Abstract
Alcohol consumption is a complex trait determined by both genetic and environmental factors, and is correlated with the risk of alcohol use disorders. While a small number of genetic loci have been reported to be associated with variation in alcohol consumption, genetic factors are estimated to explain about half of the variance in alcohol consumption, suggesting that additional loci remain to be discovered. We conducted a genome-wide association study (GWAS) of alcohol consumption in the large Genetic Epidemiology Research in Adult Health and Aging (GERA) cohort, in four race/ethnicity groups: non-Hispanic Whites, Hispanic/Latinos, East Asians, and African Americans. We examined two statistically independent phenotypes reflecting subjects’ alcohol consumption during the past year, based on self-reported information: any alcohol intake (drinker/non-drinker status), and the regular quantity of drinks consumed per week (drinks/week) among drinkers. We assessed these two alcohol consumption phenotypes in each race/ethnicity group, and in a combined trans-ethnic meta-analysis comprising a total of 86 627 individuals. We observed the strongest association between the previously-reported single nucleotide polymorphism (SNP) rs671 in ALDH2 and alcohol drinker status (OR=0.40, p=2.28×10−72) in East Asians, and also an effect on drinks/week (beta=−0.17, p=5.42×10−4) in the same group. We also observed a genome-wide significant association in non-Hispanic Whites between the previously-reported SNP rs1229984 in ADH1B and both alcohol consumption phenotypes (OR=0.79, p=2.47×10−20 for drinker status and beta=−0.19, p=1.91×10−35 for drinks/week), which replicated in Hispanic/Latinos (OR=0.72, p=4.35×10−7 and beta=−0.21, p=2.58×10−6, respectively). While prior studies reported effects of ADH1B and ALDH2 on lifetime measures, such as risk of alcohol dependence, our study adds further evidence of the effect of the same genes on a cross-sectional measure of average drinking. Our trans-ethnic meta-analysis confirmed recent findings implicating the KLB and GCKR loci in alcohol consumption, with strongest associations observed for rs7686419 (beta=−0.04, p=3.41×10−10 for drinks/week and OR=0.96, p=4.08×10−5 for drinker status), and rs4665985 (beta = 0.04, p=2.26×10−8 for drinks/week and OR=1.04, p=5.00×10−4 for drinker status), respectively. Finally, we also obtained confirmatory results extending previous findings implicating AUTS2, SGOL1, and SERPINC1 genes in alcohol consumption traits in non-Hispanic whites.
INTRODUCTION
Alcohol consumption is a common, complex trait, and heavy alcohol use increases the risk of alcohol use disorders (abuse and dependence).1–3 Drinking above the NIAAA-recommended maximum safe limits of no more than 14 drinks per week for men and 7 drinks per week for women is associated with an increased risk of alcohol-related harm.4–6 The harms from excessive alcohol consumption include a greater risk of a number of health conditions, including liver, cardiovascular, and infectious diseases, cancer, and neuropsychiatric disorders.7–16 Excessive alcohol consumption is also a preventable risk factor for many injuries and accidents.15–17 In total, excessive alcohol consumption contributes to nearly 3.3 million deaths per year worldwide (or 5.9% of all deaths), and 9.8% of all deaths in the United States.16, 18, 19
Alcohol drinking behavior can be measured in a number of ways, including cross-sectional measures of alcohol consumption (e.g. drinks per week), and lifetime measures (e.g. alcohol dependence). There is strong evidence that individual variation in each of these measures is determined by both genetic and environmental factors, and there is strong, if incomplete genetic correlation among them.20, 21 Genetic epidemiologic studies, such as twin and family/adoption studies, have estimated that about half of the variance in these traits may be explained by genetic factors.20, 22, 23 Genetic association studies have demonstrated a pharmacogenetic effect of missense variants in genes in the alcohol metabolism pathway, specifically, alcohol dehydrogenase 1B (ADH1B) and aldehyde dehydrogenase 2 (ALDH2), that influence lifetime measures, such as risk of alcohol dependence.24, 25 While these variants play an important role in determining individual levels of alcohol use, they explain a small proportion of the genetic contribution to variation in alcohol use reported in twin studies. Recently, genome-wide association studies (GWAS) of alcohol consumption have identified potential novel susceptibility loci, however, only a few have been confirmed in independent samples.26–29 That suggests that there remain specific genetic factors that could be discovered in large and ethnically diverse populations, using genotyping platforms with improved genome-wide coverage.
To address this gap, we conducted a GWAS of alcohol consumption in the large and ethnically diverse Genetic Epidemiology Research in Adult Health and Aging (GERA) cohort (n=110 266). It has been previously noted that the use of questionnaire data in large cohorts such as ours may be an efficient and productive approach towards elucidating the genetic basis of alcohol-related traits.30 We examined two statistically independent phenotypes reflecting subjects’ alcohol consumption during the past year, based on self-reported information: any intake (drinker/non-drinker status) and the regular quantity of drinks consumed per week (drinks/week), in four race/ethnicity groups (non-Hispanic whites, Hispanic/Latinos, East Asians, and African Americans) analyzed individually and also combined in a trans-ethnic meta-analysis. Further, we assessed genetic variants that had previously been reported to be associated with alcohol consumption and related traits in our cohort.
MATERIALS AND METHODS
Study Population
We analyzed participants from the Genetic Epidemiology Research in Adult Health and Aging (GERA) cohort which includes 110 266 adult men and women members of the Kaiser Permanente Medical Care Plan, Northern California Region (KPNC) and has been described in detail (dbGAP at http://ncbi.nlm.nih.gov/gap (Study Accession: phs000674.v2.p2)).31, 32 In this study, we focused on subjects who were at least 21 years of age at time of the survey, were of non-Hispanic white, Hispanic/Latino, Asian, or African American race/ethnicity, and provided self-reported information regarding their alcohol consumption during the past year (N=86 627) (Table 1). All study procedures were approved by the Institutional Review Board of the Kaiser Foundation Research Institute.
Table 1.
Characteristics of the GERA subjects with alcohol consumption information by race/ethnicity group
| Characteristics | non-Hispanic whites | Hispanic/Latinos | East Asians | African Americans |
|---|---|---|---|---|
| N (%) | 71 071 (82.0) | 7 047 (8.1) | 6 034 (7.0) | 2 475 (2.9) |
|
| ||||
| Male, n (%) | 24 971 (35.1) | 2 237 (31.7) | 1 968 (32.6) | 714 (28.9) |
| Female, n (%) | 46 100 (64.9) | 4 810 (68.3) | 4 066 (67.4) | 1 761 (71.1) |
|
| ||||
| Age at Survey (years) | ||||
| Mean ± SD | 62.3 ± 13.5 | 55.3 ± 14.9 | 55.3 ± 14.8 | 58.2 ± 14.0 |
|
| ||||
| Lean Body Mass (kg) | ||||
| Mean ± SD | 53.5 ± 10.2 | 51.6 ± 9.8 | 47.0 ± 8.6 | 53.9 ± 9.5 |
|
| ||||
| Nondrinker, n (%) | 23 104 (32.5) | 2 673 (37.9) | 3 288 (54.5) | 1 165 (47.1) |
| Drinker, n (%) | 47 967 (67.5) | 4 374 (62.1) | 2 746 (45.5) | 1 310 (52.9) |
| Alcohol Drinks per Week | ||||
| Mean ± SD | 7.3 ± 6.9 | 6.1 ± 6.4 | 4.9 ± 5.2 | 5.6 ± 6.2 |
Abbreviations: N, number; SD, standard deviation
Phenotype Definitions
Two statistically independent alcohol consumption phenotypes were examined: any intake (drinker/non-drinker status) and the regular quantity of alcoholic drinks consumed per week as a quantitative trait (drinks/week). Both phenotypes were assessed based on the Research Program on Genes, Environment, and Health (RPGEH) survey. On this survey, participants were asked regarding the past year: “On average, how many days a week do you have a drink containing alcohol?” (no days, 1 day, 2 days, 3 days, 4 days, 5 days, 6 days, or every day). Further, participants were asked: “On a typical day that you drink, how many drinks do you have?” (none, 1, 2, 3, 4, 5, 6, 7, or ≥8). Individuals who reported drinking ≥ 1 day per week and ≥ 1 drink per day were defined as ‘drinkers’, whereas those who provided negative answers (“no days” and “none”) were considered as ‘non-drinkers’. For alcohol drinkers, the regular quantity of alcohol drinks consumed per week was calculated by multiplying the two answers. Because this quantitative measure of quantity of alcoholic drinks consumed per week was positively skewed, we performed a log transformation prior to conducting genetic association analyses.
Genotyping and Quality Control Procedures
DNA samples were extracted from Oragene kits (DNA Genotek Inc., Ottawa, ON, Canada) at KPNC and genotyped at the Genomics Core Facility of the Institute for Human Genetics at the University of California, San Francisco (UCSF) on four race/ethnicity-specific Affymetrix Axiom arrays (Affymetrix, Santa Clara, CA, USA) optimized for individuals of European, African American, East Asian, and Latino race/ethnicity.32 Design details and genome-wide coverage of those arrays have been previously described.33, 34 Genotype quality control (QC) procedures for the GERA cohort were performed on an array-wise basis as described in detail elsewhere.32 Briefly, we included SNPs with initial genotyping call rate ≥ 97%, allele frequency difference (≤ 0.15) between males and females for autosomal markers, and genotype concordance rate (> 0.75) across duplicate samples. Around 94% of samples and more than 98% of genetic markers assayed passed QC procedures.32 Prior to imputation, we additionally excluded genetic markers with a minor allele frequency (MAF) < 1%, or a genotype call rate < 90%.
Imputation
Imputation was also conducted on an array-wise basis and has been described elsewhere.35 Following the pre-phase of the genotypes with Shape-IT v2.5,36 genetic markers were imputed from the cosmopolitan reference panel of the 1000 Genomes Project (phase I integrated release) using IMPUTE2 v2.3.1.37–39 As a QC metric, we used the info r2 from IMPUTE2, which estimates the correlation between the true and imputed genotype.40 Herein, we reported imputed markers with info-metric r2 ≥ 0.9 and MAF ≥ 1%; all reported genotyped markers exceeded a genotype call rate ≥ 98%, and a p-value ≥ 0.001 for Hardy-Weinberg equilibrium deviation.
Single Nucleotide Polymorphisms (SNPs) Selection for Extending Prior GWAS Findings
SNPs previously reported to be associated at the genome-wide level of significance (p<5×10−8) with alcohol consumption-related traits, including the maximum quantity of drinks consumed in a 24 hour period (MaxDrinks), were examined for association in this sample. When several SNPs were reported at a genome-wide level of significance at the same locus/gene, we chose the SNP with the most significant p-value. In total, 14 SNPs other than our top GWA findings were selected (Supplementary Table 1), including 7 SNPs previously associated in European ancestry populations,26–29 4 SNPs in Asian ancestry populations,41–43 and 3 SNPs in African ancestry populations.26 As two candidate SNPs (rs2309169 and rs200475889) were not in 1000 Genomes Project (phase I) and so could not be imputed, 12 SNPs remained for association analysis.
Statistical Analysis
GWAS Analyses and Covariate Adjusment
GWA analyses were conducted using PLINK44 v1.07 (http://pngu.mgh.harvard.edu/purcell/plink/) and R45 (https://www.R-project.org). Regional Manhattan plots were performed using LocusZoom v1.1 (http://locuszoom.sph.umich.edu/locuszoom/) including SNP association values ± 1Mb upstream and downstream of our peak significant SNPs.46 We assessed single-marker associations with drinker status and with the quantity of alcoholic drinks consumed per week using logistic and linear regression, respectively. We assumed an additive genetic model using allele counts (i.e., 0, 1, or 2 copies of the minor allele) for typed markers or additive dosages for imputed markers. We first analyzed each of the four race/ethnicity groups separately, adjusting for age, and sex, and lean body mass, which serves as a proxy for body size.47 Lean body mass was estimated using the James equation which relies on sex, height (cm), and total body weight (kg).48 We conducted sensitivity analyses without lean body mass as a covariate, and those analyses without lean body mass produced relatively similar results (Supplementary Table 2). To correct for differences in genetic ancestry, we include ancestry principal components (PCs) in our GWA analyses. To calculate the PCs, we used Eigenstrat49 v4.2 on each of the four race/ethnicity groups as previously described.31 For the non-Hispanic Whites, the first 10 ancestry PCs were included in each regression model, while for the 3 other race/ethnicity groups, the first 6 ancestry PCs were included. For the non-Hispanic whites, the percentage of Ashkenazi ancestry was also used as a covariate in the GWA analyses to adjust for genetic ancestry, as described previously.31 Further, the genomic inflation factor λ was calculated for each GWA analysis to assess inflation due to population stratification and was found to be modest (all ≤ 1.07).
Trans-Ethnic Meta-Analysis
For the combined meta-analysis of the two alcohol consumption phenotypes across the 4 race/ethnicity groups, fixed effects and random effects summary estimates were calculated for an additive model using R package “meta”. Heterogeneity index, I2 (0–100%) as well as P-value for Cochrane’s Q statistic were assessed among groups. We report fixed effects results, except when there was significant heterogeneity between race/ethnicity groups, in which case we report values for a random effects model.
Association Analysis of Previously Reported SNPs
To determine whether the 12 SNPs previously-reported as genome-wide significant in GWAS for alcohol consumption-related traits (e.g. MaxDrinks) were associated with alcohol consumption in the current study, we used a nominal significance level of 0.05. A more stringent multiple testing correction accounting for the 2 phenotypes tested and for the number of variants tested for each ethnic group is also presented (Bonferroni-corrected alpha level of 0.05/(2×6) for European population, of 0.05/(2×4) for Asian population, and of 0.05/(2×2) for African ancestry population).
Conditional Analysis
To assess whether the GCKR, and KLB SNPs recently reported by Schumann et al.,29 contribute to alcohol consumption independently from our most strongly associated SNPs in our trans-ethnic meta-analysis, we adjusted for the effect of our top SNPs (rs4665985, and rs7686419) in our non-Hispanic white sample. In addition, to assess whether previously reported SNPs in the CCDC63, MYL2, C12orf51, and OAS3 genes in the chromosome 12q24 region, that also reached genome-wide significance in our sample, contribute independently from the ALDH2 locus to alcohol consumption in East Asians, we reran association analysis by including rs671 (our top SNP) as a covariate in the regression model.
RESULTS
Study Population
In this study, the proportion of subjects that report consuming alcohol (drinkers) was greatest for non-Hispanic whites (67.5 %) followed by Hispanic/Latinos (62.1%), African Americans (52.9%), and East Asians (45.5%) (Table 1). Among drinkers, the average number of alcoholic drinks consumed per week during the past year followed a similar pattern, with 7.3 for non-Hispanic whites, 6.1 for Hispanic/Latinos, 5.6 for African Americans, and 4.9 for East Asians. We also observed a lower proportion of drinkers and reduced number of drinks per week in women compared to men in all race/ethnicity groups (Supplementary Figures 1 – 2).
Top GWAS Findings
We first conducted a GWAS analysis of the two alcohol consumption phenotypes, stratified by race/ethnicity, detecting two loci associated at a genome-wide level of significance (p<5×10−8). The strongest association was observed in East Asians between the well-known ALDH2 locus25 on 12q24 and alcohol drinker status (Figure 1. a). This association was driven by the missense SNP rs671 in ALDH2 (G>A Glu457Lys), with subjects carrying the rs671 A allele considerably less likely to report any alcohol intake (OR=0.40, p=2.28×10−72) (Table 2). Among East Asian subjects who report drinking alcohol, we also observed an association between rs671 and a reduction of 0.76 drinks/week per copy (p=5.42×10−4). rs671 was not polymorphic (MAF<0.5%) in non-Hispanic whites and African Americans, and was observed at a low frequency with poor imputation quality in Hispanic/Latinos, and so was excluded from the analysis in these race/ethnicity groups. The second strongest association was detected in non-Hispanic white subjects for the well-known rs122998425, a missense SNP in ADH1B (C>T Arg48His), which was associated with both alcohol consumption phenotypes. Subjects who carry the rs1229984 T allele were less likely to report consuming alcohol (OR=0.79, p=2.47×10−20), and among those who did report consuming alcohol, a reduction of 1.26 drinks per week per copy was reported (p=1.91×10−35). Similarly, we found associations between ADH1B rs1229984 and both alcohol consumption phenotypes in Hispanic/Latinos (OR=0.72, p=4.35×10−7 and β=−0.21, p=2.58×10−6). ADH1B rs1229984 showed a consistent direction of effect in all race/ethnicity groups, however, the effect of this SNP in East Asians was smaller than in the other groups. To determine whether the effect of ADH1B rs1229984 was attenuated by the strong effect of ALDH2 rs671 in East Asians, we performed a sub-group analysis among those not carrying the protective allele at ALDH2 rs671 (GG homozygotes). For drinker/non-drinker status, we found similar results (OR=0.94, p=0.21) than in the non-stratified East Asian sample. However, for the quantitative trait drinks/week, we found a relatively small effect of rs1229984 in this sub-group analysis (β=−0.04, p=0.19), suggesting that the effect of AHD1B rs1229984 on alcohol consumption in East Asians may not be as important as in other race/ethnicity groups.
Figure 1.
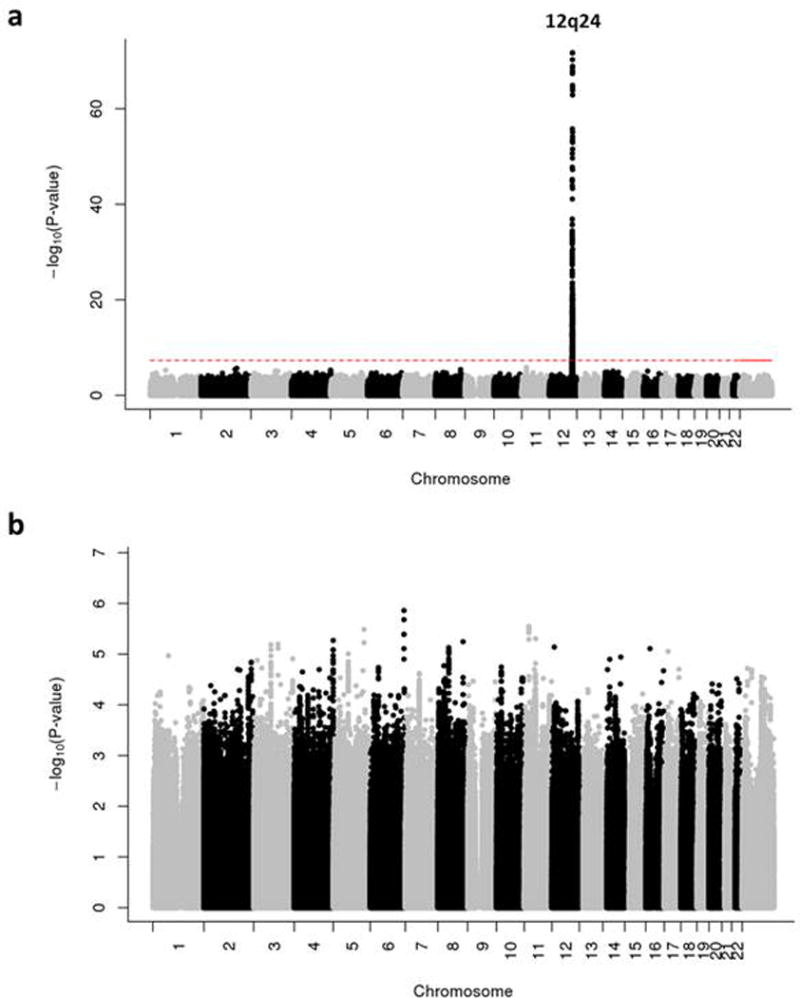
Manhattan plot of alcohol drinker status in East-Asians (a) Before and (b) After conditioning for our top SNP ALDH2 rs671.
Table 2.
Top genome-wide associations with alcohol consumption in each individual race/ethnicity group and in the trans-ethnic meta-analysis
|
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Drinker Status | Drinks/Week | ||||||||||
|
| |||||||||||
| SNP | Chr | Position (bp) | Type | Gene | EA | Ethnicity | EAF | OR (95%CI) | P | Beta (95%CI) | P |
| NHW | 0.28 | 1.05 (1.03–1.08) | 6.74×10−5 | 0.04 (0.02–0.05) | 1.05×10−6 | ||||||
| H/L | 0.38 | 0.97 (0.90–1.04) | 0.41 | 0.05 (0.002–0.09) | 0.04 | ||||||
| rs4665985 | 2 | 27,753,878 | Intergenic | GCKR | C | EAS | 0.44 | 1.02 (0.94–1.09) | 0.68 | 0.02 (−0.04–0.08) | 0.58 |
| AA | 0.12 | 1.05 (0.86–1.79) | 0.63 | 0.21 (0.08–0.33) | 0.002 | ||||||
|
|
|||||||||||
| Meta | 1.04 (1.02–1.06) | 5.0×10−4 | 0.04 (0.03–0.06) | 2.26×10−8 | |||||||
|
| |||||||||||
| NHW | 0.47 | 0.96 (0.94–0.99) | 0.0014 | −0.03 (−0.05–−0.02) | 8.95×10−8 | ||||||
| H/L | 0.48 | 0.92 (0.85–0.98) | 0.01 | −0.05 (−0.09–−0.01) | 0.02 | ||||||
| rs7686419 | 4 | 39,406,370 | Upstream | KLB | A | EAS | 0.46 | 0.94 (0.87–1.02) | 0.13 | −0.10 (−0.15–−0.04) | 7.67×10−4 |
| AA | 0.45 | 0.97 (0.85–1.73) | 0.61 | 0.01 (−0.08–0.09) | 0.91 | ||||||
|
|
|||||||||||
| Meta | 0.96 (0.94–0.98) | 4.08×10−5 | −0.04 (−0.05–−0.02) | 3.41×10−10 | |||||||
|
| |||||||||||
| NHW | 0.05 | 0.79 (0.74–0.84) | 2.47×10−20 | −0.19 (−0.22–−0.16) | 1.91×10−35 | ||||||
| H/L | 0.07 | 0.72 (0.59–0.85) | 4.35×10−7 | −0.21 (−0.30–−0.12) | 2.58×10−6 | ||||||
| rs1229984 | 4 | 100,239,319 | Missense | ADH1B | T | EAS | 0.69 | 0.94 (0.86–1.02) | 0.15 | −0.07 (−0.12–−0.01) | 0.02 |
| AA | 0.03 | 0.61 (0.26–0.69) | 0.004 | −0.11 (−0.39–0.16) | 0.41 | ||||||
|
|
|||||||||||
| Meta | *0.79 (0.69–0.91) | *8.0×10−4 | *−0.15 (−0.23–−0.07) | *3.04×10−4 | |||||||
|
| |||||||||||
| NHW | 0.0005 | NA | NA | NA | NA | ||||||
| H/L | 0.03 | NA | NA | NA | NA | ||||||
| rs671 | 12 | 112,241,766 | Missense | ALDH2 | A | EAS | 0.21 | 0.40 (0.29–0.50) | 2.28×10−72 | −0.17 (−0.26–−0.07) | 5.42×10−4 |
| AA | 0.004 | NA | NA | NA | NA | ||||||
|
|
|||||||||||
| Meta | NA | NA | NA | NA | |||||||
Abbreviations: Chr, chromosome; bp, base pair (based on UCSC Genome Browser Assembly February 2009 (GRCh37/hg19)); EA, Effect Allele; EAF, Effect Allele Frequency; NWW, non-Hispanic whites; H/L, Hispanic/Latinos; EAS, East Asians; AA, African Americans; Meta, combined trans-ethnic meta-analysis. The combined meta-analysis includes 71 071 non-Hispanic Whites, 7 047 Hispanic/Latinos, 6 034 East Asians, and 2 475 African Americans.
values for random effects model are reported when significant heterogeneity between race/ethnicity groups was detected (I2≥60%; Q≤0.05)
Bold numbers are p-values <5.0×10−8; NA: ALDH2 rs671 was not polymorphic (MAF<0.5%) in non-Hispanic whites and African Americans, and was observed at a low frequency and imputation quality in Hispanic/Latinos, and so was excluded from the analysis in these race/ethnicity groups.
Trans-Ethnic Meta-Analysis
We then conducted a combined meta-analysis across the 4 race/ethnicity groups (non-Hispanic Whites, Hispanic/Latinos, East Asians, and African Americans) for the two alcohol consumption phenotypes. We identified two loci that exceeded genome-wide significance (p<5.0×10−8) with drinks/week in the regions of GCKR (rs4665985, beta=0.04, p=2.26×10−8), and KLB (rs7686419, beta=−0.04, p=3.41×10−10) (Table 2). Consistently, GCKR rs4665985 and KLB rs7686419 were also associated with drinker status (p=5.0×10−4 and 4.08×10−5, respectively). Regional Manhattan plots on chromosome 2 and 4 near GCKR and KLB, respectively, are shown in Figure 2. Results for each individual race/ethnicity group are also presented in Table 2, showing no evidence of heterogeneity.
Figure 2.
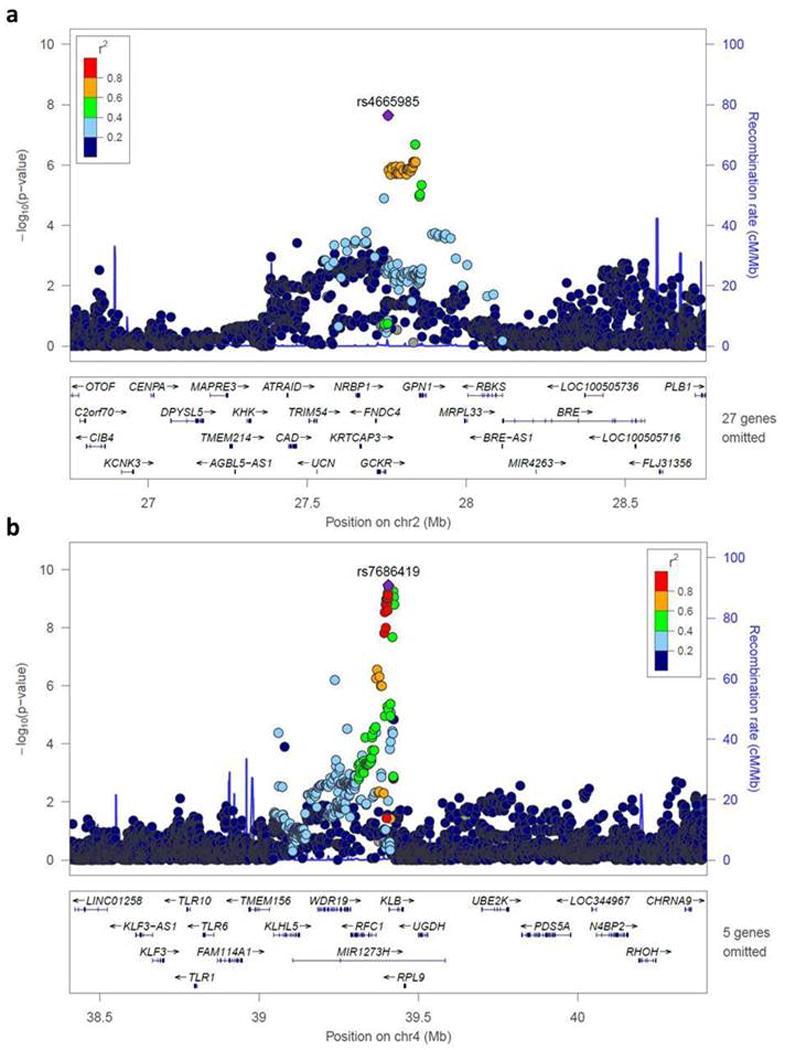
Regional locus zoom plots for the alcohol consumption-associated loci in the trans-ethnic meta-analysis. Regional plots for the loci near (a) GCKR, and (b) KLB which reached genome-wide significance in the combined meta-analysis.
Extending Findings from Prior GWAS
Finally, we investigated a total of 12 SNPs (6 SNPs for European ancestry subjects, 4 SNPs for East Asian ancestry subjects, and 2 SNPs for African ancestry subjects) associated with alcohol consumption-related traits (e.g. MaxDrinks) at a genome-wide significance level in previous studies (Supplementary Table 1).26–29, 41–43 In non-Hispanic whites, the strongest evidence of association was obtained for GCKR rs780094 and KLB rs11940694 (the same two loci identified in our meta-analysis) with drinks/week (beta=−0.034, Bonferroni-corrected p=2.86 × 10−6 and beta=−0.035, p=1.64 × 10−6, respectively) (Table 3). The same two SNPs were also associated with drinker status in a consistent direction (OR=0.96, Bonferroni-corrected p=0.013 and OR=0.95, p=0.002, respectively). These findings are consistent with the recent report from Schumann et al., who observed genome-wide significant associations for these two SNPs, in their discovery GWAS and in their combined discovery and replication data of European descent, respectively.29 However, when we repeated the analyses, conditioning on our most strongly associated SNPs from our trans-ethnic meta-analysis (rs4665985 at GCKR and rs7686419 at KLB), GCKR rs780094 and KLB rs11940694 did not remain significant (p=0.10 and 0.38, respectively), suggesting that our top associated SNPs and theirs represent the same signals at those two loci. We also confirmed the association between AUTS2 rs6943555 and drinks/week (beta=0.03, Bonferroni-corrected p=0.005) and a consistent but not significant association with drinker status. This SNP was previously associated with MaxDrinks in a meta-analysis combining 26,316 European-ancestry individuals.27 In non-Hispanic whites, we further detected weak suggestive associations of drinks/week (but not drinker status) with rs1799876 in SERPINC1 and drinker status (but not drinks/week) with rs11128951 at the SGOL1 locus, both of which were previously associated with MaxDrinks.26, 28 In East Asians, we observed genome-wide significant associations with alcohol drinker status for the 4 SNPs tested in the region of 12q24. None of these association signals remained significant, however, after conditioning on rs671 (our most strongly associated SNP), indicating that these SNPs do not contribute independently to variation in alcohol consumption in East Asians (Figure 1. b). In African Americans, the LOC100507053/ADH1B loci with SNPs rs28864441 and rs2066702 previously associated with MaxDrinks26, showed marginal evidence of association with drinks/week (but not drinker status) in our sample (Table 3). However, we note that the African Americans are the smallest subgroup in our study. Taken together, 3 of the 12 SNPs tested (25.0%) remained significant after Bonferroni correction and conditioning on the most significant SNP in each region. Thus, we confirmed and extended previous findings implicating AUTS2, SGOL1, and SERPINC1 genes in alcohol consumption-related traits in non-Hispanic whites.
Table 3.
Results in our GERA cohort of SNPs previously associated with alcohol consumption and related phenotypes at genome-wide level significance (p<5×10−8)
|
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-Hispanic Whites | ||||||||||||||
|
|
||||||||||||||
| Drinker Status | Drinks/Week | |||||||||||||
|
| ||||||||||||||
| Chr | Position (bp) | SNP | Gene | Previous Study | MA | MAF | OR | P | Padjusted | Pcondi | Beta | P | Padjusted | Pcondi |
| 1 | 173878471 | rs1799876 | SERPINC1 | Xu et al. 2015 | G | 0.33 | 0.99 | 0.43 | 1.0 | – | 0.02 | 0.0026 | 0.031 | – |
| 2 | 27741237 | rs780094 | GCKR | Schumann et al. 2016 | T | 0.41 | 0.96 | 0.0011 | 0.013 | 0.52 | −0.034 | 2.38 × 10−7 | 2.86 × 10−6 | 0.10 |
| 3 | 20375546 | rs11128951 | SGOL1 | Pan et al. 2013 | G | 0.19 | 1.05 | 0.0016 | 0.019 | – | 0.003 | 0.75 | 1.0 | – |
| 4 | 39414993 | rs11940694 | KLB | Schumann et al. 2016 | A | 0.42 | 0.95 | 1.93×10−4 | 0.002 | 0.17 | −0.035 | 1.37 × 10−7 | 1.64 × 10−6 | 0.38 |
| 7 | 50531885 | rs11575537 | DDC | Pan et al. 2013 | T | 0.02 | 1.03 | 0.48 | 1.0 | – | 0.03 | 0.33 | 1.0 | – |
| 7 | 69806023 | rs6943555 | AUTS2 | Schumann et al. 2011 | A | 0.25 | 1.02 | 0.095 | 1.0 | – | 0.03 | 4.0×10−4 | 0.005 | – |
|
| ||||||||||||||
| East Asians | ||||||||||||||
|
|
||||||||||||||
| Drinker Status | Drinks/Week | |||||||||||||
|
| ||||||||||||||
| Chr | Position (bp) | SNP | Gene | Previous Study | MA | MAF | OR | P | Padjusted | Pcondi | Beta | P | Padjusted | |
|
| ||||||||||||||
| 12 | 111333622 | rs10849915 | CCDC63 | Baik et al. 2011 | C | 0.23 | 1.70 | 4.21×10−28 | 3.37×10−27 | 0.79 | 0.04 | 0.26 | 1.0 | |
| 111414461 | rs12229654 | MYL2 | Baik et al. 2011 | G | 0.18 | 2.31 | 1.75×10−48 | 1.40×10−47 | 0.06 | 0.10 | 0.041 | 0.33 | ||
| 112645401 | rs2074356 | C12orf51 | Baik et al. 2011 | A | 0.14 | 2.41 | 5.48×10−48 | 4.38×10−47 | 0.19 | 0.11 | 0.045 | 0.36 | ||
| 113409176 | rs2072134 | OAS3 | Baik et al. 2011 | A | 0.16 | 1.58 | 3.24×10−16 | 2.59×10−15 | 0.34 | 0.06 | 0.22 | 1.0 | ||
|
| ||||||||||||||
| African-Americans | ||||||||||||||
| Drinker Status | Drinks/Week | |||||||||||||
|
| ||||||||||||||
| Chr | Position (bp) | SNP | Gene | Previous Study | MA | MAF | OR | P | Padjusted | Beta | P | Padjusted | ||
|
| ||||||||||||||
| 4 | 100190805 | rs28864441 | LOC100507053 | Xu et al. 2015 | T | 0.16 | 1.00 | 1.0 | 1.0 | 0.12 | 0.027 | 0.11 | ||
| 4 | 100229017 | rs2066702 | ADH1B | Xu et al. 2015 | A | 0.17 | 1.00 | 0.97 | 1.0 | 0.12 | 0.026 | 0.10 | ||
Abbreviations: Chr, chromosome; bp, base pair (based on UCSC Genome Browser Assembly February 2009 (GRCh37/hg19)); MA, Minor Allele; MAF, Minor Allele Frequency; Bold numbers are p-values <0.05; P adjusted: p-values adjusted for Bonferroni correction (for 2 phenotypes and the number of SNPs tested for each ethnic group); P condi: p-values adjusted for GCKR rs4665985, KLB rs7686419, or ALDH2 rs671
DISCUSSION
In this study we observed substantial differences in alcohol drinking behavior across race/ethnicity groups, particularly in East Asians who have a larger proportion of non-drinkers than other groups, and fewer drinks per week among those who do drink. This appears to be driven by ALDH2 rs671, which is the strongest effect SNP in our GWAS analyses and seen almost exclusively in the East Asian group. Our findings are consistent with previous genetic association studies which have reported strong evidence for association with alcohol consumption-related traits in the region of 12q24 in Asian populations, most likely due to linkage disequilibrium with ALDH2 rs671.41–43, 50 Our results support that rs671 is a single hit with no other independently associated risk variants nearby, as our conditional analysis completely attenuated the result for previously reported SNPs at 12q24. Our second strongest SNP association, between ADH1B rs1229984 and alcohol consumption in non-Hispanic whites, is consistent with previous findings in European Americans.26, 51 However, ADH1B rs1229984 had a relatively smaller effect in our East Asian sample in comparison to other race/ethnicity groups, suggesting that its effect might be attenuated by ALDH2 rs671 in East Asians. Previous studies have found that both genetic polymorphisms ADH1B rs1229984 and ALDH2 rs671 are especially common in Asian populations,52, 53 and that they significantly influence drinking behavior in a synergistic manner.54 To determine whether the relatively small effect of rs1229984 in East Asians was due to the strong effect of ALDH2 rs671, we examined the association of ADH1B rs1229984 among those East Asians who do not carry the non-drinking allele of rs671. The association for rs1229984 was not increased in this sub-group analysis, suggesting that the effect of AHD1B rs1229984 on alcohol consumption in East Asians may not be as important as in other race/ethnicity groups.
ADH1B and ALDH2 play a central role in the metabolism of alcohol in humans, and the functional impact of their two missense polymorphisms rs1229984 and rs671 has been extensively reviewed elsewhere.24, 25 In vivo, individuals who carry at least one copy of rs1229984 eliminate ethanol more quickly after heavy drinking and have lower blood alcohol concentrations in comparison to homozygous wild-type individuals.55, 56 Thus, individuals who carry at least one copy of rs1229984 are less exposed to circulating ethanol and might be less likely to develop a tolerance and dependence to alcohol, as previously speculated.55, 56 Our findings provide additional strong evidence of ADH1B rs1229984 as the variant in the ADH gene cluster with the largest impact on alcohol consumption, as previously reported.57, 58 On the other hand, rs671 impairs the processing of ALDH2, causing an accumulation in the body of acetaldehyde, which is toxic and leads to undesirable reactions, including nausea.56, 59 Early work demonstrated the strong effect of a single non-functional allele at this locus, as heterozygotes and homozygotes for rs671 (GA and AA genotypes) are deficient in ALDH2 activity, suggesting that rs671 allele A is dominant.60 Therefore, individuals who carry at least one copy of rs671 drink typically less, and are protected against heavy alcohol use and alcohol use disorders.50, 61 Consistently, we found that GERA subjects who carried even a single deficient metabolizing allele of ALDH2 rs671 were less likely to consume alcohol. Thus, the current study adds further evidence of an effect on average alcohol drinking of ADH1B and ALDH2, whereas much of the extensive prior literature is on effects on lifetime measures, such as risk of alcohol dependence.25, 61–64
In this study, we also identified two loci for alcohol consumption in our trans-ethnic meta-analysis. One of these loci was near KLB on chromosome 4 and showed the strongest association with alcohol consumption in our meta-analysis. The second locus with genome-wide significance was near GCKR on chromosome 2. Our findings are consistent with a recent study reporting genome-wide associations at those two loci with daily alcohol intake (log grams per day) in a large meta-analysis of Europeans,29 even though the strongest SNPs reported by Schumann et al. were different than ours. However, our lead SNPs at KLB and GCKR loci were relatively close to theirs (8.6 and 12.6 kb apart) and were moderately correlated in European ancestry populations (r2 ranged from 0.4 to 0.6). Our conditional analyses also indicated that our lead SNPs and theirs represent the same signals at those two loci, suggesting that all the identified SNPs (rs780094 and rs4665985 at GCKR, and rs11940694 and rs7686419 at KLB) are most likely proxies of causal variants influencing alcohol consumption.
Our results also extend findings of previous studies implicating other genes in determining variation in alcohol consumption and related phenotypes (e.g. MaxDrinks), especially in European-ancestry populations, including AUTS2, SGOL1, and SERPINC1. Schumann et al. previously reported a genome-wide association with alcohol consumption for AUTS2 rs6943555 in a meta-analysis combining 26,316 European-ancestry individuals.27 Two other studies conducted in European-ancestry populations identified potential novel loci associated with MaxDrinks, including SGOL1 rs11128951,28 and SERPINC1 rs1799876.26 We obtained modestly consistent results for these loci with alcohol consumption during the past year in our independent non-Hispanic white sample.
In contrast, other SNPs that were reported as genome-wide significant in previous GWAS of MaxDrinks, were not associated with either of our alcohol consumption traits in the current study. Specifically, several SNPs in DDC at 7p12.2 showed significant associations with MaxDrinks in a previous study in a European ancestry population,28 however DDC rs11575537 was not associated in our European ancestry sample with either of our traits. This lack of association at DDC is consistent with prior results of Xu et al. where no significant association with MaxDrinks was observed at this locus.26 In addition, two genome-wide associations for MaxDrinks previously reported in African Americans for ADH1B rs2066702, and rs28864441 at LOC100507053,26 were not associated with alcohol consumption in our African ancestry sample. However, we note that the African American subgroup is the smallest in our study, and we may have been underpowered to detect effects with statistical significance.
Finally, lack of an association may be influenced by variation in genotyping array coverage, because coverage affects the power to detect individual variants.65–67 Differences in coverage may explain why previous studies did not report a significant association between alcohol consumption-related traits and ALDH2 rs671 in Asian populations or ADH1B rs1229984 in European ancestry populations.
We recognize several potential limitations of our study. First, the alcohol consumption phenotypes were based on self-reported information, which may result in underestimates of the effects of individual SNPs due to phenotype misclassification. Nonetheless, self-reported measures of alcohol consumption, including drink frequency and regular quantity at the time of heaviest drinking, have been shown to correlate strongly with the genetic risk for alcohol use disorders such as dependence.20 Second, the GERA cohort members are older on average than the general population, which could affect both the generalizability of our findings and our power to observe genetic effects which may be strongest in young adulthood. For instance, older subjects may consume alcohol in a different manner (e.g. better quality of alcoholic beverages, low-risk pattern of drinking while eating) in comparison to younger subjects.16 Further, older subjects may reduce their alcohol consumption due to comorbid conditions or medication use. In addition, participants included in the present study may be healthier and/or more affluent than the general population, as they are all members of the KPNC health plan. We recognize that our restriction to study common variants (MAF ≥ 1%) cannot exclude the possibility that rare functional variants may contribute to variation in alcohol consumption. Despite these limitations, our study is based on a unique and very large cohort of individuals, who were all members of a single integrated delivery system. Participants were recruited in a similar manner, and were assessed for their alcohol consumption using a single questionnaire providing more consistency, in contrast to consortia which often include different questions across studies. Samples were genotyped on a single platform, overall allowing us to confirm previous associations with the most established alcohol consumption-related risk variants in terms of effect size and direction.
It is also important to note phenotypic differences between this study and previous ones. Most previous GWAS studies have focused on lifetime measures related to alcohol use, such as alcohol dependence, while others have used a single time measure of maximum drinks in a 24-hour period. By comparison, ours focused on two measures of alcohol consumption over the past year (drinker status and drinks/week). All of these traits have been shown to be highly correlated genetically, but there are also differences. For example, alcohol use disorders (abuse or dependence) are reflective of lifetime exposure – but also other potential factors not directly related to consumption. That could explain why some genetic findings associated with alcohol use disorders might not be found associated with measures of alcohol consumption. However, our findings have generally confirmed prior associations, suggesting that many of the previously reported genetic variants for alcohol use disorders are directly related to consumption, measured either over a short or long time period.
In conclusion, we identified 4 loci associated with alcohol consumption at a genome-wide significance level, including ALDH2, ADH1B, KLB, and GCKR. Our results also extend previous findings implicating AUTS2, SGOL1, and SERPINC1 genes in alcohol consumption-related traits in non-Hispanic whites. Our study of 86 627 individuals had 80% power to detect effects of common SNPs (MAF>0.10) of 1.10 on drinker status and a proportion of variance of 0.05% in drinks/week. This suggests that future genetic association studies of alcohol consumption traits will need to include hundreds of thousands if not millions of subjects to identify and confirm the small effects of individual SNPs on these phenotypes.
Supplementary Material
Acknowledgments
The study was supported by the National Institute of Alcohol Abuse and Alcoholism (NIAAA) R21 no. R21AA021223.
Footnotes
CONFLICT OF INTEREST
The authors declare no competing financial interests in relation to the work described.
Supplementary information is available at Molecular Psychiatry’s website
References
- 1.Dawson DA. The link between family history and early onset alcoholism: earlier initiation of drinking or more rapid development of dependence? J Stud Alcohol. 2000;61(5):637–646. doi: 10.15288/jsa.2000.61.637. [DOI] [PubMed] [Google Scholar]
- 2.Grant BF, Stinson FS, Harford TC. Age at onset of alcohol use and DSM-IV alcohol abuse and dependence: a 12-year follow-up. J Subst Abuse. 2001;13(4):493–504. doi: 10.1016/s0899-3289(01)00096-7. [DOI] [PubMed] [Google Scholar]
- 3.Sartor CE, Lynskey MT, Heath AC, Jacob T, True W. The role of childhood risk factors in initiation of alcohol use and progression to alcohol dependence. Addiction. 2007;102(2):216–225. doi: 10.1111/j.1360-0443.2006.01661.x. [DOI] [PubMed] [Google Scholar]
- 4.Dawson DA. US low-risk drinking guidelines: an examination of four alternatives. Alcohol Clin Exp Res. 2000;24(12):1820–1829. [PubMed] [Google Scholar]
- 5.Dawson DA, Grant BF, Li TK. Quantifying the risks associated with exceeding recommended drinking limits. Alcohol Clin Exp Res. 2005;29(5):902–908. doi: 10.1097/01.alc.0000164544.45746.a7. [DOI] [PubMed] [Google Scholar]
- 6.Willenbring ML, Massey SH, Gardner MB. Helping patients who drink too much: an evidence-based guide for primary care clinicians. Am Fam Physician. 2009;80(1):44–50. [PubMed] [Google Scholar]
- 7.Irving HM, Samokhvalov AV, Rehm J. Alcohol as a risk factor for pancreatitis. A systematic review and meta-analysis Jop. 2009;10(4):387–392. [PMC free article] [PubMed] [Google Scholar]
- 8.Lonnroth K, Williams BG, Stadlin S, Jaramillo E, Dye C. Alcohol use as a risk factor for tuberculosis - a systematic review. BMC Public Health. 2008;8:289. doi: 10.1186/1471-2458-8-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson DE, Jarman DW, Rehm J, Greenfield TK, Rey G, Kerr WC, et al. Alcohol-attributable cancer deaths and years of potential life lost in the United States. Am J Public Health. 2013;103(4):641–648. doi: 10.2105/AJPH.2012.301199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patra J, Taylor B, Irving H, Roerecke M, Baliunas D, Mohapatra S, et al. Alcohol consumption and the risk of morbidity and mortality for different stroke types–a systematic review and meta-analysis. BMC Public Health. 2010;10:258. doi: 10.1186/1471-2458-10-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehm J, Parry C. Alcohol consumption and infectious diseases in South Africa. Lancet. 2009;374(9707):2053. doi: 10.1016/S0140-6736(09)62150-4. [DOI] [PubMed] [Google Scholar]
- 12.Samokhvalov AV, Irving H, Mohapatra S, Rehm J. Alcohol consumption, unprovoked seizures, and epilepsy: a systematic review and meta-analysis. Epilepsia. 2010;51(7):1177–1184. doi: 10.1111/j.1528-1167.2009.02426.x. [DOI] [PubMed] [Google Scholar]
- 13.Samokhvalov AV, Irving HM, Rehm J. Alcohol consumption as a risk factor for pneumonia: a systematic review and meta-analysis. Epidemiol Infect. 2010;138(12):1789–1795. doi: 10.1017/S0950268810000774. [DOI] [PubMed] [Google Scholar]
- 14.Stein E, Cruz-Lemini M, Altamirano J, Ndugga N, Couper D, Abraldes JG, et al. Heavy daily alcohol intake at the population level predicts the weight of alcohol in cirrhosis burden worldwide. J Hepatol. 2016 doi: 10.1016/j.jhep.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 15.Taylor B, Irving HM, Kanteres F, Room R, Borges G, Cherpitel C, et al. The more you drink, the harder you fall: a systematic review and meta-analysis of how acute alcohol consumption and injury or collision risk increase together. Drug Alcohol Depend. 2010;110(1–2):108–116. doi: 10.1016/j.drugalcdep.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.WHO. Global Status Report on Alcohol and Health 2014. Geneva: 2014a. [Google Scholar]
- 17.Korcha RA, Cherpitel CJ, Ye Y, Bond J, Andreuccetti G, Borges G, et al. Alcohol use and injury severity among emergency department patients in six countries. J Addict Nurs. 2013;24(3):158–165. doi: 10.1097/JAN.0b013e3182a04b47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shield KD, Gmel G, Kehoe-Chan T, Dawson DA, Grant BF, Rehm J. Mortality and potential years of life lost attributable to alcohol consumption by race and sex in the United States in 2005. PLoS One. 2013;8(1):e51923. doi: 10.1371/journal.pone.0051923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stahre M, Roeber J, Kanny D, Brewer RD, Zhang X. Contribution of excessive alcohol consumption to deaths and years of potential life lost in the United States. Prev Chronic Dis. 2014;11:E109. doi: 10.5888/pcd11.130293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kendler KS, Myers J, Dick D, Prescott CA. The relationship between genetic influences on alcohol dependence and on patterns of alcohol consumption. Alcohol Clin Exp Res. 2010;34(6):1058–1065. doi: 10.1111/j.1530-0277.2010.01181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dick DM, Meyers JL, Rose RJ, Kaprio J, Kendler KS. Measures of current alcohol consumption and problems: two independent twin studies suggest a complex genetic architecture. Alcohol Clin Exp Res. 2011;35(12):2152–2161. doi: 10.1111/j.1530-0277.2011.01564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansell NK, Agrawal A, Whitfield JB, Morley KI, Zhu G, Lind PA, et al. Long-term stability and heritability of telephone interview measures of alcohol consumption and dependence. Twin Res Hum Genet. 2008;11(3):287–305. doi: 10.1375/twin.11.3.287. [DOI] [PubMed] [Google Scholar]
- 23.Agrawal A, Grant JD, Littlefield A, Waldron M, Pergadia ML, Lynskey MT, et al. Developing a quantitative measure of alcohol consumption for genomic studies on prospective cohorts. J Stud Alcohol Drugs. 2009;70(2):157–168. doi: 10.15288/jsad.2009.70.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30(1):5–13. [PMC free article] [PubMed] [Google Scholar]
- 25.Edenberg HJ, Foroud T. Genetics and alcoholism. Nat Rev Gastroenterol Hepatol. 2013;10(8):487–494. doi: 10.1038/nrgastro.2013.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, et al. Genomewide Association Study for Maximum Number of Alcoholic Drinks in European Americans and African Americans. Alcohol Clin Exp Res. 2015;39(7):1137–1147. doi: 10.1111/acer.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumann G, Coin LJ, Lourdusamy A, Charoen P, Berger KH, Stacey D, et al. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc Natl Acad Sci U S A. 2011;108(17):7119–7124. doi: 10.1073/pnas.1017288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan Y, Luo X, Liu X, Wu LY, Zhang Q, Wang L, et al. Genome-wide association studies of maximum number of drinks. J Psychiatr Res. 2013;47(11):1717–1724. doi: 10.1016/j.jpsychires.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, et al. KLB is associated with alcohol drinking, and its gene product beta-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci U S A. 2016;113(50):14372–14377. doi: 10.1073/pnas.1611243113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mbarek H, Milaneschi Y, Fedko IO, Hottenga JJ, de Moor MH, Jansen R, et al. The genetics of alcohol dependence: Twin and SNP-based heritability, and genome-wide association study based on AUDIT scores. Am J Med Genet B Neuropsychiatr Genet. 2015;168(8):739–748. doi: 10.1002/ajmg.b.32379. [DOI] [PubMed] [Google Scholar]
- 31.Banda Y, Kvale MN, Hoffmann TJ, Hesselson SE, Ranatunga D, Tang H, et al. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1285–1295. doi: 10.1534/genetics.115.178616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kvale MN, Hesselson S, Hoffmann TJ, Cao Y, Chan D, Connell S, et al. Genotyping Informatics and Quality Control for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1051–1060. doi: 10.1534/genetics.115.178905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann TJ, Kvale MN, Hesselson SE, Zhan Y, Aquino C, Cao Y, et al. Next generation genome-wide association tool: design and coverage of a high-throughput European-optimized SNP array. Genomics. 2011;98(2):79–89. doi: 10.1016/j.ygeno.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffmann TJ, Zhan Y, Kvale MN, Hesselson SE, Gollub J, Iribarren C, et al. Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm. Genomics. 2011;98(6):422–430. doi: 10.1016/j.ygeno.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann TJ, Van Den Eeden SK, Sakoda LC, Jorgenson E, Habel LA, Graff RE, et al. A large multiethnic genome-wide association study of prostate cancer identifies novel risk variants and substantial ethnic differences. Cancer Discov. 2015;5(8):878–891. doi: 10.1158/2159-8290.CD-15-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9(2):179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 37.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1(6):457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 41.Baik I, Cho NH, Kim SH, Han BG, Shin C. Genome-wide association studies identify genetic loci related to alcohol consumption in Korean men. Am J Clin Nutr. 2011;93(4):809–816. doi: 10.3945/ajcn.110.001776. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi F, Isono M, Nabika T, Katsuya T, Sugiyama T, Yamaguchi S, et al. Confirmation of ALDH2 as a Major locus of drinking behavior and of its variants regulating multiple metabolic phenotypes in a Japanese population. Circ J. 2011;75(4):911–918. doi: 10.1253/circj.cj-10-0774. [DOI] [PubMed] [Google Scholar]
- 43.Yang X, Lu X, Wang L, Chen S, Li J, Cao J, et al. Common variants at 12q24 are associated with drinking behavior in Han Chinese. Am J Clin Nutr. 2013;97(3):545–551. doi: 10.3945/ajcn.112.046482. [DOI] [PubMed] [Google Scholar]
- 44.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Computing RFfS, editor. Team RC. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2008. [Google Scholar]
- 46.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones AW. Pharmacokinetics of Ethanol - Issues of Forensic Importance. Forensic Sci Rev. 2011;23(2):91–136. [PubMed] [Google Scholar]
- 48.Tahari AK, Chien D, Azadi JR, Wahl RL. Optimum lean body formulation for correction of standardized uptake value in PET imaging. J Nucl Med. 2014;55(9):1481–1484. doi: 10.2967/jnumed.113.136986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 50.Quillen EE, Chen XD, Almasy L, Yang F, He H, Li X, et al. ALDH2 is associated to alcohol dependence and is the major genetic determinant of “daily maximum drinks” in a GWAS study of an isolated rural Chinese sample. Am J Med Genet B Neuropsychiatr Genet. 2014;165B(2):103–110. doi: 10.1002/ajmg.b.32213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bierut LJ, Goate AM, Breslau N, Johnson EO, Bertelsen S, Fox L, et al. ADH1B is associated with alcohol dependence and alcohol consumption in populations of European and African ancestry. Mol Psychiatry. 2012;17(4):445–450. doi: 10.1038/mp.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li H, Mukherjee N, Soundararajan U, Tarnok Z, Barta C, Khaliq S, et al. Geographically separate increases in the frequency of the derived ADH1B*47His allele in eastern and western Asia. Am J Hum Genet. 2007;81(4):842–846. doi: 10.1086/521201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Osier MV, Pakstis AJ, Soodyall H, Comas D, Goldman D, Odunsi A, et al. A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet. 2002;71(1):84–99. doi: 10.1086/341290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuchihashi-Makaya M, Serizawa M, Yanai K, Katsuya T, Takeuchi F, Fujioka A, et al. Gene-environmental interaction regarding alcohol-metabolizing enzymes in the Japanese general population. Hypertens Res. 2009;32(3):207–213. doi: 10.1038/hr.2009.3. [DOI] [PubMed] [Google Scholar]
- 55.Yokoyama A, Tsutsumi E, Imazeki H, Suwa Y, Nakamura C, Yokoyama T. Contribution of the alcohol dehydrogenase-1B genotype and oral microorganisms to high salivary acetaldehyde concentrations in Japanese alcoholic men. Int J Cancer. 2007;121(5):1047–1054. doi: 10.1002/ijc.22792. [DOI] [PubMed] [Google Scholar]
- 56.Yokoyama A, Kamada Y, Imazeki H, Hayashi E, Murata S, Kinoshita K, et al. Effects of ADH1B and ALDH2 Genetic Polymorphisms on Alcohol Elimination Rates and Salivary Acetaldehyde Levels in Intoxicated Japanese Alcoholic Men. Alcohol Clin Exp Res. 2016;40(6):1241–1250. doi: 10.1111/acer.13073. [DOI] [PubMed] [Google Scholar]
- 57.Park BL, Kim JW, Cheong HS, Kim LH, Lee BC, Seo CH, et al. Extended genetic effects of ADH cluster genes on the risk of alcohol dependence: from GWAS to replication. Hum Genet. 2013;132(6):657–668. doi: 10.1007/s00439-013-1281-8. [DOI] [PubMed] [Google Scholar]
- 58.Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, et al. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2009;18(3):580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng GS, Chen YC, Wang MF, Lai CL, Yin SJ. ALDH2*2 but not ADH1B*2 is a causative variant gene allele for Asian alcohol flushing after a low-dose challenge: correlation of the pharmacokinetic and pharmacodynamic findings. Pharmacogenet Genomics. 2014;24(12):607–617. doi: 10.1097/FPC.0000000000000096. [DOI] [PubMed] [Google Scholar]
- 60.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant J Clin Invest. 1989;83(1):314–316. doi: 10.1172/JCI113875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, et al. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48(4):677–681. [PMC free article] [PubMed] [Google Scholar]
- 62.Chen CC, Lu RB, Chen YC, Wang MF, Chang YC, Li TK, et al. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet. 1999;65(3):795–807. doi: 10.1086/302540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Higuchi S, Matsushita S, Murayama M, Takagi S, Hayashida M. Alcohol and aldehyde dehydrogenase polymorphisms and the risk for alcoholism. Am J Psychiatry. 1995;152(8):1219–1221. doi: 10.1176/ajp.152.8.1219. [DOI] [PubMed] [Google Scholar]
- 64.Li D, Zhao H, Gelernter J. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biol Psychiatry. 2011;70(6):504–512. doi: 10.1016/j.biopsych.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jorgenson E, Witte JS. Coverage and power in genomewide association studies. Am J Hum Genet. 2006;78(5):884–888. doi: 10.1086/503751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jorgenson E, Witte JS. A gene-centric approach to genome-wide association studies. Nat Rev Genet. 2006;7(11):885–891. doi: 10.1038/nrg1962. [DOI] [PubMed] [Google Scholar]
- 67.Lindquist KJ, Jorgenson E, Hoffmann TJ, Witte JS. The impact of improved microarray coverage and larger sample sizes on future genome-wide association studies. Genet Epidemiol. 2013;37(4):383–392. doi: 10.1002/gepi.21724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


