63.Trusina A, Papa FR, Tang C. Rationalizing translation attenuation in the network architecture of the unfolded protein response. PNAS. 2008;105:20280–85. doi: 10.1073/pnas.0803476105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, et al. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–76. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 65.Scheuner D, Vander Mierde D, Song B, Flamez D, Creemers JW, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11:757–64. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 66.Laybutt DR, Preston AM, Akerfeldt MC, Kench JG, Busch AK, et al. Endoplasmic reticulum stress contributes to beta cell apoptosis in type 2 diabetes. Diabetologia. 2007;50:752–63. doi: 10.1007/s00125-006-0590-z. [DOI] [PubMed] [Google Scholar]
- 67.Oyadomari S, Yun C, Fisher EA, Kreglinger N, Kreibich G, et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–39. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 68.Ladiges WC, Knoblaugh SE, Morton JF, Korth MJ, Sopher BL, et al. Pancreatic β-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54:1074–81. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- 69.Riggs AC, Bernal-Mizrachi E, Ohsugi M, Wasson J, Fatrai S, et al. Mice conditionally lacking the Wolfram gene in pancreatic islet beta cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia. 2005;48:2313–21. doi: 10.1007/s00125-005-1947-4. [DOI] [PubMed] [Google Scholar]
- 70.Fonseca SG, Fukuma M, Lipson KL, Nguyen LX, Allen JR, et al. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic β-cells. J Biol Chem. 2005;280:39609–15. doi: 10.1074/jbc.M507426200. [DOI] [PubMed] [Google Scholar]
- 71.Fonseca SG, Ishigaki S, Oslowski CM, Lu S, Lipson KL, et al. Wolfram syndrome 1 gene negatively regulates ER stress signaling in rodent and human cells. J Clin Investig. 2010;120:744–55. doi: 10.1172/JCI39678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 73.Tersey SA, Nishiki Y, Templin AT, Cabrera SM, Stull ND, et al. Islet β-cell endoplasmic reticulum stress precedes the onset of type 1 diabetes in the nonobese diabetic mouse model. Diabetes. 2012;61:818–27. doi: 10.2337/db11-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Karam JH, Grodsky GM, Forsham PH. Excessive insulin response to glucose in obese subjects as measured by immunochemical assay. Diabetes. 1963;12:197–204. doi: 10.2337/diab.12.3.197. [DOI] [PubMed] [Google Scholar]
- 75.Weir GC, Bonner-Weir S. Five stages of evolving β-cell dysfunction during progression to diabetes. Diabetes. 2004;53(Suppl 3):S16–21. doi: 10.2337/diabetes.53.suppl_3.s16. [DOI] [PubMed] [Google Scholar]
- 76.Stefan Y, Orci L, Malaisse-Lagae F, Perrelet A, Patel Y, Unger RH. Quantitation of endocrine cell content in the pancreas of nondiabetic and diabetic humans. Diabetes. 1982;31:694–700. doi: 10.2337/diab.31.8.694. [DOI] [PubMed] [Google Scholar]
- 77.Knowler WC, Barrett-Connor E, Fowler SE, Hamman RF, Lachin JM, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med. 2002;346:393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang L, Lovejoy NF, Faustman DL. Persistence of prolonged C-peptide production in type 1 diabetes as measured with an ultrasensitive C-peptide assay. Diabetes Care. 2012;35:465–70. doi: 10.2337/dc11-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 80.Wang L, Popko B, Roos RP. The unfolded protein response in familial amyotrophic lateral sclerosis. Hum Mol Genet. 2011;20:1008–15. doi: 10.1093/hmg/ddq546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vidal R, Caballero B, Couve A, Hetz C. Converging pathways in the occurrence of endoplasmic reticulum (ER) stress in Huntington’s disease. Curr Mol Med. 2011;11:1–12. doi: 10.2174/156652411794474419. [DOI] [PubMed] [Google Scholar]
- 82.Xu K, Zhu XP. Endoplasmic reticulum stress and prion diseases. Rev Neurosci. 2012;23:79–84. doi: 10.1515/rns.2011.062. [DOI] [PubMed] [Google Scholar]
- 83.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–10. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 84.Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, et al. Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. PNAS. 1998;95:6448–53. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taylor JP, Hardy J, Fischbeck KH. Toxic proteins in neurodegenerative disease. Science. 2002;296:1991–95. doi: 10.1126/science.1067122. [DOI] [PubMed] [Google Scholar]
- 86.Lindholm D, Wootz H, Korhonen L. ER stress and neurodegenerative diseases. Cell Death Differ. 2006;13:385–92. doi: 10.1038/sj.cdd.4401778. [DOI] [PubMed] [Google Scholar]
- 87.Scheper W, Hoozemans JJ. Endoplasmic reticulum protein quality control in neurodegenerative disease: the good, the bad and the therapy. Curr Med Chem. 2009;16:615–26. doi: 10.2174/092986709787458506. [DOI] [PubMed] [Google Scholar]
- 88.Roussel BD, Kruppa AJ, Miranda E, Crowther DC, Lomas DA, Marciniak SJ. Endoplasmic reticulum dysfunction in neurological disease. Lancet Neurol. 2013;12:105–18. doi: 10.1016/S1474-4422(12)70238-7. [DOI] [PubMed] [Google Scholar]
- 89.Hamos JE, Oblas B, Pulaski-Salo D, Welch WJ, Bole DG, Drachman DA. Expression of heat shock proteins in Alzheimer’s disease. Neurology. 1991;41:345–50. doi: 10.1212/wnl.41.3.345. [DOI] [PubMed] [Google Scholar]
- 90.Hoozemans JJ, Veerhuis R, Van Haastert ES, Rozemuller JM, Baas F, et al. The unfolded protein response is activated in Alzheimer’s disease. Acta Neuropathol. 2005;110:165–72. doi: 10.1007/s00401-005-1038-0. [DOI] [PubMed] [Google Scholar]
- 91.Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Eikelenboom P, Scheper W. The unfolded protein response is activated in pretangle neurons in Alzheimer’s disease hippocampus. Am J Pathol. 2009;174:1241–51. doi: 10.2353/ajpath.2009.080814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Unterberger U, Hoftberger R, Gelpi E, Flicker H, Budka H, Voigtlander T. Endoplasmic reticulum stress features are prominent in Alzheimer disease but not in prion diseases in vivo. J Neuropathol Exp Neurol. 2006;65:348–57. doi: 10.1097/01.jnen.0000218445.30535.6f. [DOI] [PubMed] [Google Scholar]
- 93.Hoozemans JJ, van Haastert ES, Eikelenboom P, de Vos RA, Rozemuller JM, Scheper W. Activation of the unfolded protein response in Parkinson’s disease. Biochem Biophys Res Commun. 2007;354:707–11. doi: 10.1016/j.bbrc.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 94.Wang HQ, Takahashi R. Expanding insights on the involvement of endoplasmic reticulum stress in Parkinson’s disease. Antioxid Redox Signal. 2007;9:553–61. doi: 10.1089/ars.2006.1524. [DOI] [PubMed] [Google Scholar]
- 95.Atkin JD, Farg MA, Walker AK, McLean C, Tomas D, Horne MK. Endoplasmic reticulum stress and induction of the unfolded protein response in human sporadic amyotrophic lateral sclerosis. Neurobiol Dis. 2008;30:400–7. doi: 10.1016/j.nbd.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 96.Holtz WA, O’Malley KL. Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. J Biol Chem. 2003;278:19367–77. doi: 10.1074/jbc.M211821200. [DOI] [PubMed] [Google Scholar]
- 97.Atkin JD, Farg MA, Turner BJ, Tomas D, Lysaght JA, et al. Induction of the unfolded protein response in familial amyotrophic lateral sclerosis and association of protein-disulfide isomerase with superoxide dismutase 1. J Biol Chem. 2006;281:30152–65. doi: 10.1074/jbc.M603393200. [DOI] [PubMed] [Google Scholar]
- 98.Kikuchi H, Almer G, Yamashita S, Guegan C, Nagai M, et al. Spinal cord endoplasmic reticulum stress associated with a microsomal accumulation of mutant superoxide dismutase-1 in an ALS model. PNAS. 2006;103:6025–30. doi: 10.1073/pnas.0509227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nishitoh H, Kadowaki H, Nagai A, Maruyama T, Yokota T, et al. ALS-linked mutant SOD1 induces ER stress– and ASK1-dependent motor neuron death by targeting Derlin-1. Genes Dev. 2008;22:1451–64. doi: 10.1101/gad.1640108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoglinger GU, Melhem NM, Dickson DW, Sleiman PM, Wang LS, et al. Identification of common variants influencing risk of the tauopathy progressive supranuclear palsy. Nat Genet. 2011;43:699–705. doi: 10.1038/ng.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stutzbach LD, Xie SX, Naj AC, Albin R, Gilman S, et al. The unfolded protein response is activated in disease-affected brain regions in progressive supranuclear palsy and Alzheimer’s disease. Acta Neuropathol Commun. 2013;1:31. doi: 10.1186/2051-5960-1-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ilieva EV, Ayala V, Jove M, Dalfo E, Cacabelos D, et al. Oxidative and endoplasmic reticulum stress interplay in sporadic amyotrophic lateral sclerosis. Brain. 2007;130:3111–23. doi: 10.1093/brain/awm190. [DOI] [PubMed] [Google Scholar]
- 103.Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12:627–36. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- 104.Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, et al. Suppression of eIF2α kinases alleviates Alzheimer’s disease–related plasticity and memory deficits. Nat Neurosci. 2013;16:1299–305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moreno JA, Halliday M, Molloy C, Radford H, Verity N, et al. Oral treatment targeting the unfolded protein response prevents neurodegeneration and clinical disease in prion-infected mice. Sci Transl Med. 2013;5:206ra138. doi: 10.1126/scitranslmed.3006767. [DOI] [PubMed] [Google Scholar]
- 106.Minamino T, Komuro I, Kitakaze M. Endoplasmic reticulum stress as a therapeutic target in cardiovascular disease. Circ Res. 2010;107:1071–82. doi: 10.1161/CIRCRESAHA.110.227819. [DOI] [PubMed] [Google Scholar]
- 107.Myoishi M, Hao H, Minamino T, Watanabe K, Nishihira K, et al. Increased endoplasmic reticulum stress in atherosclerotic plaques associated with acute coronary syndrome. Circulation. 2007;116:1226–33. doi: 10.1161/CIRCULATIONAHA.106.682054. [DOI] [PubMed] [Google Scholar]
- 108.Zhou J, Lhotak S, Hilditch BA, Austin RC. Activation of the unfolded protein response occurs at all stages of atherosclerotic lesion development in apolipoprotein E–deficient mice. Circulation. 2005;111:1814–21. doi: 10.1161/01.CIR.0000160864.31351.C1. [DOI] [PubMed] [Google Scholar]
- 109.Seimon TA, Nadolski MJ, Liao X, Magallon J, Nguyen M, et al. Atherogenic lipids and lipoproteins trigger CD36-TLR2-dependent apoptosis in macrophages undergoing endoplasmic reticulum stress. Cell Metab. 2010;12:467–82. doi: 10.1016/j.cmet.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yamaguchi O, Higuchi Y, Hirotani S, Kashiwase K, Nakayama H, et al. Targeted deletion of apoptosis signal–regulating kinase 1 attenuates left ventricular remodeling. PNAS. 2003;100:15883–88. doi: 10.1073/pnas.2136717100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tajiri S, Oyadomari S, Yano S, Morioka M, Gotoh T, et al. Ischemia-induced neuronal cell death is mediated by the endoplasmic reticulum stress pathway involving CHOP. Cell Death Differ. 2004;11:403–15. doi: 10.1038/sj.cdd.4401365. [DOI] [PubMed] [Google Scholar]
- 112.Ma Y, Hendershot LM. The role of the unfolded protein response in tumour development: friend or foe? Nat Rev Cancer. 2004;4:966–77. doi: 10.1038/nrc1505. [DOI] [PubMed] [Google Scholar]
- 113.Lee AS, Hendershot LM. ER stress and cancer. Cancer Biol Ther. 2006;5:721–22. doi: 10.4161/cbt.5.7.3120. [DOI] [PubMed] [Google Scholar]
- 114.Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr Mol Med. 2006;6:55–69. doi: 10.2174/156652406775574604. [DOI] [PubMed] [Google Scholar]
- 115.Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–34. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- 116.Carrasco DR, Sukhdeo K, Protopopova M, Sinha R, Enos M, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–60. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fernandez PM, Tabbara SO, Jacobs LK, Manning FC, Tsangaris TN, et al. Overexpression of the glucose-regulated stress gene GRP78 in malignant but not benign human breast lesions. Breast Cancer Res Treat. 2000;59:15–26. doi: 10.1023/a:1006332011207. [DOI] [PubMed] [Google Scholar]
- 118.Shuda M, Kondoh N, Imazeki N, Tanaka K, Okada T, et al. Activation of the ATF6, XBP1 and grp78 genes in human hepatocellular carcinoma: a possible involvement of the ER stress pathway in hepatocarcinogenesis. J Hepatol. 2003;38:605–14. doi: 10.1016/s0168-8278(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 119.Song MS, Park YK, Lee JH, Park K. Induction of glucose-regulated protein 78 by chronic hypoxia in human gastric tumor cells through a protein kinase C-ε/ERK/AP-1 signaling cascade. Cancer Res. 2001;61:8322–30. [PubMed] [Google Scholar]
- 120.Chen X, Ding Y, Liu CG, Mikhail S, Yang CS. Overexpression of glucose-regulated protein 94 (Grp94) in esophageal adenocarcinomas of a rat surgical model and humans. Carcinogenesis. 2002;23:123–30. doi: 10.1093/carcin/23.1.123. [DOI] [PubMed] [Google Scholar]
- 121.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–58. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pyrko P, Schonthal AH, Hofman FM, Chen TC, Lee AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 2007;67:9809–16. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 123.Luo B, Lee AS. The critical roles of endoplasmic reticulum chaperones and unfolded protein response in tumorigenesis and anticancer therapies. Oncogene. 2013;32:805–18. doi: 10.1038/onc.2012.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jamora C, Dennert G, Lee AS. Inhibition of tumor progression by suppression of stress protein GRP78/BiP induction in fibrosarcoma B/C10ME. PNAS. 1996;93:7690–94. doi: 10.1073/pnas.93.15.7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Park HR, Tomida A, Sato S, Tsukumo Y, Yun J, et al. Effect on tumor cells of blocking survival response to glucose deprivation. J Natl Cancer Inst. 2004;96:1300–10. doi: 10.1093/jnci/djh243. [DOI] [PubMed] [Google Scholar]
- 126.Romero-Ramirez L, Cao H, Nelson D, Hammond E, Lee AH, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64:5943–47. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 127.Austgen K, Oakes SA, Ganem D. Multiple defects, including premature apoptosis, prevent Kaposi’s sarcoma–associated herpesvirus replication in murine cells. J Virol. 2012;86:1877–82. doi: 10.1128/JVI.06600-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Auf G, Jabouille A, Guerit S, Pineau R, Delugin M, et al. Inositol-requiring enzyme 1α is a key regulator of angiogenesis and invasion in malignant glioma. PNAS. 2010;107:15553–58. doi: 10.1073/pnas.0914072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ghosh R, Lipson KL, Sargent KE, Mercurio AM, Hunt JS, et al. Transcriptional regulation of VEGF-A by the unfolded protein response pathway. PLOS ONE. 2010;5:e9575. doi: 10.1371/journal.pone.0009575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, et al. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–7. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 131.Zhang K, Wong HN, Song B, Miller CN, Scheuner D, Kaufman RJ. The unfolded protein response sensor IRE1α is required at 2 distinct steps in B cell lymphopoiesis. J Clin Investig. 2005;115:268–81. doi: 10.1172/JCI21848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Voorhees PM, Orlowski RZ. The proteasome and proteasome inhibitors in cancer therapy. Annu Rev Pharmacol Toxicol. 2006;46:189–213. doi: 10.1146/annurev.pharmtox.46.120604.141300. [DOI] [PubMed] [Google Scholar]
- 133.Rajkumar SV, Richardson PG, Hideshima T, Anderson KC. Proteasome inhibition as a novel therapeutic target in human cancer. J Clin Oncol. 2005;23:630–39. doi: 10.1200/JCO.2005.11.030. [DOI] [PubMed] [Google Scholar]
- 134.Papandreou I, Denko NC, Olson M, Van Melckebeke H, Lust S, et al. Identification of an Ire1α endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117:1311–14. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mimura N, Fulciniti M, Gorgun G, Tai YT, Cirstea D, et al. Blockade of XBP1 splicing by inhibition of IRE1α is a promising therapeutic option in multiple myeloma. Blood. 2012;119:5772–81. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gu JL, Li J, Zhou ZH, Liu JR, Huang BH, et al. Differentiation induction enhances bortezomib efficacy and overcomes drug resistance in multiple myeloma. Biochem Biophys Res Commun. 2012;420:644–50. doi: 10.1016/j.bbrc.2012.03.056. [DOI] [PubMed] [Google Scholar]
- 137.Ling SC, Lau EK, Al-Shabeeb A, Nikolic A, Catalano A, et al. Response of myeloma to the proteasome inhibitor bortezomib is correlated with the unfolded protein response regulator XBP-1. Haematologica. 2012;97:64–72. doi: 10.3324/haematol.2011.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Leung-Hagesteijn C, Erdmann N, Cheung G, Keats JJ, Stewart AK, et al. Xbp1s-negative tumor B cells and pre-plasmablasts mediate therapeutic proteasome inhibitor resistance in multiple myeloma. Cancer Cell. 2013;24:289–304. doi: 10.1016/j.ccr.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Boyce M, Bryant KF, Jousse C, Long K, Harding HP, et al. A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science. 2005;307:935–39. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- 141.Lu PD, Jousse C, Marciniak SJ, Zhang Y, Novoa I, et al. Cytoprotection by preemptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–79. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Han D, Upton JP, Hagen A, Callahan J, Oakes SA, Papa FR. A kinase inhibitor activates the IRE1α RNase to confer cytoprotection against ER stress. Biochem Biophys Res Commun. 2008;365:777–83. doi: 10.1016/j.bbrc.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 143.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, et al. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318:944–49. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Papa FR, Zhang C, Shokat K, Walter P. Bypassing a kinase activity with an ATP-competitive drug. Science. 2003;302:1533–37. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- 145.Wang L, Perera BG, Hari SB, Bhhatarai B, Backes BJ, et al. Divergent allosteric control of the IRE1α endoribonuclease using kinase inhibitors. Nat Chem Biol. 2012;8:982–89. doi: 10.1038/nchembio.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Atkins C, Liu Q, Minthorn E, Zhang SY, Figueroa DJ, et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res. 2013;73:1993–2002. doi: 10.1158/0008-5472.CAN-12-3109. [DOI] [PubMed] [Google Scholar]
- 147.Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, et al. Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science. 2006;313:1137–40. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Engin F, Yermalovich A, Nguyen T, Hummasti S, Fu W, et al. Restoration of the unfolded protein response in pancreatic β cells protects mice against type 1 diabetes. Sci Transl Med. 2013;5:211ra156. doi: 10.1126/scitranslmed.3006534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McManus LM, Mitchell RN, editors. Pathobiology of Human Disease. San Diego, CA: Academic; 2014. In press. [Google Scholar]