Abstract
Transmission of metabolic diseases from mother to child is multifactorial and includes genetic, epigenetic, and environmental influences. Evidence in rodents, humans and non-human primates support the scientific premise that exposure to maternal obesity or high-fat diet during pregnancy creates a long-lasting metabolic signature on the infant innate immune system and the juvenile microbiota which predisposes the offspring to obesity and metabolic diseases. In neonates, gastrointestinal microbes introduced through the mother are noted for their ability to serve as direct inducers/regulators of the infant immune system. Neonates have a limited capacity to initiate an immune response. Thus, disruption of microbial colonization during the early neonatal period results in disrupted postnatal immune responses that highlight the neonatal period as a critical developmental window. Although the mechanisms are poorly understood, increasing evidence suggests that maternal obesity or poor diet influences the development and modulation of the infant liver and other end-organs through direct communication via the portal system, metabolite production, alterations in gut barrier integrity, and the hematopoietic immune cell axis. This review will focus on how maternal obesity and dietary intake influence the composition of the infant gut microbiota and how an imbalance or maladaptation in the microbiota, including changes in early pioneering microbes, might contribute to the programming of offspring metabolism with special emphasis on mechanisms that promote chronic inflammation in the liver. Comprehension of these pathways and mechanisms will elucidate our understanding of developmental programming, and may expand the avenue of opportunities for novel therapeutics.
Keywords: Obesity, Metabolism, Pregnancy, Neonatal, NAFLD
Introduction
Nearly two-thirds of American women of childbearing age are overweight or obese and greater than half these women, once pregnant, have excess gestational weight gain (GWG) (Deputy et al. 2015; Nicklas and Barbour 2015). Alterations in maternal microbiota composition, associated with both diet and obesity status, have been widely reported (Collado et al. 2008; Collado et al. 2012; Paul et al. 2016; Santacruz et al. 2010). Disruption of the mother’s gut microbiota can influence early pioneering bacteria in the newborn and may be a prime candidate for intergenerational transmission of metabolic disease risk given the considerable influence these microbes have on gut function and immune system development. The newborn gut begins as an aerobic environment, allowing only certain microbes to temporarily take up residence, followed by dramatic fluctuations in microbe composition with introduction of breast milk and solid foods (Voreades et al. 2014). Recent studies have proposed that colonization of the gastrointestinal tract begins in utero (Aagaard et al. 2014; Collado et al. 2016; Jimenez et al. 2008); however, this idea remains controversial, as discussed in detail by Perez-Muñoz, et al. (Perez-Muñoz et al. 2017). Regardless, the first major seeding of the gut microbiota is thought to occur during the birthing process following exposure to bacteria residing in the mother’s vaginal canal and gut during vaginal birth or the mother’s skin during a cesarean section (Dominguez-Bello et al. 2010). The mother’s influence on neonatal acquisition of gut microbes continues by direct contact with the mother’s skin and through breastfeeding, which provides critical nutrients to the colonizing bacteria and is a source of new bacteria. The diversity and density of bacteria continue to expand in response to new environmental exposures throughout the first few years of life until reaching a stable adult-like microbiota.
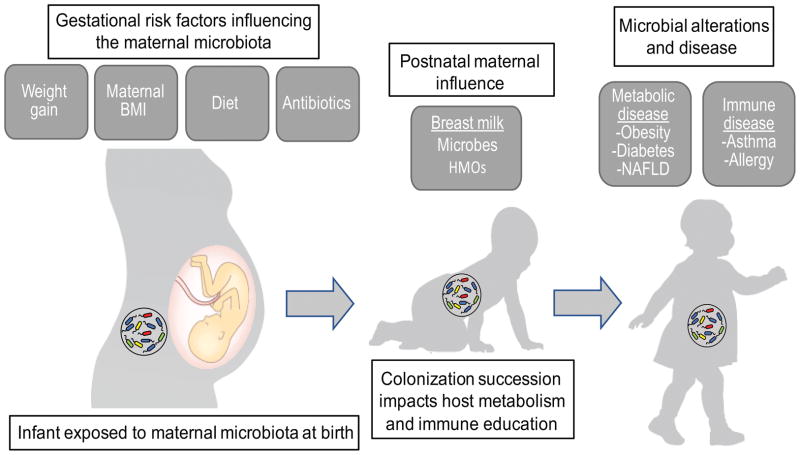
Several essential functions for human health develop in parallel with gut microbe expansion, including vitamin biosynthesis, energy extraction from the diet, gut barrier function, and immune system maturation/education. The immune system of neonates is immature and requires the exposure of gut bacteria to develop properly. Depending on the health of the mother, maternal microbe communities may already be imbalanced when passed on to the infant. In children, metabolic diseases, including obesity, insulin resistance and nonalcoholic fatty liver disease (NAFLD), along with other related immune-based diseases, are associated with alterations in infant gut microbe composition (Fig 1); however, the mechanisms remain unclear. This review will focus on maternal obesity, excessive GWG, and high-fat diet consumption during pregnancy and their impact on the microbiota of both mother and infant, including their links to early gut colonization and innate immunity in the infants that drive an increased risk for common metabolic diseases.
Figure 1.
Influential factors on the maternal and infant microbiota. Maternal factors significantly contribute to the initial colonization and succession of the infant gut microbiota. Alterations in this process may have long term health consequences related to host metabolism and immune education. HMOs, human milk oligosaccharides; NAFLD, nonalcoholic fatty liver disease.
Development of the infant microbiota and immune tolerance
The gut is the most densely populated of all the sites of microbial colonization, containing over 10 trillion bacteria cells, represented by more than 1000 different bacterial species. Mammals have evolved a mutual relationship with gut microbes, commonly referred to as “commensal”. Colonization of the infant gut by these commensals during the first few years of life is a time of significant fluctuation and maturation. Early alterations in the normal succession of the gut microbiota may have an impact on future colonization (Eggesbø et al. 2011), which could have lasting health consequences.
Commensal colonization progresses through a choreographed succession of bacterial species, and evolves rapidly during the first months of life (Del Chierico et al. 2015; Hill et al. 2017). Systemic responses to commensal microbiota are essential for the maintenance of beneficial host-microbiota interactions. For example, maternally acquired IgA and IgG in neonatal mice leads to dampened T cell–dependent immune responses against commensal bacteria (Koch et al. 2016). Additionally, systemic IgG responses to Gram-negative bacteria, acquired early and over the course of life, were shown to provide cross-protection against Gram-negative pathogens, such as Escherichia coli and Salmonella, in mice (Zeng et al. 2016). These observations reinforce the concept that microbial composition and the timing of host-commensal interactions provide the foundation for balanced immunity in the intestine.
Under healthy conditions, vaginally-delivered infants born to normal-weight mothers are initially colonized by mostly facultative anaerobic bacteria including Staphylococcus, Streptococcus, E. coli, and Enterobacteria. These anaerobes make the habitat suitable for colonization by strict anaerobes including Bacteroides, Bifidobacterium, and Clostridium (Palmer et al. 2007; Penders et al. 2006), by consuming oxygen, altering the pH, lowering the redox potential, and producing carbon dioxide and nutrients. Jakobsson, et al. (Jakobsson et al. 2014) reported a similar succession of colonization, including an initial blooming of Proteobacteria that gradually declined from 1 week to 24 months, Firmicutes that expanded from 3 months onward and a peak of Actinobacteria at 3 months. This pattern of colonization was similar in infants born by cesarean section, except the notably higher proportion of Bacteroidetes in vaginally-delivered infants during the first 12 months of life. E. coli is a member of the Proteobacteria phylum and is the most abundant facultative anaerobe in the mammalian gut microbiota, but under homeostatic conditions, this species represents only a minor fraction of the ecosystem. Nevertheless, Proteobacteria abundance may explain significant functional variability in the human gut microbiome (Bradley and Pollard 2017). Importantly, E. coli (like many other members of the family Enterobacteriaceae) has a short doubling time and a highly flexible metabolic capacity. These attributes make this bacterium highly adaptable and allow it to bloom in the presence of oxygen and nitrogen.
The importance of Proteobacteria to immune education in early life
An emerging paradigm, originally known as “The Restaurant Hypothesis” (Leatham-Jensen et al. 2012), posits that nutritional stress may alter the original colonizing bacteria, including E. coli; therefore, the signaling pathways controlled by microbial mediators may cause fine-tuning in the gut, potentially altering the immune system in early life. Several studies suggest that the critical immune mechanisms controlling the maintenance of homeostasis and tolerance to environmental exposures are determined by microbial-host interactions occurring during a narrow time frame contained within the earliest days of life (Koch et al. 2016; Zeng et al. 2016). The early colonization of Proteobacteria and its subsequent reduction in abundance over time is thought to be an important part of this process, whereby a disruption of this progression has been linked to an increased risk of neonatal diseases, particularly in pre-term infants (Neu 2015, 2016). The Proteobacteria bloom in the neonatal gut is likely under maternal control. Human milk oligosaccharides (HMOs) (De Leoz et al. 2015) and secretory IgA production (Mirpuri et al. 2014) are involved in the selective suppression of Proteobacteria during establishment of the early gut flora.
Proteobacteria are believed to be important contributors to inflammation associated with metabolic disease in adults, and their role in infant immunity is critical for early priming of the innate and adaptive immune system. A primary structural component of Gram-negative bacteria residing in the gut is the endotoxin lipopolysaccharide (LPS). Translocation of LPS from the gut into the systemic circulation has been widely reported in metabolic disease (Cani et al. 2007; Lassenius et al. 2011; Ruiz et al. 2007). A common theme that has recently emerged is that variability in the lipid A component of LPS can tip the balance of innate immune responses towards homeostatic tolerance or pro-inflammatory signaling, affecting adaptive immune responses that either alter or inhibit protective signaling normally induced by toll-like receptors (TLRs). Lipid A, the biologically active moiety of LPS, can be expressed in variant forms by many human pathogens, allowing for evasion of the host innate immune system and establishment of a chronic infection. Strikingly, several of these host-adapted Gram-negative bacteria that express immune-evasive lipid A are associated with increased risk of autoimmune disease (Vatanen et al. 2016), type I diabetes, atherosclerosis (Erridge et al. 2007b; Slocum et al. 2014), and allergic disorders in childhood and adulthood (Grönlund et al. 2000; Ogra and Welliver 2008). A role of lipid A variants is to allow a bacterium to evade TLR4 and promote chronic inflammation and these variants appear to act through dysregulation of both innate and adaptive immune responses (Kramer and Genco 2017).
While a close relationship exists between microbial flora and the intestinal epithelium, the mechanisms whereby the mucosal surface senses the presence of colonizing microbial organisms and allows for innate immune receptor-mediated responses is largely unknown (Hooper et al. 1999). Recognition of microbial structures by mammalian cells occurs through various transmembrane receptor molecules that are part of the innate immune system. TLRs recognize conserved microbial structures, such as LPS, mediate cellular activation, and thereby provide the costimulatory signal required for an efficient immune response (Medzhitov 2001). Initial exposure to bacteria and LPS in early infancy has been shown to contribute to the development of the immune system by educating it on how to appropriately response to bacteria and their components. Different bacterial species have variations in their LPS structure that impact their ability to elicit an innate immune response (Vatanen et al. 2016; Whitfield and Trent 2014).
The immunological phenomenon wherein prior exposure to TLR ligands renders macrophages refractory to LPS-elicited pro-inflammatory cytokine secretion is known as endotoxin tolerance (Biswas et al. 2007). Endotoxin tolerance is classically induced by exposure to low levels of LPS, thereby rendering the cells “tolerant” to subsequent endotoxin challenge, characterized functionally by a marked inhibition of inflammatory cytokine (e.g. TNF-α, IL-1, and IL-6) production upon re-challenge with endotoxin (Erroi et al. 1993; Granowitz et al. 1993). This immune-regulatory process of induction is also associated with a unique metabolic phenotype in macrophages that facilitates persistent improvements in antimicrobial function (phagocytosis) against Gram-negative bacteria (Coveney et al. 2015; Fensterheim et al. 2017). Immediately following TLR stimulation, macrophages shift their metabolic profile to one that favors glycolysis over oxidative metabolism (Everts et al. 2014) promoting a pro-inflammatory phenotype. In contrast, anti-inflammatory macrophages exhibit a metabolic program of augmented oxidative phosphorylation (Kelly and O’Neill 2015). The lack of appropriate alterations in early immune education may affect the overall functionality of the microbiota and macrophages thereby altering inflammation and key pathogens that can drive shifts in the community composition into dysbiosis later in life.
We have recently shown in a prospective study on vaginally-delivered, exclusively breastfed infants with no neonatal or postnatal exposure to antibiotics that neonates born to mothers with obesity showed a significant 50% reduction in Gammaproteobacteria at 2 weeks of age compared with infants of normal-weight mothers (Lemas et al. 2016). A striking relative depletion in Proteobacteria species was recently found in 2-day-old neonates delivered vaginally, but not by cesarean section, to overweight/obese mothers (Mueller et al. 2016), suggesting the differences in relative abundance of Gram-negative bacteria may stem from vertical transmission of the maternal microbiota. It is also important to note that maternal overweight/obesity could be due to a mix of environmental as well as genetic factors not accounted for, such as excess GWG or poor diet, both of which likely has its own effect on microbial community structure. Based on studies in mice (Mirpuri et al. 2014), induction of a Gammaproteobacteria-specific IgA response partially regulated the transition from a neonatal to a mature microbiota. Experiments in germ-free mice showed that colonization with microbes from mice lacking IgA had persistent increased colonization with Gammaproteobacteria that resulted in sustained intestinal inflammation and increased susceptibility to neonatal and adult models of intestinal injury. Thus, the effects of a relative depletion of intestinal Proteobacteria species in neonates of mothers with obesity could cause persistent alterations in immune development and increase an infant’s risk of developing inflammatory and metabolic diseases, but the literature on this topic in full-term human neonates is scarce.
One theory for why mothers with obesity have infants with reduced Proteobacteria and potentially more inflammation could be as simple as micronutrient deficiency. Although not universal, mothers with obesity and those with excess GWG are iron-deficient (Dosch et al. 2016; Jones et al. 2016), as well as their infants. A wide range of host immune responses are intricately linked with both systemic and cellular iron homeostasis. Perturbations to the labile iron pool have been linked with modulating various forms of innate and adaptive immune functions including proliferation, differentiation and secretion of inflammatory mediators (Ellermann and Arthur 2017). Processes associated with inflammation, including cytokine production and engagement of TLRs by microbial ligands, stimulate the production of the hormone hepcidin, which in turn modulates systemic iron homeostasis by limiting dietary iron absorption and increasing intracellular retention of iron (Drakesmith and Prentice 2012; Nemeth et al. 2004a; Nemeth et al. 2004b). Given that bacteria can secrete small molecules known as siderophores that can solubilize iron for host cells (Deriu et al. 2013), the opportunity is present for resident bacteria to modulate host immune responses at the mucosal interface. Iron has also been shown to modulate butyrate-producing bacteria (Tang et al. 2016), as well as pathogenic Proteobacteria (Kamada et al. 2013). Whether iron deficiency has a direct effect on early microbiota colonization and inflammation is a hypothesis worthy of further study.
Maternal gestational weight status and dietary intake impacts the development of the infant microbiota
The mother has a direct role in initially colonizing the infant microbiota as is evidenced by the fact that infants born vaginally have different microbiota compositions than infants born by cesarean sections (Dominguez-Bello et al. 2010). While it is unclear whether these early differences in microbiota composition affect the subsequent commensal colonization into childhood and beyond, it is clear the mother’s microbes do contribute to the colonization process. The gut microbiota has been shown to be altered with advancing gestation in normal-weight mothers, and when these microbes were transferred into germ-free mice, the mice receiving microbes from mothers in the 3rd trimester showed increased weight gain, inflammation, and insulin resistance (Koren et al. 2012). It should come as no surprise that conditions like obesity, weight gain, and diet also alter the maternal microbiota composition during pregnancy, but whether this has a significant impact on metabolic disease risk is still under investigation.
To date there are only a few studies that have explored the impact of maternal obesity and diet on maternal and infant microbiota, but the existing data suggests there is a significant impact. Collado, et al. (Collado et al. 2008) found that women that were obese prior to pregnancy had significantly different gut microbiota compositions compared with normal-weight pregnant women during both the 1st and 3rd trimesters of pregnancy. Furthermore, women that gained excessive gestational weight regardless of pre-pregnancy BMI had significant differences in their microbiota composition compared with women who had normal weight gain. Specifically, Bacteroides and Staphylococcus were found to be significantly higher in obese women and Bacteroides species were elevated in all women with excessive GWG. Santacruz, et al. (Santacruz et al. 2010) also found a difference in gut microbiota composition between normal-weight and overweight women. Overweight women were found to have significantly more Staphylococcus, Enterobacteriaceae and E. coli and fewer Bifidobacterium and Bacteroides compared with normal-weight pregnant women. They also reported that women with excessive GWG had significantly more E. coli and fewer Bifidobacterium and Akkermansia muciniphila than in women with normal weight gain during pregnancy. While there are some inconsistencies in the results of these studies, it is important to note that the samples were taken at different times during pregnancy and that one study compared normal-weight women to women with obesity and the other compared normal-weight and overweight women, which could account for the different findings. While the exact changes that occur in the microbiota of pregnant women with obesity are unclear, the important observation is that their microbiota is different from normal-weight pregnant women and that these differences (whether due to diet or body habitus) could influence the microbial colonization of the infant, as noted earlier. Collado, et al. (Collado et al. 2010) compared the microbiota of infants at 1 and 6 months of age that were born to overweight mothers (based on pre-pregnancy BMI) versus mothers that were normal weight. Infants born to overweight mothers were found to have increased Bacteroides and Staphylococcus at 1 and 6 months of age. This observation coincides with what this group showed in mothers during pregnancy (Collado et al. 2008), providing evidence for the mother’s role in directing the early infant microbiota composition. Maternal obesity has also been associated with long term changes in the offspring microbiota. Galley, et al. (Galley et al. 2014) found that toddlers born to mothers with obesity had significantly elevated levels of Oscillibacter, Parabacteroides, and an unclassified Bacteroidales genus, and reduced levels of Eubacterium and Blautia compared with toddlers born to normal-weight mothers; however, these studies did not control for mode of delivery, antibiotic use, or breastfeeding exposure.
Maternal diet has also been shown to have a significant impact on the microbiota composition of the offspring. Recently, Chu, et al. (Chu et al. 2016) showed that a maternal high-fat diet, estimated from retrospective surveys, resulted in a depletion of Bacteroides and an enrichment of Enterococcus in the meconium and a trend for reduction of Bacteroides at 6 weeks of age. Evidence of the impact on maternal diet has also been reported in animal models including both rodent and non-human primate models. In a study by Myles, et al. (Myles et al. 2013), feeding pregnant mouse dams a Western-style diet resulted in an altered microbiota composition in the offspring including an increase in Clostridiales (a class of Firmicutes), despite being weaned onto a control diet. In addition to having an altered microbiota composition, these offspring also had worse outcomes in models of infection, autoimmunity, and allergic sensitization. Importantly, in non-human primates, infants born to mothers fed a high-fat diet during gestation and lactation (whether obese or not) showed significant intestinal dysbiosis that was not completely corrected by switching to a control diet after weaning. This has important clinical relevance as the early colonized bacteria, driven by maternal diet, may have a long-lasting effect on the commensal microbe population in offspring, therefore setting the stage for increased risk for immunologic and metabolic disease patterning.
The important role of breast milk in colonization of the infant gut microbiota
The impact of the mother’s microbiota on the colonization of the infant gut continues after initial colonization at birth through breastfeeding. Breast milk is a source of live bacteria and contains energy sources for colonizing bacteria. Breast milk has also been shown to play a role in preventing the colonization of pathogenic bacteria (Jantscher-Krenn et al. 2012). Assessment of the bacterial composition of breast milk in healthy women has revealed the presence of a wide variety of bacteria, including Staphylococcus, Streptococcus, Lactobacillus, Bifidobacterium, and E. coli (Heikkilä and Saris 2003; Martín et al. 2007). Conversely, breast milk from mothers with obesity has been shown to harbor a different and less diverse bacterial community than that of normal-weight subjects, including higher levels of Staphylococcus and A. muciniphila and lower levels of Bifidobacterium (Cabrera-Rubio et al. 2012; Collado et al. 2012), and a different composition of hormones, cytokines, and oligosaccharides (Andreas et al. 2014). The source of bacteria in breast milk is unclear, but there is evidence suggesting bacteria residing in the gut might be a source of the breast milk populations (Latuga et al. 2014; Rodríguez 2014).
HMOs are an important component of breast milk and given the fact that they are indigestible to the human host, they are now believed to exist for the primary purpose of providing nutrients to the microbes colonizing the infant gut. There are over 200 known HMO molecules that are present in human breast milk which vary across individuals (Ninonuevo et al. 2006). Recently, differences in HMO composition in mother’s milk have been associated with infant growth and body composition (Alderete et al. 2015).
Breastfeeding compared to formula feeding is thought to provide significant health benefits to an infant, specifically reducing the risk of obesity (Binns et al. 2016; Dieterich et al. 2013), where the risk is inversely related to the duration (Ip et al. 2009). Given the microbial and HMO differences found in the breast milk of obese women compared with normal-weight women, this benefit may not be universal, but more research is needed to determine if the benefits outweigh the risks. Support for this concern is evidenced by the fact that antibiotic use in infants born to normal-weight mothers increases the risk of obesity, whereas antibiotic use in infants born to mothers with obesity reduces the risk of obesity (Ajslev et al. 2011). This indirectly suggests that it might be advantageous to alter the composition of the gut microbiota in infants born to mothers with obesity.
Obesity and the programming of hematopoietic immune cells
The microbiota has been implicated in altering immune system function via changes in hematopoiesis. Dietary changes can have major effects on microbial composition and release of LPS that provokes inflammation, insulin resistance, and may even program bone marrow-derived stem cells (Giusti et al. 2017). The significant increase in common myeloid progenitor cells derived from the bone marrow in diet-induced obese mice is thought to be the result of an altered microbiota composition (Luo et al. 2015). Indeed, recent evidence has emerged to indicate that innate immune memory can be transferred via hematopoietic stem and progenitor cells that alter the function of their differentiated progeny (Ng et al. 2013), depending upon the composition of the microbiota (Burgess et al. 2014). Thevaranjan, et al. (Thevaranjan et al. 2017) showed that old mice had a distinct microbiota composition compared with young mice. Bone marrow-derived macrophages and whole blood from germ-free mice colonized with gut microbiota from the old mice produced increased levels of inflammatory cytokines following stimulation by LPS compared with young mice. Singer, et al. (Singer et al. 2014) found that high-fat diet increases the production of bone marrow-derived dendritic cells and peritoneal macrophages from obese mice and they produce more inflammatory cytokines including TNF-α and IL-6 following LPS stimulation than lean mice; their findings suggest that this pro-inflammatory profile persists after the obesogenic diet is removed.
Commensal bacteria at the crossroad between inflammation and childhood NAFLD
Maternal obesity and diabetes are among the most powerful predictors of childhood obesity and other adverse health outcomes such as NAFLD, that affects up to 34% of obese children (Anderson et al. 2015). A particularly alarming statistic is the prevalence of obesity in children, affecting 10% of infants and toddlers and 17% of children and adolescents in the United States (Ogden et al. 2012). With the rise of obesity, it is predicted that NAFLD will be the most common etiology for liver transplantation in the 21st century (Agopian et al. 2012). A cross-sectional study of 538 children with biopsy-proven NAFLD illustrated the critical importance of the in utero environment to development of pediatric NAFLD (Newton et al. 2017). They showed that children born with high or low birth weight had >2-fold significantly increased incidence of nonalcoholic steatohepatitis (NASH) or advanced fibrosis, respectively, compared with those of normal birth weight.
Evidence suggests that the transgenerational passage of the maternal obese phenotype with NAFLD, or developmental programming of the offspring, involves the innate immune system as well as metabolic perturbations by gut microbes (Wesolowski et al. 2017). Because early immune development is highly dependent on triggers provided by the microbiota (Ma et al. 2014), infants born to obese mothers might be exposed to products of gut dysbiosis that activate macrophages in the liver and bone marrow. However, the timing, mechanisms involved, and whether interventions that alter microbial colonization can prevent early innate immune system “programming” are unknown. Results from recent studies demonstrate that the maternal microbiota impels early postnatal innate immune development (Gomez de Agüero et al. 2016), while in adults the transmissibility of obesity and related disorders by gut bacteria is likely mediated specifically by cells of the innate immune system, a phenomenon that is observed in Rag1−/− mice lacking all cells of the adaptive immune system (Bäckhed et al. 2004).
The source of inflammation in macrophages associated with NAFLD is incompletely understood; however, increased circulating levels of the bacterial-derived endotoxin LPS is believed to play a role. Alterations in microbial composition have been associated with increased gut permeability, resulting in translocation of LPS. Binding of LPS to TLRs triggers the release of cytokines and an associated inflammatory response. LPS will bind to LPS binding protein (LBP) and that complex will bind to CD14, located in inflammatory cells. Together LPS-LBP-CD14 activates TLR4, which is present in hepatocytes, adipocytes, Kupffer cells, the most abundant macrophage in the body, and hepatic stellate cells, triggering the release of inflammatory cytokines (i.e. TNF-α, IL-1β, and IL-6). Furthermore, the translocation of LPS from the gut is not reserved to individuals with disease. Studies have shown the ingestion of meals containing fat increase systemic LPS concentrations in healthy individuals (Erridge et al. 2007a; Ghanim et al. 2009; Ghanim et al. 2010; Laugerette et al. 2011). Under normal circumstances the immune system has a normal inflammatory response to an acute endotoxin stimulus and once the toxin is neutralized and removed, the state of inflammation returns to baseline levels. If endotoxin exposure becomes chronic, a low-grade inflammatory state may be maintained, contributing to tissue damage and the development of metabolic diseases. Those microbes most successful at driving inflammatory disease processes have evolved to interfere with host gene expression and metabolism. Many of these pathogens persist inside the cells of the immune system. Miskinyte and Gordo (Miskinyte and Gordo 2013) examined how a commensal E. coli was able to develop the virulence needed to persist inside macrophages of the immune system. Working in mice, they found that when maintained in vitro under the selective pressure of host macrophages, commensal E. coli can evolve virulent clones that escape phagocytosis and macrophage killing in vitro, while increasing their pathogenicity in vivo.
A study by Cani, et al. (Cani et al. 2007) showed that an infusion of LPS for 4 weeks in mice led to increased weight gain and development of insulin resistance, similar to mice fed a high-fat diet, providing a causative role for gut microbiota in the development of metabolic disease. In our non-human primate model of maternal obesity, high-fat diet exposure increased fetal hepatic oxidative stress and apoptosis in the early 3rd trimester, perhaps priming the liver for later development of NASH (McCurdy et al. 2009). Furthermore, there is innate immune dysfunction and necro-inflammatory changes in juvenile offspring of high-fat diet-fed dams (Thorn et al. 2014). Importantly, these alterations persist even after weaning to a normal chow diet. Lastly, in this model, a maternal high-fat diet, regardless of maternal weight gain, leads to persistent gut dysbiosis in juvenile offspring switched to a healthy diet at weaning (Ma et al. 2014).
Emerging evidence suggests that the severity of NAFLD postnatally is strongly associated with gut dysbiosis (Boursier et al. 2016) and a shift in gut metabolic function, including production and utilization of short-chain fatty acids and bile acids. Given the early gut permeability engendered by a dysbiotic infant microbe community, it is tempting to speculate that certain bacteria and their outputs might trigger programming events in the liver, and in progenitor cells in the bone marrow as well (see discussion earlier). We recently found that pregnant mice fed a Western-style diet produced an early programming effect on the gut microbiota and on bone marrow-derived macrophage polarization in 3-week-old offspring, suggesting that shaping of the early gut microbiota and programming of macrophages during the early weaning period are crucial for development of NAFLD and fibrosis in adulthood (K R Jonscher & J E Friedman, unpublished observations). The early microbes and the microenvironment and mechanisms responsible for programming myeloid cells within the bone marrow and liver of obese offspring warrant further research.
Problems and future challenges in determining causation in early microbiota colonization
Early alterations in the infant microbiota composition are associated with the development of both metabolic and immune diseases, but causation is lacking. Models imploring the use of germ-free animals or antibiotics have been the primary methods for isolating and studying the impact of the microbiota on human health. Studies using germ-free animals have produced compelling evidence for the role of the microbiota in obesity development and education of the immune system (Ridaura et al. 2013; Turnbaugh et al. 2006; Weng and Walker 2013), improving our understanding of the contribution of the microbiota in disease progression. Unfortunately, these studies do not provide sufficient information about the role of the microbiota in the initiation of the disease, or whether an altered microbiota is a cause or just a consequence of disease. Another challenge that exists is that there is still significant controversy over what is considered a healthy or diseased microbiota. This controversy is highlighted in a recent review (Sze and Schloss 2016) evaluating studies that reported the microbiota in subjects with obesity, which suggests that we are still unable to conclusively define the composition of an obese microbiota. One reason it has been difficult to assign health benefits to a specific microbiota composition is because of the functional redundancy found across various bacterial groups (Moya and Ferrer 2016). Future studies are needed to explore the impact of individual bacterial species, their interactions with each other and with their host, and how they contribute to disease development.
Long term studies are needed to determine if an altered microbiota composition alone is sufficient for disease development or whether a secondary stressor like diet is needed to accelerate disease. Identifying a programming effect due to early “obese microbes” that fine tunes the infant gut may require a second hit to reveal the causes for developmental programming of the immune and metabolic systems in early life. This has been done on a limited basis in studies showing that microbial composition dictates susceptibility to disease development in humans (Spencer et al. 2011) and animals (Le Roy et al. 2013) providing support for a causal relationship, but only in response to a secondary insult (i.e. diet). Ongoing studies are currently exploring the interaction between the microbiota composition and diet to better understand the impact of this interaction in disease development. Additionally, determining whether there are legacy costs of early alterations in microbial succession that are maintained long after the microbiota has adapted to solid foods and reached maturity and stability are also important to understand the full impact of an early alteration in the infant microbiota composition. As we continue to learn more about the importance of early bacterial colonization, we can develop strategies for interventions that could reduce the future incidence of a wide variety of metabolic and immune diseases.
Acknowledgments
Funding
This work was supported by the NIH Colorado Nutrition and Obesity Research Center (NORC) grant P30 DK048520 (CMM, JEF).
This work was supported in part by NIH R24-DK102766-06 and NIH-R01-DK140443 to JF. The authors thank Rachel Janssen for her expert assistance editing the manuscript.
Footnotes
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of this review.
References
- Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra265. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agopian VG, Kaldas FM, Hong JC, Whittaker M, Holt C, Rana A, Zarrinpar A, Petrowsky H, Farmer D, Yersiz H, et al. Liver transplantation for nonalcoholic steatohepatitis: the new epidemic. Annals of Surgery. 2012;256:624–633. doi: 10.1097/SLA.0b013e31826b4b7e. [DOI] [PubMed] [Google Scholar]
- Ajslev TA, Andersen CS, Gamborg M, Sorensen TI, Jess T. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. International Journal of Obesity. 2011;35:522–529. doi: 10.1038/ijo.2011.27. [DOI] [PubMed] [Google Scholar]
- Alderete TL, Autran C, Brekke BE, Knight R, Bode L, Goran MI, Fields DA. Associations between human milk oligosaccharides and infant body composition in the first 6 mo of life. The American Journal of Clinical Nutrition. 2015;102:1381–1388. doi: 10.3945/ajcn.115.115451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EL, Howe LD, Jones HE, Higgins JP, Lawlor DA, Fraser A. The prevalence of non-alcoholic fatty liver disease in children and adolescents: A systematic review and meta-analysis. PLoS One. 2015;10:e0140908. doi: 10.1371/journal.pone.0140908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreas NJ, Hyde MJ, Gale C, Parkinson JR, Jeffries S, Holmes E, Modi N. Effect of maternal body mass index on hormones in breast milk: a systematic review. PLoS One. 2014;9:e115043. doi: 10.1371/journal.pone.0115043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns C, Lee M, Low WY. The long-term public health benefits of breastfeeding. Asia-Pacific Journal of Public Health. 2016;28:7–14. doi: 10.1177/1010539515624964. [DOI] [PubMed] [Google Scholar]
- Biswas SK, Bist P, Dhillon MK, Kajiji T, Del Fresno C, Yamamoto M, Lopez-Collazo E, Akira S, Tergaonkar V. Role for MyD88-independent, TRIF pathway in lipid A/TLR4-induced endotoxin tolerance. Journal of Immunology. 2007;179:4083–4092. doi: 10.4049/jimmunol.179.6.4083. [DOI] [PubMed] [Google Scholar]
- Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F, Guy CD, Seed PC, Rawls JF, David LA, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63:764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PH, Pollard KS. Proteobacteria explain significant functional variability in the human gut microbiome. Microbiome. 2017;5:36. doi: 10.1186/s40168-017-0244-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SL, Buonomo E, Carey M, Cowardin C, Naylor C, Noor Z, Wills-Karp M, Petri WA., Jr Bone marrow dendritic cells from mice with an altered microbiota provide interleukin 17A-dependent protection against Entamoeba histolytica colitis. MBio. 2014;5:e01817. doi: 10.1128/mBio.01817-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. The American Journal of Clinical Nutrition. 2012;96:544–551. doi: 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, Aagaard KM. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Medicine. 2016;8:77. doi: 10.1186/s13073-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. The American Journal of Clinical Nutrition. 2008;88:894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother’s weight on infant’s microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. The American Journal of Clinical Nutrition. 2010;92:1023–1030. doi: 10.3945/ajcn.2010.29877. [DOI] [PubMed] [Google Scholar]
- Collado MC, Laitinen K, Salminen S, Isolauri E. Maternal weight and excessive weight gain during pregnancy modify the immunomodulatory potential of breast milk. Pediatric Research. 2012;72:77–85. doi: 10.1038/pr.2012.42. [DOI] [PubMed] [Google Scholar]
- Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Scientific Reports. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coveney AP, Wang W, Kelly J, Liu JH, Blankson S, Wu QD, Redmond HP, Wang JH. Myeloid-related protein 8 induces self-tolerance and cross-tolerance to bacterial infection via TLR4- and TLR2-mediated signal pathways. Scientific Reports. 2015;5:13694. doi: 10.1038/srep13694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leoz ML, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, Mills DA, Lebrilla CB. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. Journal of Proteome Research. 2015;14:491–502. doi: 10.1021/pr500759e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Chierico F, Vernocchi P, Petrucca A, Paci P, Fuentes S, Praticò G, Capuani G, Masotti A, Reddel S, Russo A, et al. Phylogenetic and metabolic tracking of gut microbiota during perinatal development. PLoS One. 2015;10:e0137347. doi: 10.1371/journal.pone.0137347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deputy NP, Sharma AJ, Kim SY, Hinkle SN. Prevalence and characteristics associated with gestational weight gain adequacy. Obstetrics and Gynecology. 2015;125:773–781. doi: 10.1097/AOG.0000000000000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deriu E, Liu JZ, Pezeshki M, Edwards RA, Ochoa RJ, Contreras H, Libby SJ, Fang FC, Raffatellu M. Probiotic bacteria reduce salmonella typhimurium intestinal colonization by competing for iron. Cell Host & Microbe. 2013;14:26–37. doi: 10.1016/j.chom.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich CM, Felice JP, O’Sullivan E, Rasmussen KM. Breastfeeding and health outcomes for the mother-infant dyad. Pediatric Clinics of North America. 2013;60:31–48. doi: 10.1016/j.pcl.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosch NC, Guslits EF, Weber MB, Murray SE, Ha B, Coe CL, Auger AP, Kling PJ. Maternal obesity affects inflammatory and iron indices in umbilical cord blood. The Journal of Pediatrics. 2016;172:20–28. doi: 10.1016/j.jpeds.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakesmith H, Prentice AM. Hepcidin and the iron-infection axis. Science. 2012;338:768–772. doi: 10.1126/science.1224577. [DOI] [PubMed] [Google Scholar]
- Eggesbø M, Moen B, Peddada S, Baird D, Rugtveit J, Midtvedt T, Bushel PR, Sekelja M, Rudi K. Development of gut microbiota in infants not exposed to medical interventions. APMIS: Acta Pathologica, Microbiologica, et Immunologica Scandinavica. 2011;119:17–35. doi: 10.1111/j.1600-0463.2010.02688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermann M, Arthur JC. Siderophore-mediated iron acquisition and modulation of host-bacterial interactions. Free Radical Biology & Medicine. 2017;105:68–78. doi: 10.1016/j.freeradbiomed.2016.10.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erridge C, Attina T, Spickett CM, Webb DJ. A high-fat meal induces low-grade endotoxemia: evidence of a novel mechanism of postprandial inflammation. The American Journal of Clinical Nutrition. 2007a;86:1286–1292. doi: 10.1093/ajcn/86.5.1286. [DOI] [PubMed] [Google Scholar]
- Erridge C, Spickett CM, Webb DJ. Non-enterobacterial endotoxins stimulate human coronary artery but not venous endothelial cell activation via Toll-like receptor 2. Cardiovascular Research. 2007b;73:181–189. doi: 10.1016/j.cardiores.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Erroi A, Fantuzzi G, Mengozzi M, Sironi M, Orencole SF, Clark BD, Dinarello CA, Isetta A, Gnocchi P, Giovarelli M. Differential regulation of cytokine production in lipopolysaccharide tolerance in mice. Infection and Immunity. 1993;61:4356–4359. doi: 10.1128/iai.61.10.4356-4359.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts B, Amiel E, Huang SC, Smith AM, Chang CH, Lam WY, Redmann V, Freitas TC, Blagih J, van der Windt GJ, et al. TLR-driven early glycolytic reprogramming via the kinases TBK1-IKKε supports the anabolic demands of dendritic cell activation. Nature Immunology. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fensterheim BA, Guo Y, Sherwood ER, Bohannon JK. The cytokine response to lipopolysaccharide does not predict the host response to infection. Journal of Immunology. 2017;198:3264–3273. doi: 10.4049/jimmunol.1602106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galley JD, Bailey M, Kamp Dush C, Schoppe-Sullivan S, Christian LM. Maternal obesity is associated with alterations in the gut microbiome in toddlers. PLoS One. 2014;9:e113026. doi: 10.1371/journal.pone.0113026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanim H, Abuaysheh S, Sia CL, Korzeniewski K, Chaudhuri A, Fernandez-Real JM, Dandona P. Increase in plasma endotoxin concentrations and the expression of Toll-like receptors and suppressor of cytokine signaling-3 in mononuclear cells after a high-fat, high-carbohydrate meal: implications for insulin resistance. Diabetes Care. 2009;32:2281–2287. doi: 10.2337/dc09-0979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanim H, Sia CL, Upadhyay M, Korzeniewski K, Viswanathan P, Abuaysheh S, Mohanty P, Dandona P. Orange juice neutralizes the proinflammatory effect of a high-fat, high-carbohydrate meal and prevents endotoxin increase and Toll-like receptor expression. The American Journal of Clinical Nutrition. 2010;91:940–949. doi: 10.3945/ajcn.2009.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giusti L, Gabriele M, Penno G, Garofolo M, Longo V, Del Prato S, Lucchesi D, Pucci L. A fermented whole grain prevents lipopolysaccharides-induced dysfunction in human endothelial progenitor cells. Oxidative Medicine and Cellular Longevity. 2017;2017:1026268. doi: 10.1155/2017/1026268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez de Agüero M, Ganal-Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, Steinert A, Heikenwalder M, Hapfelmeier S, Sauer U, et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- Granowitz EV, Porat R, Mier JW, Orencole SF, Kaplanski G, Lynch EA, Ye K, Vannier E, Wolff SM, Dinarello CA. Intravenous endotoxin suppresses the cytokine response of peripheral blood mononuclear cells of healthy humans. Journal of Immunology. 1993;151:1637–1645. [PubMed] [Google Scholar]
- Grönlund MM, Arvilommi H, Kero P, Lehtonen OP, Isolauri E. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0–6 months. Archives of Disease in Childhood. Fetal and Neonatal Edition. 2000;83:F186–192. doi: 10.1136/fn.83.3.F186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkilä MP, Saris PE. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. Journal of Applied Microbiology. 2003;95:471–478. doi: 10.1046/j.1365-2672.2003.02002.x. [DOI] [PubMed] [Google Scholar]
- Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O’Shea CA, Watkins C, Dempsey E, Mattivi F, Touhy K, et al. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET Cohort. Microbiome. 2017;5:4. doi: 10.1186/s40168-016-0213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Xu J, Falk PG, Midtvedt T, Gordon JI. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9833–9838. doi: 10.1073/pnas.96.17.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip S, Chung M, Raman G, Trikalinos TA, Lau J. A summary of the Agency for Healthcare Research and Quality’s evidence report on breastfeeding in developed countries. Breastfeeding Medicine. 2009;4(Suppl 1):S17–30. doi: 10.1089/bfm.2009.0050. [DOI] [PubMed] [Google Scholar]
- Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Björkstén B, Engstrand L, Andersson AF. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut. 2014;63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- Jantscher-Krenn E, Lauwaet T, Bliss LA, Reed SL, Gillin FD, Bode L. Human milk oligosaccharides reduce Entamoeba histolytica attachment and cytotoxicity in vitro. The British Journal of Nutrition. 2012;108:1839–1846. doi: 10.1017/S0007114511007392. [DOI] [PubMed] [Google Scholar]
- Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, Fernandez L, Rodriguez JM. Is meconium from healthy newborns actually sterile? Research in Microbiology. 2008;159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Jones AD, Zhao G, Jiang YP, Zhou M, Xu G, Kaciroti N, Zhang Z, Lozoff B. Maternal obesity during pregnancy is negatively associated with maternal and neonatal iron status. European Journal of Clinical Nutrition. 2016;70:918–924. doi: 10.1038/ejcn.2015.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nature Immunology. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B, O’Neill LA. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Research. 2015;25:771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Reiner GL, Lugo KA, Kreuk LS, Stanbery AG, Ansaldo E, Seher TD, Ludington WB, Barton GM. Maternal IgG and IgA antibodies dampen mucosal T helper cell responses in early life. Cell. 2016;165:827–841. doi: 10.1016/j.cell.2016.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Bäckhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer CD, Genco CA. Microbiota, immune subversion, and chronic inflammation. Frontiers in Immunology. 2017;8:255. doi: 10.3389/fimmu.2017.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassenius MI, Pietiläinen KH, Kaartinen K, Pussinen PJ, Syrjanen J, Forsblom C, Pörsti I, Rissanen A, Kaprio J, Mustonen J, et al. Bacterial endotoxin activity in human serum is associated with dyslipidemia, insulin resistance, obesity, and chronic inflammation. Diabetes Care. 2011;34:1809–1815. doi: 10.2337/dc10-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latuga MS, Stuebe A, Seed PC. A review of the source and function of microbiota in breast milk. Seminars in Reproductive Medicine. 2014;32:68–73. doi: 10.1055/s-0033-1361824. [DOI] [PubMed] [Google Scholar]
- Laugerette F, Vors C, Géloën A, Chauvin MA, Soulage C, Lambert-Porcheron S, Peretti N, Alligier M, Burcelin R, Laville M, et al. Emulsified lipids increase endotoxemia: possible role in early postprandial low-grade inflammation. The Journal of Nutritional Biochemistry. 2011;22:53–59. doi: 10.1016/j.jnutbio.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Le Roy T, Llopis M, Lepage P, Bruneau A, Rabot S, Bevilacqua C, Martin P, Philippe C, Walker F, Bado A, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62:1787–1794. doi: 10.1136/gutjnl-2012-303816. [DOI] [PubMed] [Google Scholar]
- Leatham-Jensen MP, Frimodt-Møller J, Adediran J, Mokszycki ME, Banner ME, Caughron JE, Krogfelt KA, Conway T, Cohen PS. The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infection and Immunity. 2012;80:1716–1727. doi: 10.1128/IAI.06193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemas DJ, Young BE, Baker PR, II, Tomczik AC, Soderborg TK, Hernandez TL, de la Houssaye BA, Robertson CE, Rudolph MC, Ir D, et al. Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome. The American Journal of Clinical Nutrition. 2016;103:1291–1300. doi: 10.3945/ajcn.115.126375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Chen GL, Hannemann N, Ipseiz N, Krönke G, Bäuerle T, Munos L, Wirtz S, Schett G, Bozec A. Microbiota from obese mice regulate hematopoietic stem cell differentiation by altering the bone niche. Cell Metabolism. 2015;22:886–894. doi: 10.1016/j.cmet.2015.08.020. [DOI] [PubMed] [Google Scholar]
- Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, Blundell P, Harris RA, Frias AE, Grove KL, et al. High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nature Communications. 2014;5:3889. doi: 10.1038/ncomms4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín R, Heilig HG, Zoetendal EG, Jimenez E, Fernandez L, Smidt H, Rodriguez JM. Cultivation-independent assessment of the bacterial diversity of breast milk among healthy women. Research in Microbiology. 2007;158:31–37. doi: 10.1016/j.resmic.2006.11.004. [DOI] [PubMed] [Google Scholar]
- McCurdy CE, Bishop JM, Williams SM, Grayson BE, Smith MS, Friedman JE, Grove KL. Maternal high-fat diet triggers lipotoxicity in the fetal livers of nonhuman primates. The Journal of Clinical Investigation. 2009;119:323–335. doi: 10.1172/JCI32661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Toll-like receptors and innate immunity. Nature Reviews Immunology. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- Mirpuri J, Raetz M, Sturge CR, Wilhelm CL, Benson A, Savani RC, Hooper LV, Yarovinsky F. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microbes. 2014;5:28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miskinyte M, Gordo I. Increased survival of antibiotic-resistant Escherichia coli inside macrophages. Antimicrobial Agents and Chemotherapy. 2013;57:189–195. doi: 10.1128/AAC.01632-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends in Microbiology. 2016;24:402–413. doi: 10.1016/j.tim.2016.02.002. [DOI] [PubMed] [Google Scholar]
- Mueller NT, Shin H, Pizoni A, Werlang IC, Matte U, Goldani MZ, Goldani HA, Dominguez-Bello MG. Birth mode-dependent association between pre-pregnancy maternal weight status and the neonatal intestinal microbiome. Scientific Reports. 2016;6:23133. doi: 10.1038/srep23133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles IA, Fontecilla NM, Janelsins BM, Vithayathil PJ, Segre JA, Datta SK. Parental dietary fat intake alters offspring microbiome and immunity. Journal of Immunology. 2013;191:3200–3209. doi: 10.4049/jimmunol.1301057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. The Journal of Clinical Investigation. 2004a;113:1271–1276. doi: 10.1172/JCI20945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004b;306:2090–2093. doi: 10.1126/science.1104742. [DOI] [PubMed] [Google Scholar]
- Neu J. Preterm infant nutrition, gut bacteria, and necrotizing enterocolitis. Current Opinion in Clinical Nutrition and Metabolic Care. 2015;18:285–288. doi: 10.1097/MCO.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neu J. Intestinal microbiota studies in preterm infants. Journal of Pediatric Gastroenterology and Nutrition. 2016;62:193–194. doi: 10.1097/MPG.0000000000001068. [DOI] [PubMed] [Google Scholar]
- Newton KP, Feldman HS, Chambers CD, Wilson L, Behling C, Clark JM, Molleston JP, Chalasani N, Sanyal AJ, Fishbein MH, et al. Low and high birth weights are risk factors for nonalcoholic fatty liver disease in children. The Journal of Pediatrics. 2017 doi: 10.1016/j.jpeds.2017.1003.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng RL, Scott NM, Strickland DH, Gorman S, Grimbaldeston MA, Norval M, Waithman J, Hart PH. Altered immunity and dendritic cell activity in the periphery of mice after long-term engraftment with bone marrow from ultraviolet-irradiated mice. Journal of Immunology. 2013;190:5471–5484. doi: 10.4049/jimmunol.1202786. [DOI] [PubMed] [Google Scholar]
- Nicklas JM, Barbour LA. Optimizing weight for maternal and infant health - tenable, or too late? Expert Review of Endocrinology & Metabolism. 2015;10:227–242. doi: 10.1586/17446651.2014.991102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninonuevo MR, Park Y, Yin H, Zhang J, Ward RE, Clowers BH, German JB, Freeman SL, Killeen K, Grimm R, et al. A strategy for annotating the human milk glycome. Journal of Agricultural and Food Chemistry. 2006;54:7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity and trends in body mass index among US children and adolescents, 1999–2010. Journal of the American Medical Association. 2012;307:483–490. doi: 10.1001/jama.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogra PL, Welliver RC., Sr Effects of early environment on mucosal immunologic homeostasis, subsequent immune responses and disease outcome. Nestlé Nutrition Workshop Series Paediatric Programme. 2008;61:145–181. doi: 10.1159/000113492. [DOI] [PubMed] [Google Scholar]
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biology. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul HA, Bomhof MR, Vogel HJ, Reimer RA. Diet-induced changes in maternal gut microbiota and metabolomic profiles influence programming of offspring obesity risk in rats. Scientific Reports. 2016;6:20683. doi: 10.1038/srep20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5:48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez JM. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Advances in Nutrition. 2014;5:779–784. doi: 10.3945/an.114.007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz AG, Casafont F, Crespo J, Cayón A, Mayorga M, Estebanez A, Fernadez-Escalante JC, Pons-Romero F. Lipopolysaccharide-binding protein plasma levels and liver TNF-alpha gene expression in obese patients: evidence for the potential role of endotoxin in the pathogenesis of non-alcoholic steatohepatitis. Obesity Surgery. 2007;17:1374–1380. doi: 10.1007/s11695-007-9243-7. [DOI] [PubMed] [Google Scholar]
- Santacruz A, Collado MC, García-Valdés L, Segura MT, Martín-Lagos JA, Anjos T, Martí-Romero M, Lopez RM, Florido J, Campoy C, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. The British Journal of Nutrition. 2010;104:83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- Singer K, DelProposto J, Morris DL, Zamarron B, Mergian T, Maley N, Cho KW, Geletka L, Subbaiah P, Muir L, et al. Diet-induced obesity promotes myelopoiesis in hematopoietic stem cells. Molecular Metabolism. 2014;3:664–675. doi: 10.1016/j.molmet.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum C, Coats SR, Hua N, Kramer C, Papadopoulos G, Weinberg EO, Gudino CV, Hamilton JA, Darveau RP, Genco CA. Distinct lipid a moieties contribute to pathogen-induced site-specific vascular inflammation. PLoS Pathogens. 2014;10:e1004215. doi: 10.1371/journal.ppat.1004215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MD, Hamp TJ, Reid RW, Fischer LM, Zeisel SH, Fodor AA. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology. 2011;140:976–986. doi: 10.1053/j.gastro.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sze MA, Schloss PD. Looking for a signal in the noise: revisiting obesity and the microbiome. MBio. 2016:7. doi: 10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Frank DN, Sherlock L, Ir D, Robertson CE, Krebs NF. Effect of vitamin E with therapeutic iron supplementation on iron repletion and gut microbiome in US iron deficient infants and toddlers. Journal of Pediatric Gastroenterology and Nutrition. 2016;63:379–385. doi: 10.1097/MPG.0000000000001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevaranjan N, Puchta A, Schulz C, Naidoo A, Szamosi JC, Verschoor CP, Loukov D, Schenck LP, Jury J, Foley KP, et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host & Microbe. 2017;21:455–466. e454. doi: 10.1016/j.chom.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn SR, Baquero KC, Newsom SA, El Kasmi KC, Bergman BC, Shulman GI, Grove KL, Friedman JE. Early life exposure to maternal insulin resistance has persistent effects on hepatic NAFLD in juvenile nonhuman primates. Diabetes. 2014;63:2702–2713. doi: 10.2337/db14-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hämäläinen AM, et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell. 2016;165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voreades N, Kozil A, Weir TL. Diet and the development of the human intestinal microbiome. Frontiers in Microbiology. 2014;5:1–9. doi: 10.3389/fmicb.2014.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng M, Walker WA. The role of gut microbiota in programming the immune phenotype. Journal of Developmental Origins of Health and Disease. 2013;4:203–214. doi: 10.1017/S2040174412000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesolowski SR, El Kasmi KC, Jonscher KR, Friedman JE. Developmental origins of NAFLD: a womb with a clue. Nature Reviews Gastroenterology & Hepatology. 2017;14:81–96. doi: 10.1038/nrgastro.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield C, Trent MS. Biosynthesis and export of bacterial lipopolysaccharides. Annual Review of Biochemistry. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, Núñez G. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity. 2016;44:647–658. doi: 10.1016/j.immuni.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]