Abstract
We collected personal, dense, dynamic data for 108 individuals over 9 months, including whole genome sequence; clinical tests, metabolomes, proteomes and microbiomes at three time points; and daily activity tracking. Using these data we generated a correlation network and identified communities of related analytes that were associated with physiology and disease. We demonstrate how connectivity within these communities identified known and candidate biomarkers, e.g. gamma-glutamyltyrosine was densely interconnected with clinical analytes for cardiometabolic disease. We calculated polygenic scores from GWAS for 127 traits and diseases, and identified molecular correlates of polygenic risk, e.g. genetic risk for inflammatory bowel disease was negatively correlated with plasma cystine. Finally, behavioral coaching informed by personalized data helped participants improve clinical biomarkers. Personal, dense, dynamic data clouds will improve understanding of health and disease, especially for early transition states. This approach to “scientific wellness” represents an opportunity largely missing in contemporary health care.
Introduction
In order to understand the basis of wellness and disease, we and others have pursued a global and holistic approach termed systems medicine1. The defining feature of systems medicine involves the collection of diverse longitudinal data for each individual that help to better decipher the immense complexity of human biology and disease—assessing both genetic and environmental determinants of health and their interactions. We refer to this collection of data as personal, dense, dynamic data clouds: personal, because each data cloud is unique to an individual; dense, because of the high number of measurements; and dynamic, because we monitor longitudinally. The convergence of advances in systems medicine, big data analysis, individual measurement devices, and consumer-activated social networks leads to a vision of healthcare that is predictive, preventive, personalized and participatory (P4)2. Such personal, dense, dynamic data clouds are needed to enable this vision and indeed underlie the essence of what Precision Medicine should be3. The U.S. healthcare system invests 97% of its resources on disease care4; accordingly, wellness and disease prevention have not been broadly explored to date. We believe this should change and give rise to the in-depth study of what we call scientific wellness, which is a quantitative data-informed approach to maintaining and improving health, as well as avoiding disease.
Several recent studies have illustrated the utility of multi-omic longitudinal data to look for signs of reversible early disease or disease risk factors in single individuals. David et al. characterized the dynamics of human gut and salivary microbiota in two individuals in response to travel abroad and enteric infection, using daily stool and saliva samples5. Chen et al. followed a single individual over a 14-month period using daily multi-omic data collection, identifying signatures of respiratory infection and the onset of type 2 diabetes6. Larry Smarr has tracked the progression of Crohn’s disease using regular blood and stool measurements over many years7. What is striking is that in essentially every personal trajectory followed we gain fascinating new insights into system dynamics.
We report here on the generation and analysis of personal, dense, dynamic data clouds for 108 individuals over the course of a 9-month study that we call the Pioneer 100 Wellness Project (P100). Our study included the whole genome sequence; clinical tests, metabolomes, proteomes, and microbiomes at three-month intervals; and frequent activity measurements (i.e. wearing a Fitbit). This study takes a different approach from previous studies, in that a broad set of assays were measured at a fewer number of time points in a (comparatively) large number of people. Furthermore, we identified ‘actionable possibilities’ for each individual to enhance her/his health. Risk factors that we observed in participants’ clinical markers and genetics were used as a starting point to identify actionable possibilities for behavioral coaching. In this manuscript, we focus our analyses on the correlations across different data types and identify population-level changes in clinical markers. This project is the pilot for the 100,000 (100K) person wellness project that we proposed in 20148. At the end of the P100, we launched a consumer-facing scientific wellness company, Arivale, which implements a data collection and coaching model based on this pilot study. Through the Arivale program (over 95% have consented to research of de-identified data thus far), we are on track to reach our goal of 100,000 individuals by 2020 with the expectation to grow significantly beyond our initial goal in the years ahead. This scale of personal, dense, dynamic data clouds holds the potential to ultimately revolutionize our understanding of scientific wellness and the early warning signs for human diseases.
Results
The P100 study had four objectives as listed in our original IRB: 1) Establish cost-efficient procedures for generating, storing and analyzing multiple sources of health data obtained over time from participants and analyzed in combination with genomic data; 2) Develop and deploy analytic tools for integrating these diverse datasets and deriving actionable information from their observed interrelationships; 3) Identify novel patterns within the streams of health data that indicate either wellness, or transitions between wellness and disease; 4) Learn how to best interface with and present longitudinal health information to individuals by studying the reactions and feedback from participants as they are presented actionable information.
Individuals in Washington state and California were informally identified as interested in the P100 via personal communication and social networks of the authors. These individuals were then sent a formal email announcement of the study from Leroy Hood with an invitation to join. Procedures for the P100 were run under the Western Institutional Review Board (IRB Protocol Number 20121979) at the Institute for Systems Biology (ISB). All 108 participants gave written informed consent for analysis of their data.
Data collection in the P100
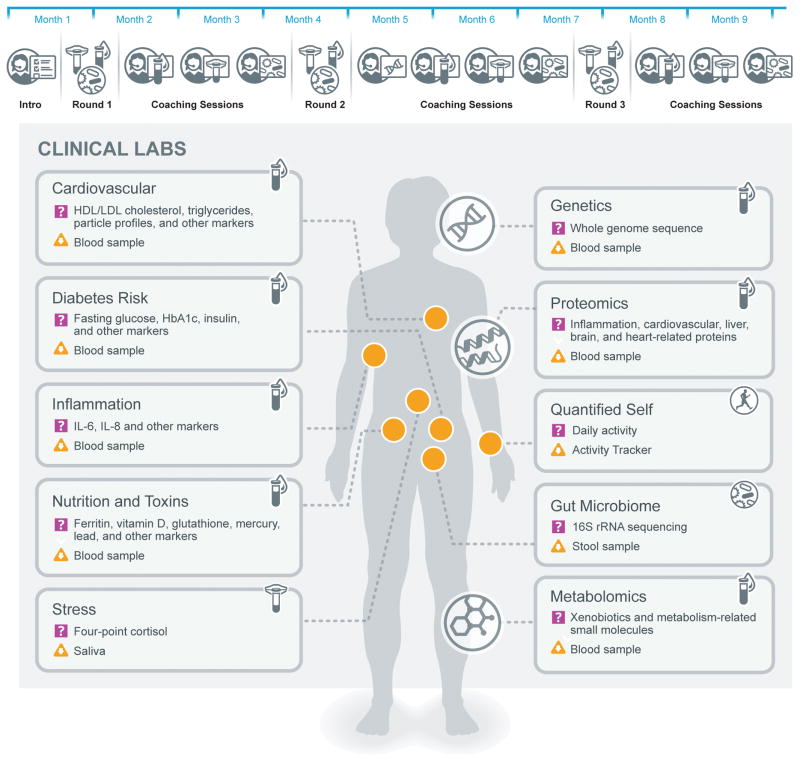
108 individuals (age 21–89+ years; 59% males, 41% females; 89% Caucasian; not recruited based on any specific phenotype) (see Supplementary Table 1) participated in an IRB-approved study from April 2014 to January 2015. Health history and behavioral assessments were performed at the beginning of the study to establish a baseline for health coaching. Each individual had the genome sequenced in full. Blood was collected in clinics every three months. Additionally, participants completed at-home collections of saliva, stool, and first morning void urine every three months. Stool and saliva samples were shipped directly to the vendor by the participant, while urine was given back to the study coordinators for distribution to the proper sample vendor (Figure 1). We named each of these three collection periods “rounds”. For each successful participant in each round we carried out 218 clinical laboratory tests, measured up to 643 metabolites and 262 proteins, and measured the abundance of 4616 operational taxonomic units (OTUs) in the gut microbiome using 16S rRNA sequencing. We used the whole genome sequence to calculate 127 polygenic scores for disease risks and quantitative traits based on previous studies selected from the NHGRI GWAS catalog9. Three common CNVs were also included as genomic features, bringing the total to 130 (see Online Methods). All vendor measurements are listed in Supplementary Table 1. Participants were asked to record weight, blood pressure and resting heart rate weekly, and to track activity and sleep daily using a wearable device (Fitbit), although compliance with this quantified-self tracking was relatively low. While data were used throughout the study for coaching, all results presented in this paper were analyzed after the conclusion of the 9-month study using uniform standardized bioinformatics pipelines. See Online Methods for details of data collection, analysis, and source code. All raw data collected as part of the P100 have been submitted to dbGaP with accession ID phs001363.v1.p1. Processed data are made available as part of the supplement to this paper.
Figure 1. Types of longitudinal data collected.
(A) Timeline of important events in the P100. (B) Schematic of the data collected every three months throughout the study.
Community structure in the correlation networks
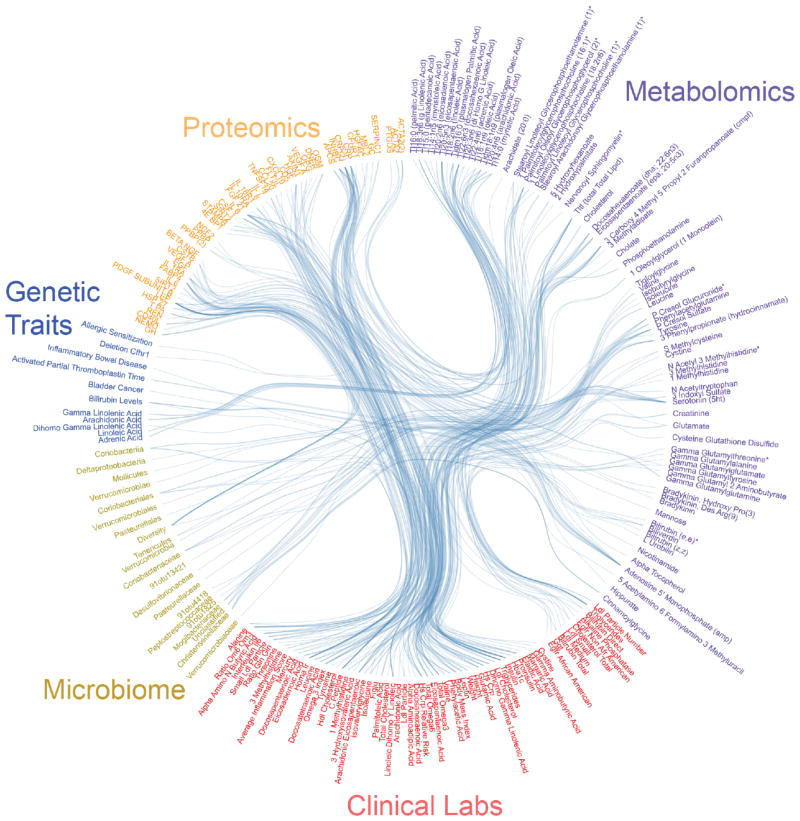
We built two age- and sex-adjusted correlation networks based on Spearman correlations across our cohort of individuals (Figure 2 and Supplementary Figure 1). Cross-sectional correlations were calculated from mean measurements of analytes calculated using all three rounds (mean A is correlated with mean B across all individuals). Delta correlations were calculated on the change in analytes between rounds (the change in A between time points is correlated with the change in B across all individuals). In these networks, vertices (V) correspond to analytes, and an edge (E) exists between two vertices if and only if a significant (padj<0.05) correlation was observed after correction for multiple hypotheses10. The inter-omic cross-sectional correlation network contains 766 nodes and 3,470 edges. The majority of edges involved a metabolite (3,309) or a clinical laboratory test (3,366), with an additional 20 edges involving the 130 tested genetic traits, 46 with microbiome taxa or diversity score, and 207 with quantified proteins. The inter-omic delta correlation network contains 822 nodes and 2,406 edges. 375 of the edges in the delta correlation network are also present in the cross-sectional network. The cross-sectional correlation network is provided in Supplementary Table 2 (inter-omic only) and Supplementary Table 3 (full). The delta correlation network is provided in Supplementary Table 4 (inter-omic only) and Supplementary Table 5 (full)).
Figure 2. Top 100 correlations per pair of data types.
Subset of top statistically-significant Spearman inter-omic cross-sectional correlations between all datasets collected in our cohort. Each line represents one correlation that was significant after adjustment for multiple hypothesis testing using the method of Benjamini and Hochberg10 at padj<0.05. The mean of all three time points was used to compute the correlations between analytes. Up to 100 correlations per pair of data types are shown in this figure. See Supplementary Figure 1 for the complete inter-omic cross-sectional network.
We identified clusters of related measurements from the cross-sectional inter-omic correlation network using community analysis, an unsupervised approach that iteratively prunes the network (removing the edges with the highest betweenness) to reveal densely interconnected subgraphs (communities)11. Seventy communities of at least two vertices (mean of 10.9 V and 34.9 E) were identified in the cross-sectional inter-omic network at the cutoff with maximum community modularity12 (Supplementary Figure 2), and are fully visualized as an interactive graph in Cytoscape13 (Supplementary File 1). 70% of the edges in the cross-sectional network remained after community edge pruning. The communities often represented a cluster of physiologically-related analytes, as described below.
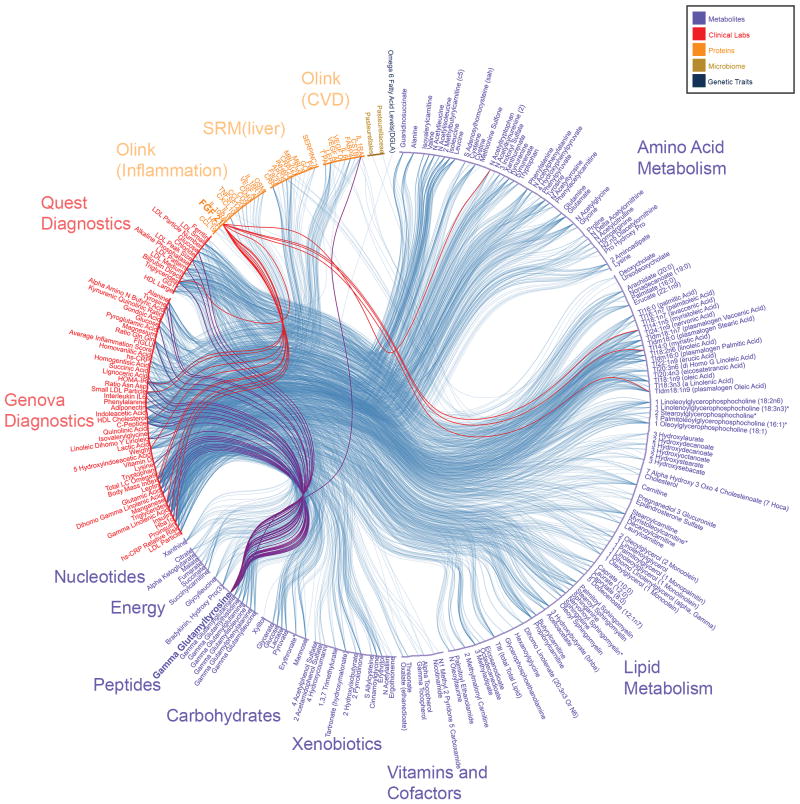
The largest community (246 V; 1645 E) contains many clinical analytes associated with cardiometabolic health, such as C-peptide, triglycerides, insulin, HOMA-IR, fasting glucose, HDL cholesterol, and small LDL particle number (Figure 3). The four most connected clinical analytes by degree (the number of edges connecting a particular analyte) are C-peptide (degree 99), insulin (88), HOMA-IR (88), and triglycerides (75). The four most connected proteins measured using targeted (SRM) mass spectrometry or Olink proximity extension assays by degree are leptin (18), C-reactive protein (15), fibroblast growth factor 21 (FGF21) (14), and inhibin beta C chain (INHBC) (10). Leptin and C-reactive protein are indicators for cardiovascular risk14,15. FGF21 is positively correlated with the clinical analytes C-peptide (Spearman’s ρ=0.51; padj=3.1e-3), triglycerides (ρ=0.50; padj=3.3e-3), HOMA-IR (ρ=0.50; padj=3.6e-3), insulin (ρ=0.47; padj=9.0e-3), and small LDL particle number (ρ=0.42; padj=4.3e-3), and is a recently reported biomarker for cardiometabolic disorders16. INHBC, a member of the TGF-beta superfamily, is positively correlated with the clinical analytes triglycerides (ρ=0.45; padj=3.0e-3), small LDL particle number (ρ=0.43; padj=6.8e-3), C-peptide (ρ=0.40; padj=1.8e-2), HOMA-IR (ρ=0.38; padj=3.4e-2), and insulin (ρ=0.38; padj=3.8e-2); it has not been reported to be a marker for cardiovascular risk and therefore represents an interesting candidate for follow-up. Serum amyloid P component (SAP) was positively correlated with LDL particle number (ρ=0.39; padj =1.8e-3). SAP is a universal constituent of amyloid deposits observed in Alzheimer’s disease17, and is associated with myocardial infarction18.
Figure 3. Cardiometabolic community.
All vertices and edges of the cardiometabolic community, with lines indicating significant (padj<0.05) correlations. Associations with FGF21 (red lines) and gamma-glutamyltyrosine (purple lines) are highlighted.
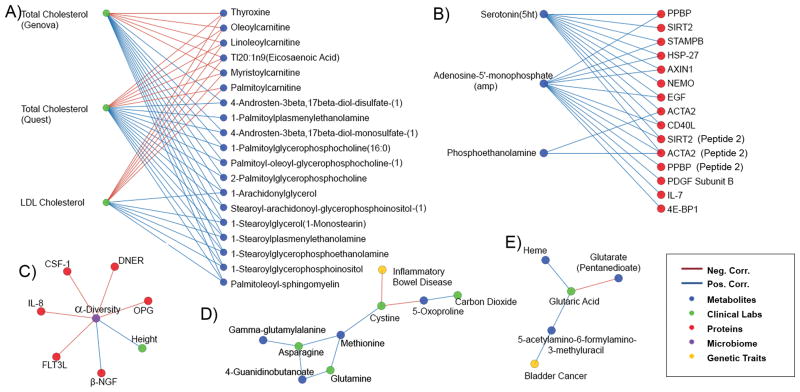
Total cholesterol and LDL cholesterol (LDL-C) segregate into a separate community from the cardiometabolic community (22 V; 48 E) with a broad array of plasma lipids (Figure 4A). Thyroid hormone L-thyroxine is also present and is negatively correlated with total cholesterol levels (ρ=−0.44; padj=5.0e-4) as well as LDL cholesterol (ρ= −0.41; padj=2.1e-3). Hypothyroidism has long been recognized clinically as a cause of elevated cholesterol values19.
Figure 4. Cholesterol, serotonin, α-diversity, IBD, and bladder cancer communities.
(A) Cholesterol community (B) Serotonin community (C) α-diversity community (D) The polygenic score for inflammatory bowel disease is negatively correlated with cystine (E) The polygenic score for bladder cancer is positively correlated with 5-acetylamino-6-formylamino-3-methyluracil (AFMU).
A community formed around plasma serotonin (18 V; 25 E) containing twelve proteins listed in Supplementary Table 6, for which the most significant enrichment identified in a STRING ontology analysis20 was platelet activation (padj=1.7e-3) (Figure 4B). Serotonin is known to induce platelet aggregation21; accordingly, selective serotonin reuptake inhibitors (SSRIs) may protect against myocardial infarction22.
We identified several communities containing microbiome taxa, suggesting that there are specific microbiome-analyte relationships. Hydrocinnamate, L-urobilin, and 5-hydroxyhexanoate clustered with the bacterial class Mollicutes and family Christensenellaceae (8 V; 8 E). Another community emerged around the Verrucomicrobiaceae and Desulfovibrionaceae families and p-cresol-sulfate (7 V; 6 E). The Coriobacteriaceae and Mogibacteriaceae families were associated (12 V; 19 E) with phenylacetic acid, eicosadienoic acid, p-cresol-glucuronide, taurine, and phenylacetylglutamine. Phenylacetylglutamine, a known microbial metabolite23, was recently identified as a risk factor for mortality and cardiovascular disease in chronic kidney disease patients24. Finally, the bile acid cholate clusters with the Peptostreptococcaceae family (2 V; 1 E).
A community formed around microbiome α-diversity (8 V; 7 E), a measure of the number of OTUs observed and the evenness of their distributions; elevated diversity is generally thought to be associated with better health in part by ameliorating inflammation25. Microbiome α-diversity was negatively correlated with inflammatory and immune-related proteins, including interleukin-8 (IL-8), FMS-related tyrosine kinase 3 (FLT3LG), and macrophage colony-stimulating factor 1 (CSF1) (Figure 4C). In contrast, β-nerve growth factor (NGF) was positively correlated with microbiome α-diversity. An analysis using STRING20 on α-diversity community members revealed a significant enrichment in the KEGG pathway cytokine-cytokine receptor interaction (padj=1.1e-4); other pathway members have been implicated in the pathogenesis of inflammatory bowel disease.26
Mining multi-omic communities for potential biomarkers
One highly interconnected metabolite in the cardiometabolic community, gamma-glutamyltyrosine (degree 27), was significantly correlated with markers of cardiometabolic disease: glucose (ρ=0.41; padj=1.6e-3), HOMA-IR (ρ=0.38; padj=6.0e-3), and insulin (ρ=0.36; padj=9.7e-3), as well as triglycerides (ρ=0.41; padj=1.5e-3), small LDL particle number (ρ=0.35; padj =1.5e-2), and HDL cholesterol (ρ= −0.35; padj=1.6e-2). Gamma-glutamyltyrosine is produced by the enzyme gamma-glutamyl transferase (GGT), a known biomarker of diabetes risk independent of BMI27,28. We carried out an ordinary least squares (OLS) regression with homeostatic risk assessment (HOMA-IR, a common marker for insulin resistance), as the dependent variable and GGT, gamma-glutamyltyrosine, age, sex, and BMI as the regressors (R2adj=0.46) (Supplementary Table 7). In this model, gamma-glutamyltyrosine has a more significant effect on HOMA-IR (p=4.3e-6) than does GGT (p=0.09). If this finding is confirmed in a larger number of unrelated samples, gamma-glutamyltyrosine could be a candidate biomarker for diabetes risk independent of BMI.
Delta correlation network identifies changes over time
Thirty-three communities of at least two vertices (mean of 24.9 V and 59.2 E) were identified in the inter-omic delta correlation network at the cutoff with maximum community modularity12 (Supplementary File 2). 81% of the edges in the delta network remained after community edge pruning. This network contains many interesting relationships not found in the cross-sectional network. For example, changes in HDL cholesterol were positively correlated with changes in galanin (ρ=0.36; padj=4.8e-3), a neuropeptide hormone with many physiological functions, including therapeutic associations with diabetes and Alzheimer’s disease29. One of the delta communities (V=15; E=28) involved the omega-3 fatty acids eicosapentaenoic acid and docosahexaenoic acid (DHA), as well as the clinical analyte omega-3 index. Also present in this delta community is the furan fatty acid metabolite 3-carboxy-4-methyl-5-propyl-2-furanpropanoate (CMPF). CMPF is elevated in the plasma of type 2 diabetes patients and directly implicated in β cell dysfunction30, and has previously been observed to increase in response to omega-3 fatty acid supplements in diabetic patients31.
Polygenic scores correlate with disease-risk analytes
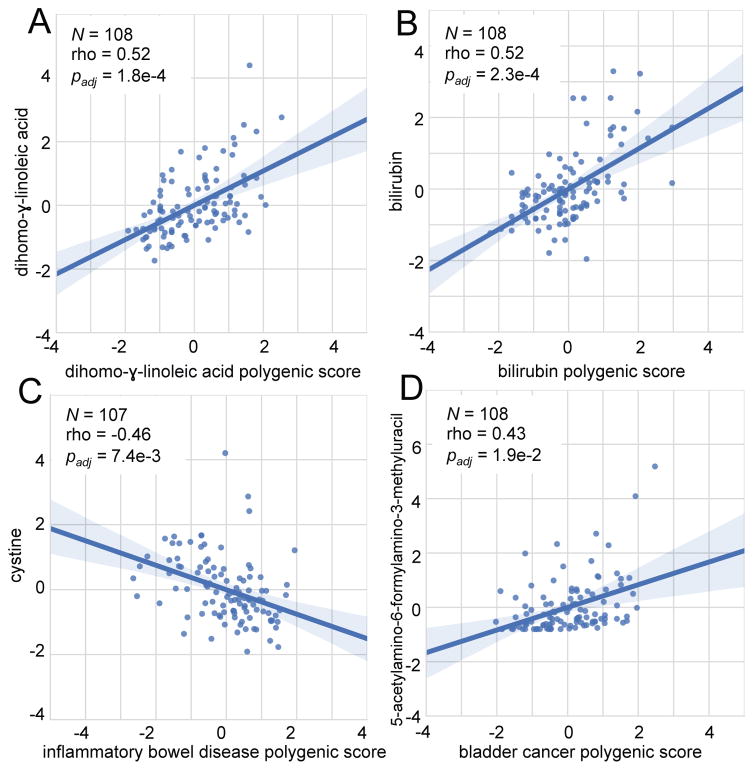
Several edges in the cross-sectional network represented correlations between genetic traits and corresponding biomarkers already identified in published studies. For example, blood levels of dihomo-γ-linolenic acid (DGLA) in our study were strongly correlated (ρ=0.52; padj=1.8e-4) with a polygenic score computed from genotypes in 6 variants that were previously associated with DGLA levels32 (Figure 5A). We observed similar results for other omega-6 fatty acids including arachidonic acid, linoleic acid, and eicosadienoic acid as well as bilirubin, a marker of liver dysfunction (ρ=0.52; padj=2.3e-4)33 (Figure 5B). All tested associations with quantitative traits are presented in Supplementary Table 8.
Figure 5. Polygenic scores correlate with blood analytes.
Spearman correlations between polygenic scores (x-axis) and analyte measurements (y-axis) from our correlation network. The number of measurements used for each pairwise comparison, correlation coefficients, and adjusted p-values are indicated on each figure. Values have been age and/or sex adjusted as described in Online Methods. The line shown is a y~x regression line, and the shaded regions are 95% confidence intervals for the slope of the line.
Although GWAS studies that model quantitative traits are most directly applicable to the measurements made in our study, other edges in the network occurred between polygenic disease risk and specific analytes. For example, the genetic risk of inflammatory bowel disease (IBD) in Europeans has been associated with 110 SNVs26. In our cohort, the polygenic score for IBD calculated from all 110 SNVs was significantly negatively correlated with plasma cystine, the disulfide form of cysteine (ρ= −0.46; padj=7.4e-3) (Figure 4D and 5C).
We computed a bladder cancer polygenic score for all of our participants from 9 SNVs previously associated with bladder cancer in a European cohort34. We identified an edge between this polygenic bladder cancer score and plasma levels of 5-acetylamino-6-formylamino-3-methyluracil (AFMU), an acetylated metabolite of caffeine, in our cohort. (ρ=0.43; padj=1.9e-2). One variant is located downstream of the gene NAT2 for the enzyme N-acetyltransferase 2 responsible for acetylating carcinogenic compounds in urine. Polymorphisms in NAT2 are known to produce ‘fast’ and ‘slow’ acetylator phenotypes, of which the latter conveys higher risk for bladder cancer35 (Figure 4E and 5D).
Coaching and clinical lab improvements
In order to help participants modify their behavior and potentially improve their health throughout the 9-month period of this study, a behavioral coach talked participants through actionable possibilities from their data. Each month the coach worked with the participant and made recommendations for lifestyle changes with the aim of altering markers of known clinical significance and/or compensating for genetic predispositions (Figure 1 and Supplementary Figure 3). Specific coaching recommendations based on personal data were customized by the coach, in consultation with the study physician. These individual recommendations typically fell into one of several major categories: diet, exercise, stress management, dietary supplements, or physician referral. Coaching focused on four primary health areas: Cardiovascular, Diabetes, Inflammation, and Nutrition. The clinical tests used to quantify these health areas are provided in Supplementary Table 9. We used generalized estimating equations (GEE) to estimate the average population change in each clinical analyte by round while controlling for the effects of age, sex, and self-reported ancestry. Complete results are shown in Table 1 and Supplementary Table 10. The most significant improvements for those who began the study out-of-range were observed in vitamin D (+7.2 ng/mL/round), mercury (−0.002 mcg/g/round), and HbA1c (−0.085 %/round). We observed consistent improvements in total cholesterol measured by both Quest and Genova (−6.4 mg/dL/round and −5.4 mg/dL/round, respectively). LDL cholesterol, measured only with Genova, significantly decreased (−4.8 mg/dL/round), while HDL cholesterol significantly increased (+4.5 mg/dL/round). Other significant improvements were observed in other diabetes risk factors (fasting glucose, fasting insulin, and HOMA-IR), and inflammation (IL-8 and TNF-alpha). Lipoprotein fractionation, performed by both lab companies, produced significant but discordant results for LDL particle number.
Table 1. Longitudinal analysis of clinical changes by round.
Generalized estimating equations (GEE) were used to estimate average changes in clinical labs over time. The Δ per round is an estimate of the average change in the population for that analyte by round adjusted for age, sex, and self-reported ancestry. ‘Out-of-range at baseline’ indicates the estimates using only those participants who were out-of-range for that analyte at the beginning of the study. Green rows indicate statistically-significant improvement, while red rows indicate statistically-significant worsening. N/A values are present where no participants were out-of-range at baseline. For example, the mean improvement in vitamin D for the 95 participants that began the study out-of-range was +7.2 ng/mL per round. Several analytes are measured by both Quest and Genova; with the exception of LDL particle number, the direction of effect for significantly changed analytes was concordant across the two labs. An independence working correlation structure was used in the GEE. See Supplementary Table 10 for the complete results.
| Clinical Laboratory Test | Out-of-range at Baseline Participants | |||
|---|---|---|---|---|
| Quadrant | Name | N | Δ per round | P-value |
| Nutrition | Vitamin D | 95 | +7.2 ng/mL/round | 7.1E-25 |
| Nutrition | Mercury | 81 | −0.002 mcg/g/round | 8.9E-09 |
| Diabetes | HbA1c | 52 | −0.085 %/round | 9.2E-06 |
| Cardiovascular | LDL particle number (Quest) | 30 | +130 nmol/L/round | 9.3E-05 |
| Nutrition | Methylmalonic acid (Genova) | 3 | −0.49 mmol/mol creat/round | 2.1E-04 |
| Cardiovascular | LDL pattern (A or B) | 28 | −0.16 /round | 4.8E-04 |
| Inflammation | Interleukin-8 | 10 | −6.1 pg/mL/round | 5.9E-04 |
| Cardiovascular | Total cholesterol (Quest) | 48 | −6.4 mg/dL/round | 7.2E-04 |
| Cardiovascular | LDL cholesterol | 57 | −4.8 mg/dL/round | 8.8E-04 |
| Cardiovascular | LDL particle number (Genova) | 70 | −69 nmol/L/round | 1.2E-03 |
| Cardiovascular | Small LDL particle number (Genova) | 73 | −56 nmol/L/round | 3.5E-03 |
| Diabetes | Fasting glucose (Quest) | 45 | −1.9 mg/dL/round | 8.2E-03 |
| Cardiovascular | Total cholesterol (Genova) | 43 | −5.4 mg/dL/round | 1.2E-02 |
| Diabetes | Insulin | 16 | −2.3 IU/mL/round | 1.5E-02 |
| Inflammation | TNF-alpha | 4 | −6.6 pg/mL/round | 1.8E-02 |
| Diabetes | HOMA-IR | 19 | −0.56 /round | 2.0E-02 |
| Cardiovascular | HDL cholesterol | 5 | +4.5 mg/dL/round | 2.2E-02 |
| Nutrition | Methylmalonic acid (Quest) | 7 | −42 nmol/L/round | 5.2E-02 |
| Cardiovascular | Triglycerides (Genova) | 14 | −18 mg/dL/round | 1.4E-01 |
| Diabetes | Fasting glucose (Genova) | 47 | −0.98 mg/dL/round | 1.5E-01 |
| Nutrition | Arachidonic Acid | 35 | +0.24 wt%/round | 1.9E-01 |
| Inflammation | hs-CRP | 51 | −0.47 mcg/mL/round | 2.1E-01 |
| Cardiovascular | Triglycerides (Quest) | 17 | −14 mg/dL/round | 2.4E-01 |
| Nutrition | Glutathione | 6 | +11 micromol/L/round | 2.5E-01 |
| Nutrition | Zinc | 4 | −0.82 mcg/g/round | 3.0E-01 |
| Nutrition | Ferritin | 10 | −14 ng/mL/round | 3.1E-01 |
| Inflammation | Interleukin-6 | 4 | −1.1 pg/mL/round | 3.8E-01 |
| Cardiovascular | HDL large particle number | 8 | +210 nmol/L/round | 4.9E-01 |
| Nutrition | Copper | 10 | +0.006 mcg/g/round | 6.0E-01 |
| Nutrition | Selenium | 6 | +0.035 mcg/g/round | 6.2E-01 |
| Cardiovascular | Medium LDL particle number | 20 | +2.8 nmol/L/round | 8.5E-01 |
| Cardiovascular | Small LDL particle number (Quest) | 14 | −2.3 nmol/L/round | 8.8E-01 |
| Nutrition | Manganese | 0 | N/A | N/A |
| Nutrition | EPA | 0 | N/A | N/A |
| Nutrition | DHA | 0 | N/A | N/A |
During the introductory coaching call one participant, a 65-year old male, reported decreased mobility during hiking trips with his family and that his primary care physician had identified cartilage damage in his ankle. The baseline blood collection revealed that he had ferritin levels of 399 ng/mL, above the clinical reference range, and subsequent genetic analysis revealed he was homozygous for HFE C282Y, the primary genetic risk factor for hereditary hemochromatosis (HH). Given his reported ferritin levels and genetic risk factors, our clinical team referred him to a hematologist, who diagnosed HH and prescribed therapeutic phlebotomy. At the next blood draw, ferritin levels had dropped to 175 ng/mL and remained normal throughout the remainder of the study (Supplementary Figure 4). HH leads to excessive accumulation of dietary iron in various tissues and can be associated with serious complications later in life, including cartilage damage, liver disease, diabetes, and cardiac decompensation. Post diagnosis, this individual’s primary care physician attributed his cartilage damage to early symptoms of HH. Six other males presented with high ferritin levels but neither of the common genetic risk factors; four of the six were of Asian ancestry, out of only six male Asians in our study. It has previously been observed that Asians and Pacific Islanders have the highest mean population levels of ferritin despite a very low prevalence of risk factors for hemochromatosis36. These individuals were referred to their physicians for monitoring.
Discussion
This paper focuses on four main findings from the P100 Wellness Project. First, thousands of statistically significant inter-omic correlations were computed using personal, dense, dynamic data clouds to identify many associations that could be followed up with perturbation experiments. Second, we partitioned the correlations into data communities, which placed biomarkers in context within biological networks. This in turn led to the identification of putative biomarkers such as gamma-glutamyltyrosine, which was highly interconnected with clinical analytes for cardiometabolic disease. Third, we identified molecular correlates of polygenic disease risk scores computed from published GWAS data, revealing possible ways in which genetic predisposition is manifested through analyte changes. Finally, on average participants significantly improved their clinical biomarkers (Table 1 and Supplementary Table 10) during the course of this pilot study (e.g. type 2 diabetes and cardiovascular risk factors). These personal, dense, dynamic data clouds embody the essence of precision medicine3 and present possibilities for the discovery of important medical applications.
Data integration generated 3,470 significant (padj<0.05) cross-sectional correlations and 2,406 significant delta (change over time) correlations after multiple hypothesis correction. Two known correlations point to the potential existence of therapeutically valuable relationships. First, our analysis identified FGF21 as a potential contributor to cardiometabolic health. Indeed, obese diabetic patients treated with an FGF21 analog have shown improvements in triglycerides and other cardiovascular markers37. Second, L-thyroxine, through a negative correlation, was placed in a data community with cholesterol markers; supplementation with L-thyroxine lowered total cholesterol and LDL-C levels in patients with hypothyroidism in a clinical trial38. These two examples were identified from our data in an unsupervised manner.
A novel association we detected was for gamma-glutamyltyrosine, a metabolite of the enzyme biomarker gamma-glutamyl transferase (GGT). GGT is a clinical biomarker for liver disease, diabetes27,28, and cardiovascular disease risk39. GGT catalyzes the transfer of the gamma-glutamyl moiety of glutathione to a substrate, commonly another amino acid, producing gamma-glutamyl dipeptides40. One of these dipeptides, gamma-glutamyltyrosine, is highly interconnected within the cardiometabolic community and much more predictive of HOMA-IR (insulin resistance) than GGT; so it may be a useful diagnostic marker for diabetes risk if these findings are replicated in an unrelated larger cohort. In clinical studies gamma-glutamyl dipeptides also discriminate different forms of liver disease41 and predict 28-day mortality in intensive care unit patients42. These are just a few of our hundreds of community correlations that may provide deeper insights into known biology or reveal interesting biological associations.
We identified correlations between calculated polygenic scores derived from common GWAS variants and measured blood analytes. For several studies (see Supplementary Table 8) we were able to independently validate the cumulative associations of these variants with the expected quantitative trait (e.g. DGLA, LDL cholesterol, or bilirubin) – a nice confirmation of the ability for these polygenic scores for quantitative traits to reproduce well. We also found new genetic trait/metabolite associations. For example, the polygenic score for IBD was significantly negatively correlated with levels of cystine in plasma across our cohort. In a case-control study of IBD patients with either Crohn’s disease or ulcerative colitis, plasma cystine and cysteine levels were abnormally low in affected individuals relative to controls, with the effect increasing with disease severity43. Decreased availability of the limiting substrate cystine suggests an impairment of glutathione synthesis in the intestine. Glutathione is an important intracellular antioxidant that is depleted in IBD inflammatory episodes, leading to excess reactive oxygen species (ROS) and subsequent colonic inflammation and oxidative damage. Although Sido et al. discuss cystine deficiency as an effect rather than a cause of IBD, our preliminary data suggest that lower levels of blood cystine may be more common in individuals at higher genetic risk for IBD before the disease ever manifests itself.
Specific genetic variants have been used to explain metabolite profiles using targeted variant-pathway interactions44. Our data suggest that GWAS polygenic scores can identify analyte associations with disease risk in a non-targeted manner (e.g. AFMU vs. bladder cancer) and in the absence of direct associations between GWAS loci and plausible metabolic pathways (e.g. cystine vs. IBD). It is possible that supplementation with cystine in healthy individuals with high IBD genetic risk could avoid the long-term low grade inflammation and oxidative damage—and thus avoid the wellness to disease transition to IBD. Such hypotheses could be tested in future studies.
Most (89%) of our study participants were of Caucasian ancestry, and most (87%) of the 127 GWAS used as features for the correlation network were determined using European ancestry populations (Supplementary Table 1). We are currently evaluating approaches to control for ancestry of individuals in the computation of polygenic scores. This study was constrained to a small population of individuals living primarily in Seattle and northern California, but as we expand to other geographic areas our population diversity will increase. PCA plots of the population distribution of the P100 participants are shown in Supplementary Figure 5 and Supplementary Figure 6.
While we did provide activity trackers (Fitbits) to our participants with the goal of measuring activity and sleep, we observed only modest compliance. We required a minimum of 40 days of Fitbit usage in order to estimate the average activity for each participant; 64% of the participants met this criterion. We included mean activity calories as a feature in our correlation network, but did not observe any statistically significant correlations with this feature. We observed even lower compliance with sleep tracking (see Online Methods).
The opportunities for observing health transitions in a cohort of 108 individuals over 9 months are limited. For this reason, we are extending this pilot program to very large populations45. The main vehicle for obtaining these data will be through Arivale, an Institute for Systems Biology spin-out company that plans to scale to more than 100,000 individuals by 2020. For those who sign up for Arivale and consent for their data to be used anonymously for research (over 95% have agreed thus far), these de-identified data are shared with ISB by partnership and can be used for scientific discovery to improve understanding in biology and medicine. Critically, it is via individuals being willing to share their data that we best learn how to interpret it, and this is what maximizes the value back to each individual as well.
The broad nature of such a study as the P100 necessarily includes limitations. First, we didn’t follow a randomized clinical trial design: none of the participants were denied wellness coaching or access to personal data. While this makes statistical analysis more difficult, such models need to be built to learn from data in the real world where these kinds of approaches are going to scale very large—and individuals will expect to get best practices in every case. Such approaches are very relevant to major efforts such as those aimed at translating ‘omics-based data and building a learning healthcare system, as recently advocated by the U.S. National Academy of Medicine46. Second, while we demonstrate the usefulness of our initial correlation-based strategy, we recognize its limitations. Even after stringent multiple hypothesis correction, false discoveries are statistically inevitable. Nonetheless, many of our strongest associations recapitulated known biology, while the rest represent incipient hypotheses for follow-up studies. Conversely, in the face of human biological complexity no doubt many important interactions are obscured or missed entirely using this approach.
With a larger cohort we expect to be able to observe transitions from wellness to disease for many common diseases, as well as transitions to improved health. Analyses of these longitudinal data should enable the identification of network perturbations that result in common diseases, the design of diagnostics to detect early disease transitions, and the development of drugs and other interventions to reverse disease at its very earliest state. Personal, dense, dynamic data clouds will place on a solid foundation the emerging field of scientific wellness, drive the predictive, preventative, personalized, and participatory (P4) medicine of the 21st century, and they are the essence of what Precision Medicine should be.
Online Methods
Procedures for the P100 were run under the Western Institutional Review Board (IRB Protocol Number 20121979) at the Institute for Systems Biology (ISB). All 108 participants gave written informed consent. See Supplementary Figure 3 for a flowchart on recruitment/dropout and important events in the P100. At three time points throughout the study blood and urine samples from each participant were collected and processed using the procedures outlined by Genova Diagnostics and Quest Diagnostics and couriered to the testing facilities to maintain maximum sample stability.
Additional whole blood and plasma samples were collected from participants and shipped to BioStorage Technologies, an international CAP-accredited biorepository. Additional samples were used for metabolomics (Metabolon), SRM proteomics (ISB), Proseek Multiplex protein panels (Olink), and whole genome sequencing (Complete Genomics and the New York Genome Center). Participants collected stool samples at home for 16S rRNA sequencing (Second Genome), and were asked to provide daily activity and sleep data using personal monitoring devices (Fitbit).
Participants were asked to fast for 12 hours before all blood collections. We observed a 99.3% compliance rate in fasting. Participants were asked by the phlebotomist to confirm compliance with the 12-hour fast before each blood draw, and this was recorded on the requisition document. The P100 project manager sent out reminder emails prior to each blood draw period (‘round’) with instructions on how long to fast. Our clinical team reviewed the clinical data from each blood draw prior to its use in coaching.
Clinical Laboratory Tests
For Genova, a total of one urine tube and nine blood tubes were collected. The blood tubes consisted of two Na-Heparin Trace Element tubes, three SST tubes, three EDTA purple top tubes, and one NMR black-top LipoTube. First morning void urine was collected in the Genova-provided green-top tube by participants the morning of their blood draw. Urine was sent frozen to Genova. Both Na-Heparin tubes were spun for 15 minutes at 3000rpm. The plasma from one Na-Heparin tubes was transferred to a blue-top preservative tube provided by Genova and shaken and spun for 5 minutes at 2500rpm. Supernatant was then transferred to the yellow top transfer tube provided by Genova and shipped frozen. Plasma from the second Na-Heparin was transferred to an amber top transfer tube and shipped frozen. Each SST tube was left to clot for 15 minutes then spun for 15 minutes at 3000rpm. The plasma for all three was pipetted to transfer tubes and shipped frozen. All three EDTA-lavender top tubes were refrigerated after collection and shipped refrigerated. The single NMR black-top LipoTube was clotted for 30 minutes then spun for 15 minutes at 3000rpm. The specimen was left in the tube and shipped refrigerated.
Each saliva collection consisted of four samples within a single day (four-point cortisol test). For collection of the four saliva samples, participants were instructed to abstain from eating or drinking 30 minutes prior to each collection. All participants were given the following collection times for each of their four samples: Sample 1: Collect before breakfast, between 7am – 9am and one hour after waking up. Sample 2: Collect before lunch, between 11am – 1pm. Sample 3: Collect before dinner, between 3pm – 5pm. Sample 4: Collect before bedtime, between 10 pm–12am. All samples were frozen overnight after collection and shipped directly to Genova.
Two SST tubes were collected for Quest Diagnostics. After collection the two tubes were left to clot for 15 minutes and then spun for 15 minutes at 3000rpm. Samples were left in the tube and shipped at ambient temperature.
All clinical labs measured using both vendors are listed in Supplementary Table 1.
Whole Genome Sequencing
Participant whole blood samples were submitted to either Complete Genomics Inc. (41 participants) or the New York Genome Center (67 participants) for whole genome sequencing (WGS). Complete Genomics conducted the whole genome sequencing using their standard complete sequencing platform employing high-density DNA nanoarrays populated with DNA nanoballs for 40X average coverage. The New York Genome Center used Illumina’s 2x150bp HiSeq X technology for 30X average coverage, using TruSeq kits for library prep. Both vendors aligned sequenced reads to human reference sequence GRCh37/hg19. NYGC used BWA v0.7.8-r455.
Complete Genomics provided a vcfBeta file for each sequenced sample calculated with CGAPipeline v2.5.0.20. NYGC provided a VCF4.1 file for each sequenced sample calculated with GATK HaplotypeCaller, following duplicate marking with Picard v1.83, and indel realignment and base quality recalibration. GATK v3.1.1-g07a4bf8 was used for BAM file post-processing and variant calling. Only variants with a FILTER value of PASS were used in downstream analyses for both CGI and Illumina data. Copy number variant status was determined using Reference Coverage Profiles47. Variant frequencies were annotated using Kaviar48. For comparison of the two technologies, we used monozygotic twins sequenced using separate technologies. We observed 99.12% concordance in variant calls across technologies in 6601 distinct loci from the GWAS catalog, while 0.21% were fully observed and discordant. Supplementary Table 11 lists the full statistics of this comparison.
Gut Microbiome 16S rRNA Sequencing
Gut microbiome data in the form of 16S OTU (Operational Taxonomic Unit) read counts were provided by Second Genome. 250bp paired end MiSeq profiling of the 16S v4 region was performed as described previously49, with 50,000–150,000 reads generated per sample. 16S sequence clustering and open reference OTU picking50 were performed using USEARCH with a proprietary strain database. Each OTU was then represented as a fraction of an individual’s total microbiome composition. These OTU proportions were placed in a vendor-provided taxonomy and aggregated at the kingdom, phylum, class, order, family, genus, and species levels (Supplementary Table 1 and Supplementary Table 12). α-diversity51, a measure of the number of OTUs observed as well as the evenness of their distributions, was calculated as the within-sample Shannon diversity index:
where pij is the relative abundance of OTU i in sample j.
Second Genome performed our microbiome OTU picking using their proprietary strain database. Many microbiome studies are performed using OTU picking against the publicly available Greengenes database, but Second Genome recommended that we use their curated database. Their database is specifically customized for microbes that exist in the human gut, whereas the Greengenes database spans a broad range of microbes, for example soil and water microbes and those found in other organisms. The proprietary strain database used for microbiome analyses can be downloaded using the following URL:
http://secondgenome.com/solutions/resources/data-analysis-tools/strain-select/
We used β-diversity to assess the degree to which each participant’s microbiome composition resembled itself over time (Supplementary Figure 7). In nearly all cases, individuals’ microbiome composition was more similar to their previous sample than to other individuals. For these inter-individual comparisons, representative sequences were aligned using PyNAST 1.2.252 via QIIME 1.9.153 with the Greengenes54 85% OTU representative sequences as a template. The alignment was filtered to remove high entropy positions using the Lane mask55. A phylogeny was reconstructed using FastTree 2.1.7. Unweighted UniFrac distances56–58 were computed on the table using QIIME. scikit-bio 0.2.3 (http://scikit-bio.org) was used in a custom Jupyter Notebook59 with matplotlib60 and seaborn to process the distance matrix. Specifically, for each sample, the distance between it and the participant’s successive time point was determined (the red points in Supplementary Figure 7). All the distances from that sample to all other samples at the successive time point were then retrieved (the box-whisker plots in Supplementary Figure 7). Subsequent statistics were computed using SciPy 0.17.0.
Metabolomics
Metabolon Inc. conducted the metabolomics assays on participant plasma samples at three time points for each participant throughout the course of the study. Metabolon Inc. generated the data using their DiscoveryHD4 platform in addition to their Fatty Acid Metabolism (FAME) panel that use a combination of ultra-high-performance liquid chromatography with tandem mass spectrometry (MS) and gas chromatography (GC) in the identification of metabolites and fatty acids. The metabolite values were reported relative to their concentrations among all participants, except for lipids that were measured via GC-FID, which were reported as molar percentages of each participant’s total fatty acids. For analysis, the metabolomics data was median scaled, such that the median value for each metabolite was one and values that fell beneath the range of detection were imputed to be the minimum observed value. This scaling was performed across all samples. All time points were run as a single batch. Counts of metabolites detected using each technology are listed in Supplementary Table 13. See Supplementary Table 1 for all metabolites detected.
Olink Proximity Extension Assays
Protein levels in plasma were determined by Proximity Extension Assays using two Olink (Uppsala, Sweden) Proseek Multiplex 96×96 kits and quantified by real-time PCR using the Fluidigm (South San Francisco, California) BioMark HD system. Each kit provides a microtitre plate for measuring 92 protein biomarkers in 90 samples. Each well contains 96 pairs of DNA-labeled antibody probes. When a matched pair of probes bind to their target protein, their DNA labels are brought into close proximity and a PCR target sequence is formed by a proximity-dependent DNA polymerization. One plate contains 96 wells for processing 90 samples, 3 positive controls, and 3 negative controls to determine the lower detection limit. Each sample is also spiked with four controls to monitor variation in the three steps of the PEA process. Two non-human antigens serve as incubation controls, one DNA-labeled antibody serves as an extension control, and an oligonucleotide serves as a detection control.
The Proseek cardiovascular (CVD I) and inflammation (Inflammation I) panels target 158 different proteins with 19 overlapping measurements. Plasma samples from 80 subjects drawn at three intervals were assayed. One sample was assayed in triplicate on all plates and additional samples were replicated for a total of 270 multiplex cardiovascular and 270 multiplex inflammation assays. A total of 41,085 data points were collected. Assays were run according to the manufacturer’s instructions. In short, 1 μl of each sample was incubated with the antibody probes at 4°C overnight. After binding, the extension mix was added and the products were extended and amplified using 17 cycles of PCR (Applied Biosystems 9700, Life Technologies, Carlsbad, California). Next, 2.8 μl of each PCR product was added to the detection mix and loaded into the sample wells of a Fluidigm 96.96 Dynamic Array plate (Fluidigm Corporation) while kit primers were loaded into the primer wells. The Dynamic Array was primed in a Fluidigm HX IFC controller and then loaded into the Fluidigm Biomark imaging thermocycler for quantitative PCR. Quantification cycle (Cq) values for each measurement were determined using Fluidigm’s Real-Time PCR Analysis software and BiomarkDataCollection version 4.1.3. Data was normalized using the extension positive control and the negative control Cq values. The limit of detection was defined as three times the standard deviation of the negative controls. See Supplementary Table 1 for all proteins detected using Olink proximity extension assays.
Selected Reaction Monitoring (SRM) Analysis
SRM Assay and Method Development
SRM assays were developed for 200 peptides representing 100 proteins. See Supplementary Table 1 for all SRM peptides. For each peptide sequence the heavy isotope labeled analogue was synthesized (PEPotec SRM library Grade 1, Thermo-Fisher Scientific, Huntsville, AL) with cysteine residues carbamidomethylated and the C-terminal arginine as R[13C6, 15N4] or lysine as K[13C6, 15N2] to allow for relative quantification. The 200 synthetic peptides were individually analyzed on a 6530 accurate-mass Q-TOF liquid chromatography mass spectrometry (LC-MS) system (Agilent Technologies, Santa Clara, CA) using a ProtID-Chip-150 (II) (Agilent Technologies, Santa Clara, CA) to verify and confirm successful peptide synthesis. The 200 peptides were pooled as internal standard. Multiplexed SRM assays were established with the human SRMAtlas61 (www.srmatlas.org) and the synthetic peptides on a 6460 QQQ MS system equipped with Jet Stream ESI technology and a 1290 Series UHPLC (Agilent Technologies, Santa Clara, CA). SRM assays were optimized with regard to sensitivity and specificity, and with the aim to target 200 peptides in a single analysis. 1200 transitions were determined, 3 transitions to target each light endogenous peptide and 3 transitions to target each isotope labeled heavy peptide, and peptides separated on a reversed phase column (Zorbax SB-C18, 50mm x 2.1 mm I.D., 1.8 μm dp, Agilent Technologies, Santa Clara, CA) using a gradient from 3% to 30.5% acetonitrile / 0.1% formic acid / water over 55 min at a flow rate of 0.2 mL/min. Data were acquired in dynamic MRM mode with a fixed cycle time of 2500 ms and a minimum dwell time of 10 ms.
Plasma Sample Preparation
Plasma samples were thawed on ice and centrifuged for 10 min at 14,000 rpm to separate tissue debris or a lipid layer. 110 μL plasma were depleted from the 14 most abundant plasma proteins using the multiple affinity removal system (MARS Hu-14, 4.6x100 mm, Agilent Technologies, Santa Clara, CA) according to the manufacturer’s protocol. The depleted fraction was collected in 1.25 mL of MARS14 Buffer A and denatured by adding 600 mg urea to 8 M final concentration. Samples were reduced with 5 mM dithiothreitol for 30 min at 55°C, alkylated with 14 mM iodoacetamide for 30 min at room temperature in darkness and desalted using a GE HiPrep 26/10 column (GE HealthCare Life Sciences, Pittsburgh, PA) and 1200 HPLC system (Agilent Technologies, Santa Clara, CA). The protein concentration of the desalted samples was determined by bicinchoninic acid assay (BCA) (Thermo-Fisher Scientific, San Jose, CA). An aliquot of the pooled 200 synthetic peptides was spiked into an aliquot of each plasma sample (equal protein amounts) prior to the digestions with trypsin (Promega, Madison, WI) at 1:50 enzyme:substrate ratio for 16 h at 37°C. Digests were dried under centrifugal vacuum evaporation (Savant, Thermo-Fisher Scientific, San Jose, CA) and reconstituted to 1 μg/μL protein concentration.
Plasma Sample Analysis
20 μg of each plasma sample spiked with the 200 isotope labeled peptides was subjected to SRM analysis using the method described above. SRM data were analyzed with Skyline62. SRM traces were integrated with default settings and manual inspected to verify correct peak assignment and co-elution of endogenous and isotope labeled standard peptides. The relative peptide abundance level was reported as ratio of endogenous light to the heavy standard.
Quantified Self Tracking
Participants were asked to wear a Fitbit activity tracker throughout the 9-month study. Participants were offered either a Fitbit Flex (wrist) or a Fitbit One (clip-on). These Fitbit models measure activity using the number of steps an individual takes each day. The models available at the time of the study did not measure heart rate, as current models do, resulting in inconsistent activity measurements for e.g., running vs. cycling. Furthermore, the devices required manual entry and exit of ‘sleep mode’ for sleep tracking, for which compliance was too low to provide useful data. We required a minimum of 40 days of Fitbit usage in order to estimate the average activity for each participant; 64% of the participants met this criterion. The Fitbit device estimates user-specific ‘activity calories’ independently of basal metabolic rate (BMR). For all calculations, we used only the estimated ‘activity calories’, excluding BMR. We used these data only as a relative indicator of activity levels rather than an absolute measure of caloric burn.
Genomic traits
The National Human Genome Research Institute’s GWAS catalog lists results from more than 2000 published studies comprising over 1000 genetic traits9. We applied a strict filtering procedure to identify GWAS used for this study. First, we excluded studies which did not contain at least one SNV with p<1.0e-8. Studies which contain few SNVs are likely to produce a vector of cumulative genetic variation with low entropy, where almost all values are identical save a few. Such low entropy measurements are more likely to produce spurious correlations in our relatively small number of samples. We therefore excluded all traits associated with five or fewer SNVs. Furthermore, we required studies to have a sample size of at least 5000 individuals. In the event that multiple studies examined the same trait, we kept the study with the largest sample size. Finally, we manually excluded traits with too-generic descriptions (e.g. ‘common traits’ or ‘metabolic traits’), which did not provide a useful description of the purpose of the original study. The combination of these filters retained 127 genetic traits that we used for further analysis. Three common CNVs were included as additional genetic features computed using Reference Coverage Profiles47, bringing the total to 130. See Supplementary Table 1 for all genetic traits computed for this manuscript.
Included in the GWAS catalog are the beta-coefficients/odds ratios as well as the p-values for the predicted effect of each variant for that trait based on the association models from the original paper. We made two assumptions to simplify the calculation of the polygenic scores. First, we assumed that the beta-coefficients (or log odds ratios) combined in an additive manner based on the number of effect alleles present in each individual. Therefore, if a single effect allele were present we added the beta-coefficient for that variant into the cumulative polygenic score. If two copies of the effect allele were present we added twice the value of the beta-coefficient into the cumulative genetic effect for that individual. The second assumption was that the effects of each variant are independent of the effects of all other variants used in the model. In other words, the values of all interaction terms are zero. These two simplifying assumptions allowed us to calculate the polygenic score for each trait across each individual in our study.
There are a number of pitfalls to this approach that served to temper our expectations. First, GWAS only identify variants that occur commonly enough in the population to be associated statistically with a trait. Unless one is able to genotype a substantial fraction of the human population at risk for a particular trait, many rare variants will never rise above the level of noise in a GWAS. Furthermore, because they employ genotyping chips most GWAS ignore copy number variations (CNVs) or structural variations (SVs) that may have a significant effect on genetic traits. We included as part of our study three common CNVs as additional genomic features. Finally, many GWAS are applied to cohorts of individuals from similar ancestries to improve their likelihood of discovering associated variants; it is therefore possible that results from these studies do not generalize to individuals from differing ancestral populations.
There are other analysis options available for WGS data that would be appropriate for subsequent studies with an ‘N of 1’ focus. For example, one could perform rare or de novo variant analysis, which identify genetic variants that are either very rare in the population or unique to an individual, respectively. GWAS focus on variants which are common enough in the population to find significant associations with quantitative traits or diseases. The interpretation of rare and de novo variants can be difficult, as each variant must be interpreted in the context of functional impact. Sequencing and phenotyping relatives (e.g. family-based analysis) is a method to assist in interpreting the functional impact of de novo variants. Another possible analysis technique for WGS data is burden analysis, which calculates a cumulative burden score on each gene and attempts to associate these scores with phenotypes.
Coaching, charting, and compliance tracking
The P100 was designed as a prospective study that attempted to help participants modify their behavior to enhance their health throughout the 9-month period. Participants were assigned to a behavioral coach, who walked them through actionable possibilities from their data and made recommendations on lifestyle changes. These lifestyle changes were recommended in an attempt to alter markers of known clinical significance and/or compensate for genetic predispositions for which reliable published evidence is available. Each participant was eligible for one 30-minute coaching session per month, though participants were not penalized or excluded from the study if they chose not to participate in the coaching sessions. Participants were also able to communicate privately and securely with the coach via a website portal created specifically for this project. Participants also received their data through the website portal. The P100 collected statistics on participation in the coaching calls and compliance with sample collection.
As previously stated, clients were offered specific coaching recommendations based on their genetics and clinically actionable data. These recommendations were customized prior to each call by the study clinician and coach, in consultation with the study physician. All clinical markers and recommendations were reviewed and approved by the study physician prior to their communication to each participant. While these recommendations were specific to each individual based on their data, they typically fell into one of several major categories, including diet, exercise, stress management, dietary supplements, or physician referral. Coaching focused on four primary health areas: Cardiovascular, Diabetes, Inflammation, and Nutrition. The clinical tests we used to quantify these health areas are provided in Supplementary Table 9. We used generalized estimating equations (GEE) to estimate the average change for each clinical lab by round while controlling for the effects of age, sex, and self-reported ancestry. Coefficients, 95% confidence intervals, and p-values for all participants as well as those who began the study out-of-range are listed in Supplementary Table 10. See also Table 1.
Action items were recorded in each participant’s chart by our behavioral coach during each coaching call. These charts were used to keep participants on track and follow standard clinical practice guidelines. Post-study we reviewed each de-identified chart in detail with our behavioral coach to extract compliance data for each recommendation. We learned a great deal about how to merge standard clinical practice (e.g. charting in free-text fields, as practiced by clinicians) with the need for automated database storage of pre-defined and enumerated recommendations. Subsequent studies will investigate specific effects of recommendations and compliance on clinical data as well as other omics data with far larger N
Data preprocessing
Each dataset was transformed into comparable data vectors for statistical analysis. All measurements were mean centered and scaled by the standard deviations of the observed measurements. The microbiome measurements were compared independently at the phylum, class, order and family taxonomic levels. With the exception of the median-scaled metabolomics data, missing data were not imputed; participants that had a missing value were dropped from pairwise comparisons utilizing that value. Each analyte was age- and/or sex-corrected if a trimmed mean robust regression identified a significant relationship (p<0.01, unadjusted) between age and/or sex and the dependent variable. Age and sex correction were performed independently, so it was possible for an analyte to be age corrected but not sex corrected, and vice versa. If an analyte was corrected the residuals of the model were used in place of the original observations. If no corrections were made, the original mean centered and scaled measurements were used. See Supplemental Table 14 for statistics on age and sex correction.
Correlation network and community analysis
We created two different types of correlation networks: ‘cross-sectional’ and ‘delta’ correlations. Cross-sectional correlations were calculated from mean measurements of analytes calculated across all rounds (i.e. mean A is correlated with mean B across all individuals). Delta correlations were calculated on the change in analytes between rounds (i.e. the change in A for an individual between time points is correlated with the change in B, where the correlation is again calculated across all individuals). We used each pair of adjacent time points (r2–r1) and (r3–r2) to build the delta correlation network, where all such comparisons were used in the two vectors that were being compared. Therefore, each individual with three observations is represented twice for each calculated delta correlation. For example, while the cross-sectional correlation network was created by correlating vectors of maximum length N=108, the delta correlation network was created by correlating vectors of maximum length N=216. Our reasoning is that each pair of adjacent time points is an independent observation of a potential correlation in time, even though they are not drawn from a completely independent set of individuals. For each pairwise set of data (e.g. clinical tests vs. proteomics, clinical tests vs. metabolomics, etc.), each measurement from the first dataset was correlated with every measurement from the second dataset using Spearman’s ρ. P-values were adjusted for multiple hypothesis testing using the method of Benjamini and Hochberg10; we chose an adjusted p-value (padj) cutoff of 0.05 as our significance level. Only inter-omic correlations were used for community analysis. Both inter-omic and intra-omic (e.g. metabolomics vs. metabolomics) cross-sectional and delta correlations are reported in Supplementary Table 2, Supplementary Table 3, Supplementary Table 4, and Supplementary Table 5 and visualized in Figure 2 and Supplementary Figure 1. We assessed reproducibility of duplicate measurements across two clinical laboratories (Supplementary Figure 8). As correlations between repeat measurements do not represent physiologically relevant information, they are not included in our network or subsequent analysis.
We performed community analysis using the method of Girvan and Newman 11. This method involves iteratively calculating edge betweenness centrality on a network: the number of weighted shortest paths from all vertices to all other vertices that pass over that edge. After each iteration, the edge(s) with the highest betweenness centrality were removed and the process was repeated until only individual nodes remain. All communities can also be dynamically visualized in Cytoscape13 (Supplementary File 1 and Supplementary File 2).
Community analysis forms a dendrogram that can be analyzed at multiple hierarchical levels. For this manuscript we analyzed our network at a cut level determined using an unbiased method, the modularity of the community structure12. Briefly, modularity of community structure corresponds to an arrangement of edges which is statistically improbable when compared to an equivalent network with edges placed at random. At every iteration of the community analysis, we computed the modularity, and analyzed the communities at the iteration which maximized this quantity. A visualization of community modularity vs. iteration is shown in Supplementary Figure 2.
OLS Regression Tests
To test for heteroscedasticity in our HOMA-IR regression model, we fit the model using White’s heteroscedasticity-consistent estimator (HCE), and the results were consistent with those reported in the manuscript: gamma-glutamyltyrosine was still significantly more predictive (p=2.3e-4) of HOMA-IR than GGT (p=0.02). To test for the effects of outliers, we fit a robust regression model and again the results were consistent with those reported in the manuscript. To test for multicollinearity, we calculated the variance inflation factors (VIF) for each predictor. The maximum VIF was 1.7, indicating a low amount of correlation between the predictors of the model.
Statistical Analyses
All data types used in the cross-sectional and delta correlation networks were normalized as described in their respective method sections. Additionally, where we were able to identify a significant effect (p<0.01, unadjusted) with age and/or sex using trimmed mean robust regression we used the residuals as the comparison value. These adjustments were performed independently for age and sex. We report the calculated p-values for age and sex with every variable, as well as whether the variable was age and/or sex adjusted in Supplementary Table 14. All transformations were performed with the Python Statsmodels package (v0.6) with robust linear models using the trimmed mean norm. Unadjusted analytes were compared using the original mean centered and scaled measurements.
We created two different types of correlation networks: ‘cross-sectional’ and ‘delta’ correlations. Cross-sectional correlations were calculated from mean measurements of analytes calculated across all rounds (i.e. mean A is correlated with mean B across all individuals). Delta correlations were calculated on the change in analytes between rounds (i.e. the change in A for an individual between time points is correlated with the change in B, where the correlation is again calculated across all individuals). We used each pair of adjacent time points (r2–r1) and (r3–r2) to build the delta correlation network, where all such comparisons were used in the two vectors that were being compared. Therefore, each individual with three observations is represented twice for each calculated delta correlation. For example, while the cross-sectional correlation network was created by correlating vectors of maximum length N=108, the delta correlation network was created by correlating vectors of maximum length N=216. For each pairwise set of data (e.g. clinical tests vs. proteomics, clinical tests vs. metabolomics, etc.), each measurement from the first dataset was correlated with every measurement from the second dataset using Spearman’s ρ. P-values were adjusted for multiple hypothesis testing using the method of Benjamini and Hochberg10; we chose an adjusted p-value (padj) cutoff of 0.05 as our significance level. Only inter-omic correlations were used for community analysis. We assessed reproducibility of duplicate measurements across two clinical laboratories (Supplementary Figure 8). As correlations between repeat measurements do not represent physiologically relevant information, they are not included in our network or subsequent analysis.
We performed community analysis using the method of Girvan and Newman 11. This method involves iteratively calculating edge betweenness centrality on a network: the number of weighted shortest paths from all vertices to all other vertices that pass over that edge. After each iteration, the edge(s) with the highest betweenness centrality were removed and the process was repeated until only individual nodes remain.
For this manuscript we analyzed our network at a cut level determined using an unbiased method, the modularity of the community structure12. At every iteration of the community analysis, we computed the modularity, and analyzed the communities at the iteration which maximized this quantity (Supplementary Figure 2).
OLS regression on the dependent variable HOMA-IR, with regressors including sex, GGT, gamma-glutamyltyrosine, age, and body mass index was performed (Supplementary Table 7). To test for heteroscedasticity in our HOMA-IR regression model, we fit the model using White’s heteroscedasticity-consistent estimator (HCE), testing for the effects of outliers using a robust regression model and testing for multicollinearity by calculating the variance inflation factors (VIF) for each predictor.
Generalized estimating equations (GEE) were used to estimate the average change in each clinical analyte by round while controlling for the effects of age, sex, and self-reported ancestry (Table 1 and Supplementary Table 10). An independence working correlation structure was used in the GEE.
Software packages
NYGC used BWA v0.7.8-r455 to align sequences. Complete Genomics CGAPipeline v2.5.0.20. NYGC provided a VCF4.1 file for each sequenced sample calculated with GATK HaplotypeCaller, following duplicate marking with Picard v1.83, and indel realignment and base quality recalibration. GATK v3.1.1-g07a4bf8 was used for BAM file post-processing and variant calling. Copy number variant status was determined using Reference Coverage Profiles47. Variant frequencies were annotated using Kaviar48
OTU picking for the microbiome was performed using USEARCH with a proprietary strain database, which can be downloaded at http://secondgenome.com/solutions/resources/data-analysis-tools/strain-select/. Inter-individual comparisons were performed using PyNAST 1.2.252 via QIIME 1.9.153 with the Greengenes54 85% OTU representative sequences as a template. The alignment was filtered to remove high entropy positions using the Lane mask55. A phylogeny was reconstructed using FastTree 2.1.7. Unweighted UniFrac distances56–58 were computed on the table using QIIME. scikit-bio 0.2.3 (http://scikit-bio.org) was used in a custom Jupyter Notebook59 with matplotlib60 and seaborn to process the distance matrix.
Olink Cq values for each measurement were determined using Fluidigm’s Real-Time PCR Analysis software and BiomarkDataCollection version 4.1.3.
Multiplexed SRM assays were established with the human SRMAtlas61 (www.srmatlas.org) and analyzed with Skyline62.
All pairwise statistical tests (Spearman) were performed using the Python scipy.stats package (v0.14). All regression models (OLS regressions and GEE) were fit using the Python Statsmodels package (v0.6)63. An independence working correlation structure was used for GEEs. Variance inflation factors were calculated using Python Statsmodels (v0.6) package. Correlation network p-values were adjusted for multiple hypothesis using the Benjamini-Hochberg10 method via the Python Statsmodels (v0.6) package for each inter-datatype comparison. Community analysis and modularity calculations were performed in Python with the NetworkX64 package, the python-louvain package (v0.3), and custom code. All custom code used in this manuscript can be downloaded from Github using the link below and is also available for download as Supplementary File 3. The Github version used for the manuscript is ‘v-release’.
https://github.com/trueprint/p100-network-code/tree/v-release
Running the custom code
This code is meant to be run in a Jupyter59 notebook that has the scipy stack installed. We recommend using the datascience docker image created by the Jupyter group at
https://github.com/jupyter/docker-stacks/tree/master/datascience-notebook
The raw data are available from dbGap in a compressed tar.gz file and should be extracted to the same directory containing the code. We recommend downloading the Jupyter docker image using docker pull jupyter/datascience-notebook on a machine with docker installed. This is not required, but is recommended and all instructions will be based on the use of this image. An example shell script is provided with the custom code (startup-notebook.sh). The startup command is:
docker run -d -p [SOME LOCAL PORT]:8888 -e USE_HTTPS=yes -e GRANT_SUDO=yes -v [LOCAL PATH TO p100-network-code]:[ROOT PATH OF NOTEBOOKS ON JUPYTER]/p100-network-code -v [LOCAL PATH TO UNZIPPED data]:[ROOT PATH OF NOTEBOOKS ON JUPYTER]/data jupyter/datascience-notebook
Then, navigate in your browser to https://[your url]:[SOME LOCAL PORT]. For example, if you ran this on your localhost, with SOME LOCAL PORT = 8888, then you would navigate to https://localhost:8888. Note: it will give you a warning about an invalid certificate. The default password for datascience-notebook is empty, i.e. just hit ENTER. The notebooks provided are: Generate correlation network.ipynb which generates a correlation network of all data for the p100 project, Community Generation-DELTA.ipynb which generates a correlation network for change in data measurements for the p100 project. Community Generation.ipynb which performs community analysis using the intraomic correlation network, Community Generation-DELTA.ipynb which performs community analysis using the intraomic delta(change) correlation network and GEE longitudinal clinical changes.ipynb which uses GEE (generalized estimating equations) to calculate average change over the course of the study in clinically relevant biomarkers. We also provide the notebook Convert csvs to pickles.ipynb, which converts raw CSV files and associated meta data in JSON from the csv folder of the data into Python pickles appropriate to the currently installed pandas version.
Supplementary Material
Acknowledgments
We would like to acknowledge significant contributions to this study from our 108 Pioneers, S. Kaplan, S. Mecca, S. Bell, G. Sorensen, C. Lewis, T. Kilgallon, M. Brunkow, S. Huang, C.-Y. Huang, D. Mauldin, S. Speck, M. Raff, J. Pizzorno, J. Guiltinan, R. Green, L. Smarr, E. Lazowska, C. Witwer, M. Flores, and many others who helped us on this wellness journey. This work was supported in part by the Robert Wood Johnson Foundation (LH, NDP), the M.J. Murdock Charitable Trust (LH, NDP), NIH grants 2P50GM076547 (LH, NDP), P30ES017885 (GSO), U24CA210967 (GSO), RC2HG005805 (RLM), and Arivale.
Footnotes
Accession Codes
All raw data collected as part of the P100 have been submitted to dbGaP with accession ID phs001363.v1.p1.
Competing Financial Interests Statement
LH and NDP are co-founders of Arivale and hold stock in the company. NDP is on the Arivale Board of Directors; LH is chair and GSO a member of Arivale’s Scientific Advisory Board. ATM, JCE, KB, and JCL are employees of Arivale and have stock options in the company, as do GG and GSO.
Author Contributions
LH and NDP conceived of and led the study. JCL designed and managed the clinical and coaching aspects of the study. ATM and JCE performed most of the computational analyses. GG contributed many important ideas from the beginning of the study. GG, RL, and DTM performed additional computational analysis. NDP, ATM, JCE, GG, RL, DTM, GSO, JCL, and LH analyzed data. CL generated the Olink proteomics data. UK, CM, YZ, SQ, and RLM generated the mass spectrometry proteomics data. KB managed most of the logistics of implementing the study. ATM, NDP, and LH were the primary writers of the paper, with contributions from all authors.
References
- 1.Hood L, Flores M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. N Biotechnol. 2012;29:613–624. doi: 10.1016/j.nbt.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011;8:184–187. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- 3.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yong PL, Saunders RS, Olsen L. The Healthcare Imperative: Lowering Costs and Improving Outcomes: Workshop Series Summary. National Academies Press (US); 2010. [PubMed] [Google Scholar]
- 5.David LA, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smarr L. Quantifying your body: a how-to guide from a systems biology perspective. Biotechnol J. 2012;7:980–991. doi: 10.1002/biot.201100495. [DOI] [PubMed] [Google Scholar]
- 8.Hood L, Price ND. Promoting Wellness and Demystifying Disease: The 100K Project. Clinical OMICs. 2014;1:20–23. [Google Scholar]
- 9.Welter D, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–6. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 1995;57:289–300. [Google Scholar]
- 11.Girvan M, Newman MEJ. Community structure in social and biological networks. Proc Natl Acad Sci USA. 2002;99:7821–7826. doi: 10.1073/pnas.122653799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newman MEJ. Modularity and community structure in networks. PNAS. 2006;103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koh KK, Park SM, Quon MJ. Leptin and cardiovascular disease: response to therapeutic interventions. Circulation. 2008;117:3238–3249. doi: 10.1161/CIRCULATIONAHA.107.741645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ridker PM. Clinical application of C-reactive protein for cardiovascular disease detection and prevention. Circulation. 2003;107:363–369. doi: 10.1161/01.cir.0000053730.47739.3c. [DOI] [PubMed] [Google Scholar]
- 16.Woo YC, Xu A, Wang Y, Lam KSL. Fibroblast growth factor 21 as an emerging metabolic regulator: clinical perspectives. Clin Endocrinol (Oxf) 2013;78:489–496. doi: 10.1111/cen.12095. [DOI] [PubMed] [Google Scholar]
- 17.Duong T, Pommier EC, Scheibel AB. Immunodetection of the amyloid P component in Alzheimer’s disease. Acta Neuropathol. 1989;78:429–437. doi: 10.1007/BF00688180. [DOI] [PubMed] [Google Scholar]
- 18.Jenny NS, Arnold AM, Kuller LH, Tracy RP, Psaty BM. Serum amyloid P and cardiovascular disease in older men and women: results from the Cardiovascular Health Study. Arterioscler Thromb Vasc Biol. 2007;27:352–358. doi: 10.1161/01.ATV.0000254150.97741.fe. [DOI] [PubMed] [Google Scholar]
- 19.Althaus BU, Staub JJ, Ryff-De Lèche A, Oberhänsli A, Stähelin HB. LDL/HDL-changes in subclinical hypothyroidism: possible risk factors for coronary heart disease. Clin Endocrinol (Oxf) 1988;28:157–163. doi: 10.1111/j.1365-2265.1988.tb03651.x. [DOI] [PubMed] [Google Scholar]
- 20.Jensen LJ, et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li N, Wallén NH, Ladjevardi M, Hjemdahl P. Effects of serotonin on platelet activation in whole blood. Blood Coagul Fibrinolysis. 1997;8:517–523. doi: 10.1097/00001721-199711000-00006. [DOI] [PubMed] [Google Scholar]
- 22.Sauer WH, Berlin JA, Kimmel SE. Selective serotonin reuptake inhibitors and myocardial infarction. Circulation. 2001;104:1894–1898. doi: 10.1161/hc4101.097519. [DOI] [PubMed] [Google Scholar]
- 23.Li M, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poesen R, et al. Microbiota-Derived Phenylacetylglutamine Associates with Overall Mortality and Cardiovascular Disease in Patients with CKD. J Am Soc Nephrol. 2016;27:3479–3487. doi: 10.1681/ASN.2015121302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manichanh C, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jostins L, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradley R, Fitzpatrick AL, Jenny NS, Lee DH, Jacobs DR. Associations between total serum GGT activity and metabolic risk: MESA. Biomark Med. 2013;7:709–721. doi: 10.2217/bmm.13.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim JS, Lee DH, Park JY, Jin SH, Jacobs DR. A strong interaction between serum gamma-glutamyltransferase and obesity on the risk of prevalent type 2 diabetes: results from the Third National Health and Nutrition Examination Survey. Clin Chem. 2007;53:1092–1098. doi: 10.1373/clinchem.2006.079814. [DOI] [PubMed] [Google Scholar]
- 29.Lang R, Gundlach AL, Kofler B. The galanin peptide family: receptor pharmacology, pleiotropic biological actions, and implications in health and disease. Pharmacol Ther. 2007;115:177–207. doi: 10.1016/j.pharmthera.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Prentice KJ, et al. The furan fatty acid metabolite CMPF is elevated in diabetes and induces β cell dysfunction. Cell Metab. 2014;19:653–666. doi: 10.1016/j.cmet.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Zheng JS, et al. Serum metabolomics profiles in response to n-3 fatty acids in Chinese patients with type 2 diabetes: a double-blind randomised controlled trial. Sci Rep. 2016;6:29522. doi: 10.1038/srep29522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan W, et al. Genome-wide association study of plasma N6 polyunsaturated fatty acids within the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet. 2014;7:321–331. doi: 10.1161/CIRCGENETICS.113.000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang TW, et al. Genome-wide association of serum bilirubin levels in Korean population. Hum Mol Genet. 2010;19:3672–3678. doi: 10.1093/hmg/ddq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothman N, et al. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat Genet. 2010;42:978–984. doi: 10.1038/ng.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okkels H, Sigsgaard T, Wolf H, Autrup H. Arylamine N-acetyltransferase 1 (NAT1) and 2 (NAT2) polymorphisms in susceptibility to bladder cancer: the influence of smoking. Cancer Epidemiol Biomarkers Prev. 1997;6:225–231. [PubMed] [Google Scholar]
- 36.Adams PC, et al. Hemochromatosis and iron-overload screening in a racially diverse population. N Engl J Med. 2005;352:1769–1778. doi: 10.1056/NEJMoa041534. [DOI] [PubMed] [Google Scholar]
- 37.Gaich G, et al. The effects of LY2405319, an FGF21 analog, in obese human subjects with type 2 diabetes. Cell Metab. 2013;18:333–340. doi: 10.1016/j.cmet.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Meier C, et al. TSH-controlled L-thyroxine therapy reduces cholesterol levels and clinical symptoms in subclinical hypothyroidism: a double blind, placebo-controlled trial (Basel Thyroid Study) J Clin Endocrinol Metab. 2001;86:4860–4866. doi: 10.1210/jcem.86.10.7973. [DOI] [PubMed] [Google Scholar]
- 39.Ruttmann E, et al. Gamma-glutamyltransferase as a risk factor for cardiovascular disease mortality: an epidemiological investigation in a cohort of 163,944 Austrian adults. Circulation. 2005;112:2130–2137. doi: 10.1161/CIRCULATIONAHA.105.552547. [DOI] [PubMed] [Google Scholar]
- 40.Thompson GA, Meister A. Interrelationships between the binding sites for amino acids, dipeptides, and gamma-glutamyl donors in gamma-glutamyl transpeptidase. J Biol Chem. 1977;252:6792–6798. [PubMed] [Google Scholar]
- 41.Soga T, et al. Serum metabolomics reveals γ-glutamyl dipeptides as biomarkers for discrimination among different forms of liver disease. Journal of Hepatology. 2011;55:896–905. doi: 10.1016/j.jhep.2011.01.031. [DOI] [PubMed] [Google Scholar]
- 42.Rogers AJ, et al. Metabolomic derangements are associated with mortality in critically ill adult patients. PLoS ONE. 2014;9:e87538. doi: 10.1371/journal.pone.0087538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sido B, et al. Impairment of intestinal glutathione synthesis in patients with inflammatory bowel disease. Gut. 1998;42:485–492. doi: 10.1136/gut.42.4.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo L, et al. Plasma metabolomic profiles enhance precision medicine for volunteers of normal health. Proc Natl Acad Sci USA. 2015;112:E4901–10. doi: 10.1073/pnas.1508425112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hood L, Price ND. Demystifying disease, democratizing health care. Sci Transl Med. 2014;6:225ed5–225ed5. doi: 10.1126/scitranslmed.3008665. [DOI] [PubMed] [Google Scholar]
- 46.Micheel CM, Nass SJ, Omenn GS. Evolution of Translational Omics: Lessons Learned and the Path Forward. National Academies Press (US); 2012. [PubMed] [Google Scholar]
- 47.Glusman G, et al. Identification of copy number variants in whole-genome data using Reference Coverage Profiles. Front Genet. 2015;6:45. doi: 10.3389/fgene.2015.00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Glusman G, Caballero J, Mauldin DE, Hood L, Roach JC. Kaviar: an accessible system for testing SNV novelty. Bioinformatics. 2011;27:3216–3217. doi: 10.1093/bioinformatics/btr540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caporaso JG, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rideout JR, et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. PeerJ. 2014;2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whittaker RH. Evolution and Measurement of Species Diversity. Taxon. 1972;21:213. [Google Scholar]
- 52.Caporaso JG, et al. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lane DJ. 16S/23S rRNA sequencing. Nucleic acid techniques in bacterial systematics. 1991:115–175. [Google Scholar]
- 56.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pérez F, Granger BE. IPython: A System for Interactive Scientific Computing. Computing in Science & Engineering. 2007;9:21–29. [Google Scholar]
- 60.Hunter JD. Matplotlib: A 2D Graphics Environment. Computing in Science & Engineering. 2007;9:90–95. [Google Scholar]
- 61.Kusebauch U, et al. Human SRMAtlas: A Resource of Targeted Assays to Quantify the Complete Human Proteome. Cell. 2016;166:766–778. doi: 10.1016/j.cell.2016.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Seabold S, Perktold J. Statsmodels: Econometric and statistical modeling with python. Proceedings of the 9th Python in Science …; 2010. pp. 57–61. [Google Scholar]
- 64.Schult DA, Swart P. Exploring network structure, dynamics, and function using NetworkX. Proceedings of the 7th Python in Science …; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.