Abstract
Proteins fold into unique native structures stabilized by thousands of weak interactions that collectively overcome the entropic cost of folding. Though these forces are “encoded” in the thousands of known protein structures, “decoding” them is challenging due to the complexity of natural proteins that have evolved for function, not stability. Here we combine computational protein design, next-generation gene synthesis, and a high-throughput protease susceptibility assay to measure folding and stability for over 15,000 de novo designed miniproteins, 1,000 natural proteins, 10,000 point-mutants, and 30,000 negative control sequences, identifying over 2,500 new stable designed proteins in four basic folds. This scale—three orders of magnitude greater than that of previous studies of design or folding—enabled us to systematically examine how sequence determines folding and stability in uncharted protein space. Iteration between design and experiment increased the design success rate from 6% to 47%, produced stable proteins unlike those found in nature for topologies where design was initially unsuccessful, and revealed subtle contributions to stability as designs became increasingly optimized. Our approach achieves the long-standing goal of a tight feedback cycle between computation and experiment, and promises to transform computational protein design into a data-driven science.
The key challenge to achieving a quantitative understanding of the sequence determinants of protein folding is to accurately and efficiently model the balance between the many energy terms contributing to the free energy of folding (1–3). Minimal protein domains (30–50aa) such as the villin headpiece and WW-domain are commonly employed to investigate this balance because they are the simplest protein folds found in nature (4). The primary experimental approach used to investigate this balance has been mutagenesis (5–12), but the results are context-dependent and do not provide a global view of the contributions to stability. Molecular dynamics simulations on minimal proteins have also been employed to study folding (13–15), but these do not reveal which interactions specify and stabilize the native structure, and cannot in general determine whether a given sequence will fold into a stable structure.
De novo protein design has the potential to reveal the sequence determinants of folding for minimal proteins by charting the space of non-natural sequences and structures to define what can and cannot fold. Protein sequence space (16) is vastly larger than the set of natural proteins that currently form the basis for nearly all models of protein stability (9, 12, 17–19), and is unbiased by selection for biological function. However, only two minimal proteins (<50 a.a. and stabilized exclusively by noncovalent interactions) have been computationally designed to date (FSD-1 (20) and DS119 (21)). In part, this is due to the cost of gene synthesis, which has limited design studies to testing tens of designs at most -- a miniscule fraction of design space. Because of the small sample sizes, design experiments are typically unable to determine why some designs are stable and others are unstructured, molten globule-like, or form aggregates (22).
Here we present a new synthetic approach to examine the determinants of protein folding by exploring the space of potential minimal proteins using de novo computational protein design on a three order of magnitude larger scale. To enable this new scale, both DNA synthesis and protein stability measurements are parallelized. To encode our designs, we employ oligo library synthesis technology (23, 24), originally developed for transcriptional profiling and large gene assembly applications, and now capable of parallel synthesis of 104–105 arbitrarily specified DNA sequences long enough to encode short proteins (Fig. S1). To assay designs for stability, we express these libraries in yeast so that every cell displays many copies of one protein sequence on its surface, genetically fused to an expression tag that can be fluorescently labeled (25) (Fig. 1A). Cells are then incubated with varying concentrations of protease, those displaying resistant proteins are isolated by FACS (Fig. 1B), and the frequencies of each protein at each protease concentration are determined by deep sequencing (Fig. 1C, for reproducibility of the assay see Fig. S2). We then infer protease EC50 values for each sequence from these data by modeling the complete selection procedure (Fig. 1D, details given in Methods). Finally, each design is assigned a “stability score” (Fig. 1E): the difference between the measured EC50 and the predicted EC50 in the unfolded state, according to a sequence-based model parameterized using EC50 measurements of scrambled sequences (Fig. S3, S4). A stability score of 1 corresponds to a 10-fold higher EC50 than the predicted EC50 in the unfolded state. The complete experimental procedure applied here costs under $7,000 in reagents (mainly from DNA synthesis and sequencing), and required ~10 hours of sorting per protease for each library.
Fig. 1. Yeast display enables massively parallel measurement of protein stability.
(A) Each yeast cell displays many copies of one test protein fused to Aga2. The c-terminal c-Myc tag is labeled with a fluorescent antibody. Protease cleavage of the test protein (or other cleavage) leads to loss of the tag and loss of fluorescence. (B) Libraries of 104 unique sequences are sorted by flow cytometry. Most cells show high protein expression (measured by fluorescence) before proteolysis (blue). Only some cells retain fluorescence after proteolysis; those above a threshold (shaded green region) are collected for deep sequencing analysis. (C) Sequential sorting at increasing protease concentrations separates proteins by stability. Each sequence in a library of 19,726 proteins is shown as a gray line tracking the change in population fraction (enrichment) of that sequence, normalized to each sequence’s population in the starting (pre-selection) library. Enrichment traces for seven proteins at different stability levels are highlighted in color. (D) EC50s for the seven highlighted proteins in (C) are plotted on top of the overall density of the 46,187 highest-confidence EC50 measurements from design rounds 1–4. (E) Same data as at left, showing that stability scores (EC50 values corrected for intrinsic proteolysis rates) correlate better than raw EC50s between the proteases. (F–I) Stability scores measured in high-throughput correlate with individual folding stability measurements for mutants of four small proteins. The wild-type sequence in each set is highlighted as a red circle. Credible intervals for all EC50 measurements are provided in supplementary materials. (F) Pin1 ΔGunf data at 40°C from (31) by thermal denaturation (G) hYAP65 Tm data from (5, 10) (H) Villin HP35 ΔGunf data at 25°C from (7, 11) by urea denaturation (I) BBL ΔGunf data at 10°C from (8) by thermal denaturation.
Massively parallel measurement of folding stability
Proteolysis assays have been previously used to select for stable sequences (26–28) and to quantify stability for individual proteins (29) and proteins from cellular proteomes (30), but have not been applied to date to quantify stability for all sequences in a constructed library. To evaluate the ability of the assay to measure stability on a large scale, we obtained a synthetic DNA library encoding four small proteins (pin1 WW-domain (31), hYAP65 WW-domain (5, 10), villin HP35 (7, 11), and BBL (8)) and 116 mutants of these proteins whose stability has been characterized in experiments on purified material. The library also contained 19,610 unrelated sequences (a fourth-generation designed protein library, detailed below), and all sequences were assayed for stability simultaneously as described. Although the stability score is not a direct analog of a thermodynamic parameter, stability scores measured with trypsin and separately measured with chymotrypsin were each well-correlated with folding free energies (or melting temperatures) for all four sets of mutants, with r2 values ranging from 0.63 to 0.85 (Fig. 1F–I). Most mutants in this dataset were predicted to have similar unfolded state EC50 values to their parent sequences, so the relative stability scores of the mutants are very similar to their relative EC50 values. However, in the case of villin assayed with chymotrypsin, the unfolded state model improved the correlation between protease resistance and folding free energy from r2 = 0.46 (using raw EC50 values) to the reported r2 = 0.77 by correcting for the effect mutations such as K70M and F51L have on intrinsic chymotrypsin cleavage rates. The mutual agreement between trypsin results, chymotrypsin results, and experiments on purified protein indicate that the assay provides a robust measure of folding stability for small proteins.
Massively parallel testing of designed miniproteins
We selected four protein topologies (ααα, βαββ, αββα, and ββαββ) as design targets. These topologies have increasing complexity: the ααα topology features only two loops and exclusively local secondary structure (helices); the ββαββ fold requires four loops and features a mixed parallel/antiparallel β-sheet bridging the N- and C-termini. Of these topologies, only ααα proteins have been found in nature within the target size range of 40–43 residues; no proteins have been previously designed in any of the four topologies at this size (excluding designed ααα and βαββ proteins stabilized by multiple disulfide linkages (32)). For each topology, we first designed between 5,000 and 40,000 de novo proteins using a blueprint-based approach described in (33). Each design has its own unique three-dimensional main chain conformation and its own specific sequence predicted to be near-optimal for that conformation. We then selected 1,000 designs per topology for experimental testing by ranking the designs by a weighted sum of their computed energies and additional filtering terms (see Methods: Protein design). The median sequence identity between any pair of tested designs of the same topology ranged from 15–35%, and designs were typically no more than 40–65% identical to any other design. This diversity is due to the different backbone conformations possible within a topology, along with the vast sequence space available even for small proteins (Fig. S5). For each design, we also included two control sequences in our library: one made by scrambling the order of amino acids in that design (preserving the overall amino acid composition), and a second made by scrambling the order while preserving both the composition and the hydrophobic or polar character at each position (34–36). This library comprised 12,459 different sequences in total: 4,153 designed proteins and 8,306 control sequences. The designed proteins are named using their secondary structure topology (using H for α-helix and E for β-strand), their design round, and a design number.
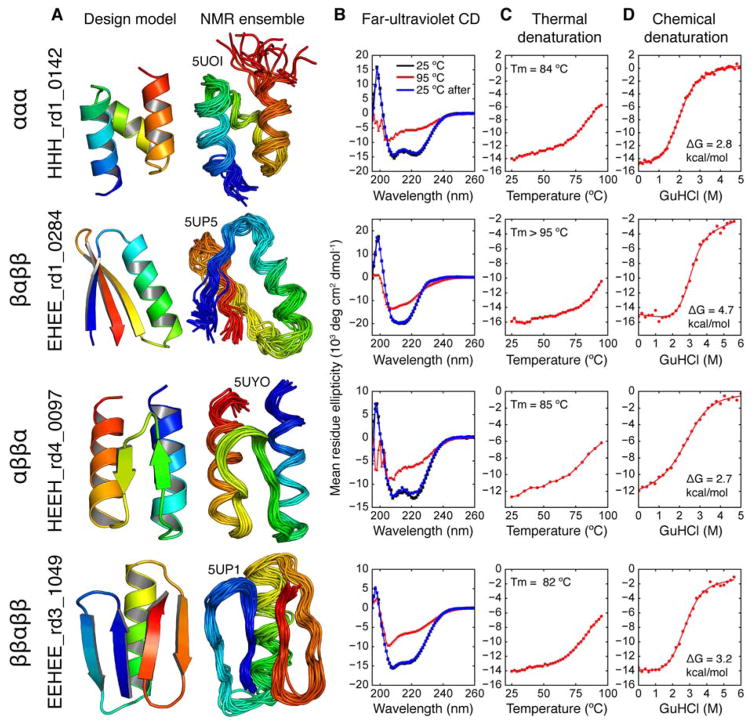
We assayed the sequence library for stability using both chymotrypsin and trypsin. To stringently identify stable designs, we ranked sequences by the lower of their trypsin or chymotrypsin stability score, referred to simply as their (overall) stability score from here on. The fully scrambled sequences and patterned scrambled sequences had similar stability score distributions; most of these controls had stability scores below 0.5, and only one had a score greater than 1.0 (Fig. 2A, Round 1). In contrast, 206 designed sequences had stability scores above 1.0 (Fig. 2A, Round 1). Most of these (195/206) were ααα designs (both left-hand and right-handed bundles); the remaining 11 were βαββ. The clustering of the 206 most stable designs around the ααα topology, and the high stability of designed sequences compared with chemically identical control sequences, strongly suggests these stable designs fold into their designed structures. To examine this further, we selected six stable designs (four ααα and two βαββ) for E. coli expression, purification, and further characterization by size-exclusion chromatography (SEC) and circular dichroism spectroscopy (CD). All six designs eluted from SEC as expected for a 5–7 kDa monomer, and the CD spectra were consistent with the designed secondary structure (Fig. S6A and Table S1). Five of the six designs had clear, cooperative melting transitions, re-folded reversibly and were highly stable for minimal proteins: all had melting temperatures above 70°C, and the βαββ design EHEE_rd1_0284 had only partially melted at 95°C (ΔGunf = 4.7 kcal/mol, Fig. 3D); the sixth design HHH_rd1_0005 did not refold and showed signs of aggregation (Fig. S6A). We determined solution structures for EHEE_rd1_0284 and the left-handed ααα design HHH_rd1_0142 by NMR; each structure closely matched the design model (average backbone root-mean-squared deviation (RMSD) 2.2 + for each NMR ensemble member against the design model, Fig. 3A; NMR data summary given in Table S2). In sum, both high-throughput control experiments and low-throughput characterization of individual proteins indicate that the protease resistant designs fold as designed.
Fig. 2. Iterative, high-throughput computational design generates thousands of stable proteins and reveals stability determinants.
(A) Stability data for designs and control sequences separated by topology (ααα, βαββ, αββα, and ββαββ) and by design round (1–4). For each round and topology, the upper plot shows the total number of designed proteins (y-axis) exceeding a given stability score threshold (x-axis, stability increases left to right). The number of designs tested (top left) may be lower than the number originally ordered (described in the text) due to removal of low-confidence data (see Methods: EC50 estimation). Lower plots show the relative amounts of the three categories of sequences (y-axis) exceeding a given stability score threshold (x-axis), as above. Round 1 categories were designed sequences (colors), fully scrambled sequences (“Scramb.”, light grey), and hydrophobic-polar pattern-preserving scrambled sequences (“Pattern”, dark grey). Round 2–4 categories were designs, patterned scrambles, and point mutants of designs with single Asp mutations expected to be destabilizing (“BuryAsp”, yellow). (B–G) Determinants of stability from Rounds 1–3 (as labeled in A). Colored histograms show the number of tested designs (left y-axis) in each bin for the structural metric on the x-axis. Black lines show the success rate (fraction of designs tested with stability score > 1.0, right y-axis) within a moving window the size of the histogram bin-width, with a shaded 95% confidence interval from bootstrapping. (B,D,E,F) Design success as a function of buried nonpolar surface area (NPSA) from hydrophobic residues. (C) Design success as a function of geometric agreement between 9-residue fragments of similar sequences in the design models and natural proteins (see text and Methods: Fragment analysis), measured in average root-mean-squared deviation (RMSD). (G) Design success as a function of Rosetta total energy. (H) Overall success rate and number of successful designs per round (stability score > 1.0 with both proteases) for all topologies across all rounds. (I) Design success as a function of predicted success according to the topology-specific logistic regression models used to select Round 4 designs for testing (trained on data from Rounds 1–3). As in B–G, colored histograms indicate the number of tested designs at each level of predicted success (left y-axis), and the black line indicates the success rate (right y-axis). Individual success rates for each topology shown in Fig. S8.
Fig. 3. Biophysical characterization of designed minimal proteins.
(A) Design models and NMR solution ensembles for designed minimal proteins. PDB codes are given above each NMR ensemble. (B) Far-ultraviolet circular dichroism (CD) spectra at 25°C (black), 95°C (red), and 25°C following melting (blue). (C) Thermal melting curves measured by CD at 220 nm. Melting temperatures determined using the derivative of the curve. (D) Chemical denaturation in GuHCl measured by CD at 220 nm and 25°C. Unfolding free energies determined by fitting to a two-state model (red solid line). CD data for all 22 purified proteins are given in Table S1 and Fig. S6.
Global determinants of stability
This unprecedentedly large set of stable and unstable minimal proteins with varying physical properties enabled us to quantitatively examine which protein features correlated with folding. We computed over 60 structural and sequence-based metrics and examined which metrics differed between the 195 most stable ααα designs (stability score > 1.0, considered to be design successes) and the 664 remaining ααα designs (considered to be failures) using the K-S 2-sample test. Significant differences indicate that a particular metric captures an important contribution to protein stability, and that this contribution was poorly optimized among the tested designs.
The dominant difference between stable and unstable ααα designs was the total amount of buried nonpolar surface area (NPSA) from hydrophobic amino acids (Fig. 2B). Stable designs buried more NPSA than did unstable designs (p < 5e–38, Fig. S7A), and none of the 95 designs below 32 Å2/residue were stable. Above this threshold, the success rate (successful designs / tested designs) steadily increased as buried NPSA increased (Fig. 2B). Stable designs also had better agreement between their sequences and their local structures as assessed by quantifying the geometric similarity (in Å of RMSD) between 9-residue long fragments of the designs and 9-residue long fragments of natural proteins similar in local sequence to the designed fragment (Fig. 2C and Methods: Fragment analysis). Fragments of stable designs were more geometrically similar to fragments of natural proteins of similar local sequence, while fragments of unstable designs were more geometrically distant from the fragments of natural proteins matching their local sequence (p < 2e–26, Fig. S7B). Other metrics were only weakly correlated with success despite substantial variability among designs, including different measures of amino acid packing density, and the total Rosetta energy itself. Although local sequence-structure agreement and especially buried NPSA are well known to be important for protein stability (1, 9), it is very challenging to determine the precise strength of these contributions at a global level in the complex balance of all the energetic contributions influencing protein structure. Our results directly demonstrate how specific imbalances (under-weighting buried NPSA and local sequence-structure agreement in the Rosetta energy model and the design procedure) led to hundreds of design failures, and our data and approach provide a new route to refining this balance in biophysical modeling.
Iterative, data-driven protein design
We sought to use these findings to increase the success rate of protein design by (1) changing the design procedure to increase buried NPSA, and (2) re-weighting the metrics used to select designs for testing (see Methods: Protein design). Using the improved design and ranking procedure, we built a second generation of 4,150 designs, along with two control sequences per design: a pattern-preserving scrambled sequence as before (now also preserving Gly and Pro positions), and a second control identical to the designed sequence, but with the most buried side chain (according to the design model) replaced with aspartate. As in Round 1, almost no scrambled sequences had stability scores above 1 (our cutoff defining success) despite the increased hydrophobicity of the scrambled sequences (Fig. 2A, Round 2). However, a much greater fraction of second-generation designs proved stable: success for ααα designs improved from 23% to 69%, βαββ designs improved from 1% to 11% successful, and we also obtained 7 stable αββα designs and one stable ββαββ design (Fig. 2H). These increases demonstrate how iterative, high-throughput protein design can make concrete improvements in design and modeling. Nearly all stable designs were destabilized via the single buried Asp substitution: the median drop in stability score for these designs was 1.1, and only 33 buried Asp controls had stability scores greater than 1.0, compared with 271 designs (Fig. 2A, Round 2). This significant destabilization from a single designed substitution provides further large-scale evidence that the stable designs fold into their designed structures. We purified and characterized seven second-generation proteins by SEC and CD, all of which (including three αββα designs and one ββαββ design) were monomeric, displayed their designed secondary structure in CD, and folded cooperatively and reversibly after thermal denaturation (Fig. S6B, Table S1). Although the αββα and ββαββ designs were only marginally stable, the second-generation βαββ design EHEE_rd2_0005 is, to our knowledge, the most thermostable minimal protein ever found (lacking disulfides or metal coordination): its CD spectrum is essentially unchanged at 95°C, and its Cm is above 5 M GuHCl (Fig. S6B).
The amount of buried NPSA was the strongest observed determinant of folding stability for second-generation βαββ designs (Fig. 2E), and continued to show correlation with stability for second-generation ααα designs (Fig. 2D). The success rate for ααα designs improved in Round 2 at all levels of buried NPSA (cf. Fig. 2D versus Fig. 2B), indicating that improving design properties unrelated to buried NPSA (mainly local sequence-structure compatibility) contributed to the increase in success rate along with the increase in NPSA. This also illustrates the coupling between different contributions to stability. Although analyzing single terms makes it possible to identify key problems with the design procedure and imbalances in the energy model, the specific success rates shown in Fig. 2 depend on the overall protein context and are not, on their own, fully general.
To improve the stability of the other two topologies, we built a third generation of designs with even greater buried NPSA, at the cost of increased exposure of hydrophobic surface. This might decrease the solubility of the designs, highlighting one of the limits of our approach aimed at optimizing stability. To increase buried NPSA in the ββαββ topology, we expanded the architecture from 41 to 43 residues. This led to a large increase in the ββαββ success rate (~0% to 13%, Fig. 2H) and 236 newly discovered stable ββαββ designs (Fig. 2A, Round 3). We purified four third-generation designs (Fig. S6C, Table S1) and found the ββαββ design EEHEE_rd3_1049 to be very stable (Fig. 3). We determined the solution structure of this design by NMR, revealing that it folds into its designed structure, which is not found in nature at this size range (average backbone RMSD 1.5 +, Fig. 3). Buried NPSA remained the dominant determinant of stability within the tested ββαββ designs (Fig. 2F). We also observed that a newly improved Rosetta energy function (optimized independently from this work (19)) provided significant discrimination between stable and unstable designs, both for the ββαββ topology (Fig. 2G) and for other topologies.
Having accumulated nearly 1,000 examples of stable designs from rounds 1–3, we asked whether more systematic utilization of this data could be used to select better designs. We designed 2,000–6,000 new proteins per topology (using the improved energy function), and then selected 1,000 designs each for experimental testing by ranking the designs using topology-specific linear regression, logistic regression, and gradient boosting regression models trained on the structural features and experimental stabilities of the 10,000 designs from rounds 1–3. Many designs selected for testing were predicted to have a low likelihood of folding, but were included to increase sequence diversity and because better designs could not be found (see Methods: Protein design). Despite this, an even larger fraction of designs proved stable than before: most notably, the success rate for βαββ designs increased from 17% to 39%, and the success rate for ββαββ designs increased from 13% to 58% (Fig. 2H). Although the success rate for designing the αββα topology remained low (as predicted by the models), five purified fourth-generation designs in this topology possessed the highest stability yet observed for the fold by CD (Fig. S6D, Table S1). We solved the structure of one of these (HEEH_rd4_0097) by NMR and found that it adopts the designed structure in solution (average backbone RMSD 1.5 +, Fig. 3). The overall increase in success across the four rounds (Fig. 2H) -- from 200 stable designs in Round 1 (nearly all in a single topology) to over 1,800 stable designs in Round 4 spread across all four topologies -- demonstrates the power of our massively parallel approach to drive systematic improvement in protein design.
Of the models used to rank designs, logistic regression was the most successful, and was quite accurate: when designs are binned according to their predicted success probability, the number of successes in each bin is close to that predicted beforehand by the logistic regressions (Fig. 2I, Fig. S8A). The accuracy of the regression models demonstrates that large-scale analysis of stable and unstable designed proteins can be used to build predictive models of protein stability. Although the models we built are limited by their training data and not fully general, the inputs to the models were global features of all proteins, such as buried NPSA and total hydrogen bonding energy. This gives these models greater potential for generality than other models used in iterative protein engineering that are typically specific to particular protein families (37, 38), although those approaches have their own advantages. Retrospectively, we found that a single logistic regression trained on data from all topologies from rounds 1–3 performed comparably to the topology-specific regressions at ranking Round 4 designs within each topology (Fig. S8B). Ultimately, continued application of our approach should greatly expand and broaden the available training data, which can be integrated with other sources of physical, chemical, and biological information (19, 39) to build a new generation of general-purpose protein energy functions (22).
Sequence determinants of stability
We next examined determinants of stability at the individual residue level by constructing a library containing every possible point mutant of 14 designs, as well as every point mutant in three paradigm proteins from decades of folding research: villin HP35, pin1 WW-domain, and hYAP65 WW-domain L30K mutant. This library of 12,834 point mutants is comparable in size to the 12,561 single mutants found in the entire ProTherm database (40) and is unbiased toward specific mutations. We assayed this library for stability using trypsin and chymotrypsin, and determined an overall stability effect for each mutation by using the independent results from each protease to maximize the dynamic range of the assay (see Methods: Mutational stability effects and Fig. S9). The mutational effects were qualitatively consistent with the designed structures for 13 of 14 designs (Fig. S10A–N). As expected, the positions on the designs that were most sensitive to mutation were the core hydrophobic residues, including many alanine residues, which indicates the designed cores are tightly packed (Fig. 4A, Fig. S10A–N). Mutations to surface residues had much smaller effects, highlighting the potential of these proteins as stable scaffolds whose surfaces can be engineered for diverse applications.
Fig. 4. Comprehensive mutational analysis of stability in designed and natural proteins.
(A) Average change in stability due to mutating each position in thirteen designed proteins, depicted on the design model structures. Positions where mutations are most destabilizing are colored yellow and shown in stick representation, positions where mutations have little effect are colored blue. Each protein’s color scale is different to emphasize the relative importance of positions; full data for all proteins is shown in Fig. S10. (B) As in (A) for villin HP35. In red, W64, K70, L75, and F76 (HP35 consists of residues 42–76) have little effect on stability but are conserved for function (F-actin binding). (C) As in (A) for pin1 WW-domain, shown bound to a doubly-phosphorylated peptide. In red, S16 is conserved and critical for function but is destabilizing compared with mutations at that position. (D) As in (A) for hYAP65 L30K, shown bound to a Smad7 derived peptide. In red, H32, T37, and W39 form the peptide recognition motif and are conserved but unimportant for stability. (E–L) Average stability effect of each amino acid at different categories of surface positions, in units of stability score (positive meaning stabilizing and negative destabilizing). The average stability of all amino acids in each panel was set to zero. The number of individual positions examined in each category is listed in parentheses with the category name. The average stability effect of the original “wild-type” designed residue (unique to each particular site within a category) is shown by a black star. Error bars indicate the 50% confidence interval for the average stability effect, calculated using bootstrapping. See Methods: Mutational stability effects for a full description of the analysis.
To examine the mutability of protein surfaces in greater detail and to probe more subtle contributions to stability, we divided the 260 surface positions in 12 of the designs into categories based on secondary structure, and calculated the average stability effect of each amino acid for each category using the ~5,000 stability measurements at these positions (Fig. 4E–L and Methods: Mutational stability effects). We observed specific, though weak, preferences for helices (Fig. 4E), helix N-caps (Fig. 4F), the first and last turns of helices (Fig. 4G,H), middle strands and edge strands (Fig. 4I,J), and loop residues (Fig. 4K,L). Amino acids that were favorable for capping helices (Asp, Ser, Thr, and Asn) were unfavorable within helices; these amino acids (except Asn) were as destabilizing as glycine when inside helices (Fig. 4E,F). Hydrophobic side chains were stabilizing even when located on the solvent-facing side of a β-sheet, and this effect was stronger at middle strand positions compared with edge strand positions (Fig. 4I,J). Most notably, we observed stabilization from charged amino acids on the first and last turns of α-helices when these charges counteract the C-to-N negative-to-positive helical dipole; charges that enhanced the dipole were destabilizing (41). We isolated this effect by comparing the average stability of each amino acid on first and last helical turns with the average stability of each amino acid at all helical sites (polar sites only in both cases, Fig. 4G,H); the effect remained significant even when we restricted the analysis to positions that were Arg or Lys in the original designs to control for any bias in the designed structures favoring original, designed residues compared with mutant residues, although no significant effect was seen at Glu positions (Fig. S11). We had not examined agreement with this dipolar preference during the four rounds of design, and after this observation, we found that the net favorable charge on first and last helical turns (stabilizing charges minus destabilizing charges summed over all helices) discriminated between stable and unstable fourth-generation ααα designs better than any other metric we examined, explaining in part why the success rate had not reached 100%.
In the three naturally occurring proteins, mutations at conserved positions were generally destabilizing, although each natural protein possessed several highly conserved positions that we experimentally determined to be unimportant or deleterious to stability. In villin HP35, these were W64, K70, L75, and F76 (villin HP35 consists of residues 42–76), which are required for villin to bind F-actin (Fig. 4B, Fig. S12, (42, 43)). In pin1, the highly conserved S16 is deleterious for stability, but directly contacts the phosphate on phosphopeptide ligands of pin1 (44), highlighting a stability-function trade-off in pin1 (6, 45) discoverable without directly assaying function (Fig. 4C, Fig. S12, (44)). In hYAP65, the conserved residues H32, T37, and W39 are relatively unimportant for stability, but these residues form the peptide recognition pocket in YAP-family WW-domains (Fig. 4D, Fig. S12, (46, 47)). These examples illustrate how our approach enables high-throughput identification of functional residues, even without a functional assay or a protein structure (as in computational approaches (48)), via comparison between stability data and residue conservation.
Stability measurement of all known small protein domains
How stable are these designed proteins compared with naturally occurring proteins? To examine this, we synthesized DNA encoding (1) all 472 sequences in the protein databank (PDB) between 20 and 50 residues in length and containing only the 19 non-Cys amino acids, and (2) one representative for all 706 domains meeting these criteria in the Pfam protein family database. These DNA sequences were prepared by reverse translation in an identical manner to the designs (see Methods: DNA synthesis). We included this DNA (and DNA for all stable designs from rounds 1–3) in the library containing our fourth-generation designs to facilitate a head-to-head comparison. The large majority of these natural proteins successfully displayed on yeast (92% each for PDB and Pfam sequences), which was comparable to the fourth generation buried aspartate mutants (also 92%) but lower than fourth generation scrambled sequences (96%) and fourth generation designs (99%). The most resistant overall sequence (measured by stability score) was a C-terminal coiled-coil domain from a TRP channel (3HRO, stability score 1.93). This protein is likely stabilized by inter-subunit interactions made possible by assembly on the yeast surface (49). Of the 100 unique, monomeric sequences with PDB structures, the most protease-resistant was a peripheral subunit binding domain (ααα topology) from the thermophile Bacillus stearothermophilus (2PDD, stability score 1.48), which has been heavily studied as an ultrafast-folding protein (4, 8). A total of 774 designed proteins had higher stability scores than this most protease-resistant natural monomeric protein. As illustrated in Fig. 5, the number of stable proteins discovered in this paper is 50-fold larger than that of natural proteins in the PDB (monomeric or not) in this size range.
Fig. 5. Comparison of naturally occurring and designed protein stability.
Designed and naturally occurring proteins are separated into bins by stability score (y-axis). The total number of designed proteins in each bin is shown by the colored bar, subdivided by topology from left to right as follows: ααα (green), βαββ (blue), αββα (violet), ββαββ (red). The total number of naturally occurring proteins with PDB structures (lacking disulfides) in each bin is shown by the black bar.
Conclusion
We have shown that proteins can be computationally designed and assayed for folding thousands at a time, and that high-throughput design experiments can provide quantitative insights into the determinants of protein stability. Large libraries can be designed in a relatively unbiased manner (as in our first generation) to maximize the protein property space examined, or properties can be tuned to increase the design success rate at the cost of diversity. The power of our iterative learning approach to progressively hone in on more subtle contributions to stability is highlighted by the progression of our ααα design sets from early rounds in which design failures were caused by insufficient buried nonpolar surface area to the last round where helix-sidechain electrostatics had the greater effect. The large numbers of folded and not folded designs will also provide stringent tests of molecular dynamics simulation approaches which have successfully reproduced structures (13, 15) and some thermodynamic measurements (14, 50) of natural proteins, but have not yet been challenged with plausible but unstable protein structures like our design failures.
The four solution structures, saturation mutagenesis data on 13 of 14 designs, and over thirty thousand negative control experiments indicate that the large majority of our stable sequences are structured as designed. These 2,788 designed proteins, stable without disulfides or metal coordination, should have numerous applications in bioengineering and synthetic biology. Many are more stable than any comparably-sized monomeric proteins found in the PDB, making them ideal scaffolds for engineering inhibitors of intracellular protein-protein interactions. Their small size may also help promote membrane translocation and endosomal escape (51, 52). As DNA synthesis technology continues to improve, high-throughput protein design will become possible for larger proteins as well, revealing determinants of protein stability in more complex structures and leading to a new era of iterative, data-driven de novo protein design and modeling.
Supplementary Material
Acknowledgments
We acknowledge support from Howard Hughes Medical Institute (to D.B.), and the Natural Sciences and Engineering Research Council of Canada (to C.H.A.). G.J.R. is a Merck Fellow of the Life Sciences Research Foundation. C.H.A. holds a Canada Research Chair in Structural Genomics. We would also like to thank Stephen Rettie for mass spectrometry support, Charlie Lee for deep sequencing support, Sergey Ovchinnikov for assistance quantifying sequence conservation, Vanessa Nguyen, Angelica Yehdego, Tessa Howard, and Kara Lau for assistance with protein purification, and Hannah Gelman and many other members of the Baker Lab for helpful discussions. This work was facilitated by the Hyak supercomputer at the University of Washington and by donations of computing time from Rosetta@Home participants. The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute [OGI-055], Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck & Co., Novartis Pharma AG, Ontario Ministry of Research, Innovation and Science (MRIS), Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and the Wellcome Trust. The RosettaScripts code and blueprint files used for protein design are provided in the supplementary materials. The data for this work (designed sequences and structures, deep sequencing counts, EC50 values, stability scores, and structural analysis of the designed models) are also provided in supplementary materials. The python code for inferring EC50 values and for fitting the unfolded state model is provided at https://github.com/asford/protease_experimental_analysis.
Footnotes
Author contributions
G.J.R. designed the research, the experimental approach, and the proteins. G. J. R., T. M. C., I. G., S. H., L. C., R. R., and A. C. performed experiments. All authors analyzed data. G. J. R., A. F., and V. M. contributed new computational tools. C. H. A. and D. B. supervised research. G. J. R. and D. B. wrote the manuscript.
Methods
Definition of scoring metrics
Explanation of supplementary datasets
Supplementary datasets and design scripts
References
- 1.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 2.Robertson AD, Murphy KP. Protein Structure and the Energetics of Protein Stability. Chem Rev. 1997;97:1251–1268. doi: 10.1021/cr960383c. [DOI] [PubMed] [Google Scholar]
- 3.Nick Pace C, Martin Scholtz J, Grimsley RG. Forces stabilizing proteins. FEBS Lett. 2014;588:2177–2184. doi: 10.1016/j.febslet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gelman H, Gruebele M. Fast protein folding kinetics. Q Rev Biophys. 2014;47:95–142. doi: 10.1017/S003358351400002X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang X, Kowalski J, Kelly JW. Increasing protein stability using a rational approach combining sequence homology and structural alignment: Stabilizing the WW domain. Protein Sci. 2001;10:1454–1465. doi: 10.1110/ps.640101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jager M, et al. Structure-function-folding relationship in a WW domain. Proceedings of the National Academy of Sciences. 2006;103:10648–10653. doi: 10.1073/pnas.0600511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao S, Bi Y, Shan B, Raleigh DP. Analysis of core packing in a cooperatively folded miniature protein: the ultrafast folding villin headpiece helical subdomain. Biochemistry. 2009;48:4607–4616. doi: 10.1021/bi8021763. [DOI] [PubMed] [Google Scholar]
- 8.Neuweiler H, et al. The folding mechanism of BBL: Plasticity of transition-state structure observed within an ultrafast folding protein family. J Mol Biol. 2009;390:1060–1073. doi: 10.1016/j.jmb.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 9.Pace CN, et al. Contribution of hydrophobic interactions to protein stability. J Mol Biol. 2011;408:514–528. doi: 10.1016/j.jmb.2011.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araya CL, et al. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci U S A. 2012;109:16858–16863. doi: 10.1073/pnas.1209751109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao S, et al. Rational modification of protein stability by targeting surface sites leads to complicated results. Proc Natl Acad Sci U S A. 2013;110:11337–11342. doi: 10.1073/pnas.1222245110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pace CN, et al. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014;23:652–661. doi: 10.1002/pro.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindorff-Larsen K, Piana S, Dror RO, Shaw DE. How Fast-Folding Proteins Fold. Science. 2011;334:517–520. doi: 10.1126/science.1208351. [DOI] [PubMed] [Google Scholar]
- 14.Piana S, Lindorff-Larsen K, Shaw DE. Protein folding kinetics and thermodynamics from atomistic simulation. Proc Natl Acad Sci U S A. 2012;109:17845–17850. doi: 10.1073/pnas.1201811109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen H, Maier J, Huang H, Perrone V, Simmerling C. Folding simulations for proteins with diverse topologies are accessible in days with a physics-based force field and implicit solvent. J Am Chem Soc. 2014;136:13959–13962. doi: 10.1021/ja5032776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang PS, Boyken SE, Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- 17.Rohl CA, Strauss CEM, Misura KMS, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 18.Magliery TJ. Protein stability: computation, sequence statistics, and new experimental methods. Curr Opin Struct Biol. 2015;33:161–168. doi: 10.1016/j.sbi.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H, et al. Simultaneous Optimization of Biomolecular Energy Functions on Features from Small Molecules and Macromolecules. J Chem Theory Comput. 2016;12:6201–6212. doi: 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dahiyat BI, Mayo SL. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 21.Liang H, et al. De Novo Design of a βαβ Motif. Angew Chem Int Ed. 2009;48:3301–3303. doi: 10.1002/anie.200805476. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Yang Y, Zhan J, Dai L, Zhou Y. Energy Functions in De Novo Protein Design: Current Challenges and Future Prospects. Annu Rev Biophys. 2013;42:315–335. doi: 10.1146/annurev-biophys-083012-130315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosuri S, Church GM. Large-scale de novo DNA synthesis: technologies and applications. Nat Methods. 2014;11:499–507. doi: 10.1038/nmeth.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun MGF, Seo MH, Nim S, Corbi-Verge C, Kim PM. Protein engineering by highly parallel screening of computationally designed variants. Sci Adv. 2016;2:e1600692. doi: 10.1126/sciadv.1600692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 26.Sieber V, Plückthun A, Schmid FX. Selecting proteins with improved stability by a phage-based method. Nat Biotechnol. 1998;16:955–960. doi: 10.1038/nbt1098-955. [DOI] [PubMed] [Google Scholar]
- 27.Finucane MD, Tuna M, Lees JH, Woolfson DN. Core-directed protein design. I. An experimental method for selecting stable proteins from combinatorial libraries. Biochemistry. 1999;38:11604–11612. doi: 10.1021/bi990765n. [DOI] [PubMed] [Google Scholar]
- 28.Park C, Zhou S, Gilmore J, Marqusee S. Energetics-based protein profiling on a proteomic scale: identification of proteins resistant to proteolysis. J Mol Biol. 2007;368:1426–1437. doi: 10.1016/j.jmb.2007.02.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park C, Marqusee S. Pulse proteolysis: a simple method for quantitative determination of protein stability and ligand binding. Nat Methods. 2005;2:207–212. doi: 10.1038/nmeth740. [DOI] [PubMed] [Google Scholar]
- 30.Leuenberger P, et al. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science. 2017;355 doi: 10.1126/science.aai7825. [DOI] [PubMed] [Google Scholar]
- 31.Jäger M, Dendle M, Kelly JW. Sequence determinants of thermodynamic stability in a WW domain--an all-beta-sheet protein. Protein Sci. 2009;18:1806–1813. doi: 10.1002/pro.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhardwaj G, et al. Accurate de novo design of hyperstable constrained peptides. Nature. 2016;538:329–335. doi: 10.1038/nature19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koga N, et al. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamtekar S, Schiffer J, Xiong H, Babik J, Hecht M. Protein design by binary patterning of polar and nonpolar amino acids. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 35.Davidson AR, Sauer RT. Folded proteins occur frequently in libraries of random amino acid sequences. Proc Natl Acad Sci U S A. 1994;91:2146–2150. doi: 10.1073/pnas.91.6.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hecht MH, Das A, Go A, Bradley LH, Wei Y. De novo proteins from designed combinatorial libraries. Protein Sci. 2004;13:1711–1723. doi: 10.1110/ps.04690804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fox RJ, et al. Improving catalytic function by ProSAR-driven enzyme evolution. Nat Biotechnol. 2007;25:338–344. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- 38.Romero PA, Krause A, Arnold FH. Navigating the protein fitness landscape with Gaussian processes. Proc Natl Acad Sci U S A. 2013;110:E193–201. doi: 10.1073/pnas.1215251110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leaver-Fay A, et al. Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol. 2013;523:109–143. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar MDS, et al. ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res. 2006;34:D204–6. doi: 10.1093/nar/gkj103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker EG, et al. Local and macroscopic electrostatic interactions in single α-helices. Nat Chem Biol. 2015;11:221–228. doi: 10.1038/nchembio.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doering DS, Matsudaira P. Cysteine scanning mutagenesis at 40 of 76 positions in villin headpiece maps the F-actin binding site and structural features of the domain. Biochemistry. 1996;35:12677–12685. doi: 10.1021/bi9615699. [DOI] [PubMed] [Google Scholar]
- 43.Meng J, et al. High-resolution crystal structures of villin headpiece and mutants with reduced F-actin binding activity. Biochemistry. 2005;44:11963–11973. doi: 10.1021/bi050850x. [DOI] [PubMed] [Google Scholar]
- 44.Verdecia MA, Bowman ME, Lu KP, Hunter T, Noel JP. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat Struct Biol. 2000;7:639–643. doi: 10.1038/77929. [DOI] [PubMed] [Google Scholar]
- 45.Shoichet BK, Baase WA, Kuroki R, Matthews BW. A relationship between protein stability and protein function. Proceedings of the National Academy of Sciences. 1995;92:452–456. doi: 10.1073/pnas.92.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chong PA, Lin H, Wrana JL, Forman-Kay JD. An expanded WW domain recognition motif revealed by the interaction between Smad7 and the E3 ubiquitin ligase Smurf2. J Biol Chem. 2006;281:17069–17075. doi: 10.1074/jbc.M601493200. [DOI] [PubMed] [Google Scholar]
- 47.Aragón E, et al. Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF-β Pathways. Structure. 2012;20:1726–1736. doi: 10.1016/j.str.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elcock AH. Prediction of functionally important residues based solely on the computed energetics of protein structure. J Mol Biol. 2001;312:885–896. doi: 10.1006/jmbi.2001.5009. [DOI] [PubMed] [Google Scholar]
- 49.Boder ET, Bill JR, Nields AW, Marrack PC, Kappler JW. Yeast surface display of a noncovalent MHC class II heterodimer complexed with antigenic peptide. Biotechnol Bioeng. 2005;92:485–491. doi: 10.1002/bit.20616. [DOI] [PubMed] [Google Scholar]
- 50.Piana S, Klepeis JL, Shaw DE. Assessing the accuracy of physical models used in protein-folding simulations: quantitative evidence from long molecular dynamics simulations. Curr Opin Struct Biol. 2014;24:98–105. doi: 10.1016/j.sbi.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 51.Appelbaum JS, et al. Arginine topology controls escape of minimally cationic proteins from early endosomes to the cytoplasm. Chem Biol. 2012;19:819–830. doi: 10.1016/j.chembiol.2012.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.LaRochelle JR, Cobb GB, Steinauer A, Rhoades E, Schepartz A. Fluorescence correlation spectroscopy reveals highly efficient cytosolic delivery of certain penta-arg proteins and stapled peptides. J Am Chem Soc. 2015;137:2536–2541. doi: 10.1021/ja510391n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang PS, et al. RosettaRemodel: a generalized framework for flexible backbone protein design. PLoS One. 2011;6:e24109. doi: 10.1371/journal.pone.0024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leaver-Fay A, et al. Methods in Enzymology. 2013:109–143. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alford RF, et al. The Rosetta all-atom energy function for macromolecular modeling and design. 2017 doi: 10.1101/106054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pedregosa F, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 57.Hoover DM, Lubkowski J. DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Res. 2002;30:e43. doi: 10.1093/nar/30.10.e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Benatuil L, Perez JM, Belk J, Hsieh CM. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng Des Sel. 2010;23:155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- 59.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nat Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patil A, Huard D, Fonnesbeck CJ. PyMC: Bayesian Stochastic Modelling in Python. J Stat Softw. 2010;35:1–81. [PMC free article] [PubMed] [Google Scholar]
- 62.The Theano Development Team et al. Theano: A Python framework for fast computation of mathematical expressions. 2016 arXiv [cs.SC] (available at http://arxiv.org/abs/1605.02688)
- 63.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 64.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 65.Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 67.Orekhov VY, Ibraghimov I, Billeter M. Optimizing resolution in multidimensional NMR by three-way decomposition. J Biomol NMR. 2003;27:165–173. doi: 10.1023/a:1024944720653. [DOI] [PubMed] [Google Scholar]
- 68.Kazimierczuk K, Orekhov VY. Accelerated NMR spectroscopy by using compressed sensing. Angew Chem Int Ed Engl. 2011;50:5556–5559. doi: 10.1002/anie.201100370. [DOI] [PubMed] [Google Scholar]
- 69.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 70.Lemak A, et al. A novel strategy for NMR resonance assignment and protein structure determination. J Biomol NMR. 2011;49:27–38. doi: 10.1007/s10858-010-9458-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lemak A, Steren CA, Arrowsmith CH, Llinás M. Sequence specific resonance assignment via Multicanonical Monte Carlo search using an ABACUS approach. J Biomol NMR. 2008;41:29–41. doi: 10.1007/s10858-008-9238-2. [DOI] [PubMed] [Google Scholar]
- 72.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 73.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 74.Linge JP, Williams MA, Spronk CAEM, Alexandre MJ, Nilges M. Refinement of protein structures in explicit solvent. Proteins: Struct Funct Bioinf. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 75.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 76.Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9:173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 77.Alva V, Nam SZ, Söding J, Lupas AN. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016;44:W410–W415. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Crooks GE. WebLogo: A Sequence Logo Generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pan Y, et al. Quantitative proteomics reveals the kinetics of trypsin-catalyzed protein digestion. Anal Bioanal Chem. 2014;406:6247–6256. doi: 10.1007/s00216-014-8071-6. [DOI] [PubMed] [Google Scholar]
- 80.Schellenberger V, Braune K, Hofmann HJ, Jakubke HD. The specificity of chymotrypsin. A statistical analysis of hydrolysis data. Eur J Biochem. 1991;199:623–636. doi: 10.1111/j.1432-1033.1991.tb16163.x. [DOI] [PubMed] [Google Scholar]
- 81.Schellenberger V, Turck CW, Hedstrom L, Rutter WJ. Mapping the S’ subsites of serine proteases using acyl transfer to mixtures of peptide nucleophiles. Biochemistry. 1993;32:4349–4353. doi: 10.1021/bi00067a026. [DOI] [PubMed] [Google Scholar]
- 82.Schellenberger V, Turck CW, Rutter WJ. Role of the S’ Subsites in Serine Protease Catalysis. Active-Site Mapping of Rat Chymotrypsin, Rat Trypsin,. alpha.-Lytic Protease, and Cercarial Protease from Schistosoma mansoni. Biochemistry. 1994;33:4251–4257. doi: 10.1021/bi00180a020. [DOI] [PubMed] [Google Scholar]
- 83.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 84.Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- 85.Kabsch W, Sander C. Dictionary of protein secondary structure: Pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 86.Lin YR, et al. Control over overall shape and size in de novo designed proteins. Proc Natl Acad Sci U S A. 2015;112:E5478–85. doi: 10.1073/pnas.1509508112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Monera OD, Sereda TJ, Zhou NE, Kay CM, Hodges RS. Relationship of sidechain hydrophobicity and α-helical propensity on the stability of the single-stranded amphipathic α-helix. J Pept Sci. 1995;1:319–329. doi: 10.1002/psc.310010507. [DOI] [PubMed] [Google Scholar]
- 88.Zheng F, Zhang J, Grigoryan G. Tertiary structural propensities reveal fundamental sequence/structure relationships. Structure. 2015;23:961–971. doi: 10.1016/j.str.2015.03.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.