Abstract
Direction switching in the flagellar motor of Escherichia coli is under the control of a complex on the rotor formed from the proteins FliG, FliM, and FliN. FliG lies at the top of the switch complex (i.e., nearest the membrane) and is arranged with its C-terminal domain (FliGC) resting on the middle domain (FliGM) of the neighboring subunit. This organization requires the protein to adopt an open conformation that exposes the surfaces engaging in inter-subunit FliGC/FliGM contacts. In a recent study, Baker and coworkers [Nat. Struct. Mol. Biol. 23(3), 197–203 (2016)] obtained evidence that FliG in the cytosol is monomeric and takes on a more compact conformation, with FliGC making intra-molecular contact with FliGM of the same subunit. In the present work, we examine the conformational preferences and interactions of FliG through in vivo crosslinking experiments in cells that lack either all other flagellar proteins or just the MS-ring protein FliF. The results indicate that FliG has a significant tendency to form multimers independently of other flagellar components. The multimerization of FliG is promoted by FliF, and also by FliM. FliM does not multimerize efficiently by itself but does so in the presence of FliG. Thus, pre-assemblies of the switch-complex proteins can form in the cytosol and might function as intermediates in assembly.
Graphical abstract
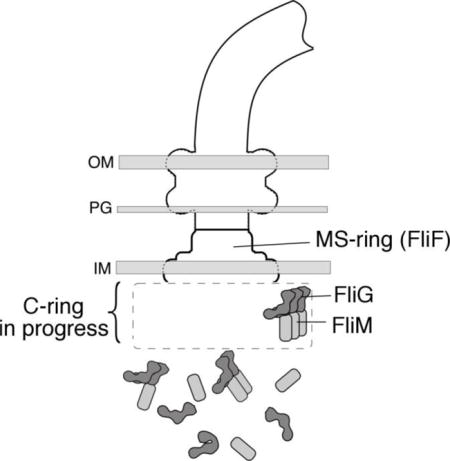
Introduction
Many species of motile bacteria direct their movement by regulating the direction of flagellar rotation [1, 2]. In E. coli and S. typhimurium, counterclockwise (CCW) rotation of the flagella produces smooth swimming and CW rotation causes rapid tumbling; as motors spontaneously switch direction, the cells alternately run and tumble, executing a largely random walk. Chemotaxis occurs when the runs that happen to be in a favorable direction are prolonged [3]. Reversals in motor direction are under the control of the ‘switch-complex’ mounted on the flagellar rotor [4, 5], which is formed, in E. coli and S. typhimurium, from the proteins FliG, FliM, and FliN. Structurally, the switch complex corresponds to the C-ring (C = Cytoplasmic) observed in electron microscopic reconstructions [6]. FliG functions most closely in the generation of torque [7] and is positioned at the top (membrane-proximal part) of the switch, where it interacts with the integral membrane protein MotA, which together with MotB forms the stator [8–11]. FliM and FliN occupy lower positions in the structure and are important for controlling the direction of rotation, presumably by regulating the position or orientation of parts of FliG.
Recent studies using pulsed dipolar ESR spectroscopy [12] and targeted crosslinking [13, 14] have clarified key aspects of switch-complex organization. A stacking interaction between the C-terminal and middle domains of FliG, first proposed by Lee et al. [15], appears well supported. In the assembled motor, each FliGC domain stacks onto FliGM of a neighboring subunit, forming an array of FliG subunits in the upper part of the complex. In a process termed “adaptive remodeling,” motors with excessive CCW bias add more copies of FliM to restore a more normal CW/CCW balance [16–19]; thus, the FliG array must be capable of adjusting to an underlying FliM array of variable size. We recently obtained evidence that a subset of the FliG subunits can adopt a more extended conformation that would allow the FliG array to enlarge as needed to conform to the number of FliM subunits present [14]. The positioning of the FliGC domain is especially important as it is the domain that contacts the stator and engages most directly in the generation of torque. Whatever the details of FliG organization, it is clear that in the motor the protein must adopt a relatively open conformation to expose the FliGC and FliGM surfaces involved in inter-subunit subunits.
In a recent study of the conformation and interactions of FliG, Baker et al. [13] obtained evidence that prior to its installation in the motor FliG exists in a more compact conformation, with FliGC stacked intra-molecularly on FliGM. Interaction of FliG with the basal body MS-ring (which is formed from the protein FliF) was hypothesized to trigger the multimerization of FliG by a ‘domain-swap’ mechanism involving the exchange of intra-molecular for inter-molecular contacts. The self-stacked conformation might serve to prevent premature and potentially detrimental polymerization of the protein in the cytoplasm [13]. In an extensive FRET-based study of flagellar assembly, Li and Sourjik [20] examined patterns of localization of several flagellar proteins in various mutant backgrounds and concluded that the MS-ring and switch complex assemble in a cooperative rather than strictly sequential fashion. They suggested, specifically, that components of the switch might facilitate assembly of the MS-ring, a direction of influence opposite that in the linear flagellar assembly pathway as it is usually depicted [21]. A subsequent fluorescence localization study by Morimoto et al. [22] further endorses this view.
In the present work, we have examined the conformation and interactions of FliG by studying crosslinking in mutant strains that are unable to synthesize flagella, owing to the absence of the MS-ring protein FliF or of all chromosomally encoded flagellar proteins. The results indicate that FliG has a significant tendency to form multimers even in the absence of any other flagellar proteins. This inherent tendency of FliG to multimerize is enhanced by both FliF and FliM. FliM shows little tendency to multimerize by itself but is effectively organized into multimers by FliG. The results support a scheme in which not only individual switch-protein subunits but a range of pre-assemblies are present in the cytosol and can contribute to construction of the switch complex. Such pre-assemblies can be quite large, judging from the cross-linked multimers observed, but do not appear to adversely affect cellular physiology.
Results
Baker and coworkers [13] probed the conformation and multimeric state of FliG by examining crosslinking through positions that give either intra- or inter-molecular disulfide bonds depending on whether the protein is in the closed (self-stacked) conformation or the open and multimeric (inter-molecularly stacked) state. Cellular localization of the crosslinked products was examined by lysing the cells, treating with oxidant to induce disulfide formation, then separating samples into membrane and soluble fractions for examination on immunoblots. The FliG in the membrane fraction, presumably representing protein installed in flagellar motors, was crosslinked into multimers, whereas the cytosolic fraction contained only monomer, in some cases including a faster-migrating band characteristic of an intra-molecular crosslink [13]. A potential concern with the approach is that the cytosolic contents were diluted by lysis of the cells prior to crosslinking, which might have caused existing multimers to dissociate and be counted as monomer in the crosslinking experiment.
As an alternative means of examining the state of association intrinsic to FliG, we carried out crosslinking experiments in intact cells lacking the MS-ring protein FliF (owing to deletion of the fliF gene) or lacking all chromosomally encoded flagellar proteins (owing to deletion of the master-regulator flhDC genes). Cells deleted for fliF were also deleted for fliG, to ensure that the plasmid-encoded FliG was the only version present. (Plasmid-based expression of FliG was necessary in these experiments because chromosomal flagellar genes are not expressed in the ΔflhDC background.) In initial experiments, expression of Cys-containing FliG proteins was induced with 30 μM IPTG. Cys replacements were made in FliGM and FliGC at positions previously shown to crosslink [13, 14].
On treatment with iodine, several of the Cys-containing FliG proteins were crosslinked into multimers even in the absence of FliF or any other flagellar proteins (Fig. 1). Cys pairs showing especially efficient crosslinking included 166/194 and 158/214, which also gave the highest yields in previous experiments using ΔfliG cells, where the protein is largely incorporated into flagella [13, 14]. Multimerization of FliG can thus occur in the absence of the MS-ring or any other flagellar components. With certain Cys pairs, a relatively weak band was also observed below the normal monomer, indicative of intra-molecular crosslinking of FliG that was in the intra-molecularly stacked conformation.
Figure 1.
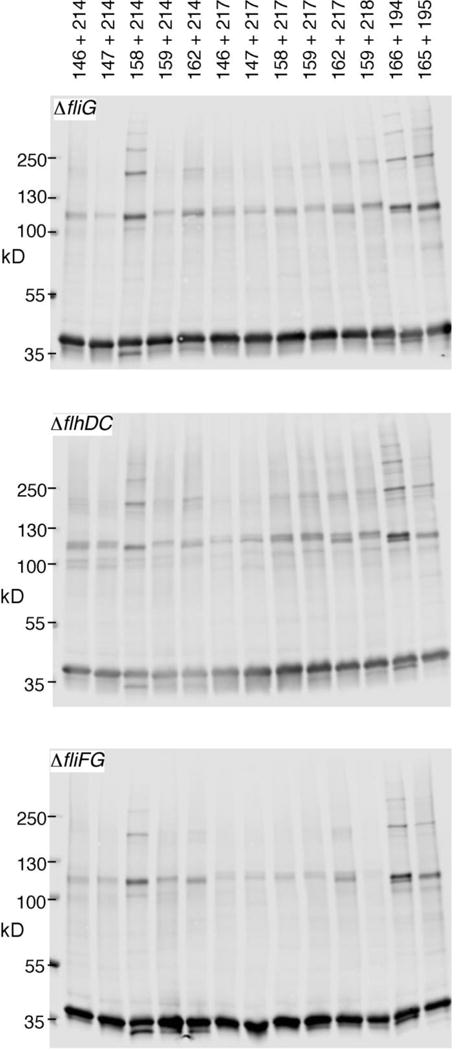
Cross-linking of FliG proteins in strains with various deletions of flagellar genes. Positions of Cys replacements are indicated. FliG expression was induced with 30 μM IPTG. Crosslinking was induced with 1 mM I2. Non-crosslinked controls are shown in Figure S1.
We previously examined the blocking of FliGC-FliGM crosslinking by the sulfhydryl reagent N-ethyl maleimide (NEM), in experiments using the 158C/214C protein expressed from the normal chromosomal locus [14]. Fairly strong reaction conditions (5 mM NEM for 30 min at 20° C) were needed to block the crosslinking, suggesting that the Cys residues are well protected from modification when the protein is present in assembled motors. We carried out a similar blocking experiment with the 158C/214C protein expressed (again from a plasmid) in the ΔflhDC background. In the ΔflhDC cells where none of the protein can be in motors, the mildest NEM treatment was able to block almost all crosslinking (Fig. 2). Thus, while the FliGM/FliGC interaction is strong enough to allow formation of multimers in the cytosol, it appears significantly looser, as judged by susceptibility to NEM, than the interaction in assembled motors. A blocking experiment using plasmid-based expression was also done in the ΔfliG background, for comparison; in this case, a significant portion of the crosslinking was resistant to block, as reported previously [14] (Figure 2).
Figure 2.
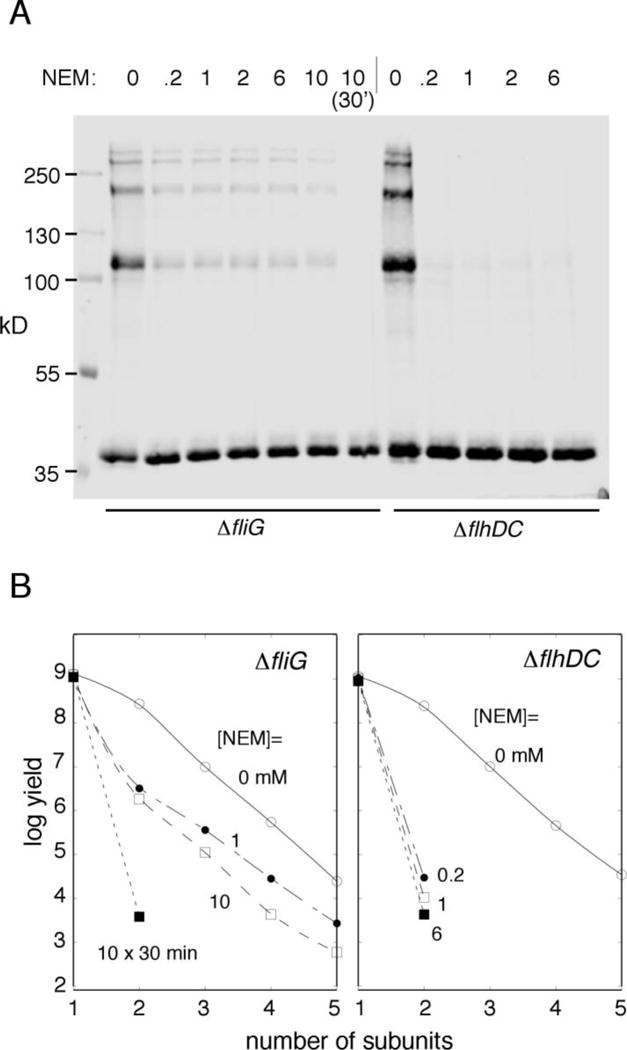
(A) Inhibition of crosslinking by pre-treatment with NEM, in cells that form flagella (ΔfliG) and nonflagellate (ΔflhDC) cells. Treatment with NEM was for 3 min except in the lane indicated (30 min). FliG expression was induced with 40 μM IPTG. (B) Quantification of the blocking experiment in panel A.
The Cys replacements at positions 158 and 214 are well tolerated in the sense of preserving nearly wild-type motility [14], but might nevertheless weaken the FliGM-FliGC interaction and thus promote the open conformation. The wild-type protein might then exist in mainly the self-stacked conformation. In this case, wild-type FliG should not readily join FliG multimers in the cytosol and should not inhibit ladder formation by the Cys-containing FliG. Inhibition was observed, however: With wild-type FliG expressed from a compatible second plasmid, total FliG level increased but crosslinking was decreased (Figure 3). Thus, wild-type FliG, like the Cys-containing protein used in crosslinking, appears able to adopt the open conformation.
Figure 3.
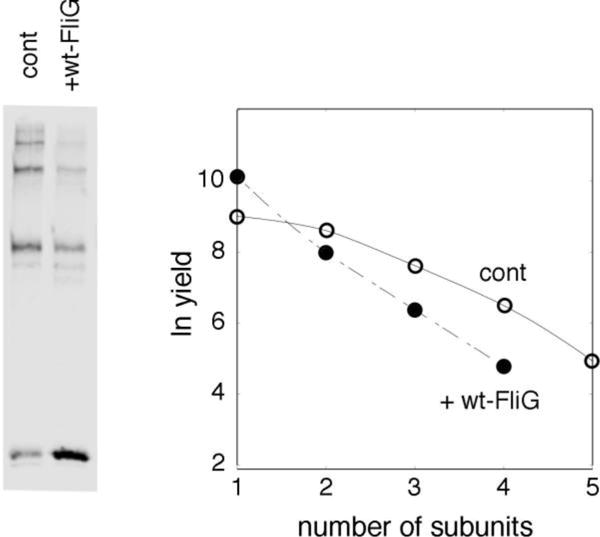
Inhibition of crosslinking of 158C/214C FliG by wild-type FliG. Cys-containing FliG was under control of the tac promoter with induction by 30 μM IPTG, and wild-type FliG was under control of a salicylate-regulated promoter with induction by 2.5 μM salicylate. Bands are quantified in the plot at the right. For quantifying yields, signals on immunoblots were assumed proportional to the total amount of protein in a band (so that, for example, one equivalent of dimer should give twice the signal of one equivalent of monomer).
While multimer formation by FliG does not strictly depend on any other flagellar proteins, it might be facilitated by other mechanisms that bring monomers into proximity, such as association with the membrane. Cross-linked products might then be found largely in the membrane fraction of ΔflhDC cells. Fractionation of cross-linked samples showed that this is not the case: Although most of the FliG ladder was found in the membrane fraction in cells that assemble flagella, in the ΔflhDC background the bulk of the cross-linked FliG was found in the soluble fraction (Fig. 4, panel A). As noted above, the absence of crosslinked products from the cytosolic fraction in previous experiments [13] might have been due to dilution-induced dissociation of FliG multimers upon lysis of the cells. To test this possibility, we carried out experiments in which cells were lysed either before or after treatment with I2, in both the ΔfliG and ΔflhDC backgrounds. In ΔfliG cells where the protein can be incorporated into flagella, crosslinking into multimers was observed using either protocol. In the ΔflhDC background, the protein crosslinked into multimers when the crosslinking was induced prior to lysis, but remained almost entirely monomeric when lysis came first (Fig. 4, panel B).
Figure 4.
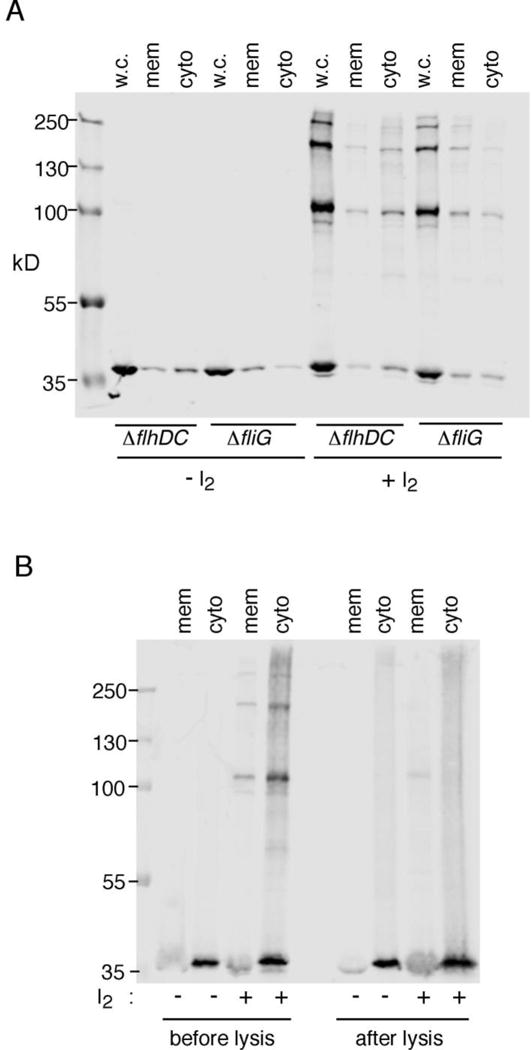
Cellular fractionation of products of FliG crosslinking. Cys residues were present at positions 158 and 214 in FliG. Expression was induced with 40 μM IPTG. Crosslinking was with 1 mM I2. (A) Fractionation experiment showing that most FliG multimer occurs in the membrane fraction in cells that make flagella, but in the soluble fraction of cells that do not. (B) Comparison of crosslinking before or after lysis of the cells.
In experiments above, the FliG induction level (30 μM IPTG) was sufficient for fair motility in liquid but lower than that needed for full complementation in a soft-agar motility assay (which required 100 μM IPTG; [23]). FliG is nevertheless somewhat overexpressed at this level of induction, relative to the levels in wild-type cells where the protein is expressed from the chromosome. Immunoblots of cells induced at various levels indicate that 10 μM IPTG suffices to give FliG levels like wild type (Fig. S2, panel A). In a crosslinking experiment in the ΔflhDC strain using induction by only 10 μM IPTG, yields were decreased but some crosslinked product was still formed (Fig. 5). The FliG level in wild-type cells thus appears poised near the threshold for multimer formation. This is in accordance with the suggestion by Baker et al. [13] that multimerization of FliG is triggered by binding to the MS-ring, formed from the protein FliF. FliF binds very strongly to the N-terminal domain of FliG [24–27]. To examine the influence of FliF, we carried out crosslinking experiments in the ΔflhDC strain with both fliF and fliG present on the plasmid, in their normal arrangement. Induction was with 15 μM IPTG, which gave FliG levels close to wild type using this plasmid (Figure S2, panel B). FliG multimer formation was modestly enhanced in the presence of FliF, with products extending to tetramer (Fig. 5, panel A). FliG also interacts with FliM [28–30]. In a crosslinking experiment with FliM expressed from a compatible second plasmid (again in the ΔflhDC background), crosslinking of FliG was modestly enhanced, to about the same extent as with FliF. FliM and FliN were also tested together but were no more effective than FliM alone in enhancing the crosslinking of FliG (not shown). The largest effect was observed with FliF and FliM together, which induced FliG to crosslink into ladders extending to at least octamer (Figure 5, panel A). To determine whether the FliM/FliG effect is reciprocal, we examined the crosslinking of FliM, through positions in its middle domain (residues 63 and 188) that were shown previously to crosslink efficiently in cells that synthesize flagella (a ΔfliM background) but not in nonflagellate ΔflhDC cells [31]. In the absence of any other flagellar proteins, FliM showed little crosslinking, confirming the earlier result. With FliG expressed from a compatible second plasmid, FliM crosslinked into products as large as tetramer (Fig. 5, panel B).
Figure 5.
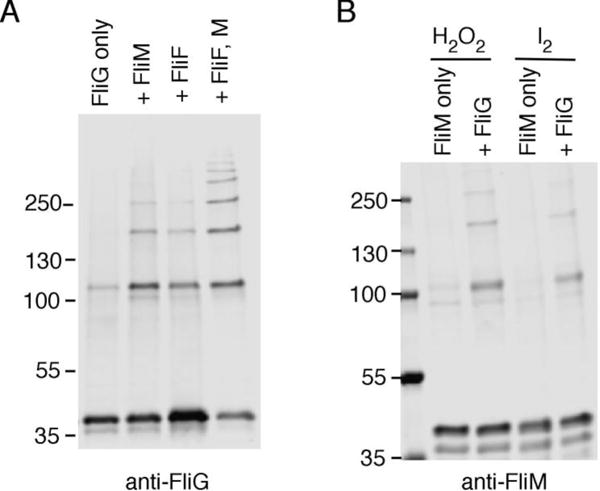
FliG and FliM pre-assemblies detected by crosslinking. (A) Comparison of FliG crosslinking in absence and presence of FliF and/or FliM. Cys residues were present at positions 158 and 214 in FliG. Experiments used the ΔflhDC strain that expresses no flagellar genes from the chromosome. Crosslinking was with 1 mM I2. In experiments that include FliF, FliF was expressed together with FliG from a plasmid encoding the genes in their normal tandem arrangement. Induction levels were those giving wild-type levels of FliG (10 μM for FliG; 15 μM for FliF + FliG; Figure S2). For experiments including FliM, FliM was expressed from a compatible second plasmid induced with 0.45 μM sodium salicylate (to give a wild-type FliM level; Figure S2, panel C). (B) Comparison of FliM crosslinking in the presence or absence of FliG. The experiment used the ΔflhDC strain. FliM had Cys replacements at positions 63 and 188 and was expressed from plasmid pEK454 with induction by 0.45 μM salicylate. FliG was expressed from plasmid pKG619 with induction by 10 μM IPTG. Crosslinking was induced with either 1 mM I2 or 48 mM H2O2, as indicated.
Discussion
The stacking of FliGC onto FliGM of the adjacent subunit has a key role in organizing the upper part of the flagellar switch complex [13–15]. FliGC can also stack intra-molecularly onto its ‘own’ FliGM domain, a conformation that was suggested to be the dominant state prior to installation, perhaps serving to prevent premature multimerization [13]. The present results indicate that FliG multimerization is indeed enhanced by FliF, as predicted [13], but also occurs to a significant extent in the absence of FliF or other flagellar proteins. FliG multimerization is promoted not only by FliF but also by FliM; multimerization of FliM is in turn assisted by FliG. The formation of FliG multimers and FliG/FliM assemblages in the cytosol does not appear to be harmful to the cells, as judged by normal growth and morphology of the ΔflhDC cells expressing the proteins. Although a substantial fraction of the cytosolic FliG is likely to be monomeric as suggested by Baker et al. [13], the finding that it is exclusively so may have been due to dilution (by cell lysis) prior to crosslinking (Figure 4).
Some pre-organization of FliG and FliM in the cytosol might facilitate the formation of the switch complex by allowing for the addition of various pre-assemblies in addition to individual protein subunits. A potential issue with such a mechanism is that the size and composition of arriving sub-complexes might not always be compatible with the gaps remaining in the structure at a given stage in assembly. Installation would fail in such cases if the sub-complexes are bound together strongly. The NEM-blocking experiment (Figure 3) indicates, however, that the inter-subunit contacts in the FliG multimers in the cytosol are less stable than those in the assembled motor. Arriving complexes should thus be able to shed excess subunits to allow installation of the parts that fit.
Our results are in accordance with the conclusions of Li and Sourjik [20], who monitored the localization of flagellar proteins in various deletion backgrounds and concluded that FliG and the other switch-complex proteins can assist in the assembly of the MS-ring, even as the MS-ring facilitates assembly of the switch complex. Such bi-directional action is not featured in the assembly sequence as typically drawn [21] but would fit with the ability of FliG and FliM to form associations independently of FliF. The early stages in flagellar assembly might then involve not just a few narrowly defined intermediates but a wide variety of sub-assemblies including partial MS-rings, FliF/FliG heterodimers, and switch-protein multimers.
Materials and Methods
Strains and media
Strains and plasmids are listed in Table 1. Strains were derivatives of Escherichia coli RP437, a gift of J.S. Parkinson. Site-directed mutations were made using the QuikChange method (Stratagene). DNA sequencing and oligonucleotide synthesis were carried out by core facilities at the University of Utah and by Genewiz, LLC (South Plainfield, NJ). TB medium contained 10 g tryptone and 5 g NaCl per liter. LB medium contained the same plus 5 g yeast extract. Ampicillin was used at 100 μg/ml and chloramphenicol at 50 μg/ml. Isopropyl-β-D-thiogalactopyranoside (IPTG) and sodium salicylate were prepared as aqueous 0.1 M stocks and used at the concentrations indicated in the figures.
Table 1.
Strains and Plasmids.
| Strain | Relevant genotype or property | source |
|---|---|---|
| RP437 | Wild-type for motility and chemotaxis | J. S. Parkinson |
| DB225 | ∆fliG in RP437 | [7] |
| RP3098 | ∆flhDC | J. S. Parkinson |
| EKS9 | ∆fliFG in RP437 | [24] |
| Plasmid | Relevant genotype or property | source |
| pRR48 | Ptac expression vector; AmpR | J.S. Parkinson |
| pKG116 | Salicylate-regulated expression vector; CmR | J. S. Parkinson |
| pKP619 | fliG in pRR48 | K.Paul |
| pEK176 | fliM in pKG116 | This study |
| pKP617 | fliG L159C/V218C in pKP619 | K.Paul |
| pEK256 | fliG L146C/V214C in pKP619 | This study |
| pEK257 | fliG A147C/V214C in pKP619 | This study |
| pEK258 | fliG M158C/V214C in pKP619 | This study |
| pEK259 | fliG L159C/V214C in pKP619 | This study |
| pEK260 | fliG A162C/V214C in pKP619 | This study |
| pEK261 | fliG L146C/A217C in pKP619 | This study |
| pEK262 | fliG A147C/A217C in pKP619 | This study |
| pEK263 | fliG M158C/A217C in pKP619 | This study |
| pEK264 | fliG L159C/A217C in pKP619 | This study |
| pEK265 | fliG A162C/A217C in pKP619 | This study |
| pEK312 | fliFG in pRR48 | This study |
| pEK349 | fliG G166C/G194C in pKP619 | This study |
| pEK353 | fliG G165C/G195C in pKP619 | This study |
| pEK454 | fliM R63C/F188C in pEK176 | This study |
Crosslinking
Cells expressing proteins (FliG or FliM) with Cys replacements were cultured overnight to saturation in LB, then diluted 100-fold into TB plus antibiotic and inducer and re-cultured at 32° C to midlog (typically 4.5 h). Cells were collected by centrifugation and washed into XL buffer (200 mM Na-phosphate, 150 mM NaCl, pH 7.5). Oxidation with I2 (1mM) iodine or H2O2 (48mM) was carried out as described previously [32]. Samples were mixed with an equal volume of 2X non-reducing loading buffer and heated at 95 °C for 10 min before loading on SDS-PAGE gels.
SDS page and immunoblotting
Proteins were resolved by SDS-PAGE (either 7.5% gels or 4%–20% gradient gels) and transferred onto nitrocellulose using a Transblot turbo or semi-dry apparatus (BioRad). Rabbit polyclonal antibodies against FliG and FliM were used at 1:1,000 dilution in a solution containing phosphate-buffered saline pH 7.4, 0.1% gelatin, and 0.01% Na-azide. Immunoblots were visualized and analyzed using the LiCor Odyssey infrared-imaging system.
Membrane Fractionation
For lysis and x-link fractionation, cells of either the ΔfliG or ΔflhDC strain expressing the M158C/V214C FliG variant (from plasmid pEK258) were cultured overnight at 32° C, diluted 100 fold into TB with 40 μM IPTG, then grown at 32° C to mid-log. OD600 was measured to adjust the cell density, then equal numbers of cells were pelleted and resuspended in 200 μl of lysis buffer (50mM Tris pH 8.0, 0.5 M sucrose, 10 mM EDTA, 0.2 mg/ml lysozyme) to an OD600 of 0.7–0.8. Cells were incubated on ice for 1 h then subjected to osmotic shock by addition of 1.8 ml of ice-cold water. Samples were sonicated briefly (Branson model 250; power 3, duty cycle 50%, 10 s). Membrane and cytosolic fractions were separated by centrifugation (16,000 × g for 30 min, 4° C). Pellets (membrane) were resuspended in 1× non-reducing dye. The supernatant (cytosol) was transferred to a new tube, mixed with 10% trichloroacetic acid solution and incubated on ice for 10 min, then centrifuged (16,000 × g for 10 min, 4° C), washed with acetone, and centrifuged once more. Precipitated protein was resuspended in 1× reducing dye and immunoblotted with anti-FliG. Cross-linking with I2 was done using the procedure described above, either before or after cell lysis as indicated in the figures.
Supplementary Material
Highlights.
Bacterial chemotaxis is regulated by a 3-protein switch complex whose assembly is imperfectly understood
The work addresses questions regarding the sequence of events in assembly
Evidence is obtained for pre-assemblies containing the proteins FliG, FliM, and FliF
FliG concentration appears poised to allow assembly when assisted by partners
This represents a revision of previous schemes in which assembly follows a strictly linear sequence
Acknowledgments
We thank Sandy Parkinson for strains and Richard Berry for comments on the manuscript. Supported by NIH grant GM-46683 to D.F.B.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Larsen SH, Reader RW, Kort EN, Tso W-W, Adler J. Change in direction of flagellar rotation is the basis of the chemotactic response in E. coli. Nature. 1974;249:74–7. doi: 10.1038/249074a0. [DOI] [PubMed] [Google Scholar]
- 2.Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nature Rev Mol Cell Biol. 2004;5:1024–37. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 3.Berg H, Brown D. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature. 1972;239:500–4. doi: 10.1038/239500a0. [DOI] [PubMed] [Google Scholar]
- 4.Yamaguchi S, Aizawa S-I, Kihara M, Isomura M, Jones CJ, Macnab RM. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–9. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisenbach M, Caplan SR. Unsolved mystery of the flagellar switch. Current Biology. 1998;8:R444–R6. doi: 10.1016/s0960-9822(98)70288-x. [DOI] [PubMed] [Google Scholar]
- 6.Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica Serovar typhimurium. J Bacteriol. 2008;188:7039–48. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lloyd SA, Tang H, Wang X, Billings S, Blair DF. Torque generation in the flagellar motor of Escherichia coli: Evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol. 1996;178:223–31. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou J, Lloyd SA, Blair DF. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1998;95:6436–41. doi: 10.1073/pnas.95.11.6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takekawa N, Kojima S, Homma M. Contribution of many charged residues at the stator-rotor interface of the Na+-driven flagellar motor to torque generation in Vibrio alginolyticus. J Bacteriol. 2014;196:1377–85. doi: 10.1128/JB.01392-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morimoto YV, Nakamura S, Kami-ike N, Namba K, Minamino T. Charged resiudes in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol Microbiol. 2010;78:1117–29. doi: 10.1111/j.1365-2958.2010.07391.x. [DOI] [PubMed] [Google Scholar]
- 11.Morimoto YV, Nakamura S, Hiraoka KD, Namba K, Minamino T. Distinct roles of highly conserved charged residues at the MotA-FliG interface in bacterial flagellar motor rotation. J Bacteriol. 2013;195:474–81. doi: 10.1128/JB.01971-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sircar R, Borbat PP, Lynch MJ, Bhatnagar J, Beyersdorf MS, Halkides C, et al. Assembly states of FliM and FliG within the flagellar switch complex. J Mol Biol. 2015;427:867–86. doi: 10.1016/j.jmb.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker MA, Hynson RM, Ganuelas LA, Mohammadi NS, Liew CW, Rey AA, et al. Domain-swap polymerization drives the self-assembly of the bacterial flagellar motor. Nat Struct Mol Biol. 2016;23:197–203. doi: 10.1038/nsmb.3172. [DOI] [PubMed] [Google Scholar]
- 14.Kim EA, Panushka J, Meyer T, Carlisle R, Ogden S, Ide N, et al. Architecture of the flagellar switch complex of Escherichia coli: Conformational plasticity of FliG and implications for adaptive remodeling. J Mol Biol. 2017 doi: 10.1016/j.jmb.2017.02.014. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature. 2010;466:996–1000. doi: 10.1038/nature09300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan J, Branch RW, Hosu BG, Berg HC. Adaptation at the output of the chemotaxis signalling pathway. Nature. 2012;484:233–6. doi: 10.1038/nature10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lele PP, Branch RW, Nathan VS, Berg HC. Mechanism for adaptive remodeling of the bacterial flagellar switch. Proc Natl Acad Sci USA. 2012;109:20018–22. doi: 10.1073/pnas.1212327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Delalez NJ, Berry RM, Armitage JP. Stoichiometry and turnover of the bacterial flagellar switch protein FliN. mBio. 2014;5 doi: 10.1128/mBio.01216-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Branch RW, Sayegh MN, Shen C, Nathan VSJ, Berg HC. Adaptive remodelling by FliN in the bacterial rotary motor. J Mol Biol. 2014;426:3314–24. doi: 10.1016/j.jmb.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Sourjik V. Assembly and stability of flagellar motor in Escherichia coli. Mol Microbiol. 2011;80:886–99. doi: 10.1111/j.1365-2958.2011.07557.x. [DOI] [PubMed] [Google Scholar]
- 21.Macnab RM. How bacteria assemble flagella. Ann Rev Microbiol. 2003;57 doi: 10.1146/annurev.micro.57.030502.090832. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 22.Morimoto YV, Ito M, KHiraoka KD, Che Y-S, Bai F, Kami-ike N, et al. Assembly and stoichiometry of FliF and FlhA in Salmonella flagellar basal body. Mol Microbiol. 2014;91:1214–26. doi: 10.1111/mmi.12529. [DOI] [PubMed] [Google Scholar]
- 23.Kim EA, Blair DF. Function of the histone-like protein H-NS in motility in Escherichia coli: Multiple regulatory roles rather than direct action at the flagellar motor. J Bacteriol. 2015;197:3110–20. doi: 10.1128/JB.00309-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kihara M, Miller GU, Macnab RM. Deletion analysis of the flagellar switch protein FliG of Salmonella. J Bacteriol. 2000;182:3022–8. doi: 10.1128/jb.182.11.3022-3028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levenson R, Zhou H, Dahlquist FW. Structural insights into the interaction between the gadterial flagellar motor prpoteinw FliF and FliG. Biochemistry. 2012;51:5052–60. doi: 10.1021/bi3004582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oosawa K, Ueno T, Aizawa S-I. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J Bacteriol. 1994;176:3683–91. doi: 10.1128/jb.176.12.3683-3691.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lynch MJ, Levenson R, Kim EA, Sircar R, Blair DF, Dahlquist FW, et al. Cofolding of a FliF-FliG split domain forms the basis of the MS:C ring interface within the bacterial flagellar motor. Structure. 2016 doi: 10.1016/j.str.2016.12.006. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang H, Braun TF, Blair DF. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–21. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 29.Toker AS, Macnab RM. Distinct regions of bacterial flagellar switch protein FliM interact with FliG, FliN and CheY. J Mol Biol. 1997;273:623–34. doi: 10.1006/jmbi.1997.1335. [DOI] [PubMed] [Google Scholar]
- 30.Vartanian AS, Paz A, Fortgang EA, Abramson J, Dahlquist FW. Structure of flagellar motor proteins in complex allows for insights into motor structure and switching. J Biol Chem. 2012;287:35779–83. doi: 10.1074/jbc.C112.378380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR. Structure of FliM provides insight into the assembly of the switch compex in the bacterial flagella motor. Proc Natl Acad Sci USA. 2006;103:11886–91. doi: 10.1073/pnas.0602811103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul K, Gonzalez-Bonet G, Bilwes AM, Crane BR, Blair DF. Architecture of the flagellar rotor. EMBO J. 2011;30:2962–71. doi: 10.1038/emboj.2011.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


