Abstract
The T-box transcription factor Eomes (also known as Tbr2) shows short-lived expression in various localized domains of the embryo, including epiblast cells during gastrulation and intermediate progenitor cells in the cerebral cortex. In these tissues Eomes fulfills crucial roles for lineage specification of progenitors. To directly observe Eomes-dependent cell lineages in the living embryo, we generated a novel dual-fluorescence reporter allele that expresses a membrane-bound tdTomato protein for investigation of cell morphology and a nuclear GFP for cell tracing. This allele recapitulates endogenous EOMES protein expression and is suitable for live imaging. We found that the allele can also be used as a short-to-medium-term lineage tracer, as GFP persists in cells longer than EOMES protein and marks Eomes-dependent lineages with a timeframe of days to weeks depending on the proliferation rate. In summary, we present a novel genetic tool for investigation of Eomes-dependent cell types by live imaging and lineage tracing.
Keywords: Mouse, Eomesodermin, Tbr2, membraneTomato, H2B:GFP
A key feature of the T-box transcription factor Eomesodermin (Eomes) is its tightly controlled, short-lived expression in confined domains during embryonic development and in few cell types during adulthood. Numerous gene-deletion studies have revealed fundamental roles of Eomes for cell lineage specification during embryogenesis (Arnold et al., 2008a; Costello et al., 2011; Nowotschin et al., 2013; Teo et al., 2011). First Eomes expression can be found during early postimplantation stages of mammalian embryogenesis in two extraembryonic tissues, namely extraembryonic ectoderm and the visceral endoderm (VE) (Nowotschin et al., 2013; Russ et al., 2000). Proper development of both tissues depends on Eomes (Nowotschin et al., 2013; Russ et al., 2000). Accordingly, the constitutive disruption of Eomes gene-function leads to early developmental arrest shortly after implantation due to failing trophectoderm development (Russ et al., 2000; Strumpf et al., 2005). Shortly later during gastrulation, the specification of two distinct cell lineages, anterior mesoderm and definitive endoderm, strictly depends on Eomes (Arnold 2008, Costello 2011). Eomes loss-of-function in the primitive streak (PS) additionally results in morphogenetic defects due to incomplete epithelial-to-mesenchymal transition (EMT) during mesoderm formation ((Arnold et al., 2008a) and reviewed by (Probst and Arnold, 2017)).
In the central nervous system (CNS) Eomes (here also referred to as Tbr2) is transiently expressed in several neurogenic progenitor populations during embryonic development and also during postnatal neurogenesis (Hodge 2012, reviewed in Mihalas and Hevner 2017, Hevner 2006, Englund 2005). Here, Eomes/Tbr2 functions were most intensively studied in the context of generating cortical excitatory pyramidal projection neurons from subventricularly located intermediate progenitor cells (IPCs) of the cerebral neocortex (Arnold et al., 2008b; Mihalas et al., 2016; Sessa et al., 2008) and within the dentate gyrus (Arnold et al., 2008b; Hodge et al., 2013; 2012). Additional sites of CNS expression include mitral cells of the olfactory bulbs, ganglion cells of the retina and unipolar brush cells of the internal granular cell layer of the cerebellum (reviewed by (Mihalas and Hevner, 2017)).
In immune cells Eomes expression is prominent in lymphocytes of the innate and adaptive immune system, including subsets of CD8+ memory T cells and conventional natural killer (cNK) cells (Gordon et al., 2012; Intlekofer et al., 2005; Pearce et al., 2003). The importance of Eomes in these cell types is best exemplified by the fact that the selective deletion of Eomes results in a reduction of memory T cells (Banerjee et al., 2010) and near complete loss of cNK cells (Klose et al., 2014).
The requirement for precise expression control of Eomes as a cell-fate defining factor is prominent during gastrulation. Two separate cell lineages, anterior mesoderm and definitive endoderm (DE), are generated from Eomes+ progenitors under the control of dynamic levels of SMAD2/3-mediated Nodal-signaling. It was proposed that these different cell types acquire their fate according to the location and the time of lineage commitment during gastrulation, reflecting a Nodal-signaling gradient within the embryo (reviewed in (Arnold and Robertson, 2009)). Similarly, during corticogenesis the generation of different neuronal subtypes follows a spatio-temporal pattern (Elsen et al. 2013).
To investigate the timing and spatial organization of specification of different cell lineages live imaging of embryos has become a powerful tool, as demonstrated for the formation of DE or analysis of EMT during gastrulation (Kwon et al., 2008; Ramkumar et al., 2016). To enable the direct observation and to follow Eomes-dependent cell lineages, we developed a novel dual-fluorescence reporter allele harboring a membrane-bound tdTomato and a nuclear H2B:GFP reporter in the Eomes gene locus. Previously established reporter lines, such as one expressing cytoplasmic GFP from the Eomes locus (EomesGFP)(Arnold et al., 2009) are less suitable for live imaging, since they do not allow for discriminating and following single cells over time.
For the generation of the novel Eomes knock-in reporter allele we followed a previously applied strategy (Arnold et al., 2009; Costello et al., 2011; Pimeisl et al., 2013). Briefly, the targeting construct including a membrane-bound tdTomato (mT) and a nuclear H2B:GFP (nG) linked by a T2A cotranslational “cleavage” peptide (Fig. 1a) was inserted into the start-codon of Eomes exon 1 by homologous recombination in embryonic stem (ES) cells, thereby deleting ~500 bp of coding exon 1 (Fig. 1b). ES cell clones carrying the EomesmTnG allele were identified by Southern blot (Fig. 1c) and used for chimera generation. Following germline transmission, the neomycin positive selection cassette was removed by crossing mice carrying the EomesmTnG allele to females carrying the Sox2::Cre transgene (Vincent and Robertson, 2003) that acts as general deleter in the female germ line. Resulting EomesmTnG reporter mice were kept at heterozygous state. The EomesmTnG allele acts as a null allele and thus no homozygous mice could be generated from heterozygous intercrosses as described previously (Arnold et al., 2009).
Figure 1. Generation of the EomesmTomatoH2B:GFP (EomesmTnG) allele.
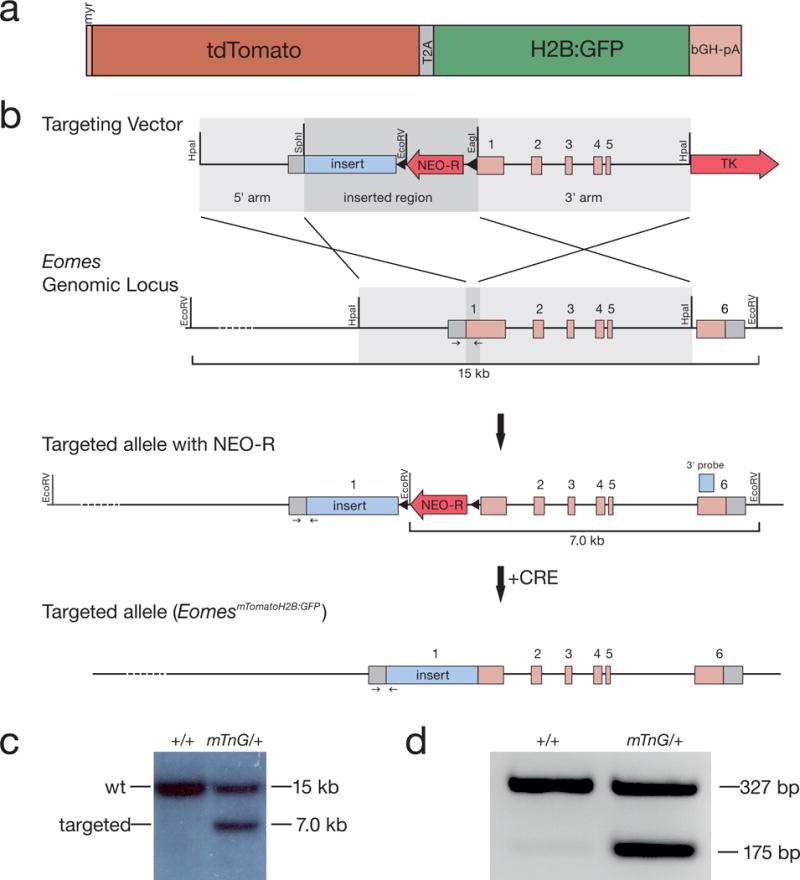
a) Illustration of the reporter sequences that were inserted into the start-codon of the Eomes gene locus shown in detail. The reporter construct consists of a membrane-targeted (myr) tandem dimer Tomato fluorescent protein and a nuclear-localized GFP (H2B:GFP) linked by the T2A “cleavable” peptide followed by the bovine growth hormone polyA signal (bGH-pA). b) Schematic of the targeting strategy to introduce the reporter cassette (insert) into the ATG of the Eomes locus by homologous recombination in ES cells. The Eomes genomic locus comprises 6 exons depicted by boxes (grey: untranslated regions (UTRs), pink: coding regions). The targeting vector includes a removable LoxP-flanked neomycin selection cassette (NEO-R, red arrow) for positive selection and the thymidine kinase negative selection cassette (TK, red arrow). After homologous recombination the insert and the NEO-R cassette are introduced into the Eomes genomic locus. The NEO-R cassette is removed by Cre recombination to generate the targeted allele. Small arrows indicate PCR primers used for genotyping. c) Southern blot analysis of targeted ES cell clones using an EcoRV restriction digest as illustrated in b) showing a wt (+/+) and a correctly targeted (mTnG/+) clone. The wt band is detected at 15 kb and the targeted band is detected at 7 kb on EcoRV digested genomic DNA using the indicated Southern blot probe (light blue box in b). d) Genotyping PCR of a wt (+/+) and a heterozygous (mTnG/+) mouse showing wt band at 327 kb and EomesmTnG band at 175 bp. Genotyping primers are indicated by small arrows in b).
Analysis of reporter expression at early gastrulation stage (E6.25) shows mTnG fluorescence in proximally located cells of the posterior half of the epiblast marking the future PS that can be observed already previous to cells leaving the streak region (Fig. 2a). More distally the epiblast is devoid of mTnG signal at this stage (Fig. 2b). Also cells of the outer VE layer show reporter expression (Fig. 2a). One day later at E7.25 the posterior half of the epiblast as well as newly generated mesoderm and DE cells show mTnG signal (Fig. 2c and d). Distally the mesoderm layer has not migrated as far to anterior as proximally and the mTnG positive domain in the epiblast is more restricted to the posterior (Fig. 2d). The maximum intensity projection of a whole mount EomesmTnG embryo at E7.5 shows strongly mTnG positive cells in the outer DE layer (Fig. 2e). These domains of expression at E6.25 and E7.5 fully correlate with those described for the cytoplasmic EomesGFP allele (Arnold et al., 2009). Time-lapse imaging of an E7.25 EomesmTnG embryo in culture shows anterior migration of mesendodermal cells. These can be observed using the membrane label to discriminate different cell morphologies and the nuclear signal for precisely following cells over time including the observation of cell divisions (Suppl. movie 1). These types of cell analyses could previously not be performed using the cytoplasmic EomesGFP allele since here single cells are difficult to distinguish (Suppl. movie 2).
Figure 2. Expression of the EomesmTnG reporter allele at gastrulation stages.
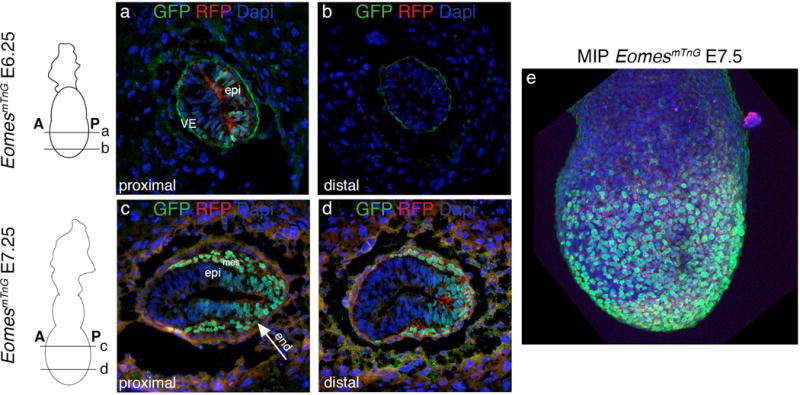
Schematics of embryos next to the images indicate the section planes for E6.25 and E 7.25 EomesmTnG embryos and anterior (A) and posterior (P) orientation of embryos. a and b) Shortly before gastrulation E6.25 embryos show nuclear H2B:GFP and membrane-localized tdTomato fluorescence in cells of half of the epiblast (epi) in the proximal domain (a) of the embryo but not in the distal domain (b). Additionally, reporter expression is found in some visceral endoderm cells (VE). c and d) At E7.25 the posterior half of the epiblast (epi) is positive for GFP and tdTomato in addition to cells of the newly generated mesoderm layer (mes) and of the endoderm layer (arrow end). c) In proximal regions mesoderm cells have migrated further anteriorly than in distal portions of the embryo (d). e) The maximum intensity projection (MIP) of an E7.5 EomesmTnG embryo shows GFP and tdTomato positive cells covering the entire surface of the embryonic region. All embryos are oriented with anterior to the left and posterior to the right.
The comparison of endogenous EOMES protein and nG reporter expression shows gross overlap at E6.5 and E7.5 in epiblast cells (Fig. 3a–g). Some differences in staining signals are observed in the extraembryonic VE such as few EOMES-positive cells that fail to show nG reporter signal (Fig. 3b, arrowhead) and faint GFP signal in anterior epiblast cells that is not always visible by IF staining (Fig. 3c,d). At E7.5 the staining-patterns of EOMES and the reporter signal entirely overlap, however differences in intensities of EOMES and nG signal are observed (Fig. 3e–g). EOMES IF shows highest intensities in the posterior epiblast and in cells that have just left the PS, whereas nuclear GFP staining is brightest in the anterior mesoderm and the DE (arrowheads Fig. 3f and g). These differences might indicate dynamic Eomes-expression and differences in reporter or EOMES protein turnover rates. The previous cytoplasmic EomesGFP allele similarly showed a discrepancy between Eomes mRNA and GFP fluorescence, which persists longer than mRNA (Arnold et al., 2009). Immunodetection of endogenous EOMES protein (Fig. 3e–g) shows that EOMES also persists considerably longer than Eomes mRNA, suggesting that both Eomes transcription and mRNA are tightly controlled. At E8.5 no more endogenous EOMES protein can be detected (Fig. 3h). In contrast nG signal is still found in the heart, gut tube, and notochord, all three tissues that derive from Eomes+ cells (Fig. 3h, arrowheads). Further reduced nG signals can be detected until E9.5 in the notochord and even weaker in the endoderm and heart (Fig. 3i and j, arrowheads and data not shown). The nG reporter thus persists longer than EOMES, which enables using this allele for short-term lineage tracing of Eomes-expressing cells, even in these highly proliferative tissues. In summary, at early embryonic stages the EomesmTnG reporter faithfully recapitulates the presence of endogenous EOMES protein. Reporter expression is found in most EOMES positive cells and is not detected at ectopic sites.
Figure 3. Comparison of endogenous EOMES protein with GFP indicates that the reporter serves as a short-term lineage tracer during gastrulation.
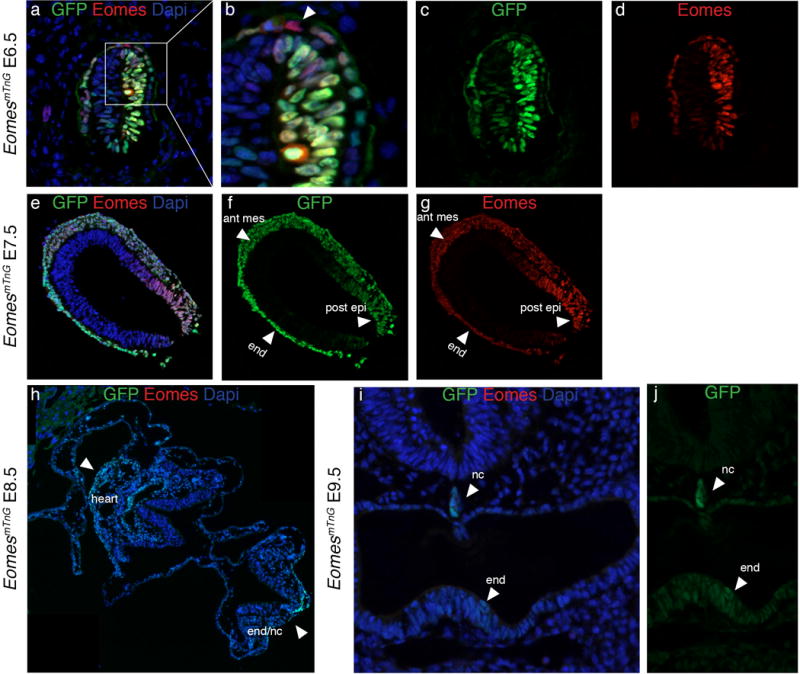
At E6.5 the expression of GFP (green) and EOMES (red) in the epiblast overlaps (a, c, and d), while some cells in the VE are positive for EOMES and not for GFP (arrowhead in b). At E7.5 GFP and EOMES proteins also show a gross overlap (e–g). Of note, the intensities of the stainings are different for GFP and EOMES. GFP is more strongly detected in the anterior mesoderm (ant mes) and endoderm (end) and EOMES protein shows strongest staining intensities in the posterior epiblast (post epi) (arrowheads in f and g). h) At E8.5 no EOMES protein can be detected by antibody staining while GFP protein is still present in the heart, the notochord, and the endoderm (end/nc) (arrowheads). i and j) Weak GFP expression remains detectable in some endoderm cells (end) and the notochord (nc) until E9.5, while EOMES protein is absent at this stage. All embryos are oriented with anterior to the left and posterior to the right, except for i and j where sections are oriented with dorsal to the top and ventral to the bottom.
To determine if the EomesmTnG allele also accurately reports expression in the CNS, we studied brain cryostat sections and retina flat mount preparations from embryonic and postnatal EomesmTnG/+ mice. In embryonic forebrain, the cerebral cortex (Ctx) was specifically labeled (Fig. 4a–c), as expected (Englund et al., 2005). Nuclear GFP was detected in scattered cells of the cortical ventricular zone (VZ) and subventricular zone (SVZ), consistent with intermediate progenitors (Fig. 4d–f) and in many neurons in the intermediate zone (IZ), cortical plate (CP), and marginal zone (MZ), consistent with lineage-traced cortical glutamatergic neurons that have already lost nuclear EOMES protein (Fig. 4b–f) (Kowalczyk et al., 2009). mT fluorescence was most prominent in cortical axons in the IZ (Fig. 4c). IF-staining for EOMES shows a few cells in the VZ that are EOMES positive but do not yet show the nG signal, most likely demonstrating slower protein maturation kinetics of H2B:GFP in comparison to EOMES. In contrast, EOMES is lost more quickly while nG signals fate-label cells for longer periods of time (Fig. 4d–f). While a delay in GFP maturation was also observed in the previous EomesGFP allele, the cytoplasmic GFP did not allow for labeling of newborn neurons in the cerebral cortex for longer time periods (Arnold et al., 2009). In contrast, nuclear H2B:GFP appears to persist longer, thus allowing for lineage labeling of newborn cortical neurons. In the postnatal (P8) olfactory bulb, green fluorescence was prominent in mitral/tufted neurons of the mitral cell layer (MCL) and in periglomerular neurons (Brill et al., 2009; Kahoud et al., 2014). Red fluorescence labeled olfactory glomeruli in the glomerular layer, containing mitral/tufted/periglomerular dendrites, and the internal plexiform layer (IPL), rich in mitral/tufted axons (Fig. 4g). Also in P8 mice, the posterior periventricle (pPV), previously identified as neurogenic source after brain injury (Nakatomi et al., 2002), was rich in nG+ cells (Fig. 4h,i), confirming previous observations that the pPV is rich in Eomes-expressing progenitors (Kowalczyk et al., 2009). The dentate gyrus (Fig. 4j) displayed nG and mT fluorescence in the subgranular zone (SGZ), consistent with IPCs (Hodge et al., 2008) and in immature neurons in the granule cell layer (GCL), presumably labeled by lineage tracing from intermediate progenitors. In the retina (Fig. k,l) the ganglion cell layer (GaCL) contained scattered nG+ nuclei (plus rare nG+ nuclei in the inner nuclear layer, presumably displaced ganglion cells), while mT fluorescence was enriched in the plexiform layers (IPL and OPL) and optic nerve (Fig. 4l, arrow). These observations in the retina matched descriptions of Eomes expression in non-image-forming visual pathways (Mao et al., 2008; Sweeney et al., 2017; 2014). Together, these observations support faithful mTnG reporter expression in Eomes+ cells, and indicate that the allele can serve as lineage tracers for periods of days to weeks in postmitotic neuronal cell types derived from EOMES+ progenitors.
Figure 4. Expression of the EomesmTnG allele in developing brain and retina.
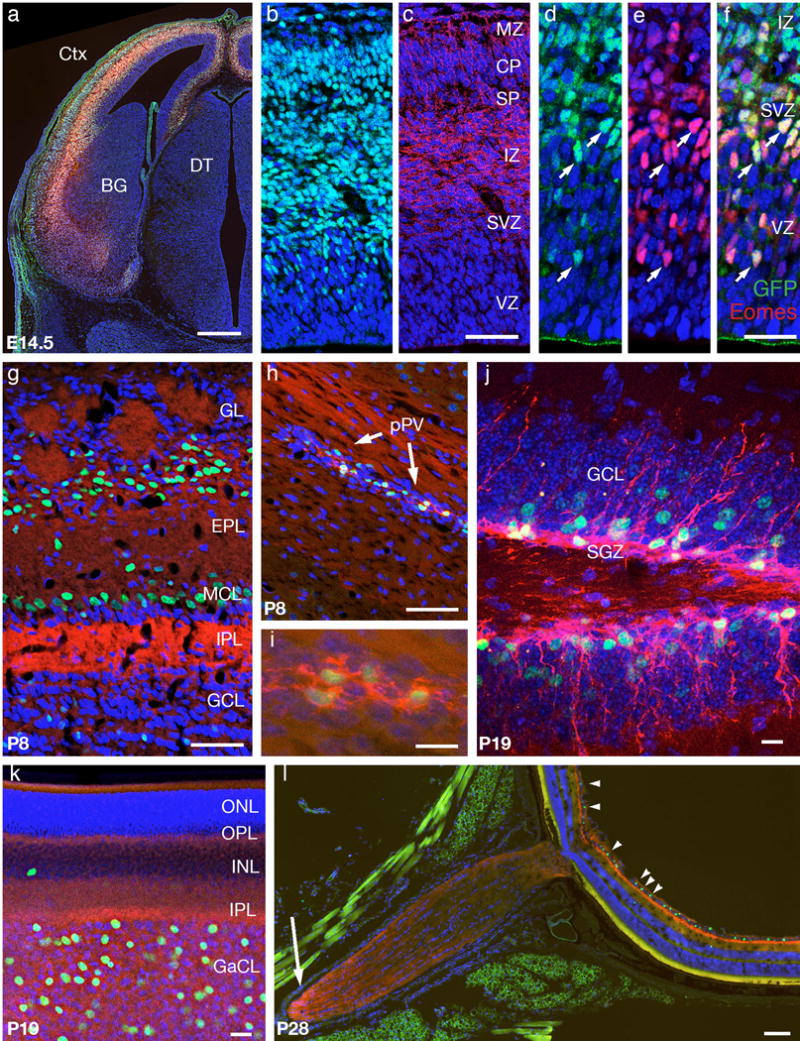
a) E14.5 forebrain, coronal section at level of thalamus. The cortex (Ctx) shows strong red and green fluorescence, while subcortical structures such as basal ganglia (BG) and dorsal thalamus (DT) do not. b and c) E14.5 cerebral neocortex comparing endogenous nG (b) and mT fluorescence (c). d-f) Immunofluorescence of E14.5 cerebral neocortex, showing overlap (arrows) of GFP (green) and EOMES (red) in the ventricular zone (VZ) and subventricular zone (SVZ) whereas in the intermediate zone in some cells only GFP is detected and not EOMES. g) P8 olfactory bulb. Many GFP+ cells were present in the mitral cell layer (MCL) and glomerular layer (GL). h) P8 forebrain with posterior periventricle (pPV) containing abundant green and red fluorescent progenitor cells. i) Higher magnification of the pPV shows small multipolar cells with green nuclei and red membranes. j) P19 dentate gyrus, with abundant red and green fluorescent cells in the subgranular zone (SGZ). Newly generated GFP+ neurons exhibited prominent red fluorescent apical dendrites that extended into the granule cell layer (GCL). k) P19 retina, flat mount, with ganglion cell layer (GaCL) viewed tangentially. A minority of DAPI+ nuclei in the GaCL express GFP; rare GFP+ nuclei are also detected in the inner nuclear layer (INL). l) P28 eye. Many GFP+ cells are seen in the retinal ganglion cell layer (GaCL; arrowheads), while red fluorescent processes are seen in segments of the optic nerve (arrow). Green fluorescence outside the retina (panel l) is non-nuclear artifact. All panels show endogenous red and green fluorescence, except (c) as indicated. All panels show cryostat sections (12 μm) except (h) as indicated. Blue fluorescence (a-l) shows DAPI nuclear counterstain. Other abbreviations: marginal zone (MZ), cortical plate (CP), subplate (SP), intermediate zone (IZ), external plexiform layer (EPL), internal plexiform layer (IPL), outer nuclear layer (ONL), outer plexiform layer (OPL), inner nuclear layer (INL), inner plexiform layer (IPL). Scale bars: a, 500 μm; b-e, 50 μm; f-g 10 μm; h, 20 μm; i, 100 μm.
Next, we wanted to test whether the dual reporter allele also marks Eomes-expressing cells of the innate immune system. cNK cells are the only innate lymphoid cells (ILCs) that express Eomes and are primarily located in the spleen and liver. EOMES+ cNK cells also express CD49b, which can be used as surrogate marker for EOMES in cNK cells (Fig. 5). In the spleen the large majority of NK1.1+ NKp46+ ILCs are EOMES+ CD49b+ cNK cells, which is faithfully reflected by the specific fluorescence expression of the dual EomesmTnG reporter in these ILCs when assessed by single cell multicolor flow cytometry (Fig. 5, left contour blot, electronically gated on NK1.1+ NKp46+ ILCs after exclusion of T cells). The liver harbors an additional NK marker positive (NK1.1+ NKp46+) ILC population that is negative for EOMES and CD49b. The dual reporter allele specifically labels only the EOMES+ CD49b+ cNK cells in the liver, whereas Eomes− CD49b− ILCs show virtually no expression of the EomesmTnG reporter (Fig. 5). Thus, the dual EomesmTnG reporter protein expression specifically marks and distinguishes different NK1.1+ NKp46+ ILC lineages at steady state in spleen and liver ILCs.
Figure 5. Faithful labeling of conventional EOMES+ NK cells by the EomesmTnG reporter allele.
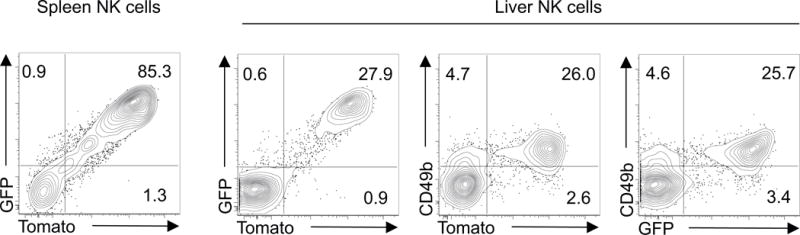
After electronic exclusion of other cells NK1.1+ NKp46+ ILC populations from spleen and liver were analyzed by flow cytometry for reporter expression. The majority of splenic NK1.1+ NKp46+ ILCs (left contour blot) are conventional EOMES+ NK cells. In the liver NK cell populations were further analyzed for the co-expression of the reporter allele and CD49b. CD49b is a surrogate marker for the expression of Eomes and distinguishes between conventional EOMES+ CD49b+ NK cells and EOMES− CD49b− NK cells. Contour blots are representative for 2 analyzed mice.
In summary we describe a novel Eomes fluorescent reporter allele, which is suited for live imaging and identification of Eomes-dependent lineages. The membrane-localized mT fluorescent protein is favorable for observing cell morphologies and clearly demarcates projections within neural tissues. The nuclear nG allows for tracking of Eomes+ cells during live imaging. In addition, this allele can serve as a short- to medium-term lineage tracer. Highly proliferative Eomes-dependent lineages can be followed for two days after endogenous EOMES protein is lost during gastrulation stages. In the brain Eomes-dependent postmitotic neurons can be traced for periods of days or weeks. This time difference is most likely due to dilution of the nG signal during cell divisions in the early embryo, while neurons are non-dividing. In conclusion, we present a novel EomesmTnG reporter allele, which is a valuable tool for studies in the live embryo of cell fate specification downstream of Eomes.
Materials and Methods
Generation of the EomesmTomatoH2BGP (EomesmTnG) allele
The targeting vector for the generation of an EomesmTnG-targeted allele comprises an 8.25 kb HpaI-HpaI fragment of the Eomes locus. A membrane_tdTomato-T2A-H2B:GFP.pA cassette and a LoxP-flanked neomycin resistance cassette were integrated between the SphI site at the translational start site and an EagI site thereby deleting ~500 bp of the coding region of exon 1. The 3′ homology region of the targeting construct was flanked with a pMCI.TK negative selection cassette. Linearized targeting vector was electroporated into CCE ES cells, and neomycin and FIAU resistant ES cell colonies were screened by Southern Blot using a 3′ external probe (wt allele: 15 kb and targeted allele: 7 kb) and a 5′ internal probe (wt allele: 15 kb and targeted allele: 11 kb) on EcoRV digested DNA. Unique insertion of the targeting vector was controlled by Southern Blot using a probe for the neomycin resistance cassette. Generation of chimeras was performed by laser-assisted injections at morula stage. F1 progeny and subsequent generations were genotyped by PCR at 61°C annealing temperature to detect wild type allele (327 bp) and the knock-in allele (175 bp) using the following primers: forward primer 5′- GAGGGAGGAAGGGGACATTA-3′; wt reverse primer 5′- AGACTGCCCGGAAACTTCTT-3′; ki reverse primer 5′- CTTTGATCACTTCCTCGCCCTTG-3′. The neomycin positive selection cassette was removed by crossing to females carrying the Sox2::Cre transgene (Vincent and Robertson, 2003). The strain was maintained on the NMRI background. To generate embryos heterozygous males or females were crossed to a wild type and embryos were dissected at the required embryonic stage. Animals carrying the EomesmTnG allele are available to the research community and can be requested from the corresponding author.
Immunofluorescence stainings in embryos
For whole mount immunofluorescent stainings (WMIF) embryos were fixed for 1 hr at 4°C with 4% PFA and washed 2x 10 min with PBS containing 0.1% Tween (PBT). Embryos were permeabilized with PBT containing 0,3% Triton-X100 at RT. Blocking was performed for 2 hrs at RT in PBT containing 1% BSA. The primary antibody was added in blocking solution over night at 4°C. Embryos were then washed 4 times in PBT and secondary antibodies were added for 3 hrs at RT in blocking solution. Nuclei were then stained with DAPI for 30 min in PBT and washed with PBT. Whole embryos were imaged with a Zeiss inverted laser-scanning microscope (LSM 510 Meta).
For IF on sectioned embryos they were processed for cryostat sectioning and sectioned at 7 μm. In brief, embryos were fixed in opened deciduae with 4% PFA for 1 hr at 4°C, processed through 15% and 30% sucrose/PBS and incubated for 1 hr in embedding medium (15% sucrose/7.5% gelatin in PBS) prior to embedding. Sections were briefly washed with PBS and then permeabilized for 20 min in PBT containing 0.2% Triton. Sections were blocked for 1 hr in blocking solution and then incubated with the primary antibodies over night in blocking solution at 4°C. After washing with PBS the slides were incubated with secondary antibodies in blocking solution for 1 hour at RT, then stained with DAPI in PBT for 5 min and mounted with ProLong Diamond Antifade Mountant (Life technologies P36970) and imaged on an inverted Zeiss Axio Observer Z1 microscope.
Primary antibodies used: GFP (1:1000, Abcam ab13970), RFP (1:500, Rockland 600-401-379), EOMES (1:300, Abcam ab23345). Secondary Alexa-Fluor-conjugated antibodies (Life Technologies) were used at a dilution of 1:1000. DNA was counterstained with DAPI.
Reporter detection and immunofluorescence in neural tissues
Insemination was assessed by the presence of a vaginal plug, with noon of that day designated embryonic day (E) 0.5. For experiments using embryos, the dam was anesthetized with Isoflurane (Patterson Veterinary) and killed by cervical dislocation. Embryos were removed, placed on ice, and decapitated; the heads were immersed in cold fixative (4% paraformaldehyde, 4% sucrose, 0.1 M sodium phosphate, pH 7.4). For experiments using postnatal (P) mice, the pups were anesthetized by hypothermia (P0.5 to P7), or isoflurane (P7.5 or older), and perfused transcardially with cold 4% paraformaldehyde, 0.1 M sodium phosphate, pH 7.4. The brains were dissected out and immersed in cold fixative for 4 hours or overnight at 4°C. Brains were then cryoprotected in 10% sucrose solution for 1 hour, 20% sucrose solution for 1 hour, then 30% sucrose for 1 hour or until the brain completely sinks (times may be longer for larger brains). Finally, brains were placed into a 50/50 mixture of 30% sucrose and O.C.T. embedding medium (Sakura Finetek, JPN) for at least 15 minutes. Brains were embedded in O.C.T. and frozen on powdered dry ice. Brains were stored at −80° C and removed when ready for cryosectioning.
Immunofluorescence of neural tissues
Brains were sectioned (12 μm) on a Leica CM1850 cryostat. Slide-mounted sections were counterstained with DAPI for direct visualization of reporter fluorescence, or treated for IF with antigen retrieval by boiling in 0.01 M sodium citrate (pH 6.0) for 1 to 5 minutes. After cooling on ice and rinsing in PBS, sections were incubated in blocking solution (10% normal goat serum, 3% bovine serum albumin, and 0.1% Triton X-100 in PBS) for 30 minutes. The sections were then incubated overnight at 4°C with primary antibodies diluted in blocking solution, PBS-T. Primary antibodies were: anti-Tbr2 (1:2000, rabbit polyclonal, R.F. Hevner, Seattle, WA), anti-Tbr2 (1:250, rat monoclonal, eBioscience 14-4875, CA), anti-RFP (1:1000, rabbit polyclonal, Clontech 632496, CA). Slides were rinsed and incubated for 2 hours at room temperature with secondary antibodies diluted in PBS-T. The Alexa Fluorescent secondary IgG antibodies conjugated to various dyes were used (Molecular Probes, OR). Slides were rinsed in PBS, counterstained with DAPI (Sigma, St. Louis, MO) to label DNA with blue fluorescence, and cover slipped in Fluoromount-G (SouthernBiotech, Birmingham, AL). After curing overnight, fluorescent images were acquired using a Carl Zeiss Axio Imager Z1 microscope equipped with a motorized stage and MosaiX software (Zeiss, USA). Confocal images were obtained using a Zeiss LSM710. Digital images were processed using Photoshop (Adobe, San Jose, CA, USA) to optimize brightness, contrast, and resolution.
Flow cytometry
Single cell suspensions of spleen and liver were obtained by filtering the tissues through a 70 μm cell strainer (BD Biosciences). For single cell suspension of liver cells 60% Percoll (Sigma-Aldrich) was overlaid with liver cells resuspended in 40% Percoll and lymphocytes were isolated at the interphase after gradient centrifugation. Erythrocytes were removed by incubation in red cell lysis buffer. After purification, single cell suspensions were incubated with FcR (CD16/CD32) blocking antibodies and stained with fluorescent-label coupled antibodies (Biolegend) in PBS (Ca2+ and Mg2+-free) supplemented with 2mM EDTA and 2% FCS. Cells were acquired on BD LSRFortessa multicolor flow cytometer (BD Bioscience). Single NK1.1+ NKp46+ ILCs were analyzed for marker expression and EomesmTnG reporter after electronic exclusion of irrelevant cell types using FlowJo software (Treestar).
Supplementary Material
Acknowledgments
This work was supported by the German Research Foundation (DFG) by the Emmy Noether Programme (AR732/1-1), project A3 of the collaborative research centre 850 (SFB 850), and BIOSS Centre of Biological Signalling Studies to SJA and (TA 436/3-1, SPP 1937 and SFB 1160, TP6) to YT; and by NIH grants R01NS092339 and R01NS085081 to RFH. SP was partially supported by a fellowship of the Novartis Foundation. We thank Mihael Pavlovic and Thilo Bass for animal husbandry, Roland Nitschke of the Life Imaging Centre (LIC) in the Center for Biological Systems Analysis (ZBSA) of the Albert-Ludwigs-University Freiburg for assistance with the confocal microscopy resources, and Carsten Schwan for time-lapse recording of gastrulation stage mouse embryos.
References
- Arnold SJ, Robertson EJ. Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat Rev Mol Cell Biol. 2009;10:91–103. doi: 10.1038/nrm2618. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Hofmann UK, Bikoff EK, Robertson EJ. Pivotal roles for eomesodermin during axis formation, epithelium-to-mesenchyme transition and endoderm specification in the mouse. Development. 2008a;135:501–511. doi: 10.1242/dev.014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AFP, Era T, Nishikawa SI, Bikoff EK, Molnár Z, Robertson EJ, Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes & Development. 2008b;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Sugnaseelan J, Groszer M, Srinivas S, Robertson EJ. Generation and analysis of a mouse line harboring GFP in the Eomes/Tbr2 locus. Genesis. 2009;47:775–781. doi: 10.1002/dvg.20562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Gordon SM, Intlekofer AM, Paley MA, Mooney EC, Lindsten T, Wherry EJ, Reiner SL. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J Immunol. 2010;185:4988–4992. doi: 10.4049/jimmunol.1002042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill MS, Ninkovic J, Winpenny E, Hodge RD, Ozen I, Yang R, Lepier A, Gascón S, Erdelyi F, Szabo G, et al. Adult generation of glutamatergic olfactory bulb interneurons. Nat Neurosci. 2009;12:1524–1533. doi: 10.1038/nn.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello I, Pimeisl IM, Dräger S, Bikoff EK, Robertson EJ, Arnold SJ. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nature Cell Biology. 2011;13:1084–1091. doi: 10.1038/ncb2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RAM, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SM, Chaix J, Rupp LJ, Wu J, Madera S, Sun JC, Lindsten T, Reiner SL. The transcription factors T-bet and Eomes control key checkpoints of natural killer cell maturation. Immunity. 2012;36:55–67. doi: 10.1016/j.immuni.2011.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Garcia AJ, Elsen GE, Nelson BR, Mussar KE, Reiner SL, Ramirez JM, Hevner RF. Tbr2 expression in Cajal-Retzius cells and intermediate neuronal progenitors is required for morphogenesis of the dentate gyrus. J Neurosci. 2013;33:4165–4180. doi: 10.1523/JNEUROSCI.4185-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Kowalczyk TD, Wolf SA, Encinas JM, Rippey C, Enikolopov G, Kempermann G, Hevner RF. Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J Neurosci. 2008;28:3707–3717. doi: 10.1523/JNEUROSCI.4280-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge RD, Nelson BR, Kahoud RJ, Yang R, Mussar KE, Reiner SL, Hevner RF. Tbr2 is essential for hippocampal lineage progression from neural stem cells to intermediate progenitors and neurons. J Neurosci. 2012;32:6275–6287. doi: 10.1523/JNEUROSCI.0532-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Kahoud RJ, Elsen GE, Hevner RF, Hodge RD. Conditional ablation of Tbr2 results in abnormal development of the olfactory bulbs and subventricular zone-rostral migratory stream. Dev Dyn. 2014;243:440–450. doi: 10.1002/dvdy.24090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CSN, Flach M, Möhle L, Rogell L, Hoyler T, Ebert K, Fabiunke C, Pfeifer D, Sexl V, Fonseca-Pereira D, et al. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 2014;157:340–356. doi: 10.1016/j.cell.2014.03.030. [DOI] [PubMed] [Google Scholar]
- Kowalczyk T, Pontious A, Englund C, Daza RAM, Bedogni F, Hodge R, Attardo A, Bell C, Huttner WB, Hevner RF. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex. 2009;19:2439–2450. doi: 10.1093/cercor/bhn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, Hadjantonakis AK. The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Developmental Cell. 2008;15:509–520. doi: 10.1016/j.devcel.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Kiyama T, Pan P, Furuta Y, Hadjantonakis AK, Klein WH. Eomesodermin, a target gene of Pou4f2, is required for retinal ganglion cell and optic nerve development in the mouse. Development. 2008;135:271–280. doi: 10.1242/dev.009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihalas AB, Hevner RF. Control of Neuronal Development by T-Box Genes in the Brain. Curr Top Dev Biol. 2017;122:279–312. doi: 10.1016/bs.ctdb.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Mihalas AB, Elsen GE, Bedogni F, Daza RAM, Ramos-Laguna KA, Arnold SJ, Hevner RF. Intermediate Progenitor Cohorts Differentially Generate Cortical Layers and Require Tbr2 for Timely Acquisition of Neuronal Subtype Identity. CellReports. 2016;16:92–105. doi: 10.1016/j.celrep.2016.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatomi H, Kuriu T, Okabe S, Yamamoto SI, Hatano O, Kawahara N, Tamura A, Kirino T, Nakafuku M. Regeneration of hippocampal pyramidal neurons after ischemic brain injury by recruitment of endogenous neural progenitors. Cell. 2002;110:429–441. doi: 10.1016/s0092-8674(02)00862-0. [DOI] [PubMed] [Google Scholar]
- Nowotschin S, Costello I, Piliszek A, Kwon GS, Mao CA, Klein WH, Robertson EJ, Hadjantonakis AK. The T-box transcription factor Eomesodermin is essential for AVE induction in the mouse embryo. Genes & Development. 2013;27:997–1002. doi: 10.1101/gad.215152.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- Pimeisl IM, Tanriver Y, Daza RA, Vauti F, Hevner RF, Arnold HH, Arnold SJ. Generation and characterization of a tamoxifen-inducible Eomes CreERmouse line. Genesis. 2013;51:725–733. doi: 10.1002/dvg.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst S, Arnold SJ. Eomesodermin-At Dawn of Cell Fate Decisions During Early Embryogenesis. Curr Top Dev Biol. 2017;122:93–115. doi: 10.1016/bs.ctdb.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Ramkumar N, Omelchenko T, Silva-Gagliardi NF, McGlade CJ, Wijnholds J, Anderson KV. Crumbs2 promotes cell ingression during the epithelial-to-mesenchymal transition at gastrulation. Nature Cell Biology. 2016;18:1281–1291. doi: 10.1038/ncb3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Sweeney NT, James KN, Nistorica A, Lorig-Roach RM, Feldheim DA. Expression of transcription factors divides retinal ganglion cells into distinct classes. J Comp Neurol. 2017;16:507. doi: 10.1002/cne.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney NT, Tierney H, Feldheim DA. Tbr2 is required to generate a neural circuit mediating the pupillary light reflex. J Neurosci. 2014;34:5447–5453. doi: 10.1523/JNEUROSCI.0035-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo AKK, Arnold SJ, Trotter MWB, Brown S, Ang LT, Chng Z, Robertson EJ, Dunn NR, Vallier L. Pluripotency factors regulate definitive endoderm specification through eomesodermin. Genes & Development. 2011;25:238–250. doi: 10.1101/gad.607311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent SD, Robertson EJ. Highly efficient transgene-independent recombination directed by a maternally derived SOX2CRE transgene. Genesis. 2003;37:54–56. doi: 10.1002/gene.10226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


