Abstract
Purpose
Interphotoreceptor retinoid-binding protein (IRBP) appears to target and protect retinoids during the visual cycle. X-ray crystallographic studies had noted a ββα-spiral fold shared with crotonases and C-terminal protein transferases. The shallow cleft formed by the fold was assumed to represent the retinol-binding site. However, a second hydrophobic site consisting of a highly restricted cavity was more recently appreciated during in silico ligand-docking studies. In this study, the ligand-binding environment within the second module of Xenopus IRBP (X2IRBP) is defined.
Methods
Pristine recombinant polypeptide corresponding to X2IRBP was expressed in a soluble form and purified to homogeneity without its fusion tag. Phenylalanine was substituted for tryptophan at each of the putative retinol-binding domains (W450F, hydrophobic cavity; W587F, shallow cleft). Binding of 11-cis and all-trans retinol were observed in titrations monitoring retinol fluorescence enhancement, quenching of tryptophan fluorescence, and energy transfer. The effect of oleic acid on retinol binding was also examined.
Results
A ligand-binding stoichiometry of ~1:1 was observed for 11-cis and all-trans with Kd in the tens of nanomolar range. The substitution mutants showed little effect on retinol binding in titrations after fluorescence enhancement. However, the W450F and not the W587F mutant showed a markedly reduced capacity for fluorescence quenching for both 11-cis and all-trans retinol. Oleic acid inhibited the binding of 11-cis and all-trans retinol in an apparent noncompetitive manner.
Conclusions
The binding site for 11-cis and all-trans retinol is a novel hydrophobic cavity that is highly restrictive and probably distinct from the long chain fatty acid–binding site.
Vision begins with the photoisomerization of 11-cis to all-trans retinal bound to rhodopsin.1,2 The 11-cis isomer is returned to the outer segments through the “visual cycle.”3–5 The cycle utilizes the retinal pigmented epithelium (RPE) and Müller cell glia to enzymatically reconvert all-trans retinol to an 11-cis isomer. The RPE returns 11-cis retinal to the rod outer segments, although the Müller cells, at least in some species, return 11-cis retinol to the cones, which are able to oxidize it to 11-cis retinal.6–10 Interphotoreceptor retinoid-binding protein (IRBP), which is the major soluble protein component of the interphotoreceptor matrix, facilitates the exchange of these retinoids between the photoreceptors, RPE, and Müller cells (reviewed in Refs. 1–3,11–15). Disruption of a phylogenetically conserved salt bridge within IRBP has been associated with a form of recessive retinitis pigmentosa.16
IRBP carries visual-cycle retinoids in a light-dependent manner, functions to remove all-trans retinol from bleached rod outer segments,17–20 delivers all-trans retinol to the RPE,21 releases 11-cis retinal from the RPE,22–24 and delivers 11-cis retinal to the rod outer segments.24 Recent studies of irbp−/− mice support a role for IRBP in the translocation of visual retinoids and cone function.25 Finally, IRBP carries endogenous 11-cis retinol, suggesting that it has a role in the cone visual cycle.6,26,27 These processes may be regulated by various fatty acids bound endogenously to IRBP.28 For example, docosahexaenoic acid (DHA) is thought to trigger the release of 11-cis retinal from IRBP to the outer segments.29,30 More recently, oleic acid was found to bind more tightly to IRBP than DHA.31 IRBP therefore functions in a complex process regulated by fatty acids, and possibly receptors, to accomplish a bidirectional transport of visual-cycle retinoids between diverse cell types while protecting their isomeric and oxidative state.32
IRBP, the largest known retinoid-binding protein, consists of two or four homologous “modules” each approximately 300 amino acids in length (four in tetrapods; two in teleosts33–36). The functional significance of this structure, unique among retinoid-binding proteins, is largely unknown. As the individual modules of IRBP appear to represent functional units of the protein, we and others have attempted to understand the structure and function of IRBP by examining the biochemical and functional properties of its individual modules.37–44 X-ray crystallographic analysis of the second module of IRBP suggests a homology with two enzyme families, the C-terminal transferases (CPTases) and crotonases. 45 Photosystem II D1 CPTase (D1P), a prototype CTPase with known structure,46 plays a role in the repair of the oxidative damage to the reaction center by renewing the photosystem II D1 protein.47 The repair involves the disassembly of the photosynthesis complex with replacement of damaged D1 protein with new protein. In a final step in this process, D1P catalyzes the C-terminal cleavage of the new D1 protein. Catalysis requires binding of DIP to the hydrophobic C-terminal of the D1 protein. Each module of IRBP is structurally similar to the D1P domains A and C which correspond to the N- and C-terminal X2IRBP domains, respectively.45 However, the PDZ domain,48 located between domains A and C conferring this enzymatic activity in DIP, is not present in IRBP. IRBP therefore retains the hydrophobic ligand-binding ability but not the proteolytic activity.
The C-terminal domain B of X2IRBP and domain C of D1P exhibit a structural homology with crotonases (enoyl Co-A hydratase/isomerases),45 as represented by the known structures of dienoyl-CoA isomerase,49 chlorobenzoyl-CoA dehydrogenase and enoyl-CoA hydratase.50 Crotonases have a low sequence identity, but adopt the same overall fold.50,51 X2IRBP shares with these enzymes a structural core composed of three helices and the large five-stranded β sheet in domain B forming a ββα-spiral fold. These elements are arranged in a topologically identical order despite peripheral insertions/deletions. However, none of the catalytic residues of the crotonase family are conserved in IRBP.45
If we are to understand the mechanism underlying the interaction between IRBP’s retinol- and fatty acid–binding sites29 and the molecular basis of retinitis pigmentosa caused by a recently described mutation in IRBP,16 it is critical to define the binding domain(s) for these physiological ligands. The structural homology among X2IRBP, crotonases, and CT-Pases suggested that at least one site corresponds to a shallow cleft associated with the ββα-spiral.45 Although results in computer docking studies have not been inconsistent with this prediction, a second site within a separate “hydrophobic cavity” was also noted15 (see Fig. 1). In the present study, we used site-directed mutagenesis to define the binding site for all-trans and 11-cis retinol in X2IRBP. The rationale for the experimental design takes advantage of the fact that X2IRBP has only two tryptophans each of which is located in one of these two putative ligand-binding domains. Tryptophan, with a fluorescence emission close to the excitation of retinol, provides a sensitive probe of the two sites. This type of tryptophan scanning has been useful in defining the ligand-binding site in cellular retinol-binding protein II52 and uncovering structure-function relationships in various protein systems (reviewed in Ref. 53). For our studies, recombinant X2IRBPs were prepared in a manner that has recently allowed preparation of diffraction-quality crystals of full length Xenopus IRBP.54 This approach, which extends earlier strategies for preparing X2IRBP,37 includes providing a reducing environment and removing the fusion tag from the final protein. We provide evidence that the hydrophobic cavity, and not the shallow cleft, represents the binding site for 11-cis and all-trans retinol in X2IRBP.
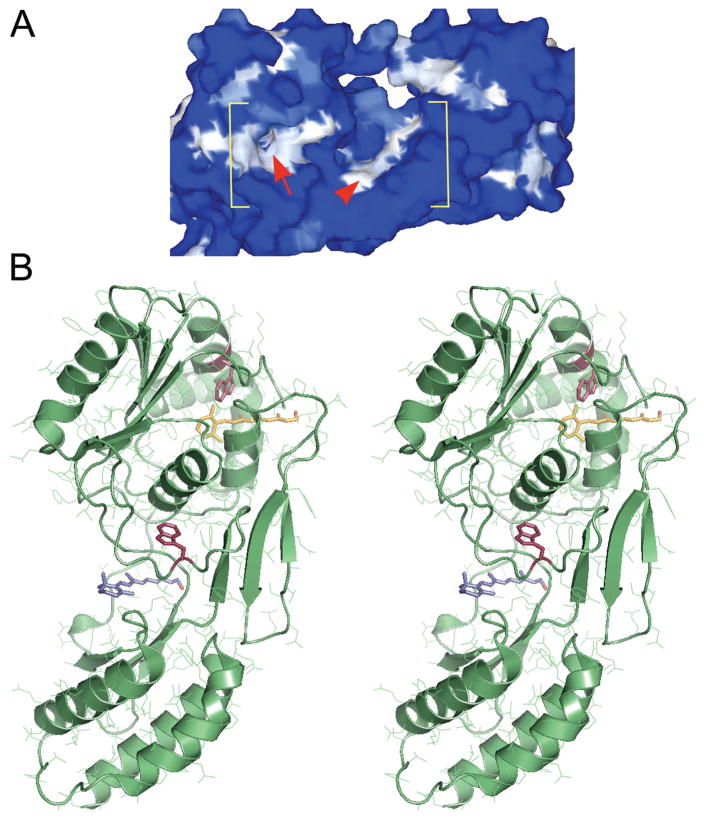
Figure 1.
Two putative retinol-binding sites within the second module of Xenopus IRBP (X2IRBP). (A) Hydrophobicity surface representation, where white represents high hydrophobicity, and blue represents high hydrophilicity. Within the bracketed area, the arrowhead identifies a shallow hydrophobic cleft. Arrow: an opening located in a hydrophobic patch that leads to a hydrophobic cavity. Modified with permission from Loew A, Gonzalez-Fernandez F. Crystal structure of the functional unit of interphotoreceptor retinoid binding protein. Structure. 2002;10(1):43–49. © Cell Press. (B) Stereoimage of computer docking of all-trans retinol in each of the two possible hydrophobic ligand-binding domains. We have previously shown that retinol-binding quenches tryptophan fluorescence in X2IRBP (Gonzalez-Fernandez et al.38). There are two tryptophan residues within X2IRBP that could support this quenching. Each of these tryptophans (red) is located in one of the putative ligand-binding domains. W450 is located in the hydrophobic cavity containing a molecule of retinol (yellow); W587 is located in the more shallow hydrophobic cleft containing a molecule of retinol (blue). Image modified with permission from Gonzalez-Fernandez F, Ghosh D. Focus on molecules: interphotoreceptor retinoid-binding protein (IRBP). Exp Eye Res. 2006;86:169–170. © Elsevier. The image may be viewed in 3-D without specialized stereo glasses. Suggestions for viewing molecular stereo images are available at http://spdbv.vital-it.ch/TheMolecularLevel/0Help/StereoView.html.
Methods
The second module of Xenopus IRBP (X2IRBP), corresponds to aminoacid residues 313 to 612 in the full-length sequence where the first residue corresponds to the initial methionine of the signaling peptide. 38 The expression construct was prepared by PCR amplifying module 2 from X10a, a full-length Xenopus IRBP cDNA that has been isolated in our laboratory.38 Ligation-independent cloning (LIC) was used to insert the amplified fragment into pET-30 Xa/LIC (Novagen, San Diego, CA).55,56 This vector provides polyhistidine and S-Tag domains, and a factor Xa cleavage site 5′ to the cloning site. During the PCR, we added a tobacco-etch virus (TEV) protease cleavage site (GluAsnLeuTyrPheGln ↓ Gly; bold in the forward primer sequence below)57 between the Xa cleavage and cloning sites (see Fig. 2A). Cleavage with TEV was efficient and added a single Gly residue to the X2IRBP N terminus. The oligonucleotide primers, synthesized by Integrated DNA Technologies (Coralville, IA), were as follows. Forward primer, GGTATTGAGGGTCGCGAAAATCTGTATTTTCAGGGCgtgcgttcatctgttacc (italic, primer extension sequence for vector compatibility; bold, TEV protease recognition site; lowercase, Xenopus-IRBP coding sequence beginning at residue 313); Reverse primer, AGAGGAGAGTTAGAGCCTTAtgcaatgatctccttggccttgtc (italic, primer extension sequence; bold, stop codon; lowercase, Xenopus-IRBP coding sequence ending with residue 612).
Figure 2.
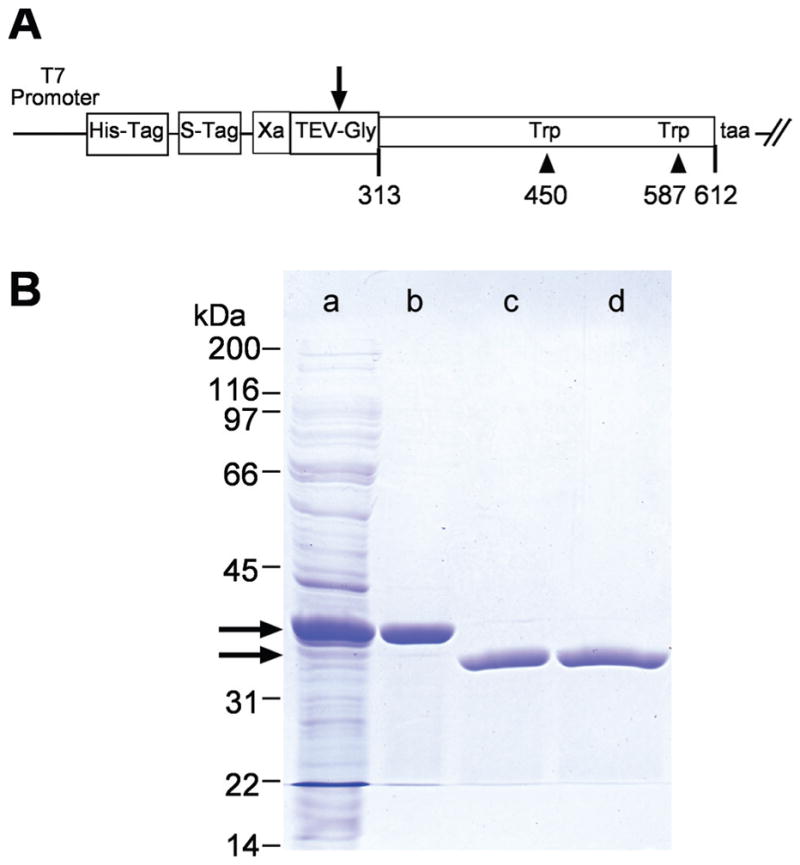
X2IRBP and the W450T and W587T mutants were expressed in a soluble form as polyhistidine-fusion proteins. The fusion domain was removed by cleavage with tobacco-etch virus (TEV) protease. (A) Map of the plasmid construct used to express the wild-type and mutant forms of X2IRBP. Arrow: cleavage site in the TEV protease recognition site (GluAsnLeuTyrPheGln ↓ Gly). Amino-acid residues are numbered according to the sequence of the full-length Xenopus IRBP. (B) Purification of X2IRBP and removal of its polyhistidine fusion tag. Lane a: total soluble E. coli lysate. Lane b: proteins binding to the HisTrap-nickel column. Lane c: Proteins not binding to a second HisTrap column after TEV cleavage. Lane d: Cleaved X2IRBP following Q-Sepharose HP ion-exchange chromatography. Arrows: positions of the fusion protein before and after TEV cleavage, respectively. Protein size standards are shown on the left of the gel. SDS (4%–20% gradient)-polyacrylamide gel stained with Coomassie blue.
Tryptophan was replaced with phenylalanine or valine at positions 450 and 587. The mutations were introduced directly into the X2IRBP-pET30 construct by using site-directed mutagenesis (QuikChange; Stratagene, La Jolla, CA). Briefly, hot-start PCR was performed on a thermocycler (model 2720; Applied Biosystems, Foster City, CA), using high-fidelity DNA polymerase (PfuUltra; Stratagene) for 18 cycles (95°C, 30 seconds; 58°C, 1 minute; 68°C, 7.5 minutes) with HPLC-purified primers: W450F: forward caccttttatagttaacacagtctttgaacccatcacaataacagaaaac, reverse gttttctgttattgtgatgggttcaaagactgtgttaactataaaaggtg; W587F: forward catagatagtaatggtgactactttttgggaggtggagttgttccag; reverse ctggaacaactccacctcccaaaaagtagtcaccattactatctatg; W450V: forward caccttttatagttaacacagtcgtggaacccatcacaataacagaaaac, reverse gttttctgttattgtgatgggttccacgactgtgttaactataaaaggtg; and W587V: forward catagatagtaatggtgactacgtgttgggaggtggagttgttccag, reverse ctggaacaactccacctcccaacacgtagtcaccattactatctatg. The final constructs were verified by automated DNA sequencing.
X2IRBP and each of the tryptophan substitution mutants were expressed in Escherichia coli BLR(DE3) (Novagen) at 22°C for 18 hours. Protein expression was induced with 1 mM IPTG at a cellular density of OD550 = 0.5. The cells were disrupted in a French pressure cell (13,000 psi; SLM-Aminco Instruments, Rochester, NY) in 250 mM NaCl, 50 mM Tris (pH 7.5), 1 mM DTT, with broad-spectrum protease inhibitors (Complete EDTA-free; Roche Diagnostics, Basel, Switzerland). The soluble fraction after centrifugation at 43,000g for 30 minutes was subjected to affinity chromatography on a 5-mL nickel column at 6°C (HisTrap; GE Healthcare, Piscataway, NJ). These columns use the tetradentate chelator Ni-NTA, which has superior Ni2+ affinity to tridentates, and is stable in DTT up to 5 mM. X2IRBP-polyhistidine fusion proteins eluted from the column at ~100 mM imidazole with a 0- to 300-mM imidazole linear gradient over 15 column volumes (pH 7.4). The imidazole was removed by ultrafiltration, and the fusion protein domain cleaved from the X2IRBP with TEV for 16 hours at 4°C. The freed X2IRBP, which had little affinity for the immobilized nickel resin, was separated from residual uncut fusion protein, by reapplication to the column. The protein was further purified by ion-exchange chromatography (resin, 20 mL bed volume; Q Sepharose HP, XK16/20 column; GE Healthcare). X2IRBP eluted from the column at 300 to 320 mM NaCl with a 1.0 M NaCl linear gradient over 12 column volumes. Protein aliquots were held at −80°C and never refrozen. The concentration of the purified protein was determined by absorbance spectroscopy (extinction 27,390 M−1 cm−1, at 280 nm), and amino acid analysis (AAA). For AAA, the samples were hydrolyzed in vacuo for 16 hours at 115°C with 6 N HCl/0.2% phenol. The acid contains 2 nanomoles norvaline per 100 μL as an internal standard. Analyses were performed with an amino acid analyzer (L-8900PH; Hitachi, Tokyo, Japan) that uses an ion-exchange column to separate the amino acids and a postcolumn reaction with ninhydrin for detection at 570 and 440 nm. AAA also allowed quantification of the number of phenylalanine residues present in the recombinant protein. Although the analysis did not support direct quantification of tryptophan, the ability to measure the increased number of phenylalanines per mole of polypeptide provides support for the presence of the intended amino acid substitutions.
Ligand-Binding Assays by Fluorescence Spectroscopy
All-trans retinol, at a purity of 99% by HPLC, was obtained from Fluka (Buchs, Switzerland). Working stocks were prepared fresh in ethanol and held under argon at −80°C for no more than 10 days. The extinction coefficient of all-trans retinol in ethanol was taken as 38,300 M−1 cm−1, 325 nM.58 11-cis Retinol was prepared from 11-cis retinal by chemical reduction.59 In brief, sodium borohydride (4.5 mg, 0.119 mmol) was added to 11-cis retinal (11.1 mg, 39.1 mmol) dissolved in 550 μL of molecular sieve–dried 2-propanol and flushed with argon. After the reaction was stirred at 21°C for 1 hour, most of the solvent was removed in a stream of argon. Water (300 μL) was added drop-wise. The product was extracted three times with benzene (1 mL each time). The three benzene phases, which contained the product, were combined. The product in the benzene was dried over MgSO4, and the solvent removed with reduced pressure under 30°C to yield 7.3 mg (65.2%) of pure 11-cis retinol. The purity of the product was confirmed by HPLC and absorption maximum (318 nm, hexane).60
The binding of all-trans and 11-cis retinol to X2IRBP was characterized in titrations using a DM 45 scanning spectrofluorimeter (On-Line Instrument Systems, Inc., Bogart, GA).61 Titrations monitoring enhancement of retinol fluorescence, quenching of intrinsic protein fluorescence, and energy transfer were performed as previously described.41 Enhancement of retinol fluorescence was determined by monitoring the increase in retinol fluorescence (all-trans retinol: excitation, 330 nm; emission, 480 nm; 11-cis retinol: excitation, 324 nm; emission, 475 nm). A fluorescence-matched solution of N-acetyl-L-tryptophanamide, which possesses a typical protein fluorescence, but is unable to interact with retinol, was used as a blank.30,62 Assays after the quenching of protein fluorescence used excitation and emission wavelengths of 280 and 340 nm, respectively. In these experiments, the inner filter effect was accounted for by graphic correction.63 Energy transfer assays monitored retinol fluorescence at 480 nm for all-trans retinol and 475 nm for 11-cis retinol (excitation, 280 nm). The dissociation constant (Kd) and number of binding sites (N) was determined by nonlinear least-squares fit to a binding equation that assumes a single type of noninteracting site(s).42 The error associated with the determination of binding parameters is given as the mean ± SE. The standard errors are estimates of the uncertainties in the estimates of the regression coefficients (analogous to the SEM). Nonlinear least-squares analysis, determination of the binding parameter SE, determination of R2 (coefficient of determination), and plotting of the fitted curves were performed (SigmaPlot Version 7; SPSS, Chicago, IL). In some experiments (see Fig. 5), each titration run was alternated. That is, during the same experiment, a titration was run with one mutant, followed by the second mutant. This was repeated a total of three times alternating between the two proteins. Collecting the data in this manner eliminates time-dependent systemic error. The data from each mutant was then combined and analyzed, as just described.
Figure 5.
Fluorescence titration of all-trans retinol binding to W450T and W587T X2IRBPs. The concentration of W450F (○) and W587F (●) were 1.0 and 0.80 μM, respectively. (A) Enhancement of retinol fluorescence (excitation at 330 nm, emission at 480 nm). (B) Quenching of tryptophan fluorescence (excitation 280 nm, emission 340 nm). The y-axis corresponds to the normalized fluorescence intensity relative to the maximum amount of quenching. Error bars are SEM for three independent titrations. The curves represent nonlinear least-squares fit of the data to the ligand-binding equation (coefficient of determination, R2 = 0.9996 for each of the curve fittings). These titrations represent protein from separate expressions and purifications from that used in the previous figure. W450F did not support sufficient quenching to allow meaningful fitting of the binding equation. Calculated number of binding sites and dissociation constants are summarized in Table 1.
Results
Crotonases and CTPases use the same fold to stabilize unique ligands. In the case of CTPases, the fold stabilizes the hydrophobic C-terminal end of a protein substrate. For dienoyl-CoA isomerase, the fold confers an ability to isomerize fatty acids, allowing their entry into the β-oxidation pathway. Superposition of X2IRBP with one of the crotonase superfamily members suggests a similar ligand-binding site.45 Of note, such comparisons place the ligand-binding site between the two domains of X2IRBP in the ββα-spiral fold. Furthermore, an equivalent superposition of 2-enoyl-CoA hydratase with D1P protease also places the ligand into the known active site of D1P.45 Although the catalytic residues, and the function between the crotonase superfamily and CTPases are not conserved, the location of the substrate-binding domain is. The structural homology between X2IRBP, crotonases and CTPases suggests that at least one binding site in X2IRBP is located in the ββα-spiral corresponding to the shallow cleft in Figure 1.
Although computer modeling studies are not inconsistent with this prediction, they also identified a second putative binding site for all-trans retinol.15 In addition to the ββα-spiral fold, retinol could also be docked within a separate “hydrophobic cavity.” Based on their size and hydrophobicity, both of these domains represent candidate retinol-binding sites. The arrow in Figure 1A identifies the entrance to the hydrophobic cavity, and the arrowhead identifies a more open cleft-like domain. The hydrophobic cavity, is more restricted, and lined by a higher percentage of hydrophobic residues compared with the larger, less restrictive cleft-like fold.38 The “shallow cleft” is large enough to provide a binding site for a long chain fatty-acid molecule like DHA. In contrast, the hydrophobic cavity is not large enough to accommodate these fatty acids, but easily docks all-trans retinol in silico15,38 (Fig. 1B).
Figure 2B illustrates the purification strategy. For X2IRBP of wild-type sequence, and both of the tryptophan-to-phenylalanine substitution mutants, 75% of the fusion protein was expressed in the soluble fraction of the crude-cell lysate. In contrast, for the tryptophan-to-valine mutants, although the overall level of expression was similar to the phenylalanine substitutions, less than 5% was expressed in a soluble form. Given this low level of soluble expression, we did not attempt to purify the valine substitution mutants. The yield for X2IRBP, W450F and W587F ranged from 45 to 55 mg/L of E. coli after TEV cleavage, and the second HisTrap column. The final yield after Q-Sepharose HP ion-exchange chromatography was 25 to 35 mg/L of E. coli. The concentration of each of the purified proteins was determined by absorbance spectroscopy and AAA. From the translated amino acid sequence, 12 phenylalanines are predicted to be present in X2IRBP of wild-type sequence. Of those, 11.9 were actually detected by AAA. For both W450F and W587F, the number of phenylalanines would be expected to increase to 13.0. Consistent with this prediction, the number of phenylalanines detected by AAA was 13.2 and 13.4, respectively (±0.15). These studies support the presence of a tryptophan-to-phenylalanine substitution in each of the mutants.
To address whether the purified X2IRBP of wild-type sequence, and the W450F and W587F mutants were in a correctly folded state, we took advantage of the fact that misfolded polypeptides often exist in solutions as protein aggregates.64 Each of the purified recombinant proteins freed of its polyhistidine fusion domain were subjected to analytical size-exclusion chromatography on a calibrated S-200 column. SDS-PAGE was used to analyze the composition of the peaks obtained. Each of the recombinant proteins migrated as a sharp peak at an elution volume consistent with the expected size of the monomer (data not illustrated). Evidence of dimers or significant multimer formation was not found. Since metastable, partially folded states generally lead to protein aggregation, the finding that the protein exists as a monomer in solution suggests that each of the proteins are present in a correctly folded state.65,66
In silico energy minimization was used to anticipate potential local effects of substituting phenylalanine for tryptophan at each site. In Figure 3, the binding pockets were energy minimized in a 4.5-Å sphere surrounding the phenylalanine. Superimposition of the X2IRBP structure with the energy-minimized mutants showed little alteration of the environment of the ligand-binding pocket. No increased steric hindrance was appreciated, suggesting that the substitution would not affect the ability of the sites to accommodate a molecule of all-trans retinol. This finding does not rule out an effect on protein folding. However, as discussed earlier, misfolding is unlikely, given the monodispersity of the mutants in solution.
Figure 3.
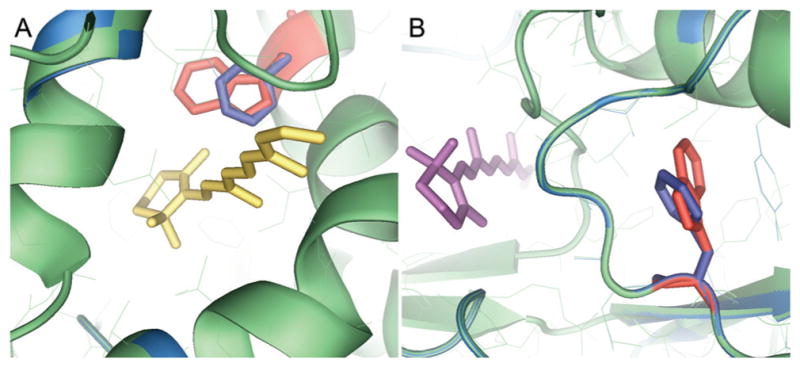
In silico energy minimization for the tryptophan-to-phenylalanine substitution in each of the two putative retinol-binding sites. The binding pockets were energy minimized in a 4.5-Å sphere surrounding the phenylalanine. (A) Hydrophobic cavity showing the W450F substitution docked with all-trans retinol (yellow). (B) Shallow cleft showing the W587F substitution docked with all-trans retinol (purple). In each panel, the native structure is shown in green with the tryptophan in red. The energy-minimized structure is shown in blue, with the orientation of the phenylalanine in blue.
Fluorescence spectroscopy allows noninvasive monitoring of ligand binding by monitoring changes in quantum yield.61 Observing the binding of all-trans or 11-cis retinol took advantage of such changes concomitant with ligand entry into the binding pocket. The amino-acid residues lining the two pockets have been tabulated.38 The hydrophobic environment of the sites support retinol fluorescence enhancement. The quenching of endogenous protein fluorescence on ligand binding is supported by the presence of tryptophan. As shown in Figure 1, the entire X2IRBP contains only two tryptophan residues. Each is located in the vicinity of one of the ligand-binding pockets. The average ring center-to-center distances between tryptophan and the docked retinol are 5.2 and 11.9 Å for the hydrophobic cavity and the shallow cleft, respectively. In contrast, W450 is 32 Å from the retinol in the shallow cleft; W587 is 20 Å from the retinol in the hydrophobic cavity.
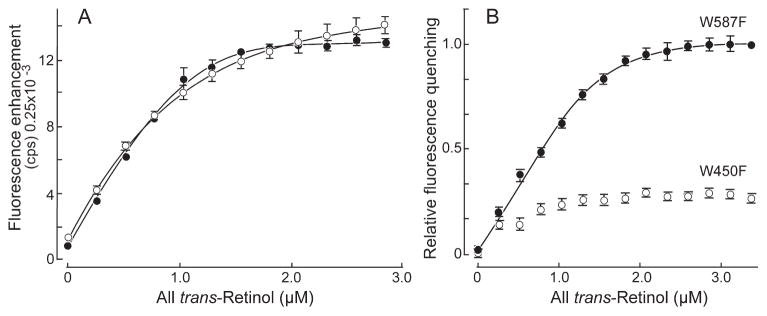
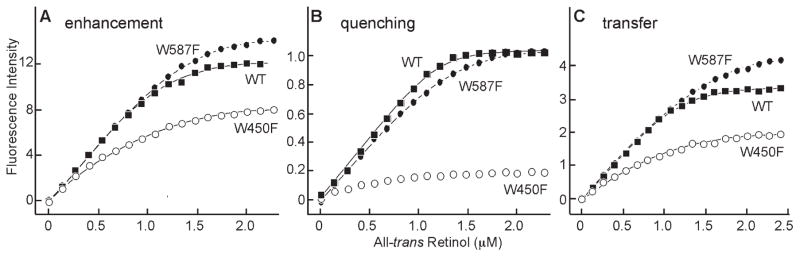
To determine whether the hydrophobic cavity, or the shallow cleft represents the principal ligand-binding site, we took advantage of the modeling indicating that the bound retinol would be near a tryptophan residue in either site. W450 is located in the hydrophobic cavity and W587 is located in the shallow cleft (Fig. 1B). In addition to the titrations with X2IRBP of wild-type sequence, Figure 4 superimposes data for the binding of all-trans retinol to the tryptophan-to-phenylalanine substitution mutants. All-trans retinol binding was observed by monitoring the enhancement of retinol fluorescence (excitation 330 nm, emission 480 nm) and quenching of tryptophan fluorescence (excitation 280 nm, emission 340 nm). The amount of enhancement was determined by subtracting the nonspecific fluorescence changes measured by titration of all-trans retinol into a fluorescence-matched NAT solution (data not shown). NAT, which has protein-like fluorescence from its indole ring, does not significantly interact with all-trans retinol, making it an ideal control solution.62 The number of binding sites per molecule of X2IRBP of wild-type (WT) sequence was calculated (see the Methods section) to be N = 1.27 ± 0.03, with Kd = 0.07 ± 0.02 μM. Figure 4B shows tryptophan fluorescence quenching, giving N = 1.26 ± 0.02 with Kd = 0.008 ± 0.003 μM (WT). Inner filter effect was accounted for as described by Mertens and Kagi.63 The plot in Figure 4C shows the energy transfer, giving N = 1.26 ± 0.02 with Kd = 0.033 ± 0.008 μM. These titrations suggest that there is at least one binding site for all-trans retinol in X2IRBP. Titrations monitoring all-trans retinol fluorescence enhancement showed significant binding for both mutants (Table 1). In contrast, titrations monitoring tryptophan quenching detected a site only in W587F (0.97 ± 0.02). No significant quenching was observed with the W450F mutant (Fig. 4B). Measurements of energy transfer detected approximately one site in both mutants.
Figure 4.
Fluorescence titrations of all-trans retinol binding to X2IRBP, and the tryptophan-to-phenylalanine substitution mutants. Each data set represents the average of three independent titrations. The curves, and binding parameters were determined by nonlinear least-squares fit of the binding equation. For each regression, the coefficient of determination (R2) was 0.9996. (A) Enhancement of retinol fluorescence (excitation 330 nm, emission 480 nm). The y-axis corresponds to the fluorescence intensity (counts per second, × 0.25 × 10−3). (B) Quenching of tryptophan fluorescence (excitation 280 nm, emission 340 nm). The y-axis corresponds to the normalized fluorescence intensity relative to the maximum amount of quenching. (C) Energy transfer (excitation 280 nm, emission 480 nm). The y-axis corresponds to the fluorescence intensity (counts per second × 0.25 × 10−3). X2IRBP of wild-type sequence (WT) (■), W450F (●), and W587F (○) mutants. The calculated number of binding sites and dissociation constants are summarized in Table 1.
Table 1.
Summary of All-trans Retinol Binding to X21RBP and Its Tryptophan to Phenylalanine Substitution Mutants
| Protein | Sites (N) Kd (μM) |
Enhancement | Quenching | Energy Transfer |
|---|---|---|---|---|
| X2IRBP | N | 1.27 ± 0.03 | 1.26 ± 0.02 | 1.26 ± 0.02 |
| Kd | 0.07 ± 0.02 | 0.008 ± 0.003 | 0.033 ± 0.008 | |
| W450F | N | 0.62 ± 0.09 | † | 0.83 ± 0.08 |
| 0.72 ± 0.06* | ND | |||
| Kd | 0.45 ± 0.11 | † | 0.29 ± 0.08 | |
| 0.48 ± 0.05* | ND | |||
| W587F | N | 1.07 ± 0.01 | 0.97 ± 0.02 | 1.20 ± 0.01 |
| 1.40 ± 0.04* | 1.31 ± 0.03* | ND | ||
| Kd | 0.07 ± 0.01 | 0.05 ± 0.01 | 0.10 ± 0.01 | |
| 0.05 ± 0.04* | 0.06 ± 0.01* | ND |
Figure 5 shows the titrations, which were performed in a side-by-side manner to directly compare the mutants during the same experiment. In the experiments depicted, each of the titration runs for W450F and W587F were alternated in the same experiment, with a total of three titrations performed for each mutant. The mutants showed a marked difference in fluorescence quenching compared to enhancement. Although the binding affinity of the two mutants was subtly altered, the enhancement titration did not detect a significant difference in the ligand-binding stoichiometry. In contrast, the W587F mutant showed a reduction in fluorescence quenching compared with W450F. As in Figure 4, the level of fluorescence quenching observed did not permit meaningful fitting of the binding equation. The calculated number of binding sites and dissociation constants are summarized in Table 1. The data suggest that all-trans retinol fluorescence quenching is markedly reduced when the tryptophan residue (W450) located in the hydrophobic cavity, but not in the shallow cleft (W587), is replaced with phenylalanine. The fact that all-trans retinol binding can still be measured by fluorescence enhancement suggests that the binding site remains intact in both mutants.
To address the possibility that one of the sites might be specific for the 11-cis and the other for the all-trans isomer of retinol, we carried out a second set of titrations shown in Figures 6 and 7. Figure 6 shows the binding of 11-cis retinol to X2IRBP of wild-type sequence. The superimposed titrations appear symmetrical in shape, and the calculated parameters were similar (by enhancement N = 0.95 ± 0.03, and Kd = 0.077 ± 0.015 μM; by quenching N = 0.93 ± 0.02, and Kd = 0.042 ± 0.009 μM). Titrations attempting to monitor energy transfer were not sensitive enough to obtain reliable data, because 11-cis retinol is not as fluorescent as all-trans retinol.
Figure 6.
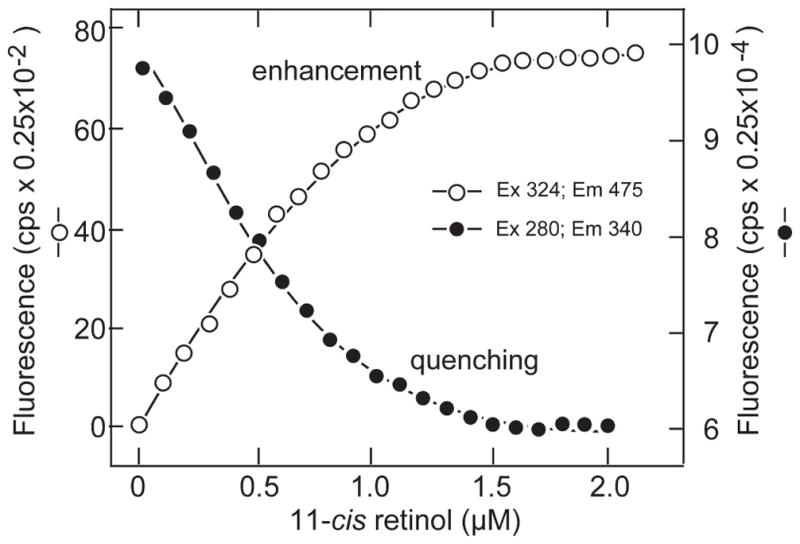
Binding of 11-cis retinol to X2IRBP of wild-type sequence. The titrations monitored the enhancement of retinol fluorescence (excitation 330 nm, emission 480 nm) and quenching of endogenous protein fluorescence (excitation 280 nm, emission 340 nm). Enhancement: N = 0.95 ± 0.03 and Kd = 0.077 ± 0.015 μM. Quenching: N = 0.93 ± 0.02 and Kd = 0.042 ± 0.009. The calculated number of binding sites and dissociation constants are summarized in Table 2.
Figure 7.
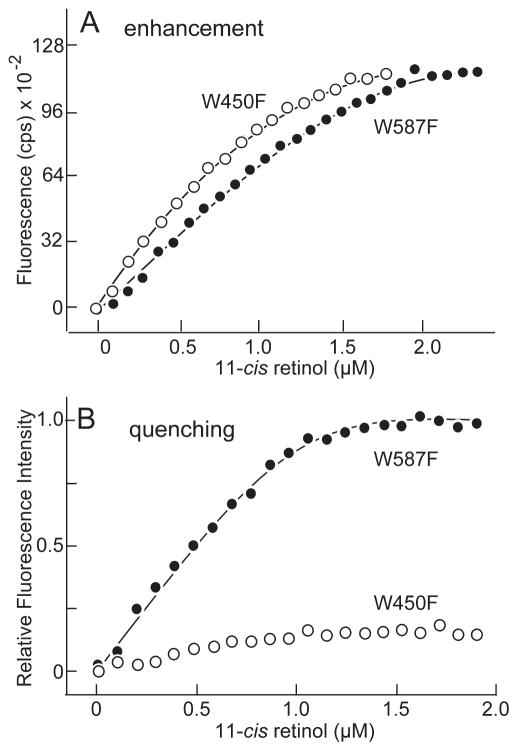
11-cis Retinol binding to mutants W450F (○) and W587F (●). (A) Binding determined by monitoring the enhancement of 11-cis fluorescence (excitation 324 nm, emission 475 nm). (B) Binding determined by monitoring quenching of tryptophan fluorescence (excitation 280 nm, emission 340 nm). Fluorescence intensity is normalized relative to the maximum amount of quenching. Calculated number of binding sites and dissociation constants are summarized in Table 2.
The results were similar for the two mutants when fluorescence enhancement was used to follow 11-cis retinol binding (Fig. 7A). One binding site with similar dissociation constants was detected in each mutant (Table 2). In contrast, when tryptophan quenching was used to monitor 11-cis retinol binding, the results were different (Fig. 7B). For W587F, values of N = 0.77 ± 0.02, with Kd = 0.024 ± 0.010 were obtained. However, for W450F, the quenching was not sufficient to obtain a meaningful fit of the ligand-binding equation. In silico docking shows that the β-ionone rings of both 11-cis and all-trans retinol are centered within the hydrophobic cavity (Fig. 8). Their side chains extend from the cavity into a less hydrophobic environment. Although the local environments of the side chain show some differences, the binding site can clearly accommodate both retinol isomers. Taken together, the results suggest that 11-cis and all-trans retinol bind in the hydrophobic cavity.
Table 2.
Summary of 11-cis Retinol Binding to X2IRBP and Its Tryptophan to Phenlyalanine Substitution Mutants
| Protein | Sites (N) Kd (μM) |
Enhancement | Quenching |
|---|---|---|---|
| X2IRBP | N | 0.95 ± 0.03 | 0.93 ± 0.02 |
| Kd | 0.077 ± 0.015 | 0.042 ± 0.009 | |
| W450F | N | 1.00 ± 0.06 | |
| Kd | 0.218 ± 0.062 | —* | |
| W587F | N | 1.67 ± 0.06 | 0.77 ± 0.02 |
| Kd | 0.132 ± 0.056 | 0.024 ± 0.010 |
No significant quenching was observed.
Figure 8.
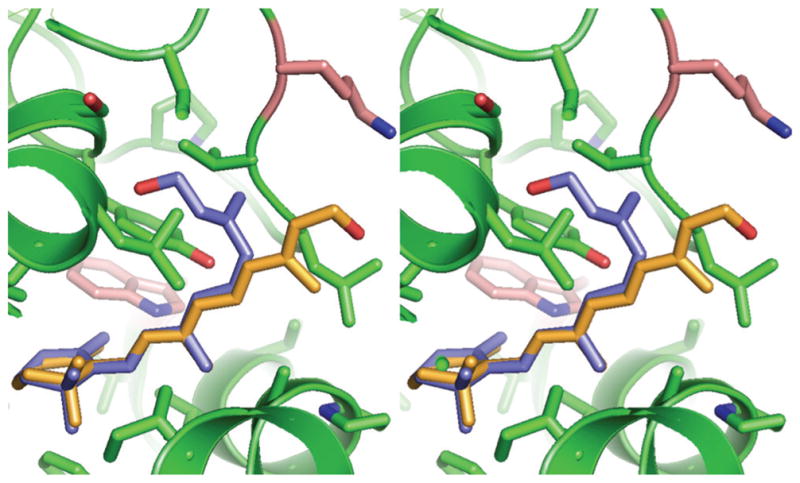
Stereo view of in silico docking of all-trans and 11-cis retinol within the hydrophobic cavity of X2IRBP. Blue: 11-cis retinol; orange: all-trans retinol. See Figure 1 for instructions for viewing stereo images.
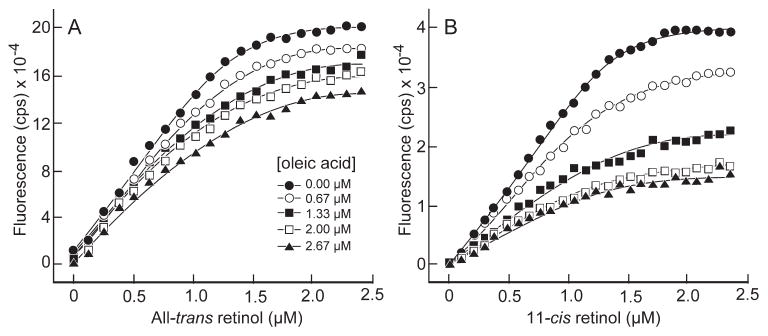
The known ability of X2IRBP to bind fatty acids is not easily explained by an interaction with the hydrophobic cavity, because the cavity is not large enough to accommodate a molecule the size of DHA or oleic acid.38 To investigate, we performed 11-cis and all-trans retinol titrations with X2IRBP in the presence of oleic acid ranging in concentration from 0.67 to 2.66 μM. The titrations, which monitored retinol fluorescence enhancement, are shown in Figure 9. For both 11-cis and all-trans retinol, the presence of oleic acid depressed the degree of fluorescence enhancement for retinol without altering significantly the inflection point of the binding–titration curve. That is, although the degree of fluorescence at saturation dropped with increasing oleic acid, the curves did not converge at high ligand concentrations. This finding suggests that oleic acid inhibits the binding of retinol to X2IRBP in a noncompetitive manner. An allosteric mechanism is consistent with the likelihood that the hydrophobic cavity, the binding site of 11-cis and all-trans retinol, is not large enough to accommodate a molecule of oleic acid.
Figure 9.
Binding of all-trans and 11-cis retinol to X2IRBP (1 μM) in the presence of oleic acid. The titrations follow retinol fluorescence enhancement (excitation 330 nm; emission 480 nm). (A, B) Titrations for all-trans and 11-cis retinol, respectively, in the presence of different concentrations of oleic acid.
Discussion
Our reductionist approach of examining an individual module has the disadvantage of not taking into account the complete quaternary structure of the protein. However, focusing on the individual module reduces the complexity of the system, allowing insight into structure–function relationships that would be difficult to obtain in the full-length protein. Recently, we established conditions to produce pristine full-length Xenopus IRBP that supports the formation of diffraction-quality crystals. 54 We extend those studies to X2IRBP, by including DTT during the purification and concentration processes and introducing a TEV protease cleavage site allowing efficient removal of the fusion tag. These improvements afforded routine preparation of soluble X2IRBP free of fusion tags. Taking advantage of the fact that there is one tryptophan within each of the two putative retinol-binding pockets, we used site-directed mutagenesis and fluorescence spectroscopy to provide evidence that the hydrophobic cavity, and not the ββα-spiral fold, as previously thought, represents the binding site for both 11-cis and all-trans retinol.
X-ray crystallographic studies of X2IRBP have pointed to two hydrophobic domains representing candidate ligand-binding sites. In the original report of the X2IRBP structure, particular attention was given to a shallow cleft (see Fig. 1). Of interest, this cleft, which is formed by the ββα-spiral fold and also used by IRBP’s relatives, the crotonases and CPTases, to bind diverse hydrophobic ligands, was considered to be the most promising candidate for the retinol-binding site. However, in silico docking sites called attention to a second more hydrophobic and restricted hydrophobic cavity.38 There appears to be only one entrance into the cavity as illustrated (Fig. 1). This cavity, which may have structural differences between the modules,38 can accommodate a molecule of all-trans retinol, but is too small to accommodate a long chain fatty acid such as DHA. In the present study, we took advantage of the fact that, according to the crystal structure, both the hydrophobic cavity and shallow cleft contain a single tryptophan residue, W450 and W587, respectively. To probe these sites we substituted phenylalanine for tryptophan at each of these position. Phenylalanine, provides a highly conservative substitution. In contrast, the less conservative valine substitutions appear to destabilize the protein promoting insoluble expression.
Substituting phenylalanine for W450 and W587 does not significantly perturb 11-cis or all-trans retinol binding when monitoring retinol-fluorescence enhancement. However, the ability to support quenching was markedly reduced in the W450F mutant. In contrast, the W587F mutant showed little change, even when binding was determined by monitoring fluorescence quenching. These findings suggest that the hydrophobic cavity, not the shallow cleft, is the primary binding site for both 11-cis and all-trans retinol. That substitution of phenylalanine for W450 and W587 does not significantly perturb the overall structure of the ligand-binding site is supported by the energy-minimization studies and the fact that X2IRBP and the mutants were expressed and purified in a soluble monodispersed form. The effect of the substitution could be further addressed in future x-ray crystallographic studies.
The shallow cleft is a good candidate for a fatty acid–binding site. Previous studies have shown that X2IRBP binds the fluorescent fatty-acid analogue 9-(9-anthroyloxy) stearic acid with stoichiometry of N = 1.49 ± 0.15.38 Caution is warranted in interpreting these data, because the effect of the fluorescent tag is unknown. A recent study has measured the binding of fatty acids particularly to purified bovine IRBP by observing protein fluorescence quenching.31 However, we were not successful in using quenching to monitor DHA or oleic acid binding to X2IRBP. This is probably related to sensitivity, as these fatty acids do not absorb well at 330 nm making quenching measurements difficult. To examine the interaction of oleic acid with X2IRBP, we monitored the binding of 11-cis and all-trans retinol in the presence of various concentrations of oleic acid. We found that oleic acid inhibits the binding of both retinols. The inability of higher retinol concentrations to overcome the inhibitory effect suggests that oleic acid inhibits retinol binding through a noncompetitive mechanism. An allosteric mechanism is consistent with the likelihood that the hydrophobic cavity is not large enough to accommodate a molecule of oleic acid. We anticipate that the hydrophobic cavity is the site of 11-cis and all-trans retinol binding and that oleic acid binds within the shallow cleft. This model of separate retinol and fatty acid–binding sites in the same module is similar to that proposed for human IRBP.67 Ongoing x-ray crystallographic studies of the holoprotein will address this question.
Acknowledgments
Supported by Award Number I01BX007080 from the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development (FG-F); National Institutes of Health Grant EY09412 (FG-F, DG) and EY04939 (RKC); an unrestricted grant from Research to Prevent Blindness to the Department of Ophthalmology at SUNY at Buffalo; a Research Support grant at MUSC; Postdoctoral fellowship awards KDscf-030228-71 Song-Cheon Foundation through Kyung-Dong Pharmaceutical Co., and BK21, Department of Molecular Biology, School of Medicine, Kyung Hee University, Seoul, South Korea (K-IL). FG-F holds the Ira Gile Ross and Elizabeth Pierce Olmsted Ross Chair in Ophthalmic Pathology, SUNY at Buffalo.
The authors thank Andreas Loew for help during the early phases of the work, Tom Loeflem for preparing the expression construct, Joseph Armenia for assistance in the initial ligand-binding studies, and Myron Crawford and Fernando Pineda (Keck Center, Yale University, New Haven, CT) for performing the amino acid analyses.
Footnotes
Disclosure: F. Gonzalez-Fernandez, None; T. Bevilacqua, None; K.-I. Lee, None; R. Chandrashekar, None; L. Hsu, None; M.A. Garlipp, None; J.B. Griswold, None; R.K. Crouch, None; D. Ghosh, None
References
- 1.Lamb TD, Pugh EN., Jr Phototransduction, dark adaptation, and rhodopsin regeneration the proctor lecture. Invest Ophthalmol Vis Sci. 2006;47(12):5138–5152. doi: 10.1167/iovs.06-0849. [DOI] [PubMed] [Google Scholar]
- 2.McBee JK, Palczewski K, Baehr W, Pepperberg DR. Confronting complexity: the interlink of phototransduction and retinoid metabolism in the vertebrate retina. Prog Retin Eye Res. 2001;20(4):469–529. doi: 10.1016/s1350-9462(01)00002-7. [DOI] [PubMed] [Google Scholar]
- 3.Lamb TD, Pugh EN. Dark adaptation and the retinoid cycle of vision. Prog Retina Eye Res. 2004;23(3):307–380. doi: 10.1016/j.preteyeres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Marmor MF, Martin LJ. 100 years of the visual cycle. Surv Ophthalmol. 1978;22(4):279–285. doi: 10.1016/0039-6257(78)90074-7. [DOI] [PubMed] [Google Scholar]
- 5.Thompson DA, Gal A. Vitamin A metabolism in the retinal pigment epithelium: genes, mutations, and diseases. Prog Retin Eye Res. 2003;22(5):683–703. doi: 10.1016/s1350-9462(03)00051-x. [DOI] [PubMed] [Google Scholar]
- 6.Mata N, Radu R, Clemmons R, Travis G. Isomerization and oxidation of vitamin a in cone-dominant retinas: a novel pathway for visual-pigment regeneration in daylight. Neuron. 2002;36(1):69–80. doi: 10.1016/s0896-6273(02)00912-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolf G. The visual cycle of the cone photoreceptors of the retina. Nutr Rev. 2004;62(7):283–286. doi: 10.1111/j.1753-4887.2004.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 8.Mata NL, Ruiz A, Radu RA, Bui TV, Travis GH. Chicken retinas contain a retinoid isomerase activity that catalyzes the direct conversion of all-trans-retinol to 11-cis-retinol. Biochemistry. 2005;44(35):11715–11721. doi: 10.1021/bi050942m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trevino SG, Villazana-Espinoza ET, Muniz A, Tsin AT. Retinoid cycles in the cone-dominated chicken retina. J Exp Biol. 2005;208:4151–4157. doi: 10.1242/jeb.01881. [DOI] [PubMed] [Google Scholar]
- 10.Muniz A, Villazana-Espinoza ET, Hatch AL, Trevino SG, Allen DM, Tsin AT. A novel cone visual cycle in the cone-dominated retina. Exp Eye Res. 2007;85(2):175–184. doi: 10.1016/j.exer.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Fernandez F. Interphotoreceptor retinoid-binding protein: an old gene for new eyes. Vision Res. 2003;43(28):3021–3036. doi: 10.1016/j.visres.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 12.Pepperberg DR, Okajima TL, Wiggert B, Ripps H, Crouch RK, Chader GJ. Interphotoreceptor retinoid-binding protein (IRBP): molecular biology and physiological role in the visual cycle of rhodopsin. Mol Neurobiol. 1993;7(1):61–85. doi: 10.1007/BF02780609. [DOI] [PubMed] [Google Scholar]
- 13.Liou GI, Geng L, Baehr W. Interphotoreceptor retinoid-binding protein: biochemistry and molecular biology. Prog Clin Biol Res. 1991;36:2115–2137. [PubMed] [Google Scholar]
- 14.Nickerson JM, Borst DE, Redmond TM, Si JS, Toffenetti J, Chader GJ. The molecular biology of IRBP: application to problems of uveitis, protein chemistry, and evolution. Prog Clin Biol Res. 1991:362139–161. [PubMed] [Google Scholar]
- 15.Gonzalez-Fernandez F, Ghosh D. Focus on molecules: interphotoreceptor retinoid-binding protein (IRBP) Exp Eye Res. 2008;86(2):169–170. doi: 10.1016/j.exer.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 16.den Hollander A, McGee TL, Ziviello C, et al. A homozygous missense mutation in the IRBP gene (RBP3) associated with autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2009;50(4):1864–1872. doi: 10.1167/iovs.08-2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Q, Blakeley LR, Cornwall MC, Crouch RK, Wiggert BN, Koutalos Y. Interphotoreceptor retinoid-binding protein is the physiologically relevant carrier that removes retinol from rod photoreceptor outer segments. Biochemistry. 2007;46(29):8669–8679. doi: 10.1021/bi7004619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qtaishat NM, Wiggert B, Pepperberg DR. Interphotoreceptor retinoid-binding protein (IRBP) promotes the release of all-trans retinol from the isolated retina following rhodopsin bleaching illumination. Exp Eye Res. 2005:81455–1463. doi: 10.1016/j.exer.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Ala-Laurila P, Kolesnikov AV, Crouch RK, et al. Visual cycle: dependence of retinol production and removal on photoproduct decay and cell morphology. J Gen Physiol. 2006;128(2):153–169. doi: 10.1085/jgp.200609557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kolesnikov AV, Ala-Laurila P, Shukolyukov SA, et al. Visual cycle and its metabolic support in gecko photoreceptors. Vision Res. 2007;47(3):363–374. doi: 10.1016/j.visres.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 21.Okajima TI, Pepperberg DR, Ripps H, Wiggert B, Chader GJ. Interphotoreceptor retinoid-binding protein: role in delivery of retinol to the pigment epithelium. Exp Eye Res. 1989;49(4):629–644. doi: 10.1016/s0014-4835(89)80059-4. [DOI] [PubMed] [Google Scholar]
- 22.Carlson A, Bok D. Promotion of the release of 11-cis-retinal from cultured retinal pigment epithelium by interphotoreceptor retinoid-binding protein. Biochemistry. 1992;31(37):9056–9062. doi: 10.1021/bi00152a049. [DOI] [PubMed] [Google Scholar]
- 23.Okajima TI, Wiggert B, Chader GJ, Pepperberg DR. Retinoid processing in retinal pigment epithelium of toad (Bufo marinus) J Biol Chem. 1994;269(35):21983–21989. [PubMed] [Google Scholar]
- 24.Okajima TI, Pepperberg DR, Ripps H, Wiggert B, Chader GJ. Interphotoreceptor retinoid-binding protein promotes rhodopsin regeneration in toad photoreceptors. Proc Natl Acad Sci U S A. 1990;87(17):6907–6911. doi: 10.1073/pnas.87.17.6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin M, Li S, Nusinowitz S, et al. The role of interphotoreceptor retinoid-binding protein on the translocation of visual retinoids and function of cone photoreceptors. J Neurosci. 2009;29(5):1486–1495. doi: 10.1523/JNEUROSCI.3882-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adler AJ, Spencer SA. Effect of light on endogenous ligands carried by interphotoreceptor: retinoid-binding protein. Exp Eye Res. 1991;53(3):337–346. doi: 10.1016/0014-4835(91)90239-b. [DOI] [PubMed] [Google Scholar]
- 27.Lin ZS, Fong SL, Bridges CD. Retinoids bound to interstitial retinol-binding protein during light and dark-adaptation. Vision Res. 1989;29(12):1699–1709. doi: 10.1016/0042-6989(89)90152-1. [DOI] [PubMed] [Google Scholar]
- 28.Bazan NG, Reddy TS, Redmond TM, Wiggert B, Chader GJ. Endogenous fatty acids are covalently and noncovalently bound to interphotoreceptor retinoid-binding protein in the monkey retina. J Biol Chem. 1985;260(25):13677–13680. [PubMed] [Google Scholar]
- 29.Chen Y, Houghton LA, Brenna JT, Noy N. Docosahexaenoic acid modulates the interactions of the interphotoreceptor retinoid-binding protein with 11-cis-retinal. J Biol Chem. 1996;271(34):20507–20515. doi: 10.1074/jbc.271.34.20507. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, Saari JC, Noy N. Interactions of all-trans-retinol and long-chain fatty acids with interphotoreceptor retinoid-binding protein. Biochemistry. 1993;32(42):11311–11318. doi: 10.1021/bi00093a007. [DOI] [PubMed] [Google Scholar]
- 31.Semenova EM, Converse CA. Comparison between oleic acid and docosahexaenoic acid binding to interphotoreceptor retinoid-binding protein. Vision Res. 2003;43(28):3063–3067. doi: 10.1016/j.visres.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Crouch RK, Hazard ES, Lind T, Wiggert B, Chader G, Corson DW. Interphotoreceptor retinoid-binding protein and alpha-tocopherol preserve the isomeric and oxidation state of retinol. Photochem Photobiol. 1992;56(2):251–255. doi: 10.1111/j.1751-1097.1992.tb02154.x. [DOI] [PubMed] [Google Scholar]
- 33.Nickerson JM, Frey RA, Ciavatta VT, Stenkamp DL. Interphotoreceptor retinoid-binding protein gene structure in tetrapods and teleost fish. Mol Vis. 2006;12:1565–1585. [PMC free article] [PubMed] [Google Scholar]
- 34.Rajendran RR, Van Niel EE, Stenkamp DL, Cunningham LL, Raymond PA, Gonzalez-Fernandez F. Zebrafish interphotoreceptor retinoid-binding protein: differential circadian expression among cone subtypes. J Exp Biol. 1996;199(12):2775–2787. doi: 10.1242/jeb.199.12.2775. [DOI] [PubMed] [Google Scholar]
- 35.Wagenhorst BB, Rajendran RR, Van Niel EE, Hessler RB, Bukelman A, Gonzalez-Fernandez F. Goldfish cones secrete a two-repeat interphotoreceptor retinoid-binding protein. J Mol Evol. 1995;41(5):646–656. doi: 10.1007/BF00175823. [DOI] [PubMed] [Google Scholar]
- 36.Dettai A, Lecointre G. New insights into the organization and evolution of vertebrate IRBP genes and utility of IRBP gene sequences for the phylogenetic study of the Acanthomorpha (Actinopterygii Teleostei) Mol Phylogenet Evol. 2008;48(1):258–269. doi: 10.1016/j.ympev.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 37.Loew A, Baer C, Gonzalez-Fernandez F. The functional unit of interphotoreceptor retinoid-binding protein (IRBP): purification, characterization and preliminary crystallographic analysis. Exp Eye Res. 2001;73(2):257–264. doi: 10.1006/exer.2001.1031. [DOI] [PubMed] [Google Scholar]
- 38.Gonzalez-Fernandez F, Baer CA, Ghosh D. Module structure of interphotoreceptor retinoid-binding protein (IRBP) may provide bases for its complex role in the visual cycle: structure/function study of Xenopus IRBP. BMC Biochem. 2007;8(1):15. doi: 10.1186/1471-2091-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez-Fernandez F, Baer CA, Baker E, et al. Fourth module of Xenopus interphotoreceptor retinoid-binding protein: activity in retinoid transfer between the retinal pigment epithelium and rod photoreceptors. Curr Eye Res. 1998;17(12):1150–1157. doi: 10.1076/ceyr.17.12.1150.5129. [DOI] [PubMed] [Google Scholar]
- 40.Baer CA, Van Niel EE, Cronk JW, et al. Arginine to glutamine substitutions in the fourth module of Xenopus interphotoreceptor retinoid-binding protein. Mol Vis. 1998;4:30–43. [PubMed] [Google Scholar]
- 41.Baer CA, Retief JD, Van Niel E, Braiman MS, Gonzalez-Fernandez F. Soluble expression in E. coli of a functional interphotoreceptor retinoid-binding protein module fused to thioredoxin: correlation of vitamin A binding regions with conserved domains of C-terminal processing proteases. Exp Eye Res. 1998;66(2):249–262. doi: 10.1006/exer.1997.0418. [DOI] [PubMed] [Google Scholar]
- 42.Baer CA, Kittredge KL, Klinger AL, Briercheck DM, Braiman MS, Gonzalez-Fernandez F. Expression and characterization of the fourth repeat of Xenopus interphotoreceptor retinoid-binding protein in E. coli. Curr Eye Res. 1994;13(6):391–400. doi: 10.3109/02713689408999866. [DOI] [PubMed] [Google Scholar]
- 43.Gross EA, Li GR, Lin ZY, et al. Prediction of structural and functional relationships of Repeat 1 of human interphotoreceptor retinoid-binding protein (IRBP) with other proteins. Mol Vis. 2000;6:30–39. [PubMed] [Google Scholar]
- 44.Nickerson JM, Li GR, Lin ZY, Takizawa N, Si JS, Gross EA. Structure-function relationships in the four repeats of human interphotoreceptor retinoid-binding protein (IRBP) Mol Vis. 1998;4:33. [PubMed] [Google Scholar]
- 45.Loew A, Gonzalez-Fernandez F. Crystal structure of the functional unit of interphotoreceptor retinoid binding protein. Structure. 2002;10(1):43–49. doi: 10.1016/s0969-2126(01)00698-0. [DOI] [PubMed] [Google Scholar]
- 46.Liao DI, Qian J, Chisholm DA, Jordan DB, Diner BA. Crystal structures of the photosystem II D1 C-terminal processing protease. 2000;7(9):749–753. doi: 10.1038/78973. [DOI] [PubMed] [Google Scholar]
- 47.Blankenship RE. Molecular Mechanisms of Photosynthesis. Williston, MA: Blackwell Science; 2002. [Google Scholar]
- 48.Jemth P, Gianni S. PDZ domains: folding and binding. Biochemistry. 2007;46(30):8701–8708. doi: 10.1021/bi7008618. [DOI] [PubMed] [Google Scholar]
- 49.Modis Y, Filppula SA, Novikov DK, Norledge B, Hiltunen JK, Wierenga RK. The crystal structure of dienoyl-CoA isomerase at 1.5 A resolution reveals the importance of aspartate and glutamate sidechains for catalysis. Structure. 1998;6(8):957–970. doi: 10.1016/s0969-2126(98)00098-7. [DOI] [PubMed] [Google Scholar]
- 50.Engel CK, Mathieu M, Zeelen JP, Hiltunen JK, Wierenga RK. Crystal structure of enoyl-coenzyme A (CoA) hydratase at 2.5 angstroms resolution: a spiral fold defines the CoA-binding pocket. EMBO J. 1996;15(19):5135–5145. [PMC free article] [PubMed] [Google Scholar]
- 51.Engel CK, Kiema TR, Hiltunen JK, Wierenga RK. The crystal structure of enoyl-CoA hydratase complexed with octanoyl-CoA reveals the structural adaptations required for binding of a long chain: fatty acid-CoA molecule. J Mol Biol. 1998;275(5):847–859. doi: 10.1006/jmbi.1997.1491. [DOI] [PubMed] [Google Scholar]
- 52.Locke BC, MacInnis JM, Qian S, et al. Fluorescence studies of rat cellular retinol binding protein II produced in Escherichia coli: an analysis of four tryptophan substitution mutants. Biochemistry. 1992;31(8):2376–2383. doi: 10.1021/bi00123a024. [DOI] [PubMed] [Google Scholar]
- 53.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2. New York: Kluwer Academic; 1999. Protein fluorescence; pp. 445–486. [Google Scholar]
- 54.Ghosh D, Griswold JB, Bevilacqua T, Gonzalez-Fernandez F. Purification of the full-length Xenopus interphotoreceptor retinoid binding protein and growth of diffraction-quality crystals. Mol Vis. 2007;13:2275–2281. [PMC free article] [PubMed] [Google Scholar]
- 55.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18(20):6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haun RS, Serventi IM, Moss J. Rapid, reliable ligation-independent cloning of PCR products using modified plasmid vectors. Biotechniques. 1992;13(4):515–518. [PubMed] [Google Scholar]
- 57.Carrington JC, Dougherty WG. A viral cleavage site cassette: identification of amino acid sequences required for tobacco etch virus polyprotein processing. Proc Natl Acad Sci U S A. 1988;85(10):3391–3395. doi: 10.1073/pnas.85.10.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Szuts EZ, Harosi FI. Solubility of retinoids in water. Arch Biochem Biophys. 1991;287(2):297–304. doi: 10.1016/0003-9861(91)90482-x. [DOI] [PubMed] [Google Scholar]
- 59.Hubbard R, Brown PK, Bownds D. Methodology of vitamin A and visual pigments. Methods Enzymol. 1971;18:615–643. [Google Scholar]
- 60.Moiseyev G, Crouch RK, Goletz P, Oatis J, Jr, Redmond TM, Ma JX. Retinyl esters are the substrate for isomerohydrolase. Biochemistry. 2003;42(7):2229–2238. doi: 10.1021/bi026911y. [DOI] [PubMed] [Google Scholar]
- 61.Ward LD. Measurement of ligand binding to protein by fluorescence spectroscopy. Methods Enzymol. 1985;117:400–414. doi: 10.1016/s0076-6879(85)17024-2. [DOI] [PubMed] [Google Scholar]
- 62.Cogan U, Kopelman M, Mokady S, Shinitzky M. Binding affinities of retinol and related compounds to retinol binding proteins. Eur J Biochem. 1976;65(1):71–78. doi: 10.1111/j.1432-1033.1976.tb10390.x. [DOI] [PubMed] [Google Scholar]
- 63.Mertens ML, Kagi JH. A graphical correction procedure for inner filter effect in fluorescence quenching titrations. Anal Biochem. 1979;96(2):448–455. doi: 10.1016/0003-2697(79)90605-5. [DOI] [PubMed] [Google Scholar]
- 64.Kiefhaber T, Rudolph R, Kohler HH, Buchner J. Protein aggregation in vitro and in vivo: a quantitative model of the kinetic competition between folding and aggregation. Biotechnology. 1991;9(9):825–829. doi: 10.1038/nbt0991-825. [DOI] [PubMed] [Google Scholar]
- 65.Ferreira ST, De Felice FG, Chapeaurouge A. Metastable, partially folded states in the productive folding and in the misfolding and amyloid aggregation of proteins. Cell Biochem Biophys. 2006;44(3):539–548. doi: 10.1385/CBB:44:3:539. [DOI] [PubMed] [Google Scholar]
- 66.Smith AV, Hall CK. Protein refolding versus aggregation: computer simulations on an intermediate-resolution protein model. J Mol Biol. 2001;312(1):187–202. doi: 10.1006/jmbi.2001.4845. [DOI] [PubMed] [Google Scholar]
- 67.Lin ZY, Li GR, Takizawa N, et al. Structure-function relationships in interphotoreceptor retinoid-binding protein (IRBP) Mol Vis. 1997;3:17. [PubMed] [Google Scholar]