Abstract
The human cerebral cortex depends for its normal development and size on a precisely controlled balance between self-renewal and differentiation of diverse neural progenitor cells. Specialized progenitors that are common in humans, but virtually absent in rodents, called ‘outer radial glia’ (ORG), have been suggested to be crucial to the evolutionary expansion of the human cortex. We combined progenitor subtype-specific sorting with transcriptome-wide RNA-sequencing to identify genes enriched in human ORG, which included targets of the transcription factor Neurogenin and previously uncharacterized, evolutionarily dynamic long noncoding RNAs. We show that activating the Neurogenin pathway in ferret progenitors promotes delamination and outward migration. Finally, single-cell transcriptional profiling in human, ferret, and mouse revealed more cells co-expressing proneural Neurogenin targets in human compared to other species, suggesting greater neuronal lineage commitment and differentiation of self-renewing progenitors. Thus, we find that the abundance of human ORG is paralleled by increased transcriptional heterogeneity of cortical progenitors.
The neurons of the cerebral cortex are generated from several diverse types of neural progenitor cells whose molecular controls and lineage relationships are still not well understood. In the embryonic mouse brain, two primary progenitor subtypes that produce the excitatory projection neurons of the neocortex are clearly distinguishable by their germinal zone location, morphology, gene expression, and lineage potential1. Radial glial cells (RGC), a progenitor subtype shared by all mammals, are highly polarized, epithelial-like progenitors whose cell bodies reside in the ventricular zone (VZ) and possess apical processes integrated into the ventricular surface, while a radial process extends basally to the pial basement membrane. RGC are multipotent and self-renewing, undergoing mitosis at the ventricular surface and sequentially producing excitatory neurons of all cortical layers as well as glial lineages2. RGC primarily produce neurons indirectly, generating intermediate progenitors (IP) which, in contrast to RGC, are multipolar, non-epithelial cells, basally located in the subventricular zone (SVZ), with limited capacity for self-renewal and fate-restricted to produce neurons3.
In contrast to rodents, primate cortical germinal zones, especially those of the human, are more complex, as exemplified by the dramatic expansion and subdivision of the SVZ into inner and outer compartments containing heterogeneous populations of progenitors with diverse morphological and molecular characteristics4–7. Most notably, basal or outer radial glial cells (ORG), which are highly abundant in the human fetal cortex but rare in the mouse, intriguingly display characteristics of both RGC and IP: although delaminated and basally located in the SVZ, ORG retain a radial process that frequently reaches the pial surface, and express many of the transcription factors and cytoskeletal markers of apical RGC5,6. Despite undergoing mitosis in the SVZ, ORG appear less restricted than multipolar IP in their self-renewal capacity and lineage potential: they can divide symmetrically to produce two daughter ORG8, and give rise to both neurons and astrocytes9. Finally, a subset of ORG co-expresses TBR26,9,10, the proneural transcription factor associated predominantly with IP in rodents. Unfortunately, although these findings highlight the need for a more detailed molecular characterization of ORG and other human progenitor subtypes, the paucity of ORG in the mouse has presented a significant barrier to better understanding their molecular and cellular identity.
Recent applications of high-throughput technologies have produced extensive transcriptome-wide atlases of gene expression in the human fetal brain, providing valuable insights into the evolution of human cortical neurogenesis and patterning11–15. Surprisingly, however, these studies so far have not uncovered a distinctive transcriptional signature of the expanded human outer SVZ, or of the ORG that reside there. The remarkable cellular heterogeneity of the human germinal zones may obscure such a signal, since comparisons of bulk tissue samples collected by microdissection are limited to producing an average gene expression profile of the many cell types present in the samples. The SVZ in particular harbors various subtypes of radial and non-radial progenitors, radially migrating projection neurons, and tangentially migrating interneurons originating from separate progenitor pools in the ventral telencephalic germinal zones.
In the present study, we dissected the cellular heterogeneity of the fetal human cortex by first isolating the cell populations of interest from dissociated tissue, and secondly applying single-cell gene expression profiling. Using this approach, we found hundreds of genes that are specifically enriched in apical RGC, and a smaller but distinct transcriptional signature of human ORG. Interestingly, the ORG transcriptional profile was dominated by proneural transcription factors of the Neurogenin pathway, indicating that at the population level, ORG represent a distinct reservoir of neuronal lineage-committed, self-renewing radial progenitors. We overexpressed the Neurogenin pathway in the developing cortex of the ferret, a carnivore with abundant ORG, and confirmed that this pathway has a conserved function in processes critical to ORG formation, including delamination from the ventricular neuroepithelium and migration into the SVZ. A single-cell comparative transcriptional analysis of human, ferret and mouse progenitors confirmed that Neurogenin pathway-expressing cells are more abundant among human progenitors, and are less common in mice, which lack a large ORG subpopulation. More generally, single-cell profiling revealed a surprising transcriptional heterogeneity of human and, to a lesser degree, ferret cortical progenitors, which we propose reflects an extended, more graded transcriptional transition from RGC to IP in these species, characterized by a large proportion of cells co-expressing classic markers of both self-renewing RGC and neuronal lineage-committed IP. Finally, comparative genomic analysis of several previously undescribed, human ORG-enriched long noncoding RNA (lncRNA) genes indicated that many of these loci, while potentially present in the common ancestor of human, ferret, and mouse, show highly distinct patterns of ORG expression accompanied by greater genomic sequence divergence in rodents.
RESULTS
Purification and RNA-seq analysis of human ORG
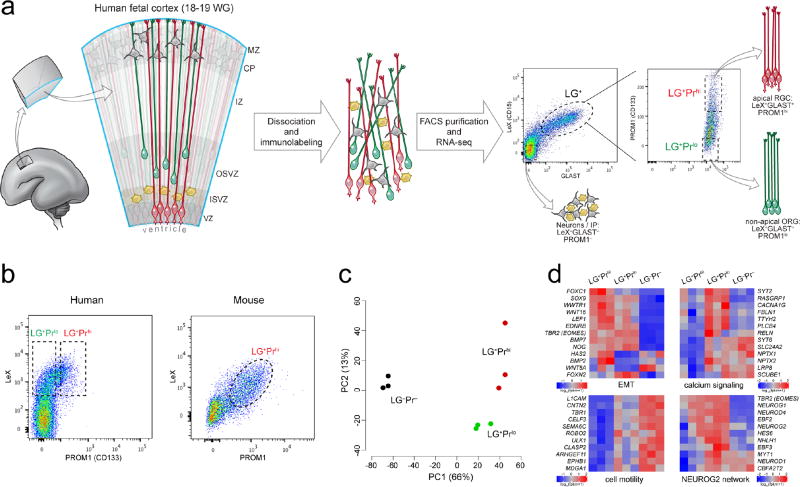
We used the differential expression of surface markers to separate cortical progenitor subtypes using fluorescence-activated cell sorting (FACS) prior to RNA-seq (Fig. 1a). Human apical RGC, the epithelial progenitor subtype, express LeX (CD15) and GLAST (SLC1A3)16–18, as well as prominin (PROM1; CD133) on their apical surface19,20. ORG express LeX and GLAST, but lack apical proteins including PROM15. Intermediate progenitors (IP) and neurons lack all three markers. Therefore we separated LeX and GLAST positive (LG+) cells showing the top (LG+Prhi) and bottom (LG+Prlo) 5–10% of PROM1 signal intensity to enrich for apical RGC and non-apical ORG, respectively, as well as cells negative for all three markers (LG−Pr−) comprising IP and neurons, among other cells, for RNA-seq analysis (Fig. 1a and Table 1).
Figure 1. Transcriptional profiling of isolated human radial glial cells distinguishes apical from non-apical subpopulations.
a, Workflow and strategy for FACS isolation of human RGC subpopulations by cell surface marker expression. LeX and GLAST are used as pan-RGC markers (LG+) while PROM1 is used to select apical (Prhi) and non-apical (Prlo) subpopulations from within the LG+ pool. WG, weeks of gestation. b, Human LG+ cells are predominantly LG+Prlo (~80%) whereas virtually all mouse LG+ cells are LG+Prhi (>95%) consistent with the relative abundance of ORG in humans and their paucity in mouse. c, Principle component (PC) analysis of transcriptome-wide gene expression estimates (FPKM) across three biological replicates of FACS-separated subpopulations reveals major gene expression differences between LG+ progenitors and LG− cells (first PC, x-axis), as well as between LG+Prhi apical and LG+Prlo non-apical radial glial subtypes (second PC, y-axis). d, Differential expression between LG+Prhi apical and LG+Prlo non-apical RGC subpopulations included genes involved in calcium signaling, epithelial-to-mesenchymal transition (EMT), and cell migration and motility, as well as several members of a proneural transcription factor network regulated by the transcription factor NEUROG2.
Table 1. Cell populations immunolabeled and collected by FACS from human fetal cortex for RNA-seq analysis.
Dissociated fetal human cortical tissue was immunolabeled for radial glial progenitor markers LeX (CD15), GLAST (SLC1A3), and PROM1 (Prominin; CD133). LeX and Glast are detected on the vast majority of radial glial cells16–18, while Prom1 is specific to the apical membrane domain found at the ventricular surface19,20.
| Sorted Pool | Antigenicity | Enriched Markers (qRT-PCR) | Cell Type |
|---|---|---|---|
| LG+Prhi | LeX+, Glast+, Prom1-high | PAX6, SOX2, VIM, NES, BLBP, PARD3 (PAR3), TJP1 (ZO1), MPP5 (PALS) | apical radial glial cells |
| LG+Prlo | LeX+, Glast+, Prom1-low | PAX6, SOX2, VIM, NES, BLBP | non-apical radial glial cells, including ORG |
| LG−Pr− | LeX−, Glast−, Prom1− | TUJ1, DCX, MEF2C, NEUN | intermediate progenitors, neurons |
Multiple lines of evidence confirm that our sorting approach enriches for RGC while separating apical from non-apical subpopulations. First, qRT-PCR confirmed that both LG+Prhi and LG+Prlo cells were enriched for markers of neural progenitors and radial glia, while being depleted for neuronal genes, compared to the LG−Pr− population (Supplementary Fig. 1a). LG+Prhi cells showed enrichment for mRNAs encoding PROM1 and other apical membrane proteins compared to LG+Prlo cells (Supplementary Fig. 1a). LeX+ cells proliferated in vitro to produce neurospheres that showed SOX2 immunoreactivity and could be serially passaged at clonal density, consistent with neural stem cell behavior16 (Supplementary Fig. 1b). Furthermore, in RGC sorted from embryonic mouse cortex, PROM1 highly overlapped with both LeX and GLAST, with few LG+Prlo cells detected, confirming the scarcity of non-apical ORG in mouse (Fig. 1b). Finally, since non-apical multipolar IP lack PROM1, the absence of a significant LG+Prlo population in the mouse also corroborates the absence of LeX and GLAST on IP, as these would appear to be LG+Prlo by FACS. Thus, our method provides an unprecedented opportunity to assay transcriptome-wide differences between, as well as heterogeneity within, human progenitor subtypes.
RNA-seq of the three FACS-enriched cell populations from three biological replicates (18–19 weeks of gestation [WG]; Supplementary Table 1) identified ~3,500 known genes with significantly different expression, as well as ~250 differentially expressed non-reference novel transcripts (FDR < 5%; FPKM > 1). Principle component analysis indicated that the greatest proportion of variability between samples reflected the differences between the LG+ RGC and LG− cells, but the second principle component highlighted differences between the LG+Prhi apical and LG+Prlo non-apical subpopulations, indicating a distinct ORG transcriptional signature (Fig. 1c). Gene set enrichment analysis further demonstrated the radial glial progenitor nature of the LG+ population: relative to the LG−Pr− pool, LG+ cells were enriched for genes involved in cell cycle regulation, DNA replication, extracellular matrix, and growth factor pathways critical for RGC maintenance and neurogenesis (Supplementary Fig. 2). Notably, LG+ enriched genes included integrin signaling and basement membrane components, such as laminins, consistent with both LG+Prhi and LG+Prlo subpopulations maintaining radial processes contacting the pial basement membrane, as has been shown for ORG5,6,9. In all, 552 genes significantly differed between LG+Prhi and LG+Prlo cells, with 79 of these genes specifically enriched or depleted in LG+Prlo non-apical ORG (Fig. 1d and Supplementary Fig. 3). Interestingly, among the genes upregulated in LG+Prlo ORG, six transcription factors (TFs) – HES6, NEUROD4 (Atoh3, Math3), NHLH1 (HEN1, NSCL1), NEUROD1, CBFA2T2 (Mtgr1), and MYT1 – are all downstream of the critical regulatory gene NEUROG221,22, which in mouse cortex and chick spinal cord initiates delamination and neuronal lineage commitment of neural precursors23–25. Interestingly, NEUROG2 itself and two additional early markers of neuronal fate commitment – TBR2 (EOMES)23 and BTG2 (Tis21)26 – were all highly expressed in both LG+Prhi and LG+Prlo subpopulations (Fig. 1d). Given that NEUROG2 expression is transient in mouse apical progenitors during their transition from RGC to TBR2+ IP23,27, we sought to validate the expression of NEUROG2 in apical and non-apical RGC, and to test its function in a model species with abundant cortical ORG.
NEUROG2 function in RGC of the gyrencephalic ferret cortex
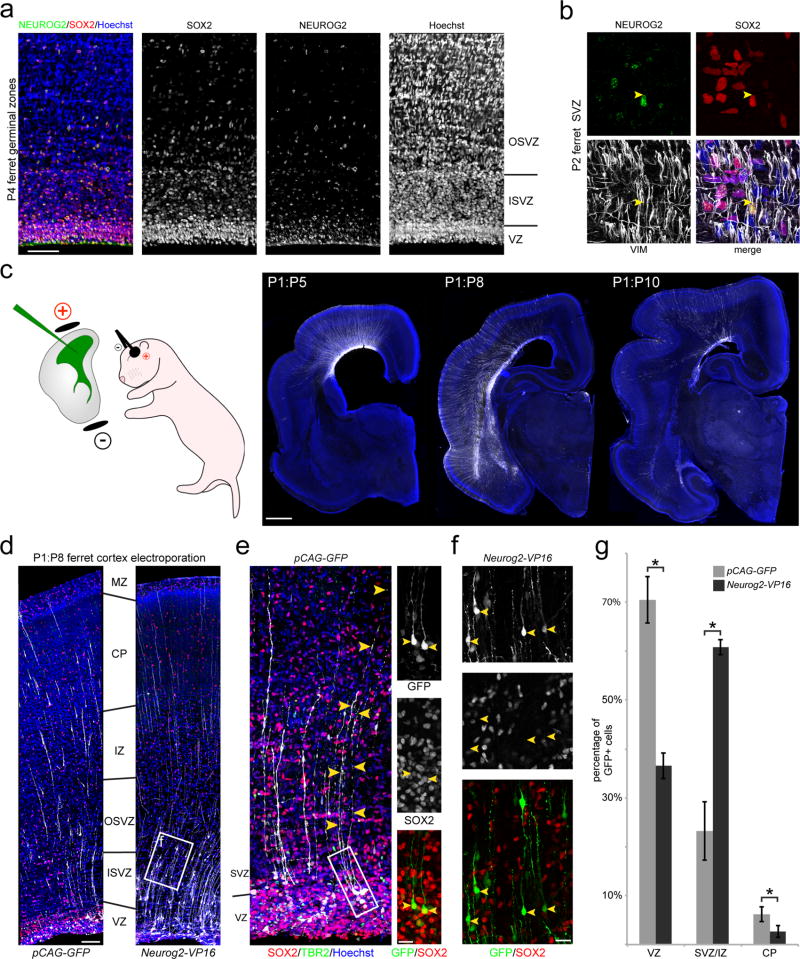
The developing ferret and human cerebral cortices share several key features including stereotyped sulci and gyri, a dramatically expanded subventricular zone (SVZ), and an abundance of ORG, making the ferret an attractive model for the study of cortical neurogenesis9,28,29. We first confirmed the expression of NEUROG2 in ferret RGC (Fig. 2a,b), identifying a marked “salt-and-pepper” pattern of NEUROG2 immunoreactivity in both the VZ and SVZ. We then used in vivo electroporation of the newborn ferret dorsal cortical VZ to express a NEUROG2-VP16 fusion protein, which links the DNA-binding domain of NEUROG2 to the VP16 constitutive transcriptional activator domain25, thus activating all direct downstream targets of NEUROG2 in ferret apical RGC. Following delivery of the Neurog2-VP16 expression construct, we allowed ferret kits to develop for up to ten days, during which time many neurons of the upper cortical layers are generated30–32 (Fig. 2c). In both control (pCAG-GFP) and Neurog2-VP16 brains, we observed numerous GFP+SOX2+ and GFP+TBR2+ cells in the VZ, including cells in the basal VZ and inner SVZ with a characteristic ORG morphology (Fig. 2d,e); as well many GFP+ cells in the SVZ and intermediate zone (IZ), with a small number reaching the cortical plate (CP) after longer survival times (up to ten days post-electroporation [DPE]) (Fig. 2f). After 7–9 DPE, NEUROG2-VP16 induced a significant shift in the proportion of GFP+ cells from the VZ to the SVZ/IZ, and a concomitant reduction in the proportion of GFP+ cells co-expressing SOX2 (Fig. 2g), with the majority of NEUROG2-VP16+ cells displaying the morphology of radially migrating postmitotic neurons in the outer SVZ and IZ. In addition, we FACS-purified electroporated cells from the ferret cortex and performed qRT-PCR analysis of ORG-enriched candidate genes identified in humans, and found that nearly all human ORG-enriched NEUROG2 downstream targets were highly upregulated by the NEUROG2-VP16 construct in ferret, relative to control GFP-expressing cells, while Sox2 was repressed (Supplementary Fig. 4). Notably, NEUROG2-VP16 expression in ferret RGC in vivo also resulted in increased expression of the ferret orthologs of several other human ORG-enriched genes, including Gadd45g and Ttyh2 (Supplementary Fig. 4), further suggesting that NEUROG2 is a critical regulator of a conserved radial progenitor development program in species with abundant ORG. Altogether, our ferret functional experiments demonstrate a conserved role for NEUROG2 transcriptional targets in driving delamination from the ventricular neuroepithelium, which is a key step in the production of ORG, while additional downstream effectors initiate repression of Sox2 and activation of a neuronal differentiation program, including radial migration, as previously described in mice. Future studies will be required to identify the specific factors downstream of NEUROG2 that regulate neuroepithelial integration, and elucidate the molecular mechanisms that permit a subset of NEUROG2-expressing ferret and human RGC to remain integrated in the VZ, while others detach and migrate into the SVZ.
Figure 2. NEUROG2 regulates progenitor morphology and molecular identity in thedeveloping cortex of the gyrencephalic ferret.
a, Co-expression of RGC marker SOX2 and proneural marker NEUROG2 in developing ferret cortex. Numerous SOX2+NEUROG2+ cells are found in both the VZ and SVZ at early postnatal ages, when these germinal zones are populated respectively by large numbers of apical and non-apical RGC producing neurons destined for the upper cortical layers. b, Higher magnification of SOX2+NEUROG2+ progenitors in the SVZ co-expressing the RGC neurofilament protein Vimentin (VIM), which labels the basal radial process (yellow arrowhead). c, Genetic manipulation of apical RGC is achieved by in vivo intraventricular injection and electroporation in neonatal ferret kits. After several days post-electroporation (DPE), GFP+ cells are observed throughout the developing cortical wall, with GFP+ radial fibers extending from the germinal zones to the pial surface and newborn neurons migrating through the SVZ and intermediate zone (IZ) into the cortical plate (CP). d, Littermates electroporated at postnatal day 1 and harvested 7 days later (P1:P8) with NEUROG2 gain-of-function (pCAG-Neurog2-VP16) or GFP control (pCAG-GFP) expression constructs, analyzed for the distribution of GFP+ cells and their co-expression of SOX2. ISVZ, inner SVZ; OSVZ, outer SVZ; MZ, marginal zone. e, At all survival time-points, we identified numerous GFP+SOX2+ apical RGC in the VZ as well as occasional GFP+SOX2+ ORG with soma at the VZ/SVZ border (insets). f, Higher magnification of the boxed area from (d) shows GFP+ cells in the SVZ/IZ that are SOX2-negative with the morphology of radially migrating newborn neurons. g, At 7 to 9 DPE, NEUROG2 gain-of-function induced a significant shift of GFP+ cells from the VZ into the SVZ/IZ compared to the control (asterisk denotes p < 0.05, paired t-test; exact p-values: VZ=0.026, SVZ/IZ=0.026, CP=0.034; n=3 animals per condition; 3–4 brain sections counted per animal; data represented as mean ± SEM), with a concomitant loss of RGC morphology and SOX2 expression, demonstrating that the NEUROG2 proneural network promotes delamination of daughter cells from the ventricular surface, migration into the SVZ, and neuronal differentiation. Scale bars: 50 µm (a), 1 mm (c), 100 µm (d), 20 µm (e,f).
Single-cell analysis of species-specific RGC heterogeneity
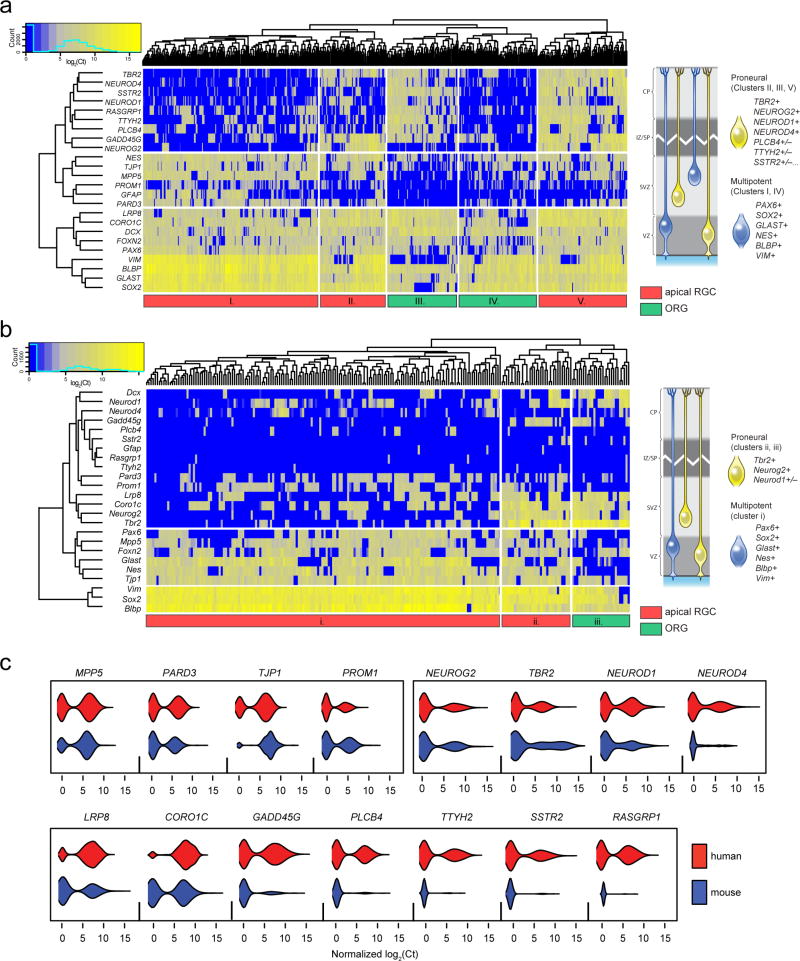
Both our human RNA-seq and ferret immunofluorescence data demonstrate NEUROG2+ RGC subpopulations in both the VZ and SVZ, intermingled with NEUROG2− progenitors, exemplifying the heterogeneity that confounds population-level transcriptome comparisons, so we turned to single cell analysis to compare the subpopulations of radial progenitors in human, ferret, and mouse. We first sorted RGC from human fetal cortex (n=6, 16–21 WG; Supplementary Table 1) into 96-well plates and performed microfluidics-based, highly multiplexed single-cell qRT-PCR to simultaneously assay several dozen genes that included markers for all RGC (PAX6, SOX2, GLAST, BLBP, VIM, NES) and apical RGC (PROM1, PARD3, MPP5, TJP1); proneural Neurogenin pathway TFs; and additional validated LG+Prlo, ORG-enriched genes (Supplementary Fig. 3). Of 546 sorted single human progenitors, PAX6, SOX2, GLAST, BLBP, and VIM were detected in virtually all cells (93±3% detection rate; Supplementary Fig. 5a), confirming their radial glial identity. Hierarchical clustering revealed several distinct transcriptional states characterized by the combinatorial expression of apical markers and proneural factors, such that cells fell into one of four main subpopulations which we refer to as apical/multipotent; apical/proneural; non-apical/multipotent; and non-apical/proneural (Fig. 3a and Supplementary Fig. 5b). Apical RGC subpopulations (clusters I, II, V in Fig. 3a; 71±8% detection rate for apical genes) could be divided into apical/multipotent (I) and apical/proneural (II and V) subpopulations based on their lesser (21±5%) or greater (81±7%) expression of proneural Neurogenin pathway TFs, respectively. ORG-like non-apical subpopulations (clusters III and IV) with lower detection rates of apical complex transcripts (31±6%) were similarly subdivided according to lower or higher rates of proneural gene expression (22±7% vs. 65±15%). Notably, most non-apical/proneural cells were NEUROG2− (cluster III), consistent with observations in the mouse that NEUROG2 represses apical identity and is then downregulated upon delamination23. Finally, both apical and non-apical proneural RGC were further subdivided by expression of other LG+Prlo-enriched genes (TTYH2, PLCB4, SSTR2, RASGRP1) that define additional transcriptional heterogeneity among human cortical progenitors. These results demonstrate significant multigenic transcriptional diversity within cortical radial glial progenitors, and are characteristic of the previously unappreciated heterogeneity recently revealed by single-cell analyses in other non-neural stem cell niches33,34.
Figure 3. Single-cell gene expression of human and mouse progenitors reveals species-specific RGC subpopulations.
a, Multiplexed gene expression profiling of 546 single human RGC reveals distinct transcriptional states defined by the presence or absence of transcripts encoding apical membrane-specific proteins, proneural transcription factors downstream of NEUROG2 such as NEUROD1 and NEUROD4, and additional LG+Prlo-enriched genes such as TTYH2 and PLCB4. Hierarchical clustering and heatmap representation of single-cell qRT-PCR data (left) indicates the co-expression patterns of these genes, and a schematic representation (right) of the four main subpopulations of RGC identified: “multipotent” RGC (blue) are found as subsets of both the apical (cluster I) and non-apical RGC (cluster IV), as are the “proneural” NEUROG2/TBR2+ RGC (clusters II, III, V). In addition, proneural RGC can be further subdivided according to their expression of downstream factors and additional LG+Prlo-enriched genes (e.g., compare clusters II and V). b, The same genes assayed in 226 RGC from E16-E17 mouse cortex yield only three subpopulations: apical multipotent (i); apical proneural (ii); and non-apical proneural (iii). Schematic representation of these subpopulations (right) highlights the major species differences, namely, the mouse has fewer non-apical cells overall; few if any multipotent (NEUROG2−TBR2−) non-apical cells, suggesting the absence of a significant subpopulation of proliferative ORG; and very few cells expressing other human subset-enriched genes (e.g., TTYH1, PLCB4). c, Violin plots of gene expression distributions for apical complex, NEUROG2 network, and ORG-enriched genes in human and mouse single RGC. Several ORG-enriched genes appear to be abundantly expressed in subsets of human but not mouse RGC including NEUROD4, GADD45G, PLCB4, TTYH2, SSTR2, RASGRP1.
In contrast to the human analysis, single RGC from the embryonic day (E)16–17 mouse cortex showed fewer distinct transcriptional states (Fig. 3b and Supplementary Fig. 5a), and rarely expressed many of the genes that defined subsets of human cells (Fig. 3b,c). Virtually all mouse single cells (n=226) expressed the RGC markers Sox2, Vim, Blbp, and Glast (93±8%), and the vast majority (88%) of cells also expressed some apical complex genes, confirming that ORG are rare in the mouse. Hierarchical clustering yielded only three subpopulations (Fig. 3b), corresponding to the apical/multipotent (cluster i), apical/proneural (ii), and non-apical/proneural (iii) subsets observed in the human cortex. Although a significant subset of mouse RGC co-expressed proneural TFs, the proportion of cells was significantly smaller than in human (27% in mouse vs. 47% in human; p=9.57E-8, Fisher’s exact test). Importantly, the absence of a significant non-apical/multipotent subpopulation (human cluster IV) suggests a critical species difference in the proliferative potential of ORG, which could underlie the paucity of ORG in the mouse. Most notably, orthologs of human ORG-enriched genes that contributed markedly to human RGC heterogeneity, including Plcb4, Gadd45g, Ttyh2, Rasgrp1, and Sstr2, were detected in a rare and uncorrelated minority of mouse RGC (<10%, compared to 45–55% in human) (Fig. 3b,c), further highlighting the species-specificity of RGC transcriptional heterogeneity.
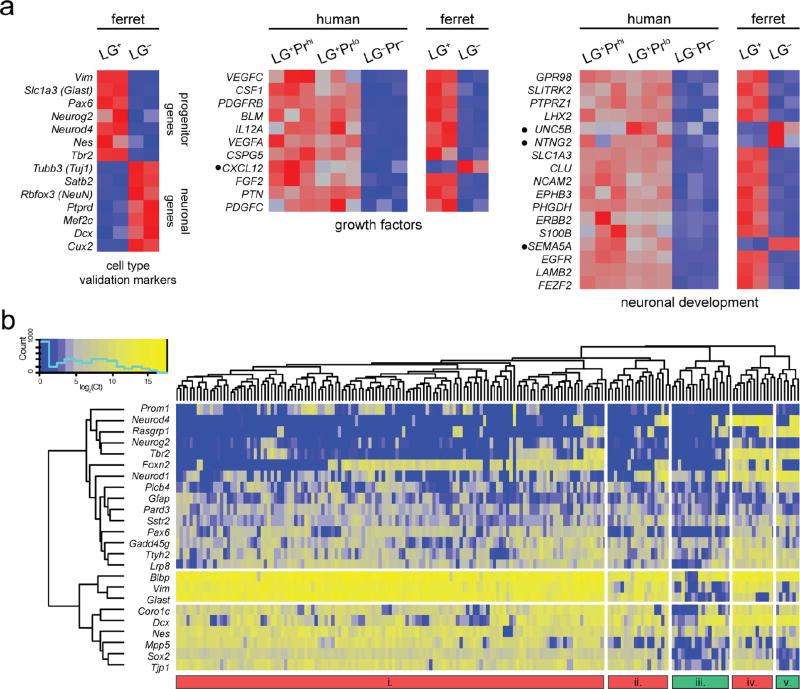
Finally, we performed RNA-seq and single-cell profiling of ferret radial glial progenitors and found that they share some of the key transcriptional states of human RGC. In the absence of working antibodies against ferret Prominin, we first validated LeX and Glast antibodies by immunohistochemistry in ferret brain sections as well as by FACS (data not shown), then collected LG+ and LG− cells from neonatal ferret cortex, at which time middle and upper layers of cortex are being generated30,31, roughly corresponding to mouse E16–17 or human 16–20 WG, and performed population-level RNA-seq. Ferret LG+ cells were enriched for most previously described RGC marker genes, and showed transcriptome-wide expression patterns similar to LG+ cells from human and mouse cortex (Fig. 4a). Having validated that LG+ cells comprise a substantial proportion of ferret RGC, we performed single-cell profiling on 185 single LG+ cells from the neonatal ferret cortex, and interestingly found an intermediate degree of heterogeneity compared to the human and mouse progenitors (Fig. 4b). The vast majority of sorted ferret cells expressed classic RGC markers, confirming the specificity of the sorting, while a subset co-expressed Tbr2 (ferret clusters iv and v, and a number of cells in cluster i), and a smaller subset of those were positive for Neurod4 and Neurod1 (clusters iv and v). Interestingly, the ferret homologs of some human ORG-enriched genes (e.g., Rasgrp1) were preferentially expressed in these proneural cells, as in human, whereas most human ORG genes were more homogeneously expressed in most ferret cells (Ttyh2, Sstr2), and conversely, some genes were heterogeneously expressed in novel subsets of cells that did not correspond to those seen in human (Foxn2). Finally, we noted that a greater proportion of ferret cells expressed the gliogenic marker Gfap compared to human cells, which is consistent with evidence that ferret ORG have astrogliogenic potential9. Taken together, our single-cell analyses from human, ferret, and mouse implicate a large number of genes acting in a coordinated network that may be responsible for the evolution of novel progenitor transcriptional states critical for human cortical development.
Figure 4. Population-level whole-transcriptome RNA-seq and single-cell expression analysis of ferret RGC.
a, Expression heatmaps of known progenitor and neuronal marker genes, as well as selected human RGC-enriched gene sets, from LG+ and LG− cells isolated by FACS from the P2 developing ferret cortex (n=2). Enrichment of classic RGC markers and a high degree of similarity between gene sets enriched in human and ferret LG+ cells validate the use of LeX and Glast to select RGC from the developing ferret cortex. Notably, however, several genes (black bullets) show distinct expression patterns between the two species (e.g., CXCL12, UNC5B, NTNG2, SEMA5A), suggesting that certain growth factor and other pathways may be expressed in a species-specific manner in RGC. b, Single-cell gene expression profiling of 185 single ferret LG+ progenitors was performed using the same gene panel as shown in Figure 3 for human and mouse RGC. As in humans, a substantial fraction of ferret cells in clusters i, iv, and v co-express both RGC markers and Tbr2/Neurog2, consistent with our immunohistochemcial analysis of NEUROG2 expression in the ferret (Fig. 2a) and suggesting this “proneural” RGC transcriptional state is conserved. Similar to human RGC, a subset of these proneural cells also express the downstream factors NEUROD1 and NEUROD4. However, some human ORG-enriched genes (e.g. Rasgrp1) are expressed in fewer ferret RGC, while others (e.g. Plcb4, Sstr2, Gadd45g, Ttyh2) appear more homogenous across all cells. Interestingly, Foxn2, which was detected in nearly all human RGC, appears to mark a distinct subpopulation of ferret apical RGC (clusters i and iv). Overall, while ferret RGC exhibit more diversity of transcriptional states than mouse and generally more similarity to human, they are nonetheless distinct in their relative proportions and composition.
Novel long noncoding transcripts enriched in human ORG
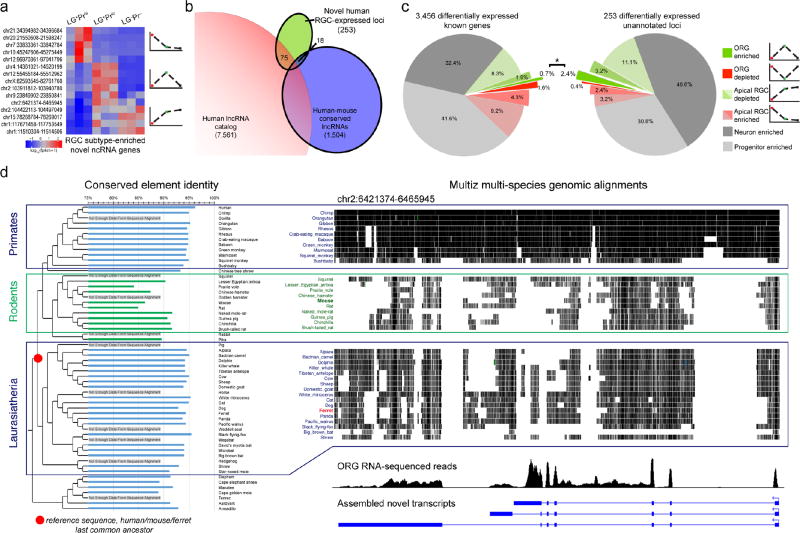
Given the species differences in RGC subpopulations revealed by our single-cell analysis, we next searched our RNA-seq data for transcriptional influences on species differences in RGC molecular identity, identifying candidate non-conserved RNA transcripts including lncRNAs, which are evolutionarily dynamic, frequently lacking human-mouse homology35, and are involved in critical neural developmental processes such as progenitor pluripotency, neurogenesis, and epithelial-mesenchymal transition36–38. We compared 253 differentially expressed unannotated loci (Fig. 5a) to two published human lncRNA catalogs39,40 and identified 75 loci overlapping putative human lncRNAs39, while only 18 loci matched reported human-mouse conserved lncRNAs40 (Fig. 5b and Table 2), suggesting that our human RGC subtype-specific novel transcripts include numerous novel lncRNAs that may lack any homologous mouse transcripts. Surprisingly, we found that a much greater proportion of differentially expressed novel loci were specifically enriched in ORG, compared to known genes (Fig. 5c; 2.4% vs. 0.7%; p = 0.012, Fisher’s exact test), suggesting that lncRNAs are especially relevant to the molecular identity and function of the ORG subpopulation in humans. By manual inspection, we determined that although a few novel transcripts reflected incomplete annotations of known genes (e.g., alternative transcription start sites or untranslated regions), the majority resemble bona fide unannotated genes, many of which show multiple exons and alternative splicing (Table 3 and Supplementary Fig. 6), and none of which have previously been reported in cortical development.
Figure 5. Novel transcripts detected by RNA-seq include previously unknown lncRNAs with distinct RGC subtype expression patterns and evolutionary conservation.
a, Differential expression patterns of selected ncRNA loci, including several novel multi-exon lncRNAs (Table 3). b, Intersection of 253 differentially expressed non-reference loci from our RNA-seq analysis with previous catalogs of human non-coding RNA genes revealed a number of reported human lncRNAs39, and a smaller number of human-mouse conserved lncRNAs40. c, A significantly greater proportion of novel differentially expressed transcripts were specifically enriched in LG+Prlo non-apical progenitors (2.4%), compared to known genes (0.7%; p=0.012, Fisher’s exact test), implicating this evolutionarily dynamic gene class in the regulation of the ORG progenitor subpopulation, which is greatly expanded in humans. d, Comparative genomics analysis of novel ORG-enriched lncRNAs was performed by comparing conserved elements from within each genomic locus from 58 species to a computed ancestral sequence for the Laurasiatherian last common ancestor (LCA) of human, ferret, and mouse, the three species examined in this study (see Table 3). Here, we show a detailed example from a human ORG-enriched lncRNA gene located on chromosome 2. At left is shown the percent identity of each species’ conserved elements from this locus to the LCA sequence, which demonstrates that rodents (highlighted in green) show a highly divergent sequence compared to both primates and other more distantly related groups, including carnivores. The panel at right illustrates multi-species genomic alignments to the same human locus for primates, rodents, and other Laurasiatherian species (top), with the human ORG RNA-seq reads (middle) and assembled transcripts (bottom), illustrating the greater sequence divergence of rodents compared to either non-human primates or other more distant Laurasiatherian species. Similar results were found for several other ORG-enriched lncRNA transcripts, as summarized in Table 3 (% ID columns). These findings are consistent with the interpretation that functional transcripts were present at these loci in the Laurasiatherian LCA, and are either highly divergent or lost in the rodent lineage. This scenario of ORG-expressed transcript divergence in rodents, which are mostly lissencephalic and lack large numbers of ORG, is consistent with the recent suggestion that gyrencephaly is an ancestral mammalian trait42,43.
Table 2. Differentially expressed known lncRNA genes.
Expression patterns of known lncRNA genes with conserved mouse orthologs, most of which show species-specific expression patterns. Human and mouse orthologous pairs were assayed in FACS-purified progenitor and neuron populations by qRT-PCR (see Supplementary Fig. 7).
| Human gene symbol (mouse ortholog) |
Locus (hg19) | Human Expression Pattern |
Mouse Expression Pattern |
Comments |
|---|---|---|---|---|
| H19 (H19) | chr11:2016405-2019065 | apical RGC-enriched | RGC-enriched | imprinted maternally-expressed tumor suppressor |
| CRNDE (Crnde) | chr16:54952777-54963101 | apical RGC-enriched | not expressed | knockout mouse made but no phenotype reported37 |
| MIR22HG (Mir22hg) | chr17:1614797-1619571 | apical RGC-enriched | no differential expression | |
| LINC00643 (1700086L19Rik) | chr14:62570095-62606691 | ORG-depleted | neuron-enriched | |
| TUNAR (Tunar) | chr14:96343108-96391908 | ORG-depleted | neuron-enriched | regulates pluripotency and neural lineage commitment in mouse |
| LINC00599(A930011O12Rik(?)) | chr8:9753778-9767085 | neuron-enriched | neuron-enriched | immediately adjacent to MIR124-1 |
| MIAT (Miat) | chr22:27042391-27176170 | neuron-enriched | no differential expression | enriched in mouse TBR2+ intermediate progenitors41 |
| LINC-PINT (linc-Pint) | chr7:130628918-130794831 | RGC-enriched | neuron-enriched | knockout mouse displays general growth defect37 |
| RMST (Rmst) | chr12:97856520-97958754 | RGC-enriched | n.d. | enriched in mouse TBR2+ intermediate progenitors41 |
n.d., no data
Table 3. Expression and conservation of novel non-coding transcripts.
Previously uncharacterized transcripts with differential expression between apical and non-apical RGC subtypes are listed with their cell type of peak expression. Some transcripts overlap previously reported putative human lncRNAs39,40 but most are completely unannotated. Manual inspection of RNA-seq reads in their genomic context reveals multiple classes of transcripts, including unannotated alternative transcription start sites (5’ UTR exons) and antisense transcripts of known protein-coding genes, as well as many multi-exon intergenic transcripts. For each lncRNA, “% ID” columns show the results of a comparative genomics analysis of each locus to a computed common ancestor of human, ferret, and mouse, indicating that for many loci, sequence homology was preferentially lost along the rodent lineage, potentially contributing to the evolutionary loss of the ORG cell subpopulation in mouse cortex (for further illustration of an example, see Fig. 5d).
| Locus (hg19) | Peak expression |
% ID primates |
% ID rodents |
% ID Laurasi atheria |
Comments | |
|---|---|---|---|---|---|---|
| chr1:11510304-11514506 | ORG and neurons | 96% | 93% | 95% | "PTCHD2-OS1", novel transcript ~25 kb upstream of PTCHD2 | |
| § | chr1:117671458-117753549 | ORG and neurons | 91% | 0% | 0% | "VTCN1-OS1", novel spliced transcript on the opposite strand overlapping the 3’UTR of VTCN1, which is not expressed |
| * | chr2:104422113-104497049 | ORG and neurons | 97% | 93% | 91% | expressed from a bidirectional promoter shared with another novel, RGC-enriched lincRNA (see Supplementary Fig. 6a) |
| chr15:78268784-78269017 | ORG and neurons | n.d. | n.d. | n.d. | single-exon transcript maps to a low-complexity region, ~20 kb downstream of apical RGC-enriched TBC1D2B | |
| * | chr2:6421374-6465945 | ORG | 95% | 89% | 94% | novel multi-exon, alternatively spliced lncRNA locus (see Supplementary Fig. 6b) |
| chr2:103911812-103940780 | ORG | 96% | 90% | 95% | novel spliced transcript, ~475 kb from nearest RefSeq gene (TMEM182) | |
| * | chr4:14361021-14520199 | ORG | 92% | 91% | 92% | opposite strand overlapping the 3’ end of another lncRNA, LINC00504, which is not expressed |
| chr9:23849902-23850841 | ORG | 94% | 89% | 94% | likely ELAVL2 alternative TSS | |
| chr12:55455184-55512962 | ORG | 97% | 90% | 95% | novel spliced transcript ~30 kb downstream of ORG-enriched NEUROD4 | |
| chrX:82593545-82761760 | ORG | 96% | 91% | 95% | "POU3F4-OS1", novel transcript ~1.5 kb upstream on the opposite strand | |
| * | chr7:33833361-33842784 | apical RGC | n.d. | n.d. | n.d. | apical-specific, intergenic (>100 kb) spliced transcript |
| chr10:45247606-45275449 | apical RGC | 94% | 86% | 94% | apical-specific, intergenic (>30 kb) spliced transcript (see Supplementary Fig. 6c) | |
| * | chr12:96970361-97041796 | apical RGC | 96% | 91% | 96% | overlaps a locus now called CFAP54; coding potential uncertain |
| * # | chr20:21550608-21598247 | apical RGC | 97% | 92% | 96% | "NKX2-2OS" (LOC101929625) |
| chr21:34394982-34396684 | apical RGC | 95% | 92% | 91% | unspliced transcript ~1.5 kb upstream of OLIG2 TSS; possible novel OLIG2 alternative TSS |
Few known lncRNAs that are functionally essential37 or have been transcriptionally profiled41 in mouse brain development were detected in human, and those that were conserved displayed species-specific expression patterns, further illustrating the dynamic evolutionary changes in lncRNAs. Of 18 lncRNAs recently knocked out in mice37 we identified only two orthologs with appreciable expression in human developing cortex (Table 2). Similarly, of 15 mouse IP-enriched lncRNAs41, only two – MIAT and RMST – showed appreciable expression in human fetal cortex but with cell-type enrichment patterns distinct from mouse (Table 2). Within conserved lncRNAs between human and mouse, we find several – including LINC-PINT, TUNAR, CRNDE, and MIR22HG – which are depleted in mouse RGC but enriched in human apical and outer RGC suggesting potentially distinct functions in cortical development (Supplementary Fig. 7). Thus, the dynamic patterns of lncRNA expression in RGC subtypes and their notable lack of conservation are consistent with the highly species- and also cell type-specific expression of lncRNAs in other contexts, and suggest that this transcript class is unusually dynamic in its evolutionary relationship to cortical development.
To probe the evolutionary history of ORG-enriched lncRNAs, we performed comparative genomic analysis, specifically evaluating their presence in a common mammalian ancestor and their conservation in gyrencephalic mammals such as the ferret and nonhuman primates compared to rodents. We extracted conserved elements within the novel lncRNA genomic loci and compared their percent identity to a reconstructed last common ancestor (LCA) of human, mouse and ferret, and indeed found that both primates and more distant non-rodent species shared greater identity to the LCA conserved sequences than rodents (Fig. 5d and Table 3). This analysis suggests that many of these newly described human ORG-enriched lncRNAs show comparative patterns of sequence conservation that parallel levels of gyrification, being more highly conserved in many larger-brained gyrencephalic species, including other primates and ferrets, and more highly divergent in non-gyrencephalic rodents42,43.
Discussion
Using a combined FACS enrichment and transcriptional profiling strategy, we identified for the first time a molecular signature of human ORG comprising hundreds of known genes and novel transcripts. Most notably, we observed a highly significant enrichment of a well-known transcription factor network, regulated by the critical regulatory factor NEUROG2, in ORG, and used ferrets to confirm the role of this transcription factor network in regulating key steps in ORG production, specifically delamination from the ventricular neuroepithelium and migration into the SVZ. On the other hand, both our human RNA-seq and our ferret immunohistochemical data indicated heterogeneity of expression of NEUROG2 itself within both apical RGC and ORG, and our human single-cell data showed remarkably diverse transcriptional states within both apical RGC and ORG, characterized by the combinatorial expression patterns of classic progenitor markers, proneural transcription factors, and novel ORG-enriched candidates such as RASGRP1, TTYH2, and SSTR2. This heterogeneity was markedly simplified in mouse, consistent with the paucity of ORG in that species, but was more evident in ferret single progenitors, which included a substantial subpopulation of NEUROG2 target-expressing RGC. Finally, we describe novel gene loci, putatively encoding lncRNAs, including several loci with enriched expression in human ORG. Several of these ORG-enriched lncRNA loci show comparative genomic evidence of having been present in the LCA of humans and ferrets, which also possess abundant ORG and are gyrencephalic, but greatly diverged during rodent evolution, suggesting that these transcripts may be expressed in other species with expanded SVZ progenitor populations. Altogether, our population level and single-cell transcriptional data intriguingly show a correlation between mature cortical size and structure and the heterogeneity of the progenitors that create this structure during development.
Recent studies have provided evidence for or against functional heterogeneity within mouse RGC, particularly with respect to the transcription factors CUX2 and FEZF2, respectively44,45. Our human RGC subtype-specific RNA-seq data confirm that FEZF2 is highly enriched in both LG+Prhi and LG+Prlo RGC subpopulations relative to LG− neurons. In contrast, CUX2 shows a highly significant, >50-fold enrichment in LG− neurons relative to LG+Prhi apical RGC, with a more modest but still highly significant ~8-fold enrichment in LG+Prlo ORG relative to LG+Prhi apical RGC. These patterns are consistent with the interpretation that FEZF2 is expressed in both apical and outer RGC, with no significant difference between the progenitor subsets illustrated in Fig. 3; whereas CUX2 is most likely enriched in the NEUROG2+ proneural subsets, consistent with this factor’s role in upper layer neuronal morphogenesis. It is important to note however that the human specimens available for our studies were from the second half of the second trimester, during the later stages of upper-layer neurogenesis, and that earlier human fetal cortical samples would be required to specifically contrast the expression or function of these two transcription factors in early versus late human radial glial progenitors.
Two previous studies have reported gene expression profiles of the human VZ, ISVZ, and OSVZ, using laser capture-assisted microdissection to separate the germinal zones from each other and from the postmitotic IZ and CP compartments13,14, with one of these studies also directly contrasting human and mouse germinal zones13, and another recent study explored the transcriptional signature of human RGC and the differences in gene expression between human and mouse progenitors15. The genes reported by these studies as OSVZ-enriched or human RGC-enriched we find to be expressed either in apical RGC or in both apical RGC as well as ORG (Supplementary Table 2), consistent with their being radial glial markers. On the other hand, few of the ORG-enriched genes we find by our methods were captured by previous studies, highlighting the ability of sorted cell populations to reveal cell type-specific expression patterns. One prior transcriptional analysis that also identified a number of the human ORG-enriched genes found in our study was performed on single progenitors from the embryonic mouse cortex (Supplementary Table 2)46. Amazingly, those authors showed by in situ hybridization that a number of human ORG-enriched genes, which they described as labeling a novel progenitor subpopulation intermediate between classic RGC and IP, were expressed in a thin band at the VZ/SVZ border in E14 mouse cortex, suggesting that these cells were indeed transitioning from RGC to IP. These data are consistent with our interpretation that the human ORG transcriptional signature reflects an abundance of cells within the ORG population persisting in just such a transitional state. Remarkably, the comparison of our data with those of Kawaguchi et al.46 suggests that the embryonic mouse cortex may have an analogous cell type to the human ORG, but which differs in both morphology, having already retracted its radial fiber, and in position, residing between the VZ and the SVZ rather than superficial to the zone of classic TBR2+ multipolar IP.
Overall, our data show that human radial glial progenitors, and to a lesser extent those of the gyrencephalic ferret, differ most strikingly from mouse RGC in the “gradedness” of their transition from NEUROG2-negative neuroepithelial RGC to delaminated, multipolar, neuronal lineage-committed IP. Live-imaging studies of the embryonic mouse cerebral cortex have consistently shown that daughter cells from the abventricular mitoses of classic RGC concurrently delaminate, retract their radial fibers, lose PAX6 expression, gain TBR2 expression, and migrate into the SVZ. In contrast, our transcriptional analysis of human ORG and our unbiased single-cell profiling of hundreds of RGC from human and ferret show that these cells exist in a surprising number of transcriptional transitional states between classic RGC and IP.
Methods
Human Tissue Specimens and Processing
Research performed on samples of human origin was conducted according to protocols approved by the institutional review boards of Beth Israel Deaconess Medical Center and Boston Children’s Hospital. Fetal brain tissue was received two to four hours following elective pregnancy termination and after release from clinical pathology. Cases with known anomalies were excluded. Gestational ages were determined using fetal foot length. Tissue was transported in HBSS medium on ice to the laboratory for research processing.
Purification of Cortical Progenitors
Cortical tissue was separated from remaining brain tissue in ice-cold HBSS medium and manually disrupted using a sterile razor blade down to ~1-mm3 pieces. The tissue was then dissociated into a single cell suspension using the trypsin Neural Dissociation Kit (Miltenyi Biotec) according to manufacturer’s instructions. Cells were placed into FACS “pre-sort” media (Neurobasal media, 0.25% HEPES, 0.5% FBS, rhEGF, rhFGF) for labeling with cell surface antibodies. Cells were labeled in aliquots of 500ul containing up to 40 million cells with anti-CD15-FITC (BD Biosciences 560997) at 1:10,000; anti-GLAST-PE (Miltenyi Biotec 130-098-804) at 1:10,000; and anti-CD133-APC (Miltenyi Biotec 130-098-829) at 1:1,000 for 30 minutes at +4°C and washed twice with pre-sort media before FACS. Alternatively, minced tissue was cryopreserved prior to enzymatic dissociation by storing in HBSS + 10% DMSO, cooled gradually in a cryochamber to −80°C overnight, and transferred to −150°C for long-term storage.
RNA Isolation, Processing, and RNA sequencing
Cells were sorted directly into RNA stabilizing lysis buffer followed by total RNA extraction (Qiagen). Next-generation sequencing libraries were prepared using Illumina TruSeq v2 according to manufacturer’s instructions and sequencing was performed on an Illumina HighSeq 2000. Data were analyzed primarily with the Tuxedo software suite (bowtie/tophat/cufflinks/cummerbund)47 using the hg19 genome and UCSC KnownGene transcriptome references. Additional R/Bioconductor packages were used for principle component analysis, clustering, and the generation of heatmaps. Gene set enrichment analyses were performed using DAVID (http://david.abcc.ncifcrf.gov/) and comparison of non-reference cufflinks transcripts to published lncRNA catalogs was done in Galaxy (http://main.g2.bx.psu.edu/).
Ferret Electroporation
Timed-pregnant ferrets (Mustela putorius furo) were obtained from Marshall Bioresources. Neonatal ferret kits were anesthetized with 5% and maintained at 3% isoflurane utilizing a nose cone during the entire procedure. A small incision was made on the skin at the dorsomedial part of the head using a surgical blade and a hole was opened anterior to the bregma on the left side of the skull, above the lateral ventricle, using an insulin syringe needle. 3–5µl of DNA construct (1µg/µl) was injected into the lateral ventricle using a pulled glass micropipette inserted through the craniotomy and the overlying cortical wall. 150V electric pulses were passed 5 times at 1s intervals using paddle electrodes positioned outside the animal’s head. The skin incision was closed using VetBond (3M) tissue adhesive and kits were returned to the nest after recovering from anesthesia. Kits were deeply anesthetized prior to transcardial perfusion with cold PBS and 4% PFA, and brains were extracted and placed in 4% PFA at +4° overnight prior to processing for immunohistochemistry.
Immunohistochemistry
Ferret brains were embedded in 4% low-melting-point agarose and sectioned at 70µm on a vibrating microtome. Sections were washed in cold 0.1M PB followed by antigen retrieval in 10mM citric acid (pH 6.0) + 0.05% Tween-20 at 80°C for 30 minutes. Sections were then cooled to room temperature and washed in cold 0.1M PB. Sections were blocked for at least 1 hour at room temperature (10% Normal Donkey Serum, 0.1% Triton X-100, 0.2% gelatin in PBS). Primary antibodies were incubated at 4°C in 0.2X blocking buffer for at least 48 hours. Primary antibodies included chicken anti-Vimentin 1:250 (Abcam ab24525), chicken anti-Tbr2 1:250 (Millipore AB15894), rabbit anti-GFP 1:1,000 (Abcam ab290), goat anti-Sox2 1:250 (Santa Cruz sc-17320). Sections were washed in PBS and then incubated for two hours in 0.2X blocking buffer containing AlexaFluor secondary antibodies (Life Technologies). Slices were then rinsed and coverslipped with Fluoromount-G (Southern Biotech) containing Hoechst 1:1,000 (Roche). Images were obtained with Zeiss LSM700 confocal microscope and Leica MZ16 F fluorescence stereomicroscope. For quantification, tiled confocal images spanning the entire cortical wall were captured at 20x, stitched, and exported to Photoshop. Three to four pairs of coronal sections from three control (GFP) and three NEUROG2-VP16 electroporated hemispheres, matched for the level of section and spatial extent of the electroporation, were imaged and the images were then coded and quantified blind to experimental condition. The Hoechst nuclear counterstain was used to demarcate the borders between VZ, SVZ, IZ, and CP, and the numbers of GFP+ cell bodies in each zone counted. The percentages of GFP+ cells in each zone were calculated separately for each image and averaged across the images for each brain. The averages for each replicate were then compared across experimental conditions using a paired student’s t-test.
Single Cell mRNA Expression Profiling
Following cell labeling, single cells were sorted by FACS into skirted 96 well PCR plates containing Pre-Amplification solution (Cells Direct Kit, Life Technologies) and appropriate mixtures of Taqman assays (for human and mouse) or validated primer pairs (for ferret). Plates were transported on ice and spun down before pre-amplification (94°C 10 minutes, 50°C 60 minutes, 94°C 30 seconds, 50°C 3 minutes × 28 cycles). Target-specific cDNA from single cells was harvested, screened for expression of housekeeping genes ACTB and GAPDH, and then loaded onto a Biomark chip (Fluidigm) for expression profiling with the panel of qRT-PCR assays. Expression data was processed and analyzed using the Singular Analysis Toolset (Fluidigm) and gplots packages in R. Hierarchical clustering was performed using complete linkage based on Euclidean distance and clusters of cells were defined by cutting the single-cell dendrogram at the same height for all three species.
Comparative Genomics Analysis of Novel lncRNA Loci
Comparative evolutionary analysis of lncRNAs was performed using a modified version of the recently published “Forward Genomics” approach48. Briefly, multi-sequence fasta files were generated for all conserved regions located within novel lncRNAs using existing 100-way vertebrate multiple alignment files available from the UCSC genome browser. Next we generated ancestral sequences for the common ancestor of human, mice, and ferrets using the prequel algorithm (--keep-gaps --no-probs --msa-format PHYLIP), part of the PHAST tools49. The percent identities of sequences from all species were determined by alignment to the corresponding ancestral sequence using Needleall, part of the EMBOSS tools50. Species with low quality or missing sequence information were excluded from the analysis. Finally, the number of identical bases from all regions within each lncRNA were calculated to yield the %ID to the common ancestor.
Statistics
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those generally employed in the field and are comparable to those reported in previous publications11,13. For parametric analyses, data distribution was assumed to be normal but this was not formally tested. A supplementary methods checklist is available.
Supplementary Material
Acknowledgments
We thank J. Partlow for coordinating human tissue protocols; D. Gonzalez for animal protocol and technical experimental assistance; Suzan Lazo-Kallanian for single-cell FACS assistance; and all members of the Walsh lab for comments and discussion. The Neurog2-VP16 construct was a generous gift from Carol Schuurmans (University of Calgary). This work was supported by grants to C.A.W. from the National Institutes of Neurological Disease and Stroke (R01 NS032457) and the Paul G. Allen Family Foundation. M.B.J. was supported by a fellowship from the Nancy Lurie Marks Family Foundation. P.P.W. is a Stuart H.Q. & Victoria Quan Fellow at Harvard Medical School. Single-cell expression profiling experiments were performed at the Molecular Genetics Core at Boston Children’s Hospital (BCH IDDRC, P30 HD18655). Transcriptome analysis was performed using Harvard Medical School’s Orchestra high-performance computing cluster, which is partially supported by NIH grant NCRR 1S10RR028832-01. C.A.W. is a Distinguished Investigator of the Paul G. Allen Family Foundation, and an Investigator of the Howard Hughes Medical Institute.
Footnotes
Accession Codes
RNA sequencing data are available from the Gene Expression Omnibus, GSE66217.
Author Contributions
M.B.J., P.P.W., R.N.D. designed and conducted experiments and analyzed data. K.D.A. and E.A.M. performed experiments and analyzed data. J.H. procured and examined human tissue samples. M.B.J., P.P.W., C.A.W. interpreted the data and wrote the manuscript.
References
- 1.Florio M, Huttner WB. Neural progenitors, neurogenesis and the evolution of the neocortex. Development. 2014;141:2182–2194. doi: 10.1242/dev.090571. [DOI] [PubMed] [Google Scholar]
- 2.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kowalczyk T, et al. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smart IHM, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fietz SA, et al. OSVZ progenitors of human and ferret neocortex are epithelial-like and expand by integrin signaling. Nat Neurosci. 2010;13:690–699. doi: 10.1038/nn.2553. [DOI] [PubMed] [Google Scholar]
- 6.Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 7.Betizeau M, et al. Precursor Diversity and Complexity of Lineage Relationships in the Outer Subventricular Zone of the Primate. Neuron. 2013;80:442–457. doi: 10.1016/j.neuron.2013.09.032. [DOI] [PubMed] [Google Scholar]
- 8.Gertz CC, Lui JH, LaMonica BE, Wang X, Kriegstein AR. Diverse behaviors of outer radial glia in developing ferret and human cortex. J Neurosci. 2014;34:2559–2570. doi: 10.1523/JNEUROSCI.2645-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reillo I, de Juan Romero C, García-Cabezas MÁ, Borrell V. A role for intermediate radial glia in the tangential expansion of the mammalian cerebral cortex. Cereb Cortex. 2011;21:1674–1694. doi: 10.1093/cercor/bhq238. [DOI] [PubMed] [Google Scholar]
- 10.Martínez-Cerdeño V, et al. Comparative analysis of the subventricular zone in rat, ferret and macaque: evidence for an outer subventricular zone in rodents. PLoS ONE. 2012;7:e30178. doi: 10.1371/journal.pone.0030178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson MB, et al. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fietz SA, et al. Transcriptomes of germinal zones of human and mouse fetal neocortex suggest a role of extracellular matrix in progenitor self-renewal. Proceedings of the National Academy of Sciences. 2012;109:11836–11841. doi: 10.1073/pnas.1209647109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JA, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lui JH, et al. Radial glia require PDGFD-PDGFRβ signalling in human but not mouse neocortex. Nature. 2014;515:264–268. doi: 10.1038/nature13973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capela A, Temple S. LeX is expressed by principle progenitor cells in the embryonic nervous system, is secreted into their environment and binds Wnt-1. Dev Biol. 2006;291:300–313. doi: 10.1016/j.ydbio.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 17.Mo Z, et al. Human cortical neurons originate from radial glia and neuron-restricted progenitors. J Neurosci. 2007;27:4132–4145. doi: 10.1523/JNEUROSCI.0111-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibata T, et al. Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J Neurosci. 1997;17:9212–9219. doi: 10.1523/JNEUROSCI.17-23-09212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weigmann A, Corbeil D, Hellwig A, Huttner WB. Prominin, a novel microvilli-specific polytopic membrane protein of the apical surface of epithelial cells, is targeted to plasmalemmal protrusions of non-epithelial cells. Proc Natl Acad Sci USA. 1997;94:12425–12430. doi: 10.1073/pnas.94.23.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uchida N, et al. Direct isolation of human central nervous system stem cells. Proc Natl Acad Sci USA. 2000;97:14720–14725. doi: 10.1073/pnas.97.26.14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seo S, Lim J-W, Yellajoshyula D, Chang L-W, Kroll KL. Neurogenin and NeuroD direct transcriptional targets and their regulatory enhancers. EMBO J. 2007;26:5093–5108. doi: 10.1038/sj.emboj.7601923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gohlke JM, et al. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 2008;6:15. doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochiai W, et al. Periventricular notch activation and asymmetric Ngn2 and Tbr2 expression in pair-generated neocortical daughter cells. Molecular and Cellular Neuroscience. 2009;40:225–233. doi: 10.1016/j.mcn.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Rousso DL, et al. Foxp-Mediated Suppression of N-Cadherin Regulates Neuroepithelial Character and Progenitor Maintenance in the CNS. Neuron. 2012;74:314–330. doi: 10.1016/j.neuron.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kovach C, et al. Neurog2 simultaneously activates and represses alternative gene expression programs in the developing neocortex. Cereb Cortex. 2013;23:1884–1900. doi: 10.1093/cercor/bhs176. [DOI] [PubMed] [Google Scholar]
- 26.Iacopetti P, et al. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. Proc Natl Acad Sci USA. 1999;96:4639–4644. doi: 10.1073/pnas.96.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaguchi A, et al. Differential expression of Pax6 and Ngn2 between pair-generated cortical neurons. Journal of Neuroscience Research. 2004;78:784–795. doi: 10.1002/jnr.20347. [DOI] [PubMed] [Google Scholar]
- 28.Reid CB, Tavazoie SF, Walsh CA. Clonal dispersion and evidence for asymmetric cell division in ferret cortex. Development. 1997;124:2441–2450. doi: 10.1242/dev.124.12.2441. [DOI] [PubMed] [Google Scholar]
- 29.Ware ML, Tavazoie SF, Reid CB, Walsh CA. Coexistence of widespread clones and large radial clones in early embryonic ferret cortex. Cereb Cortex. 1999;9:636–645. doi: 10.1093/cercor/9.6.636. [DOI] [PubMed] [Google Scholar]
- 30.Jackson CA, Peduzzi JD, Hickey TL. Visual cortex development in the ferret. I. Genesis and migration of visual cortical neurons. J Neurosci. 1989;9:1242–1253. doi: 10.1523/JNEUROSCI.09-04-01242.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noctor SC, Scholnicoff NJ, Juliano SL. Histogenesis of ferret somatosensory cortex. J Comp Neurol. 1997;387:179–193. [PubMed] [Google Scholar]
- 32.Borrell V. In vivo gene delivery to the postnatal ferret cerebral cortex by DNA electroporation. J. Neurosci. Methods. 2010;186:186–195. doi: 10.1016/j.jneumeth.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 33.Treutlein B, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature. 2014;509:371–375. doi: 10.1038/nature13173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 36.Ng S-Y, Bogu GK, Soh BS, Stanton LW. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol. Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 37.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu P, et al. LncRNA expression signatures of twist-induced epithelial-to-mesenchymal transition in MCF10A cells. Cell. Signal. 2014;26:83–93. doi: 10.1016/j.cellsig.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Cabili MN, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aprea J, et al. Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J. 2013;32:3145–3160. doi: 10.1038/emboj.2013.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelava I, et al. Abundant occurrence of basal radial glia in the subventricular zone of embryonic neocortex of a lissencephalic primate, the common marmoset Callithrix jacchus. Cereb Cortex. 2012;22:469–481. doi: 10.1093/cercor/bhr301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewitus E, Kelava I, Kalinka AT, Tomancak P, Huttner WB. An Adaptive Threshold in Mammalian Neocortical Evolution. PLoS Biol. 2014;12:e1002000. doi: 10.1371/journal.pbio.1002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franco SJ, et al. Fate-Restricted Neural Progenitors in the Mammalian Cerebral Cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo C, et al. Fezf2 Expression Identifies a Multipotent Progenitor for Neocortical Projection Neurons, Astrocytes, and Oligodendrocytes. Neuron. 2013;80:1167–1174. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawaguchi A, et al. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- 47.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hiller M, et al. A ‘forward genomics’ approach links genotype to phenotype using independent phenotypic losses among related species. Cell Rep. 2012;2:817–823. doi: 10.1016/j.celrep.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hubisz MJ, Pollard KS, Siepel A. PHAST, RPHAST: phylogenetic analysis with space/time models. Brief. Bioinformatics. 2011;12:41–51. doi: 10.1093/bib/bbq072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.