Abstract
Purpose
Developing a DNA dot hybridization model for diagnosing parasitic keratitis.
Methods
Newly designed oligonucleotide probes for detecting Acanthamoeba and microsporidia were tested with target reference strains of Acanthamoeba (n = 20) and microsporidia (n = 3), and non-target microorganisms, including bacteria (n = 20) and fungi (n = 20). These probes, which had passed the preliminary tests, were then assembled as a parasite dot hybridization (PDH) model for assessing 33 clinical samples from patients with clinically suspected Acanthamoeba and microsporidia keratitis, including eight positives for Acanthamoeba, 13 positives for microsporidia, and 12 negatives for both pathogens.
Results
Two probes for detecting Acanthamoeba and two for detecting microsporidia passed the tests using target and non-target strains and then were assembled in the PDH model. For clinical samples, one Acanthamoeba-positive sample (proved with pathology) was falsely negative according to the PDH assay. The sensitivity and specificity of the PDH assay for diagnosing Acanthamoeba keratitis were 87.5% and 100%, respectively, while the sensitivity and specificity for diagnosing microsporidia keratitis were 100%. The infectious agent of all clinical samples of microsporidia keratitis was identified as Vittaforma corneae with DNA sequencing, while those of Acanthamoeba keratitis were caused by four species of Acanthamoeba, with Acanthamoeba castellanii found in four samples (50%, 4/8).
Conclusions
The PDH model has the potential to be a molecular assay for diagnosing Acanthamoeba and microsporidia keratitis. However, a prospective clinical study might be needed before the model is adopted in routine clinical practice.
Introduction
Parasitic keratitis, primarily caused by Acanthamoeba and microsporidia, is largely underreported [1]. Acanthamoeba keratitis (AK) and microsporidia keratitis (MK) may be easily overlooked because they usually present with non-specific symptoms masquerading as viral or noninfectious keratitis [2,3]. Early AK or MK presenting coarse punctate epithelial keratitis can be misdiagnosed as herpetic or adenoviral keratitis or toxic keratitis, whereas late forms of AK or MK that present necrotizing stromal keratitis are easily confused with bacterial or fungal keratitis. Despite the recent outbreaks of AK [4-6] and MK [3,7,8], the two diseases remain rare compared to other forms of microbial keratitis, and this rarity may lead to misdiagnosis by most eye-care practitioners [3,9]. Misdiagnosis and treatment with topical corticosteroid for AK and MK may cause medically refractory stromal keratitis leading to disastrous ocular complications and permanent visual loss.
Routine microbiological examination focusing on bacterial and fungal keratitis can result in underdiagnoses of AK and MK. Culturing in special media is the conventional standard method for diagnosing AK, but the sensitivity can be less than 50% [10], and the incubation time is long (3–7 days) [11]. Although the taxonomic affiliation of microsporidia is closely related to that of fungi [3], microsporidia have evolved as obligate intracellular parasites that require special cell culture systems for isolation. Direct microscopic examination enables rapid diagnosis of AK [2] and MK [3], but the technique is not sensitive enough for diagnosing light infections, owing to the requirements for large corneal scrapes and expertise in ocular microbiology. Therefore, some researchers have proposed different diagnostic tests for rapid and sensitive detection of the two types of parasitic keratitis [12-15].
DNA-based molecular techniques are useful for diagnosing infections caused by Acanthamoeba and microsporidia [12,15-17]. In previous studies, we developed different dot hybridization models to resolve different clinical scenarios, which were found to be sensitive and specific for diagnosing bacterial keratitis [18] and fungal keratitis [19], and for differentiating AK from herpes keratitis [20]. Therefore, the aim of the present study was to develop a parasite dot hybridization (PDH) model as an alternative strategy for diagnosing AK and MK.
Methods
Reference strains and clinical isolates
To assess the candidate oligonucleotide probes for detecting Acanthamoeba and microsporidia, 20 reference strains (12 species) of Acanthamoeba and three strains (three species) of microsporidia were used as the target strains for the sensitivity test, while 20 reference strains of bacteria (ten species) and 20 strains (ten species) of fungi were used as non-target strains for the specificity test (Table 1). The probes, which had passed the preliminary tests, were then assembled in the PDH model (Table 2, Figure 1) for further clinical assessments.
Table 1. Target and non-target microorganisms used for testing candidate oligonucleotide probes.
| Microorganism |
Species and strain no.a |
|---|---|
| Target | |
|
Acanthamoeba
(n=20) |
Acanthamoeba castellanii ATCC 30,010, ATCC 50,370, ATCC 50,374
Acanthamoeba culbertsoni ATCC 30,171
Acanthamoeba griffini ATCC 30,731, ATCC 50,702
Acanthamoeba hatchetti ATCC 30,730, ATCC 50,672
Acanthamoeba jacobsi ATCC 30,732
Acanthamoeba lugdunensis ATCC 50,240
Acanthamoeba mauritaniensis ATCC 50,676
Acanthamoeba palestinensis ATCC 30,870, ATCC 50,708
Acanthamoeba polyphaga ATCC 30,461, ATCC 30,487, ATCC 30,873
Acanthamoeba pustulosa ATCC 50,252
Acanthamoeba quina ATCC 50,241
Acanthamoeba rhysodes ATCC 30,973, ATCC 50,368 |
| Microsporidia (n=3) | Encephalitozoon cuniculi ATCC 50,789 Encephalitozoon hellem ATCC 50,504 Encephalitozoon intestinalis ATCC 50,651 |
| Non-target | |
|---|---|
| Bacteria
(n=20) |
Escherichia coli BCRC 13,095, BCRC 15,481
Klebsiella pneumoniae BCRC 11,644, CCUG 15,938
Mycobacterium chelonae ATCC 35,752, CCUG 37,827
Mycobacterium fortuitum ATCC 6841, ATCC 19,542
Nocardia farcinica BCRC 13,364, BCRC 13,380
Pseudomonas aeruginosa ATCC 27,853, BCRC 10,944
Serratia marcescens BCRC 10,768, BCRC 10,948
Staphylococcus aureus BCRC 10,780, BCRC 14,957
Staphylococcus epidermidis BCRC 14,976, BCRC 14,988
Streptococcus pneumoniae BCRC 10,794, BCRC 14,733 |
| Fungi (n=20) | Alternaria alternata BCRC 32,888, CBS 109,455 Aspergillus flavus BCRC 30,006, BCRC 30,009 Aspergillus fumigatus BCRC 30,502, BCRC 32,120 Candida albicans BCRC 20,511, BCRC 20,512 Candida parapsilosis BCRC 20,515, BCRC 21,253 Curvularia pallescens CBS 156.35, CBS 102,694 Curvularia senegalensis CBS 149.71, CBS 102,171 Fusarium oxysporum ATCC 26,225, CBS 798.95 Fusarium solani BCRC 32,446, BCRC 32,448 Penicillium lilacinum BCRC 31,616, CBS 100,229 |
aATCC, American Type Culture Collection, Manassas, Va., USA; BCRC: Bioresources Collection and Research Center, Hsinchu, Taiwan; CBS, Centraalbureau voor Schimmelcultures, Utrech, The Netherlands; CCUG, Culture Collection, University of Göteborg, Sweden.
Table 2. The oligonucleotide probes used in the PDH model.
| Target microorganism | Probe codea | Sequence (5′ to 3′) | Length (nucleotide) | GC (%) | Tmb (°C) | Locationc | GenBank accession number |
|---|---|---|---|---|---|---|---|
| Acanthamoeba | AC1 | CTGCCACCGAATACATTAGCATGGttttttttttd | 24 | 50.0 | 59.3 | 1145–1168 | KT185626 |
| AC2 | GATTAACTTCTGCGAAAGCATCTtttttttttt | 23 | 39.1 | 51.7 | 1302–1324 | KT185626 | |
| Microsporidia | MS1 | GATGAAGGACGAAGGCTGGAGtttttttttt | 21 | 57.1 | 55.2 | 594–614 | XR552277 |
| MS2 | TCTGGGGATAGTATGCTCGCAAGtttttttttt | 23 | 52.2 | 57.2 | 712–734 | XR552277 |
aOligonucleotide probes are positioned on the PDH model as indicated in Figure 1. bTm, melting temperature. cThe corresponding locus of the probe to the specified GenBank accession number dSeveral bases of thymine were added to the 5′ end of the probe to increase hybridization signal.
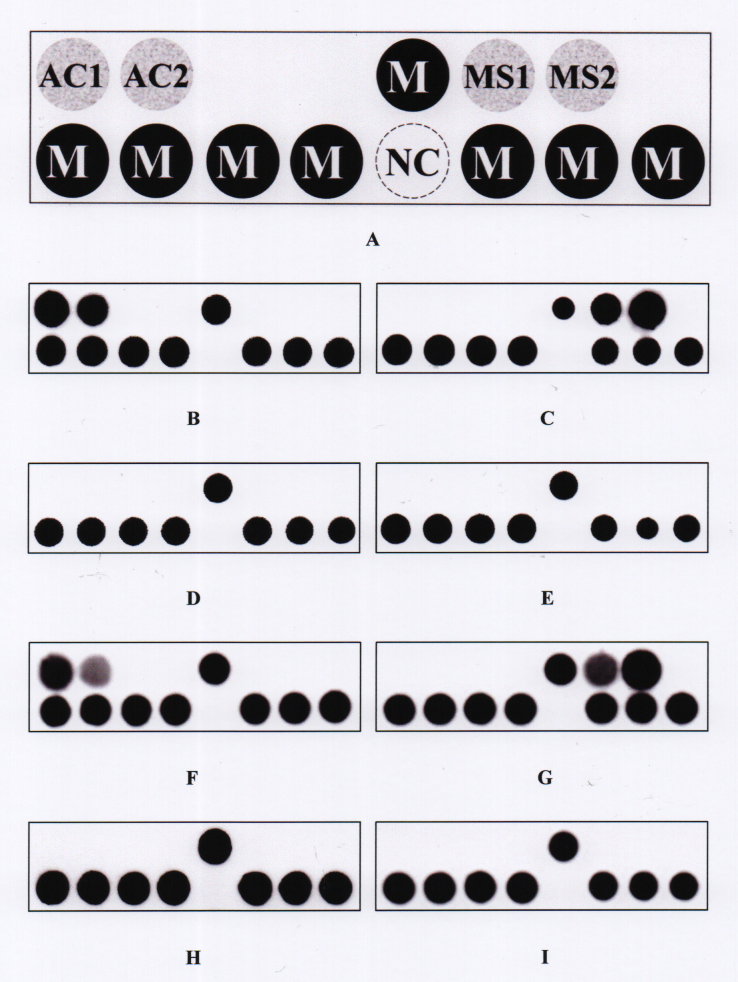
Figure 1.
The PDH model. A: Layout of oligonucleotide probes on the model (0.8 × 0.2 cm). The probes “AC1” and “AC2” were used to identify Acanthamoeba spp. The probes “MS1” and “MS2” were used to identify microsporidia. The dot “NC” is a negative control (tracing dye only). The probe “M” is a position marker probe, i.e., an irrelevant digoxigenin-labeled oligonucleotide probe (5ʹ-digoxigenin-GCA TAT CAA TAA GCG GAG GA-3ʹ). All probe sequences are listed in Table 2. B−E: Representative hybridization patterns for Acanthamoeba castellanii ATCC 30,010, Encephalitozoon cuniculi ATCC 50,789, Pseudomonas aeruginosa BCRC 10,944, and Fusarium solani BCRC 32,446, respectively. F−I: Hybridization patterns for the represented clinical samples that were positive for Acanthamoeba (sample no. 1e), positive for microsporidia (sample no. 1a), false negative for Acanthamoeba (sample no. 1f), and true negative (sample no. 1g).
Clinical specimens
A scraping procedure for corneal debridement was performed for patients with clinically suspected Acanthamoeba and microsporidia keratitis using a #15 sterilized knife under biomicroscopy. One portion of the scrape was sent to the laboratory for standard microbiological analyses, including direct microscopy (Gram stain and acid fast stain) [3,19] and culture (blood agar and Escherichia coli enriched non-nutritional agar for cultivation of amebae). In addition, PCR was performed for detection of Acanthamoeba and microsporidia [12,21]. The remaining corneal scrape on a knife was put into a 1.5-ml sterile Eppendorf tube containing 1 ml saline and stored at −20 °C before DNA extraction. Corneal biopsy for pathological examination was performed only for the patient refractory to medical treatment. For evaluation of the clinical samples with the PDH model, 33 corneal scrapes from patients with clinically suspected Acanthamoeba and microsporidia keratitis were consecutively collected from July 25, 2012, to November 25, 2015, with approval from the Institutional Review Board (IRB)/the Committee of Medical Ethics and Human Experiments of National Cheng Kung University Hospital. All procedures adhered to the Declaration of Helsinki and the ARVO statement on human subjects. Among the 33 consecutively collected samples, eight were AK positive as diagnosed with direct microscopy, culture, PCR, or pathology, and 13 were MK positive as diagnosed with direct microscopy or PCR (Table 3). Control negatives included 12 scrapes in which AK and MK were excluded with standard microbiological analyses and PCR. None of the 12 scrapes had reports of pathological examination.
Table 3. Positive clinical samples determined by the standard microbiological methods, PCR, and the PDH model, and identification of the infectious agents by DNA sequencing.
| Samples no. | Microscopy |
Culture | Pathology | PCRa | PDH model | Species of the infectious microorganismb | Sequence similarity, % | |
|---|---|---|---|---|---|---|---|---|
| GSc | AFSd | |||||||
| Acanthamoeba (+) | ||||||||
| 1e | − | − | + | NDe | + | + | Acanthamoeba quina | 99.3 |
| 1f | − | − | − | + | − | − | NAf | NA |
| 1h | − | − | − | + | + | + | Acanthamoeba polyphaga | 99.3 |
| 2a | − | − | + | ND | + | + | Acanthamoeba sp. | 92.5 |
| 2d | − | − | + | ND | + | + | Acanthamoeba castellanii | 100 |
| 2g | − | − | + | ND | + | + | Acanthamoeba castellanii | 98.8 |
| 4g | − | − | + | ND | + | + | Acanthamoeba castellanii | 98.8 |
| 4h | − | − | + | ND | + | + | Acanthamoeba castellanii | 98.6 |
| Microsporidia (+) | ||||||||
| 1a | − | − | − | ND | + | + | Vittaforma corneae | 99.2 |
| 1b | + | + | − | ND | + | + | Vittaforma corneae | 100 |
| 1c | + | − | − | ND | + | + | Vittaforma corneae | 100 |
| 1d | + | − | Streptococcus pneumoniae | ND | + | + | Vittaforma corneae | 99.0 |
| 2f | − | − | − | ND | + | + | Vittaforma corneae | 99.3 |
| 2h | − | − | − | ND | + | + | Vittaforma corneae | 99.5 |
| 2j | + | − | − | ND | + | + | Vittaforma corneae | 99.9 |
| 3a | ND | + | − | ND | + | + | Vittaforma corneae | 99.9 |
| 4b | + | − | − | ND | + | + | Vittaforma corneae | 99.9 |
| 4c | + | − | − | ND | + | + | Vittaforma corneae | 99.9 |
| 4d | + | − | − | ND | + | + | Vittaforma corneae | 97.0 |
| 4e | − | − | − | ND | + | + | Vittaforma corneae | 99.6 |
| 4l | + | + | − | ND | + | + | Vittaforma corneae | 99.7 |
aPCR, polymerase chain reaction. bThe species names of the infectious agents were determined by gene sequencing. cGS, Gram stain. dAFS, acid fast stain. eND, not done. fNA, not available.
DNA extraction and duplex PCR
The thawed corneal scrape in normal saline was transferred to a 1.5-ml Eppendorf tube and centrifuged at 13,200 ×g in a microfuge for 10 min. DNA in the precipitate was extracted using a commercial kit (DNeasy Blood & Tissue Kit, Qiagen, Valencia, CA). The extracted DNA was amplified with a duplex PCR using two pairs of primers: One pair was used to amplify the 18S rRNA gene of Acanthamoeba (JDP1, 5′-digoxigenin-GGC CCA GAT CGT TTA CCG TGA A-3′; JDP2, 5′-digoxigenin-TCT CAC AAG CTG CTA GGG GAGTCA-3′) [21], and the other pair was used to amplify the small subunit rRNA gene of microsporidia (V1, 5′-digoxigenin-CAC CAG GTT GAT TCT GCC TGA C-3′ [22], and a primer Mco807R, 5′-digoxigenin-CGC GTT GAG TCA AAT TAA G-3′ newly designed in this study). Each primer was labeled with a digoxigenin molecule at the 5′ end. The PCR mixture (25 μl) consisted of 2.5 μl template DNA, 0.2 μM each primer, and other necessary reagents from a PCR kit (KAPA2G Fast HotStart ReadyMix; Kapa Biosystems, Boston, MA). The cycling conditions were as follows: initial denaturation (95 °C, 3 min), ten cycles of denaturation (95 °C, 15 s) and annealing (60 °C, 50 s), and 36 cycles of denaturation (95 °C, 15 s), annealing (55 °C, 30 s), and extension (72 °C, 20 s). Positive controls were performed in each run by using template DNAs of Acanthamoeba castellanii ATCC 30,010 and Encephalitozoon cuniculi ATCC 50,789 (a microsporidia strain), respectively. A negative control was performed in each run by replacing the template DNA with sterile water.
Immobilization of Acanthamoeba- and microsporidia-specific oligonucleotide probes on a nylon membrane
The universal Acanthamoeba probes were designed from a conserved sequence in the 18S rRNA gene, while the universal microsporidia probes were designed from a conserved sequence in the small subunit rRNA gene (Table 2). The procedure for the immobilization of oligonucleotide probes on a nylon membrane is described elsewhere [23]. In brief, each probe in the PDH model (Figure 1A) was diluted 1:1 (final concentration, 10 μM) with a tracking dye solution and spotted on a positively charged nylon membrane (Roche, Mannheim, Germany) using a pin (400 μm in diameter) and a spotter (SR-A300; EZlife Technology, Taipei, Taiwan) to form an array (0.8 × 0.2 cm). A digoxigenin-labeled irrelevant oligonucleotide probe (code M, 5ʹ-digoxigenin-GCA TAT CAA TAA GCG GAG GA-3ʹ) was used as a position marker. The dot NC was a negative control (tracking dye only). Once all the probes had been spotted, the membrane was exposed to a shortwave ultraviolet (UV) light (Stratalinker 1800; Stratagene, La Jolla, CA) for 30 s to fix the probes on the membrane.
Experimental procedures of the PDH model
A 10-μl aliquot of the PCR product was used for the PDH model. The procedures for prehybridization, hybridization, and color development have been described elsewhere [23]. In brief, the PDH model was prehybridized at room temperature for 1 h with 1 ml of hybridization solution (5×saline sodium citrate (SSC) [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 1% [wt/vol] blocking reagent, 0.1% N-laurylsarcosine, and 0.02% sodium dodecyl sulfate). Hybridization was conducted at 55 °C for 90 min. After removing the nonhybridized PCR products and blocking solution, alkaline phosphatase-conjugated anti-digoxigenin antibodies (Fab fragments; Roche, Mannheim, Germany) and phosphatase substrates (nitroblue tetrazolium chloride-5-bromo-4-chloro-3-indolylphosphate; Roche, Mannheim, Germany) were used for color development. Acanthamoeba was identified if at least one of the two probes (codes AC1 and AC2) was hybridized, and microsporidia was identified if at least one of the two probes (codes MS1 and MS2) was hybridized. Images of hybridized arrays were captured with a scanner (PerfectionTM V600 Photo; Epson, Nagano, Japan).
Detection limits of the PDH model
The detection limits of the PDH model for Acanthamoeba and microsporidia were determined by testing tenfold serial dilutions of the prequantified DNA samples of A. castellanii ATCC 30,010 and E. cuniculi ATCC 50,789, respectively. For a sample that was positive for Acanthamoeba or microsporidia, the purified DNA was amplified with the respective PCR for Acanthamoeba and microsporidia, the amplicon was sequenced, and the determined sequence was used to search for homologous sequences of the infectious agents in GenBank using the BLASTN program.
Statistical analysis
Using the results obtained with standard microbiological methods, PCR, and/or pathological examination as the reference method, the performance indices, including sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV), for diagnosis of AK and MK were calculated. The 95% confidence intervals for the performance indices were estimated with an online calculator (Causascientia).
Results
Assessment of the candidate oligonucleotide probes with target and non-target strains
All target strains of Acanthamoeba (n = 20) and microsporidia (n = 3) listed in Table 1 were correctly identified by the two Acanthamoeba probes (AC1 and AC2) and the two microsporidia probes (MS1 and MS2), respectively. No PCR product was amplified from any microorganism of non-target bacteria (n = 20) and fungi (n = 20; Table 1), and thus, no cross-hybridization of the four probes was found. In addition, all target strains of Acanthamoeba did not cross-hybridize with the microsporidia probes, and vice versa. The four candidate probes were then assembled in the PDH model for the following assessments with clinical specimens. The representative hybridization patterns of the PDH model are shown in Figure 1.
Detection limits of the PDH model
The serial tenfold diluted DNA samples of A. castellanii ATCC 30,010 and E. cuniculi ATCC 50,789 were assessed with the PDH model. The detection limits for both pathogens were 0.25 ng DNA per test. However, both detection limits decreased to 2.5 pg DNA if a single-plex PCR was used for A. castellanii and E. cuniculi, respectively.
Species of Acanthamoeba and microsporidia causing keratitis in clinical samples
The species causing AK (n = 7) and MK (n = 13) were further determined with DNA sequencing of the PCR amplicons followed by BLASTN search in GenBank. The microorganism in four (no. 2d, 2g, 4g, and 4h) of the seven AK positive samples was identified as Acanthamoeba castellanii, and the remaining three samples were identified as A. quina (no. 1e), A. polyphaga (no. 1h), and Acanthamoeba sp. (no. 2a), respectively (Table 3). Microsporidia in all 13 cases of MK was identified as Vittaforma corneae (Table 3).
Performance of the PDH model for diagnosing AK and MK
The PDH model was then used to analyze 33 clinical samples. For AK diagnosis, 32 concordant (seven positives and 25 negatives) and one discordant (negative according to the PDH model but positive according to the standard microbiological methods) results were obtained. One sample (sample no. 1d) had a positive bacterial culture (Streptococcus pneumoniae), but the presence of a streptococcal microorganism in this sample did not influence the diagnosis of MK by the PDH model. For diagnosis of MK, all 33 samples produced concordant results (13 positives and 20 negatives). The sensitivity, specificity, PPV, and NPV of the PDH model for diagnosis of AK were 87.5%, 100%, 100%, and 96.2%, respectively, while the respective values for MK diagnosis were all 100% (Table 4). If AK and MK were considered together, the respective performance values of the PDH model were 95.2%, 100%, 100%, and 92.3%.
Table 4. Performance of the PDH model for the diagnosis of Acanthamoeba and microsporidia keratitis.
| Positive clinical samplesa (n=21) | Results of the PDH model |
Performance of the PDH model, % (95% confidence interval) |
|||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Sensitivity | Specificity | PPVb | NPVc | ||
| Acanthamoeba keratitis (n=8) | 7 | 1 | 87.5 (56.7–99.1) | 100 (89.1–100) | 100 (68.8–100) | 96.2 (83.5–99.8) | |
| Microsporidia keratitis (n=13) | 13 | 0 | 100 (80.1–100) | 100 (86.7–100) | 100 (80.7–100) | 100 (86.7–100) | |
aPositive sample as determined by the standard microbiological methods or PCR; bPositive predictive value; cNegative predictive value.
Discussion
This is the first study to describe a molecular test for diagnosing AK and MK simultaneously. The PDH model, based on specific oligonucleotide probes targeting the 18S rRNA gene of Acanthamoeba and the small subunit rRNA gene of microsporidia, had high sensitivity and specificity (Table 4). This model requires minimal instrumentation and can be completed within one working day. Although a duplex PCR was performed in this study, a single-plex PCR targeting either Acanthamoeba or microsporidia can be used on demand. The detection limits (2.5 pg/assay) were 100 times lower if single-plex PCR was adopted.
The PDH model produced one false negative (sample no. 1f) for the diagnosis of AK (Table 3); this might be caused by a low cell number of amebic cells present in the specimen or even no amebic cells sampled due to a wrong sampling locus [15]. In addition, this sample (no. 1f) was positive only with pathology; the sample was negative according to the standard microbiological analyses and PCR. Therefore, a deeper infection was highly suspected, and this might result in a sampling failure. The detection rates of direct microscopy for AK were highly variable [24,25]. No sample was found to have amebic cells by Gram stain in this study (Table 3). In general, a relatively large tissue sample and expertise in ocular microbiology are required for direct microscopy.
In a previous study, we developed a pair of PCR primers and an oligonucleotide probe to detect Acanthamoeba in clinical samples [20]. However, the two primers had low efficiency when an additional pair of primers for microsporidia was included in the duplex PCR described in this study. This might be caused by an interaction (such as dimer formation) between the primers used to amplify Acanthamoeba and microsporidia. Therefore, the primers described by Schroeder et al. [21] were used in the duplex PCR, and two new probes were designed for the diagnosis of Acanthamoeba. The sensitivity (87.5%) of the array for Acanthamoeba detection was slightly lower than that (93.3%) of the previous study [20].
Compared to the results of Joseph et al. [12] (PCR using pan-microsporidian primers had a sensitivity of >83% and a specificity of 92%), the current PDH model displayed a better performance for MK diagnosis (Table 4). In addition, this PDH model being able to differentiate AK from MK might have a benefit under confusing or atypical presentations [7]. Streptococcus pneumoniae was isolated from one (sample no. 1d) of the microsporidia-positive samples (Table 3); the result indicated a complication of polymicrobial keratitis for this patient with MK.
The amplicons of all MK-positive samples were successfully sequenced, and a single microorganism (Vittaforma corneae) was identified (Table 3); the results were in agreement with those of previous studies [7,8,16]. However, at least three species of Acanthamoeba (A. castellanii, A. polyphaga, and A. quina) and an undermined species (Acanthamoeba sp.) were found in the eight AK samples. This indicates a variety of Acanthamoeba species can infect the human eye [26].
Most AK cases are caused by wearing contact lens [6,9]. However, exposure to topical corticosteroids, contaminated soil, and water are predisposing factors of AK [27,28] and MK [7,8]. Immunocompetent and immunocompromised patients with AK and MK are at risk of developing secondary infections with other microorganisms and can progress to severe recalcitrant stromal keratitis if patients are not diagnosed early and properly treated [1,3]. As the sensitivity and specificity of the PDH model are high for diagnosis of AK and MK (Table 4), currently a multicenter study is being conducted to assess the clinical impact of this molecular technique. However, this study was unable to correlate diagnostic results with the clinical outcomes of patients because all clinical samples had been delinked with their identifiable clinical information, except their microbiological diagnostic data. In addition, this limitation also prohibits us from determining the definite final diagnoses for these eyes with negative microbiological results.
In conclusion, the PDH model developed here is a potential diagnostic tool for AK and MK. The current model can provide an alternative molecular assay for the most common parasitic keratitis. We believe this diagnostic model will facilitate early treatment, rescue vision, and minimize ocular complications due to AK and MK. However, a prospective clinical study might be needed before the model is adopted in routine clinical practice.
Acknowledgments
The authors acknowledge the Genomic & Proteomic Core Laboratory, Department of Medical Research, Chang Gung Memorial Hospital at Kaohsiung for supplying the arrayer. This work was supported by Chang Gung Research Proposal (CMRPG8C0762 and CMRPG8C0763), and the Ministry of Science and Technology (Grant No. MOST 103–2314-B-182A-044 and MOST 104–2314-B-182A-101-MY3). The sponsors or funding organizations had no role in the design or conduct of this research. A part of the study results had been presented in the Asia-Pacific Academy of Ophthalmology Congress 2017.
References
- 1.Tu EY, Joslin CE. Microsporidia and Acanthamoeba: the role of emerging corneal pathogens. Eye (Lond) 2012;26:222–7. doi: 10.1038/eye.2011.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammersmith KM. Diagnosis and management of Acanthamoeba keratitis. Curr Opin Ophthalmol. 2006;17:327–31. doi: 10.1097/01.icu.0000233949.56229.7d. [DOI] [PubMed] [Google Scholar]
- 3.Sharma S, Das S, Joseph J, Vemuganti GK, Murthy S. Microsporidial keratitis: need for increased awareness. Surv Ophthalmol. 2011;56:1–22. doi: 10.1016/j.survophthal.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Verani JR, Lorick SA, Yoder JS, Beach MJ, Braden CR, Roberts JM, Conover CS, Chen S, McConnell KA, Chang DC, Park BJ, Jones DB, Visvesvara GS, Roy SL. AcanthamoebaKeratitis Investigation Team. National outbreak of Acanthamoeba keratitis associated with use of a contact lens solution, United States. Emerg Infect Dis. 2009;15:1236–42. doi: 10.3201/eid1508.090225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser MN, Wong Q, Shah L, Holland SP, Morshed M, Isaac-Renton J, Chong M, Kibsey P, Patrick DM. Characteristics of an Acanthamoeba keratitis outbreak in British Columbia between 2003 and 2007. Ophthalmology. 2012;119:1120–5. doi: 10.1016/j.ophtha.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 6.Por YM, Mehta JS, Chua JL, Koh TH, Khor WB, Fong AC, Lim JW, Heng WJ, Loh RS, Lim L, Tan DT. Acanthamoeba keratitis associated with contact lens wear in Singapore. Am J Ophthalmol. 2009;148:7–12. doi: 10.1016/j.ajo.2009.02.030. [DOI] [PubMed] [Google Scholar]
- 7.Fan NW, Wu CC, Chen TL, Yu WK, Chen CP, Lee SM, Lin PY. Microsporidial keratitis in patients with hot springs exposure. J Clin Microbiol. 2012;50:414–8. doi: 10.1128/JCM.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwok AK, Tong JM, Tang BS, Poon RW, Li WW, Yuen KY. Outbreak of microsporidial keratoconjunctivitis with rugby sport due to soil exposure. Eye (Lond) 2013;27:747–54. doi: 10.1038/eye.2013.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robaei D, Carnt N, Minassian DC, Dart JK. The impact of topical corticosteroid use before diagnosis on the outcome of Acanthamoeba keratitis. Ophthalmology. 2014;121:1383–8. doi: 10.1016/j.ophtha.2014.01.031. [DOI] [PubMed] [Google Scholar]
- 10.Radford CF, Minassian DC, Dart JK. Acanthamoeba keratitis in England and Wales: incidence, outcome, and risk factors. Br J Ophthalmol. 2002;86:536–42. doi: 10.1136/bjo.86.5.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maubon D, Dubosson M, Chiquet C, Yera H, Brenier-Pinchart MP, Cornet M, Savy O, Renard E, Pelloux H. A one-step multiplex PCR for Acanthamoeba keratitis diagnosis and quality samples control. Invest Ophthalmol Vis Sci. 2012;53:2866–72. doi: 10.1167/iovs.11-8587. [DOI] [PubMed] [Google Scholar]
- 12.Joseph J, Sharma S, Murthy SI, Krishna PV, Garg P, Nutheti R, Kenneth J, Balasubramanian D. Microsporidial keratitis in India: 16S rRNA gene-based PCR assay for diagnosis and species identification of microsporidia in clinical samples. Invest Ophthalmol Vis Sci. 2006;47:4468–73. doi: 10.1167/iovs.06-0376. [DOI] [PubMed] [Google Scholar]
- 13.Hau SC, Dart JK, Vesaluoma M, Parmar DN, Claerhout I, Bibi K, Larkin DF. Diagnostic accuracy of microbial keratitis with in vivo scanning laser confocal microscopy. Br J Ophthalmol. 2010;94:982–7. doi: 10.1136/bjo.2009.175083. [DOI] [PubMed] [Google Scholar]
- 14.Toriyama K, Suzuki T, Inoue T, Eguchi H, Hoshi S, Inoue Y, Aizawa H, Miyoshi K, Ohkubo M, Hiwatashi E, Tachibana H, Ohashi Y. Development of an immunochromatographic assay kit using fluorescent silica nanoparticles for rapid diagnosis of Acanthamoeba keratitis. J Clin Microbiol. 2015;53:273–7. doi: 10.1128/JCM.02595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda Y, Miyazaki D, Yakura K, Kawaguchi A, Ishikura R, Inoue Y, Mito T, Shiraishi A, Ohashi Y, Higaki S, Itahashi M, Fukuda M, Shimomura Y, Yagita K. Assessment of real-time polymerase chain reaction detection of Acanthamoeba and prognosis determinants of Acanthamoeba keratitis. Ophthalmology. 2012;119:1111–9. doi: 10.1016/j.ophtha.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 16.Reddy AK, Balne PK, Gaje K, Garg P. PCR for the diagnosis and species identification of microsporidia in patients with keratitis. Clin Microbiol Infect. 2011;17:476–8. doi: 10.1111/j.1469-0691.2010.03152.x. [DOI] [PubMed] [Google Scholar]
- 17.Ge Z, Qing Y, Zicheng S, Shiying S. Rapid and sensitive diagnosis of Acanthamoeba keratitis by loop-mediated isothermal amplification. Clin Microbiol Infect. 2013;19:1042–8. doi: 10.1111/1469-0691.12149. [DOI] [PubMed] [Google Scholar]
- 18.Fang PC, Chien CC, Yu HJ. Ren-Wen Ho, Tseng SL, Lai YH, Kuo MT. A dot hybridization assay for the diagnosis of bacterial keratitis. Mol Vis. 2017;23:306–17. [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo MT, Chang HC, Cheng CK, Chien CC, Fang PC, Chang TC. A highly sensitive method for molecular diagnosis of fungal keratitis: a dot hybridization assay. Ophthalmology. 2012;119:2434–42. doi: 10.1016/j.ophtha.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 20.Kuo MT, Fang PC, Yu HJ, Chao TL, Chien CC, Chen SH, Wang JR, Tseng SL, Lai YH, Hsiao CC, Chang TC. A multiplex dot hybridization assay for detection and differentiation of Acanthamoeba and herpes keratitis. Invest Ophthalmol Vis Sci. 2016;57:2158–63. doi: 10.1167/iovs.15-17741. [DOI] [PubMed] [Google Scholar]
- 21.Schroeder JM, Booton GC, Hay J, Niszl IA, Seal DV, Markus MB, Fuerst PA, Byers TJ. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J Clin Microbiol. 2001;39:1903–11. doi: 10.1128/JCM.39.5.1903-1911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller A, Stellermann K, Hartmann P, Schrappe M, Fätkenheuer G, Salzberger B, Diehl V, Franzen C. A powerful DNA extraction method and PCR for detection of microsporidia in clinical stool specimens. Clin Diagn Lab Immunol. 1999;6:243–6. doi: 10.1128/cdli.6.2.243-246.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao CR, Huang L, Bouchara JP, Barton R, Li HC, Chang TC. Identification of medically important molds by an oligonucleotide array. J Clin Microbiol. 2005;43:3760–8. doi: 10.1128/JCM.43.8.3760-3768.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bharathi MJ, Ramakrishnan R, Meenakshi R, Mittal S, Shivakumar C, Srinivasan M. Microbiological diagnosis of infective keratitis: comparative evaluation of direct microscopy and culture results. Br J Ophthalmol. 2006;90:1271–6. doi: 10.1136/bjo.2006.096230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boggild AK, Martin DS, Lee TY, Yu B, Low DE. Laboratory diagnosis of amoebic keratitis: comparison of four diagnostic methods for different types of clinical specimens. J Clin Microbiol. 2009;47:1314–8. doi: 10.1128/JCM.00173-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Souza Carvalho FR, Carrijo-Carvalho LC, Chudzinski-Tavassi AM, Foronda AS, de Freitas D. Serine-like proteolytic enzymes correlated with differential pathogenicity in patients with acute Acanthamoeba keratitis. Clin Microbiol Infect. 2011;17:603–9. doi: 10.1111/j.1469-0691.2010.03252.x. [DOI] [PubMed] [Google Scholar]
- 27.Mascarenhas J, Lalitha P, Prajna NV, Srinivasan M, Das M, D’Silva SS, Oldenburg CE, Borkar DS, Esterberg EJ, Lietman TM, Keenan JD. Acanthamoeba, fungal, and bacterial keratitis: a comparison of risk factors and clinical features. Am J Ophthalmol. 2014;157:56–62. doi: 10.1016/j.ajo.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Zhang Y, Li R, Wang Z, Luo S, Gao M, Deng S, Chen W, Jin X. Acanthamoeba keratitis: clinical characteristics and management. Ophthalmology. 2006;113:412–6. doi: 10.1016/j.ophtha.2005.10.041. [DOI] [PubMed] [Google Scholar]