ABSTRACT
We evaluated the expression and function of the microglia‐specific growth factor, Progranulin‐a (Pgrn‐a) during developmental neurogenesis in the embryonic retina of zebrafish. At 24 hpf pgrn‐a is expressed throughout the forebrain, but by 48 hpf pgrn‐a is exclusively expressed by microglia and/or microglial precursors within the brain and retina. Knockdown of Pgrn‐a does not alter the onset of neurogenic programs or increase cell death, however, in its absence, neurogenesis is significantly delayed—retinal progenitors fail to exit the cell cycle at the appropriate developmental time and postmitotic cells do not acquire markers of terminal differentiation, and microglial precursors do not colonize the retina. Given the link between Progranulin and cell cycle regulation in peripheral tissues and transformed cells, we analyzed cell cycle kinetics among retinal progenitors following Pgrn‐a knockdown. Depleting Pgrn‐a results in a significant lengthening of the cell cycle. These data suggest that Pgrn‐a plays a dual role during nervous system development by governing the rate at which progenitors progress through the cell cycle and attracting microglial progenitors into the embryonic brain and retina. Collectively, these data show that Pgrn‐a governs neurogenesis by regulating cell cycle kinetics and the transition from proliferation to cell cycle exit and differentiation. © 2017 The Authors. Developmental Neurobiology Published by Wiley Periodicals, Inc. Develop Neurobiol 77: 1114–1129, 2017
Keywords: Progranulin‐a, retinal development, zebrafish, cell cycle, CNS development
INTRODUCTION
Progranulin (PGRN), also known as granulin‐epithelin precursor, acrogranin, proepithelin and PC cell‐derived growth factor, is an evolutionarily conserved secreted growth factor/pleiotropic glycoprotein precursor with a well‐established role in embryogenesis, tumorigenesis, and wound healing (He and Bateman, 2003; He et al., 2003; Ong and Bateman, 2003). In peripheral tissues and tumor cells, PGRN is mitogenic, stimulates cell proliferation and migration, and promotes cell survival (He et al., 2002; He and Bateman, 2003; Ong and Bateman, 2003). In the mammalian CNS, PGRN is expressed by neurons and microglia (Daniel et al., 2000). In humans, diminished PGRN production due to heterozygous mutations in the PGRN gene (hGRN) is a major cause of the neurodegenerative disease, frontotemporal lobar degeneration (Baker et al., 2006; Cruts et al., 2006). In contrast, malignancy of glioblastomas is correlated with elevated PGRN levels (Liau et al., 2000; Wang et al., 2011). In vivo, PGRN increases exercise‐induced neurogenesis in the hippocampus of adult mice (Asakura et al., 2011). In vitro, PGRN enhances neuronal survival, increases neural progenitor proliferation, and promotes neurite outgrowth and neuronal differentiation (Van Damme et al., 2008; Ryan et al., 2009; Gao et al., 2010; Nedachi et al., 2011). Modulation of PGRN levels is a molecular signature of activated microglia (De Muynck and Van Damme, 2011); CNS injury induces a microglia‐specific upregulation of progranulin mRNA and protein synthesis (Craig et al., 2008; Moisse et al., 2009; Naphade et al., 2009; Philips et al., 2010). Interestingly, in vitro PGRN acts as a microglial chemoattractant (Pickford et al., 2011). Much is known about PGRN activity in non‐neuronal tissues and the injured CNS, however the function of PGRN within the developing CNS is not well understood.
We use the zebrafish retina as a model tissue for studying brain development. The vertebrate retina is a well‐established and tractable model for investigating the cellular and molecular mechanisms that regulate both developmental and regenerative neurogenesis (Agathocleous and Harris, 2009). Among vertebrates, the cytoarchitecture of the retina is precise and evolutionarily very highly conserved. Even subtle changes in developmental programs are easily detected as structural changes (Cepko et al., 1996). Zebrafish have four protein‐encoding progranulin genes, two of which, pgrn‐a and pgrn‐b, are co‐orthologues of mammalian gene (GRN) (Cadieux et al., 2005). pgrn‐a is syntenically conserved, making it the true orthologue of hGRN. We first identified pgrn‐a as a microglia‐specific growth factor in the retina of the adult zebrafish, where it is strongly upregulated following photoreceptor death and during photoreceptor regeneration (Craig et al., 2008).
To test the hypothesis that the microglia‐specific growth factor, Pgrn‐a, regulates neurogenesis in the vertebrate retina, we evaluated its developmental expression and used protein knockdown to test its function. At 24 h postfertilization (hpf), pgrn‐a is expressed throughout the forebrain, but beginning about 36 hpf pgrn‐a expression becomes limited to macrophages/microglial precursors in the yolk sac, brain and retina. Knockdown of Pgrn‐a using morpholino oligonucleotides (MOs) results in marked developmental changes. In morphants, retinal development is delayed; retinal progenitors remain in the cell cycle at times when they are normally postmitotic, and there is a corresponding absence of neuronal differentiation. Further, depletion of Pgrn‐a prevents microglial precursors from migrating into the retina. Given that PGRN governs the cell cycle in peripheral tissues and transformed cells (Ong and Bateman, 2003), we evaluated cell cycle kinetics in retinal progenitors following Pgrn‐a knockdown. Depleting Pgrn‐a significantly increases the duration of the G2‐ and M‐phases, and this results in an overall increase in the total length of the cell cycle. We also found that diminished Pgrn‐a results in a significant increase in the expression of genes that promote cell cycle progression, and a significant reduction in the expression of genes that promote cell cycle exit, demonstrating that Pgrn‐a signaling regulates the duration of the cell cycle by governing the expression of genes that directly control cell cycle progression. From these data, we conclude that the microglia‐specific molecule, Pgrn‐a, plays fundamental role in governing developmental neurogenesis. Further, we conclude that Pgrn‐a also functions to recruit microglial precursors to the embryonic CNS.
MATERIALS AND METHODS
Animals
Adult AB wild type (WT) zebrafish (Danio rerio; ZIRC, University of Oregon, Eugene, OR) were maintained at 28.5°C on a 14/10‐h light/dark cycle. Embryos were collected immediately following natural spawns, incubated at 28.5°C on a 14/10‐h light/dark cycle and staged by hours postfertilization (hpf). The Institutional Animal Care and Use Committee at the University of Michigan approved all protocols and procedures.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as previously described (Luo et al., 2012). Briefly, embryos were fixed overnight in 4% paraformaldehyde (PFA) in 100 mM phosphate buffer at 4°C, cryoprotected with 20% sucrose in 100 mM phosphate buffer, and embedded in frozen Tissue‐Tek optical cutting temperature (OCT; Sakura Finetek USA, Torrance, CA) compound. Frontal sections (10 µm thick), immediately adjacent to or including the optic nerve, were mounted on slides, washed, incubated in heat inactivated normal sheep serum (NSS; Sigma‐Aldrich Corp., St. Louis MO), and incubated overnight at 4˚C with primary antibodies. The following day, sections were washed and incubated in secondary antibodies for 1 h at room temperature. Nuclei were stained with DAPI. The antibodies used and their concentrations are listed in Supporting Information Table S1.
S‐Phase Labels
Cells in S‐phase of the cell cycle were labeled with either 5‐Bromo‐2′deoxyuridine (BrdU; Sigma‐Aldrich Corp., St. Louis, MO) or 5‐ethynyl‐2′deoxyuridine (EdU; Invitrogen, Carlsbad, CA) as previously described (Ochocinska and Hitchcock, 2007; Luo et al., 2012). Briefly, dechorionated embryos at 24 and 48 hpf were incubated for 20 min in ice‐cold 10 mM BrdU or 1.5 mM EdU dissolved in E3 containing 15% DMSO. 72 hpf embryos were incubated for 20 min in 10 mM BrdU or 1.5 mM EdU dissolved in E3 containing 15% DMSO at room temperature (RT). 8 dpf larvae were incubated for 20 min in 2 mM EdU dissolved in 10% PBS in E3 at RT. Following the labeling step, embryos/larvae were returned to RT E3 for 10 min prior to fixation. For BrdU staining, sections were incubated in 100°C sodium citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0) for 30 min, cooled to RT for 60 min, and then immunolabeled using standard protocols described above.
In Situ Hybridization
Double in situ hybridization (ISH) on retinal sections (Hitchcock and Kakuk‐Atkins, 2004) and whole mount ISH was performed as previously described (Hitchcock and Kakuk‐Atkins, 2004; Ochocinska and Hitchcock, 2007; Craig et al., 2008; Luo et al., 2012). Sense and antisense digoxigenin (DIG)‐ or fluorescein‐labeled riboprobes (Supporting Information Table S2) were synthesized from full‐length cDNA clones for pgrn‐a (NM001001949; a gift from Dr. Hugh Bennett), fms (NM131672; a gift from Dr. Philippe Herbomel), and atoh7 (NM131632; a gift from Dr. Deborah Stenkamp) using in vitro transcription (Roche Diagnostic Corp., Indianapolis, IN). Embryos were incubated with hybridization solution containing approximately 100 ng of probe. For double fluorescent ISH, the fms and pgrn‐a probes were hybridized simultaneously.
Pgrn‐a Knockdown with MOs
For Pgrn‐a knockdown, two independent antisense MOs (Gene Tools, LLC, Philomath, OR), targeting either the 5′UTR (0.25 ng/embryo) or the splice site between exon 3 and intron 3 (SS, 1 ng/embryo) of zebrafish pgrn‐a, were diluted in 1X Danieau buffer (Nasevicius and Ekker, 2000) containing 2.5% phenol red and injected into the yolk of AB WT embryos at the 1–2 cell stage. Five‐base pair mismatch (MM) and standard control (SC) MOs (Gene Tools, LLC, Philomath, OR) were used as negative controls. To suppress potential off target effects induced by experimental MOs, p53 MO (Gene Tools, LLC, Philomath, OR) was coinjected (1.5 fold) to block nonspecific cell death (Robu et al., 2007). The MO sequences are listed in Supporting Information Table S3.
mRNA Rescue
The coding sequence of zebrafish (zf) pgrn‐a (NM_001001949.2; in pSPORT1 Vector) and the pgrn‐a paralog, zf pgrn‐b (NM_212738.1; in pBK‐CMV Image Clone from Open Biosystems, GE Dharmacon, Lafayette, CO), were PCR amplified starting from the ATG (primer sequences listed in Supporting Information Table S4), therefore removing the 5′UTR MO recognition sequence, using Platinum Taq DNA Polymerase HF (Invitrogen, Carlsbad, CA). cDNAs were subcloned into the pGEM‐T Easy Vector (Promega, Madison, WI) using standard protocols, linearized with SacI‐HF (New England Biolabs, Ipswich, MA), and capped RNAs were transcribed using the mMESSAGE mMACHINE T7 kit (Ambion, Thermo Fisher Scientific, Halethorp, MD). Human progranulin (hGRN; GE Healthcare clone ID 3457813 in pCMV‐SPORT6) and eGFP (pCS2+‐EGFP) were linearized with NotI‐HF and transcribed with SP6. For rescue experiments, zf pgrn‐a (25 pg/embryo), zf pgrn‐b (25 pg/embryo), or human progranulin (hGRN; 400 pg/embryo) was coinjected with 5′UTR MO (.25 ng/embryo) at the one cell stage. As a negative control, eGFP mRNA (25 pg/embryo) was coinjected with the 5′UTR MO (.25 ng/embryo). At 72 hpf Embryos were treated with EdU, sacrificed, and processed for IHC.
Western Blot
To confirm knockdown of Pgrn‐a, Western blotting was performed as previously described (Gramage et al., 2015). Briefly, for each condition, protein was isolated from 50 embryo heads and separated by gel electrophoresis. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (GenHunter Corp., Nashville, TN) and incubated with rabbit anti‐Pgrn antibodies (1:2000, a gift from Jen Leih Wu), see Li et al., 2010). The polyclonal anti‐PgrnA antibodies were raised against a peptide that corresponds to residues 244–264, which are unique to PgrnA (Li et al., 2010). Immunolabeled proteins were detected using enhanced chemiluminescence assay (ECL detection system, Amersham Biosciences, Arlington Heights, IL). Anti‐Actin (1:1000, Calbiochem, EMD Millipore, Billerica, MA) was used as a loading control.
PCR
To verify SS‐targeting MOs altered processing of pgrn‐a pre‐mRNA, PCR was performed on RNA isolated from SS morphant and uninjected control embryos at 48 hpf. cDNA was reverse transcribed using the SuperScript kit (Invitrogen, Carlsbad, CA). Primers flanking the SS MO target sequence (F: AGAATGTTGTGAGGACCATC and R: CTGTGCTACTGGACAGCAG) were used to amplify the pgrn‐a locus following manufacturer's protocol (SuperScript One‐Step RT‐PCR system; Invitrogen). PCR products were separated by gel electrophoresis, purified and sequenced.
Cell Cycle Kinetics
Mitotic Index
The proportion of cells in the M‐phase of the cell cycle was determined for embryos at 28 hpf Retinal sections were immunolabeled with anti‐phosphorylated histone H3 (pH3), marking cells in the M‐phase, and the number of M‐phase cells as a function of retinal area (μm2) was calculated. The spatial pattern of pH3 labeled cells was also assessed.
Percent Labeled Mitosis
The length of the G2‐phase of the cell cycle was determined using the percent labeled mitosis technique (Quastler and Sherman, 1959; Luo et al., 2012). Embryos at 28 hpf were incubated in BrdU for 20 min to label all cells in the S‐phase of the cell cycle. Starting 10 min after BrdU exposure, embryos were collected at 30‐min intervals through 35 hpf, sectioned and immunolabeled with anti‐pH3 and anti‐BrdU antibodies. The proportion of double‐labeled retinal cells ((pH3+BrdU+)/(pH3+)) was plotted as a function of time following BrdU exposure.
Pulse‐Chase‐Pulse
To determine the relative length of S‐phase (T s) and estimate the total cell cycle length (T c), a pulse chase pulse method was used as previously described (Rachel et al., 2002). At 26 hpf, embryos were incubated in 10 mM BrdU dissolved in E3 containing 15% DMSO for 20 min on ice, rinsed in E3 at RT, incubated in 10 mM thymidine at 28.5°C for 20 min (Otteson et al., 2001), washed in E3 for 1 h 40 min at 28.5°C and incubated in 1.5 mM EdU dissolved in E3 containing 15% DMSO for 20 min on ice. Embryos were then rinsed for an additional 10 min, sacrificed and processed sequentially to label BrdU and EdU. Nuclei were counterstained with DAPI. For each embryo, an optical section from the middle of a z‐stack taken of one central retina section was selected and cells labeled with BrdU (nBrdU), EdU (nEdU), and DAPI (nDAPI) were counted. The length of the S‐phase and total length of the cell cycle were determined using the following formulae, T s=2/(n BrdU+/n total EdU+) and T c=T s/(n EdU+/n DAPI+), respectively.
Reverse Transcriptase Quantitative Real‐Time PCR for Cyclins (B, D, and E) and Cyclin‐Dependent Kinase Inhibitors (p27kip and p57kip)
To determine the expression levels of genes that regulate cell cycle, Quantitative Real‐Time PCR was performed as previously described (Taylor et al., 2015). For each condition, three biological replicates of 50 embryo heads were collected. Total RNA was isolated from pooled heads using the Aurum Total RNA Mini Kit (Bio‐Rad Laboratories Inc., Hercules, CA). cDNA was synthesized using the QuantiTect Reverse Transcription kit (Qiagen, Hilden, Germany). Three technical replicates of each biological replicate were run in a Bio‐Rad CFX384 Touch Real Time PCR Detection System using 6 ng cDNA and Bio‐Rad IQ SYBR Green Supermix. Primers are listed in Supporting Information Table S5. Gene expression was calculated using Bio‐Rad CFX Manager Software and normalized to β‐actin.
Statistical Analysis
Quantitative data was represented by means and standard deviations. To calculate statistical significance, a one‐way ANOVA or Student's t‐test was used (GraphPad Prism Software, La Jolla, CA). A p‐value ≤ 0.05 was considered significant, unless otherwise noted. With the exception of the data presented in Figure 5(C,F,G), 95% confidence intervals (CI) were calculated and reported here for all statistical comparisons.
Figure 5.
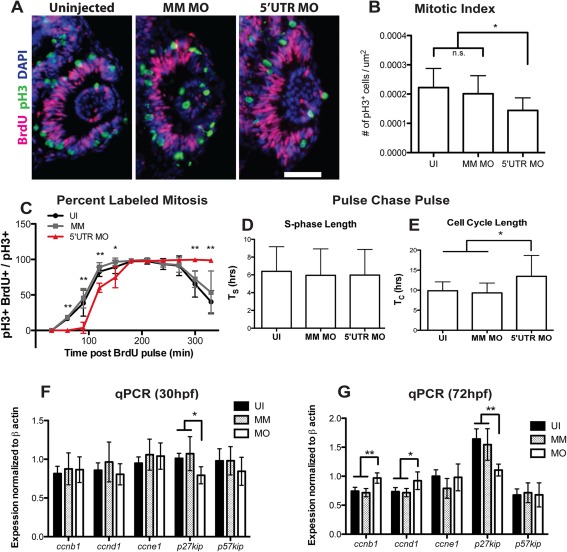
Knockdown of pgrn‐a alters cell cycle kinetics in retinal progenitors. (A) Representative retinal sections from 28 hpf uninjected (left panel), MM MO (center panel), and 5′UTR MO (right panel) embryos stained with antibodies against pH3 (green), BrdU (fusia), and DAPI (blue) used for quantification of mitotic index. (B) Histogram showing the mitotic index at 28 hpf. The number of pH3+ cells/unit area in UI (0.0002 ± 1.973e‐005 cells per μm2; n = 11; 95% CI: .0002 (−0.00018–0.00058)), MM MO (0.0002 ± 1.947e‐005 cells per μm2; n = 10; 95% CI: .0002 (0.000162–0.000238)), and 5′UTR MO retinas (0.0001 ± 1.366e‐005 cells per μm2; n = 10; 95% CI: .0001 (0.000073–0.000127)); *p≤0.01. (C) Graph showing the percent labeled mitosis for UI, MM MO and 5′UTR MO embryos between 28 and 35 hpf. (D) Histogram showing average S‐phase length (TS) in 26‐28 hpf UI (6.4 ± 2.8 hrs; n = 16; 95% CI: 6.4 (5–7.8)), MM MO (5.9 ± 3.0 hrs; n = 10; 95% CI: 6.0 (4.2–7.8)), and 5′UTR MO (6.0 ± 2.9 hrs; n = 7; 95% CI: 6.0 (3.9–8.1)) embryos. (E) Histogram showing average total cell cycle length (TC) at 26‐28 hpf in UI (9.9 ± 2.2 hrs; n = 16; 95% CI: 9.9 (8.8–11)), MM MO (9.3 ± 2.5 hrs; n = 10; 95% CI: 9.3 (7.8–10.8)), and 5′UTR MO (13.5 ± 5.2 hrs; n = 7; 95% CI: 13.5 (9.6‐17.4)); *p≤0.05. (F–G).
Imaging
All sectioned material was sealed with glass coverslips and mounting media (Electron Microscopy Sciences, Hatfield, PA). Fluorescence images were captured using a Leica TCS SP5 confocal microscope (Leica, Wetzler, Germany). Bright field images were captured using either a Leica DM6000 CFS or Leica M165FC microscope (Leica, Wetzler, Germany).
RESULTS
Developmental Expression of Pgrn‐a
We identified pgrn‐a in the zebrafish retina through an unbiased screen for genes modulated during neuronal regeneration (see http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13999), and determined that in the adult retina pgrn‐a is expressed exclusively by microglia (Craig et al., 2008). Based on these data and the observation that, in vitro, Pgrn is a chemoattractant to microglia (Pickford et al., 2011), we hypothesized that Pgrn‐a plays a fundamental role in developmental neurogenesis, and may also regulate the migration of microglial precursors from their origins in the yolk sack into neuroepithelial tissues. We first established the developmental cellular pattern of pgrn‐a expression using ISH (Fig. 1 and Supporting Information Fig. S1). At 24 hpf, pgrn‐a is expressed throughout the forebrain and retina [Fig. 1(A) and Supporting Information Fig. S 1(A)]. Between 24 and 48 hpf, however, pgrn‐a expression is down regulated in the neuroepithelium and becomes restricted to putative macrophages/microglial precursors in the yolk sac, forebrain and retina (arrowheads) [Fig. 1(B–D) and Supporting Information Fig. S1(B–E)]. There are no antibodies that detect zebrafish Pgrn‐a in tissue sections, however, Western blot analysis showed that Pgrn‐a is present at 24 hpf [Supporting Information Fig. S1(E)], suggesting that the protein is reliably expressed when transcript is present. To confirm that pgrn‐a expressing cells in the embryonic retina are microglia, pgrn‐a ISH was combined with immunstaining using the microglial markers, L‐plastin (Herbomel and Levraud, 2005) and 4C4 (Raymond et al., 2006), and double ISH was perfomed using a probes for pgrn‐a and fms, which encodes the macrophage colony stimulating factor 1 receptor (CSF1R). In the retina, all cells that express pgrn‐a [Fig. 1(E,G)] are colabeled with antibodies to L‐plastin [Fig. 1(F)] or 4C4 [Fig. 1(H)] and coexpress fms [Fig. 1(I–L)]. Equivalent labeling patterns were observed in the brain (data not shown).
Figure 1.
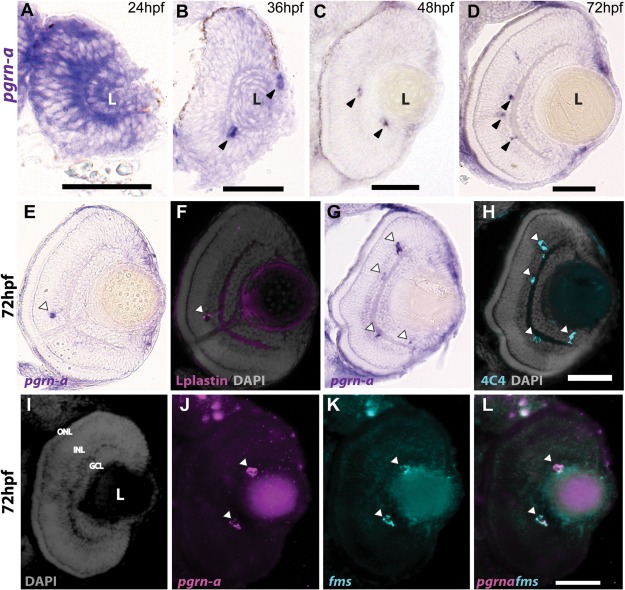
Expression of pgrn‐a in the developing retina. (A–D) Cross‐sections of the retina following whole mount in situ hybridizations (ISHs) showing pgrn‐a expression in WT at 24 (A), 36 (B), 48 (C), and 72 hpf (D). (E–L) pgrn‐a is microglia‐specific by 72 hpf. pgrn‐a ISH (E + G) followed by immunohistochemistry of L‐plastin (F) and 4C4 (H) showing colocalization of pgrn‐a and microglial markers (arrowheads). Double ISH in the same section showing DAPI (I), pgrn‐a (J), and fms (K) expression, and overlay (L) showing colocalization of pgrn‐a and fms in the retina (arrowheads). In panel D, it appears there is transcript expression in the CMZ. However, there is no evidence for this following whole mount ISH (see Supporting Information Fig. S1), and we infer that in sections this apparent labeling is spurious. Outer nuclear layer (ONL); inner nuclear layer (INL); and ganglion cell layer (GCL); (L) lens. Scale bar equals 50 µm.
Knockdown of Pgrn‐a Results in Diminished Neuronal Differentiation and a Paucity of Microglia
Two independent antisense MOs were used to block Pgrn‐a synthesis. The ability to knockdown Pgrn‐a with the 5′UTR‐targeting MO was confirmed with Western blot analysis [Fig. 2 (A,B)]. Diagnostic PCR showed that the SS‐targeting MO resulted in intron retention and a premature stop codon (data not shown). There were no qualitative changes in the appearance of morphant retinas between 48 and 72 hpf, so only data from 72 hpf embryos are described here. At 72 hpf, the overall body length and shape of control embryos [Fig. 2(D–F)] and pgrn‐a morphants [Fig. 2(G,H)] are comparable to uninjected WT embryos [Fig. 2(C)]. In contrast to controls, pgrn‐a morphants have markedly small forebrains and microphthalmia.
Figure 2.

pgrn‐a‐targeted morpholino oligonucleotides inhibit Pgrn‐a translation, which results in microphthalmia. (A) Western blot showing Pgrn‐a and Actin expression in 72 hpf uninjected (UI) and morphant (5′UTR MO) embryos. (B) Histogram showing quantification of Pgrn‐a expression in UI controls (26.2 ± 2.05%) and morphants (2.7 ± 0.9%); **p ≤ 0.01. Quantitative data from three biological and technical replicates is normalized to Actin and represented as mean; error bars represent the standard deviation. (C–H) Representative whole embryos from control (C–F) and experimental (G–H) groups at 72 hpf. 5′UTR MM MO and SS MM MO, embryos injected with 5‐nucleotide mismatch control morpholinos; 5′UTR MO, embryos injected with pgrn‐a 5′UTR‐targeted morpholinos; SS MO, embryos injected with pgrn‐a e3i3 splice site‐targeted morpholinos.
Compared to warm blooded vertebrates, retinal morphogenesis in zebrafish is very rapid. By 24 hpf, eyecups have formed from the anterior neural plate, and the retina consists of proliferating neuroepithelial cells (Schmitt and Dowling, 1994, 1996; Li et al., 2000). At approximately 28 hpf, neuronal differentiation begins and ganglion cells are the first to exit the cell cycle in a small ventronasal patch (Hu and Easter, 1999). Neuronal differentiation and lamination then progress sequentially from the ventronasal patch through each cellular lamina in circumferential waves that move dorsally, then temporally (Schmitt and Dowling, 1996, 1999; Hitchcock and Raymond, 2004). By 72 hpf, the initial phase of neurogenesis is largely complete, and the retina is fully laminated and functional (Easter and Nicola, 1996). Beyond 72 hpf and into adulthood, the retina continues to grow by expansion and the addition of new neurons from the ciliary marginal zone (CMZ) (Hitchcock et al., 2004; Raymond et al., 2006).
To determine the functional consequences of Pgrn‐a knockdown, qualitative and quantitative measures of cell proliferation, neuronal differentiation and retinal size were compared between control and experimental groups at 72 hpf (Fig. 3). At 72 hpf, in retinas from uninjected embryos, EdU‐positive progenitors are relatively few in number [Fig. 3(A–C,T)] and restricted to the CMZ (brackets) [Fig. 3(A)]. In the central retina, the absence of EdU reflects exit from the cell cycle prior to EdU exposure by neurons in the ganglion cell layer and the inner and outer nuclear layers [Fig. 3(A–C)]. Nuclear staining with DAPI reveals the retina is fully laminated, and, reflecting its functional state, markers of mature neurons label differentiated ganglion cells [Fig. 3(A)], amacrine cells [Fig. 3(B)], and photoreceptors [Fig. 3(C)]. All embryos injected with control MOs [Fig. 3(D–L)] appear identical to uninjected embryos [Fig. 3(A–C)]. In marked contrast to uninjected and control embryos, blocking the translation of Pgrn‐a results in striking changes in the retinal phenotype. Compared to controls [Fig. 3(A–L)], morphant retinas contain significantly more EdU‐positive progenitors, which are present throughout the retina, and they lack well‐defined laminae and differentiated neurons [Fig. 3(M–R)]. The microphthalmia observed in the whole mount embryos [Fig. 2(A–F)] is readily observable in retinal sections [Fig. 3(M–R)]. As is also evident in the whole embryos, the retinal phenotype in the 5′UTR morphants is slightly more severe than the SS morphants. We interpret this to be a dose‐dependent effect resulting from the SS MO targeting zygotic transcripts only, whereas the 5′UTR MO targets both maternal and zygotic transcripts (Cadieux et al., 2005; Bill et al., 2009). (All data presented below use the 5′UTR‐targeting MO). Collectively, these data show that the absence of Pgrn‐a results in a significant temporal delay in retinal neurogenesis. Progenitors fail to exit the cell cycle at the appropriate developmental time, and among postmitotic neurons there is diminished differentiation. Interestingly, the number of retinal microglia was also significantly decreased in pgrn‐a morphants compared to controls [Fig. 3(U)].
Figure 3.
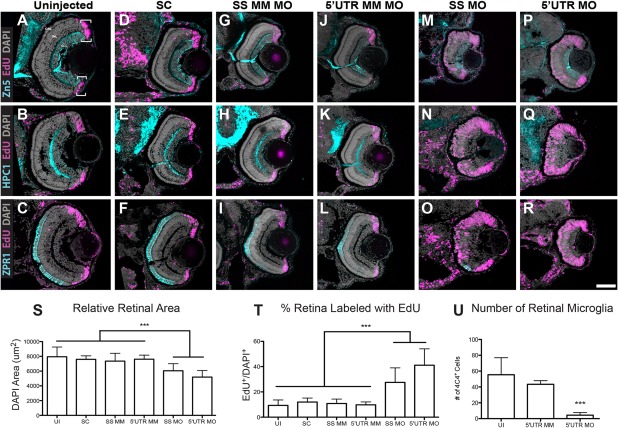
Pgrn‐a knockdown results in microphthalmia and diminished neuronal differentiation. (A–R) Sections through central retina of uninjected (UI, A–C); standard control morpholino‐injected (SC, D–F); SS MM (G–I) and 5′UTR MM (J–L), embryos injected with 5‐nucleotide mismatch control morpholinos; SS MO (M–O) and 5′UTR MO (P–R), embryos injected with e3i3 splice site‐targeted or 5′UTR‐targeted morpholinos, respectively, at 72 hpf. Sections are immunolabeled (cyan) with markers of differentiated ganglion cells (Zn5, top row), amacrine cells (HPC1, middle row), and red–green double cone photoreceptor cells (ZPR1, bottom row), EdU (fusia), and DAPI (gray). (S) Histogram showing relative retinal area in UI (7945.8 ± 1319.6 μm2; n = 11; CI, 7945 ± 886), SC‐injected (7594.3 ± 475.2 μm2; n = 9; 95% CI: 7594 (7229–7959)), SS MM MO‐injected (7354 ± 1062.6 μm2; n = 11; 95% CI: 7353 (6640‐8066)), and 5′UTR MM MO‐injected (7610.8 ± 547.6 μm2; n = 10; 95% CI: 7610 (7219–8001)), SS MO (6044.7 ± 956.2 μm2; n = 10; 95% CI: 6044 (5360–6782)), and 5′UTR MO (5188.7 ± 891.8 μm2; n = 16; 95% CI: 5188 (4713–5663)) retinas at 72 hpf; ***p≤0.001. (T) Histogram showing the percent of the retina labeled with EdU in UI (9.3 ± 4.3%; n = 11; 95% CI: 9.3 (6.4–12.2)), SC‐injected (12 ± 3.1%; n = 9; 95% CI: 12.0 (8.7–15.3)), SS MM MO‐injected (10.8 ± 3.5%; n = 11; 95% CI: 10.8 (8.7–12.9)), 5′UTR MM MO‐injected (9.8 ± 2.3%; n = 10; 95% CI: 9.8 (7.3–12.3)), SS MO‐injected (27.5 ± 11.5%; n = 10; 95% CI: 27.6 (25.9–29.3)), and 5′UTR MO‐injected (41.1 ± 13%; n = 16; 95% CI: 41.1 (35–47.2)) retinas at 72 hpf; ***p≤0.001. (U) Histogram showing the number of microglia in uninjected (UI; 55.4 ± 21.5 cells; n = 20; 95% CI: 62.4 (51.7–73.1)), 5′UTR mismatch morpholino injected (5′UTR MM; 43.4 ± 4.6 cells; n = 8; 95% CI: 43.4 (39.6–47.2)), and Pgrn‐a morphant (5′UTR MO; 4.4 ± 3.4 cells; n = 11; 95% CI: 4.4 (2.1–6.7)) retinas at 72 hpf; ***p≤0.001. Quantitative data are represented as mean; error bars represent the standard deviation. Outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL); ciliary marginal zone (CMZ, brackets). Scale bar equals 50 µm.
MO concentrations decrease with each cell division, and, as a result, MO‐dependent inhibition of protein translation lasts approximately 3‐4 dpf (Nasevicius and Ekker, 2000). Therefore, we also evaluated the potential for the recovery of retinal development in pgrn‐a morphants. When morphants are allowed to survive to 8 dpf, Pgrn‐a synthesis recovers and the cellular differentiation and cytoarchitecture of the retina follow normal developmental patterns [Supporting Information Fig. S2(A)], however, the retinal size does not recover to control values [Supporting Information Fig. S2(B)]. This indicates that the recovery of Pgrn‐a synthesis does not produce a compensatory acceleration of cell division or neuronal differentiation that would produce a retina of normal size. So too, the number of microglia in 8 dpf larvae remains significantly below control values [Supporting Information Fig. S2(C)].
Neurogenic Competence and Cell Death Is Not Altered in Morphants
MOs have the potential to create spurious results from off‐target effects (Eisen and Smith, 2008). As a first of several approaches to assess this, we determined whether or not the retinal changes observed here were due to a non‐specific developmental delay. atoh7 is required to initiate neurogenesis and serves as an indicator of the onset of neurogenic competence (Masai et al., 2000; Kay, 2005). Pgrn‐a knockdown does not alter the timing or spatial restriction of the initial atoh7 expression (Supporting Information Fig. S3), although the apparent level of atoh7 expression in morphant larvae is diminished (cf. Supporting Information S3 A–C), indicating that the changes in retinal development are not a consequence of a simple, systemic developmental delay. Cell death is another potential off target affect. In embryos at 24 and 48 hpf, counts of apoptotic cells marked by terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) [Supporting Information Fig. S3(D)] or pyknotic nuclei labeled with acridine orange (data not shown) show no significant differences between control and experimental embryos, indicating that cell death cannot account for the changes in retinal development. In addition, these data show that the presence of the p53 MO in both the MM and 5′UTR MOs does not artificially depress neuronal apoptosis between 28 and 72 hpf.
Coinjection of zf pgrn‐a/b or hGRN mRNA Rescues Pgrn‐a Knockdown
To validate the specificity of the Pgrn‐a knockdown, mRNA encoding egfp, zf pgrn‐a, zf pgrn‐b, or hGRN was coinjected with the 5′UTR MO. To determine if exogenous mRNA rescued the knockdown phenotype, comparisons were made between embryos that were uninjected [Fig. 4(A–C)], injected with 5′UTR MO alone [Fig. 4(D–F)], and coinjected with 5′UTR MO and one of the mRNA constructs [Fig. 4(G–O)]. As a control for the mRNA injections, the 5′UTR MO and egfp mRNA were coinjected, and this uniformly failed to rescue the developmental defects resulting from Pgrn‐a knockdown (data not shown) [Fig. 4(P–R)]. In contrast, both zebrafish pgrn paralogs and the human pgrn mRNA rescued the morpholino‐induced developmental defects in retinal growth, cell proliferation and neuronal maturation. In these coinjected embryos, EdU‐labeled retinal progenitors were present only in the CMZ, retinal laminae were fully formed, and retinal size was equal to controls [Fig. 4(G–O,P,Q)]. These data validate the specificity of the morpholinos targeted to pgrn‐a, and demonstrate that Pgrn‐a regulates both cell proliferation and neuronal differentiation. Finally, these experiments highlight the conserved function of progranulin protein between fish and humans. Interestingly, coinjecting mRNA only partially rescues the migration of macrophages/microglial precursors into the retina [Fig. 4(R)]. In embryos coinjected with Pgrn mRNA, the total number of retinal microglia is significantly increased compared to morphants, however, there are significantly fewer retinal microglia in coinjected embryos relative to uninjected controls [Fig. 4(R)].
Figure 4.
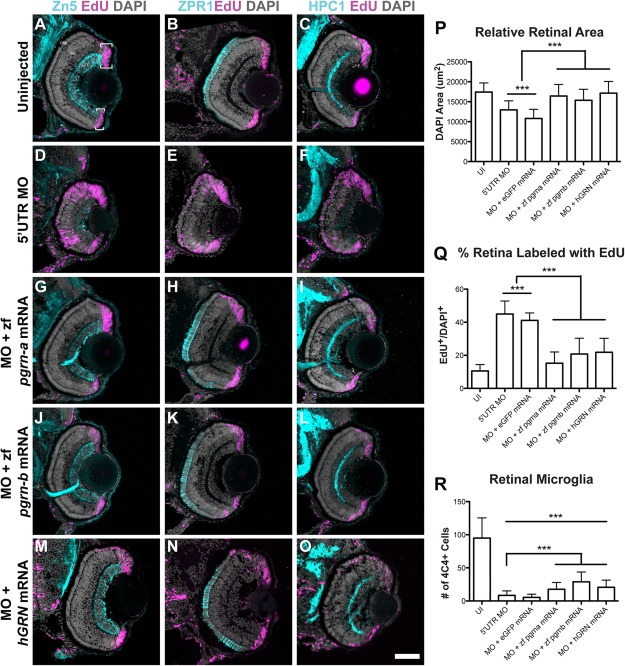
Coinjection of 5′UTR MO and zf pgrn‐a, zf pgrn‐b, or hGRN mRNA rescues most aspects of knockdown retinal phenotype. (A–O) Cross‐sections through central retina of 72 hpf uninjected (UI, A–C), 5′UTR MO‐injected (D–F), 5′UTR MO and zf pgrn‐a mRNA coinjected (G–I), 5′UTR MO and zf pgrn‐b mRNA coinjected (J–L), and 5′UTR MO and hGRN mRNA coinjected (M–O) embryos at 72 hpf. Sections are immunolabeled (cyan) with markers for ganglion cells (Zn‐5, left column), red‐green double cone photoreceptors (Zpr1, middle column), and amacrine cells (HPC1, right column), EdU (fusia), and DAPI (gray). (P) Histogram showing relative retinal area of UI (17437.4 ± 2286.1 μm2; n = 23; 95% CI: 16970 (14744–19196)), 5′UTR MO‐injected (12961.2 ± 2251.8 μm2; n = 21; 95% CI: 11329 (10561–12097)), 5′UTR MO and eGFP mRNA co‐injected (10809 ± 2282.5 μm2; n = 8; 95% CI: 14374 (12599–16149)), 5′UTR MO and zf pgrn‐a mRNA co‐injected (16435.7 ± 2882.2 μm2; n = 12; 95% CI: 16860 (15697–18023)), 5′UTR MO and zf pgrn‐b mRNA co‐injected (15357.8 ± 2750.2 μm2; n = 13; 95% CI: 14834 (13529–16139)), and 5′UTR MO and hGRN mRNA co‐injected (17146.1 ± 2930.7 μm2; n = 12; 95% CI: 18579 (17174–19984)) embryos at 72 hpf; ***p<0.001. (Q) Histogram showing the percent of the retina labeled with EdU in UI (10.5 ± 3.9%; n = 7; 95% CI: 11.0 (7–15)), 5′UTR MO‐injected (45 ± 7.9%; n = 18; 95% CI: 45 (41–49)), 5′UTR MO and eGFP mRNA co‐injected (41.1 ± 4.5%; n = 6; 95% CI: 41.0 (36–46)), 5′UTR MO and zf pgrn‐a mRNA co‐injected (15.2 ± 6.8%; n = 22; 95% CI: 15.0 (12–18)), 5′UTR MO and zf pgrn‐b mRNA co‐injected (20.8 ± 9.6%; n = 18; 95% CI: 21 (16‐26)), and 5′UTR MO and hGRN mRNA co‐injected (21.8 ± 8.5%; n = 12; 95% CI: 15 (9–21)) embryos at 72 hpf; ***p<0.001. (R) Histogram showing the number of retinal microglia (4C4+ cells) at 72 hpf in UI (95 ± 30.5 cells; n = 23; 95% CI: 95.0 (82.5–107.5)), 5′UTR MO‐injected (8.4 ± 6.8 cells; n = 21; 95% CI: 8.4 (5.5–11.3)), 5′UTR MO and eGFP mRNA co‐injected (5.4 ± 4.8 cells; n = 8; 95% CI: 5.4 (2.1–8.7)), 5′UTR MO and zf pgrn‐a mRNA co‐injected (17.7 ± 10.2 cells; n = 12; 95% CI: 17.7 (11.9–23.5)), 5′UTR MO and zf pgrn‐b mRNA co‐injected (28.9 ± 14.7 cells; n = 13; 95% CI: 28.9 (20.9′36.9)), and 5′UTR MO and hGRN mRNA co‐injected (20.6 ± 10.8; n = 12; 95% CI: 20.6 (14.5′26.7)) embryos; ***p<0.001. Quantitative data are represented as mean; error bars represent the standard deviation. Outer nuclear layer (ONL), inner nuclear layer (INL), and ganglion cell layer (GCL); ciliary marginal zone (CMZ, brackets). Scale bar equals 50 µm.
Pgrn‐a Knockdown Alters Cell Cycle Kinetics
In morphants, the persistence of retinal progenitors at a stage when these cells normally exit the cell cycle, suggests that Pgrn‐a functions to promote progression through the cell cycle. Therefore, we determined the consequences of Pgrn‐a knockdown on cell cycle kinetics using three approaches to directly measure the duration of individual components of the cell cycle and to estimate the total length of the cell cycle. First, the mitotic index was assayed at 28 hpf. Sections through central retina were immunolabeled with anti‐pH3, a marker of cells in M‐phase, and the proportion of pH3‐positive cells was determined. In the retina, cells divide at the apical surface of the neuroepithelium (Das et al., 2003; Baye and Link, 2007). Following Pgrn‐a knockdown, the location of pH3‐positive cells was similar in morphants and controls [Fig. 5(A)], indicating that the fundamental polarization of the neuroepithelium and interkinetic nuclear migration is unchanged following knockdown of Pgrn‐a (see Baye and Link, 2007). In contrast, the number and proportion of M‐phase cells was significantly less in the morphant retinas [Fig. 5(B)].
Second, the length of the G2‐phase of the cell cycle was determined using the Percent Labeled Mitosis method (Quastler and Sherman, 1959). This assay measures the interval between the S‐phase, when cells can be labeled with BrdU, and when BrdU‐positive cells undergo mitosis and become positive for pH3. The resulting curve represents the percentage of BrdU‐labeled cells that are colabeled with pH3 at each of the time points sampled. In control embryos, the percentage of double‐labeled nuclei increases sigmoidally over time [Fig. 5(C)] (see Luo et al., 2012). For control groups, the curves plateau at 100% at about 2.5 h, thereby defining the duration of the G2‐phase. In contrast, in Pgrn‐a morphants, the curve is shifted to the right. There were significantly fewer double‐labeled cells at 60, 90, 120, 150, 300, and 330 min, indicating a G2‐phase of about 3 h. Additionally, whereas the control curves begin to return to zero as BrdU‐positive cells exit the M‐phase and migrate away from the apical surface of the retina (Baye and Link, 2007), in morphant retinas the plateau at 100% persists, indicating that morphant progenitors are delayed in M. These cells eventually exit the M‐phase and re‐enter the cell cycle, because the number of progenitors does accumulate over time [cf. Figs. 3(P–R) and 5(A)]. Together, these data show that knockdown of Pgrn‐a results in a slower G2‐phase and a prolongation of the M‐phase.
Third, a pulse chase pulse paradigm was used to determine the relative length of S‐phase (T S) and estimate the total cell cycle length (T C) (Rachel et al., 2002). This analysis showed the duration of the S‐phase (T S) is 6 h and is invariant in morphants and controls [Fig. 5(D)]. The total length of the cell cycle (T C) in uninjected controls is approximately 10 h [Fig. 5(E)], which recapitulates previous findings in WT embryos (see Li et al., 2000). However, the TC in morphant retinas is approximately 14 h, which is significantly greater than controls [Fig. 5(E)]. These and the aforementioned PLM data reveal that the G1‐phase is also longer in morphants (∼5 h) than in controls (∼1.5 h).
To gain insight into the proximal molecular mechanisms that underlie control of the cell cycle by Pgrn‐a, qRTPCR was performed at 30 and 72 hpf to measure the expression levels of genes that promote cell cycle exit, for example, p27kip and p57kip2 and genes that promote cell cycle progression, for example, cyclin B, D1, and E. Compared to controls, at both 30 and 72 hpf the expression level of p27kip is significantly lower in morphants [Fig. 5(F)], whereas at 72 hpf the expression levels of cyclinB and cyclinD are significantly higher [Fig. 5(G)].
DISCUSSION
In this study, we evaluated the function of the microglia‐specific growth factor, Pgrn‐a. Knockdown of Pgrn‐a significantly altered neurogenesis; retinal progenitors do not exit the cell cycle at the appropriate developmental time, and neuronal differentiation and early retinal and ocular growth is delayed. Further, knockdown of Pgrn‐a results in a significant lengthening the cell cycle. These data serve as the mechanistic explanation for the delay in retinal development, and demonstrate that Pgrn‐a plays a fundamental role in governing early neurogenic events by regulating the brain's intrinsic timing of the cell cycle.
Progranulin is a soluble growth factor that has multiple functions in developing and adult vertebrates. In vitro, PGRN promotes migration and invasiveness of transformed cells (Tangkeangsirisin and Serrero, 2004; Monami et al., 2006; Swamydas et al., 2011; Dong et al., 2016), stimulates proliferation and migration of endothelial and fibroblast cells (Bateman and Bennett, 2009), and acts as a chemoattractant to microglia (Pickford et al., 2011). In vivo, application of PGRN to a cutaneous wound results in an increase in macrophage, neutrophil, and blood vessel accumulation in the wound (He et al., 2003). In humans, macrophage infiltration into adipose tissues correlates with circulating PGRN, further suggesting PGRN acts as a macrophage chemotactic factor (Youn et al., 2009). Our data are the first to demonstrate Pgrn‐a regulates the migration of microglial precursors, originating in the rostral blood island (RBI) (Xu et al., 2015a, 2015b), into neuroepithelial tissues and, therefore, acts as a chemoattractant to microglia in vivo. We hypothesize that during early zebrafish embryogenesis, the developing forebrain serves as a source of Pgrn‐a, thereby establishing a morphogenic gradient between the brain and yolk sac. Microglial precursors migrate up the Pgrn‐a gradient. Consistent with our hypothesis, depleting Pgrn‐a significantly decreases the number of microglia that invade the retina. In the mRNA rescue experiments, exogenous pgrn‐a (or pgrn‐b) is ubiquitously expressed in every cell. While this restores the function of Pgrn‐a during retinal neurogenesis, there is no discrete source of Pgrn‐a protein that is sufficient to recreate a gradient and, thereby, fully rescue the migration of microglia into the retina. Together, these data suggest that, similar to in vitro results (Pickford et al., 2011), in vivo, Pgrn‐a acts as a chemoattractant and directs microglial migration into neuroepthelial tissues.
Progranulin was first identified as a novel autocrine epithelial cell growth factor in peripheral tissues (Review by Bateman and Bennett, 1998). Subsequent studies have shown that in humans Progranulin is involved in both neurodegeneration and glioblastoma (De Muynck and Van Damme, 2011). PGRN was first identified as a mitogen in the tumorigenic PC cell line (Zhou et al., 1993). Later studies established that the level of PGRN expression is directly proportional to proliferation rate of cells in vitro and tumorigenicity in vivo, such that increased PGRN levels accelerates cell division (He and Bateman, 1999), and decreased PGRN levels attenuates tumor growth (Zhang and Serrero, 1998). Acting as both a competence and progression factor, PGRN promotes mitosis in embryonic and adult epithelial cells by stimulating classic growth factor signal transduction cascades, such as mitogen‐activated protein kinase (MAPK)/extracellular‐signal‐related kinase (ERK) and phosphatidylinositol 3‐kinase (PI3K) pathways (Reviewed by Ong and Bateman, 2003; Bateman and Bennett, 2009). Modulation of PGRN levels within the CNS appears similarly to affect cell cycle activity. Our data demonstrates that among retinal progenitors in vivo Pgrn‐a promotes progression through the cell cycle. Pgrn‐a knockdown results in a significant lengthening of the cell cycle. Further, when Pgrn‐a is reduced, the number of retinal progenitors exiting the cell cycle and undergoing terminal differentiation is significantly decreased. In Pgrn‐a morphants, mitotically active cells take longer to transit G1 and G2 and undergo mitosis. Overall, these data indicate that in retinal progenitors Pgrn‐a governs the duration of the G1‐, G2‐, and M‐phases of the cell cycle. Therefore, Pgrn‐a normally functions to promote cell cycle progression and neuronal differentiation in the developing retina. This illustrates that Pgrn‐a functions similarly in peripheral tissues and in the CNS by driving cells through the cell cycle. The cell cycle data were analyzed at a stage when all neural progenitors express pgrn‐a, and suggest that among these cells Pgrn‐a may function as either an autocrine or paracrine growth factor. In contrast, starting at 36–48 hpf, when Pgrn‐a expression can be ascribed solely to microglia, the changes in cell cycle kinetics likely reflect the consequence of the absence of Pgrn‐a in microglia.
We found that Pgrn‐a also governs migration of microglial precursors into the embryonic brain. Although the ontogeny of microglia was debated for many years (Reviewed in Cuadros and Navascués, 2001; Ginhoux and Prinz, 2015), it is now established in fish (Herbomel, 2001), birds (Cuadros et al., 1993), and rodents (Ginhoux et al., 2010) that embryonic microglia originate as primitive yolk sac macrophages. In zebrafish, macrophages/microglial precursors migrate from the embryonic RBI (Xu et al., 2015a, 2015b) into head mesenchyme between 22 and 40 hpf, colonize neuroepithelial tissues, including the retina, between 30 and 48 hpf, then undergo a phenotypic transformation around 60 hpf to adopt their final microglial state (Herbomel et al., 1999; Herbomel, 2001). The migration of macrophages/microglial precursors into the developing CNS is also highly conserved across vertebrate species (fish: Herbomel, 2001; rodents: Swinnen et al., 2013; humans: Verney et al., 2010). Although the mechanisms that regulate microglial migration and maturation are not completely understood, these processes depend on multiple factors and are critical for normal CNS development and function (Reviewed in Nayak et al., 2014; Prinz and Priller, 2014). In vertebrates, microglia govern early neurogenic events. When microglial precursors fail to colonize the CNS, developmental neurogenesis is aberrant. Mice that completely lack CNS microglia have multiple developmental brain abnormalities, including small brains, increased neuronal density, and decreased cortical thickness (Ginhoux et al., 2010; Erblich et al., 2011; Nandi et al., 2012). These animals do not live beyond one month of age (Dai et al., 2004).
There are numerous factors that regulate microglial migration in the developing zebrafish. Cell autonomous factors, such as the Leucine/Arginine transporter, slc7a7 (Rossi et al., 2015b), phosphate exporter XPR1 orthologue, xpr1b (Meireles et al., 2014), and noncanonical NOD‐like receptor, nlrc3‐like (Shiau et al., 2013), are required for primitive macrophage migration to and colonization of embryonic neuroepithelial tissues. Zebrafish slc7a7, xpr1b, and nlrc3‐like mutants all lack microglia (Shiau et al., 2013; Meireles et al., 2014; Rossi et al., 2015a,b), although the embryonic retinal and brain phenotypes among these mutants remain to be determined. Non‐cell autonomous factors produced by neuroepithelial tissues, such as cell death signals and chemokines, also promote macrophage chemotaxis during development of the brain and retina (Reviewed in Polazzi and Contestabile, 2002; Schwarz and Bilbo, 2014). Two recent studies demonstrate that in zebrafish apoptotic neurons and the molecules they release mediate the entry of microglial precursors into the optic tectum and their in situ proliferation (Xu et al., 2015a; Casano et al., 2016). These studies show that the tectal apoptosis that guides microglial colonization begins around 48 hpf and peaks at 72 hpf. Interestingly, this developmental cell death follows the expression of Pgrna in neural progenitors. This suggests a complex of signaling cascades (see above), where Pgrna provides signals that elicit the initial migration of microglia precursors into the brain, and neuronal cell death then sustains microglia colonization and mediates their spatial position and initial number.
Members of the zebrafish community have recently expressed concerns regarding the use of MOs as experimental tools (Schulte‐Merker and Stainier, 2014), and some have declared data based on MOs untrustworthy (Kok et al., 2015). Nonetheless, we are confident in the data presented here that are based on MO‐induced knockdown of Pgrn‐a. First, we used two independent MOs that targeted different regions of pgrn‐a, and each MO produced essentially identical results. Second, we confirmed specific knockdown of Pgrn‐a protein by Western blot analysis. Third, we used low, minimum doses of MOs: 0.25 ng and 1 ng for the 5′UTR and SS MO, respectively. Fourth, we coinjected each experimental MO with a p53‐targeting MO to suppress p53 activity and minimize off target/nonspecific effects, including ectopic cell death. Fifth, we characterized the retinal phenotype in morphants at 72 hpf, a time at which the MO‐induced knockdown and the concentration of MOs in individual cells is nearing its effective limit (unpublished observations; Bill et al., 2009). Finally, coinjecting Pgrn mRNA uniformly rescued the morphant phenotype, which validates the specificity of the MOs and the relationship between Pgrn‐a knockdown and the retinal phenotype. Together, these observations support the data presented here and suggest that, when used at limiting doses and rigorously controlled, MOs have a place in the repertoire of zebrafish biologists.
Finally, zebrafish mutant for pgrn‐a and pgrn‐b were recently generated using zinc finger nuclease genome editing technology (Solchenberger et al., 2015). Neither single or double pgrn mutants recapitulated the spinal motor neuron axonopathy observed (Laird et al., 2010) or the diminished proliferation among myogenic progenitor cells reported (Li et al., 2013) following MO‐induced knockdown of Pgrn‐a. (Retinal development was not evaluated in the pgrn‐a mutants.) The absence of concordance between knockdown and mutant phenotypes may be explained by the recent observation that targeted genetic mutations in zebrafish can result in genetic compensation by signaling molecules in related pathways that selectively mask loss‐of‐function mutations (Rossi et al., 2015a). Although the field awaits a fuller description of the pgrn mutants, the morphant data presented here can safely be interpreted to show that Pgrn‐a protein plays a fundamental role in governing developmental neurogenesis by regulating cell cycle kinetics and microglial precursor migration. Further assessment of the function of Pgrn‐a in governing microglia behavior during retinal development and regeneration will require transgenic report lines that faithfully label both microglia precursors and mature microglia.
The authors would like to thank Dilip Pawar and Laura Kakuk‐Atkins for technical assistance, and Ryan Thummel, Pamela Raymond, and Hitchcock lab members for feedback on manuscript drafts. The authors declare that they have no conflict of interest.
Supporting information
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Figure 3.
Supporting Information Table 1.
Supporting Information Table 2.
Supporting Information Table 3.
Supporting Information Table 4.
Supporting Information Table 5.
REFERENCES
- Agathocleous M, Harris WA. 2009. From progenitors to differentiated cells in the vertebrate retina. Annu Rev Cell Dev Biol 25:45–69. [DOI] [PubMed] [Google Scholar]
- Asakura R, Matsuwaki T, Shim J‐H, Yamanouchi K, Nishihara M. 2011. Involvement of progranulin in the enhancement of hippocampal neurogenesis by voluntary exercise. Neuroreport 22:881–886. [DOI] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering‐Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, et al. 2006. Mutations in progranulin cause tau‐negative frontotemporal dementia linked to chromosome 17. Nature 442:916–919. [DOI] [PubMed] [Google Scholar]
- Bateman A, Bennett HP. 1998. Granulins: The structure and function of an emerging family of growth factors. J Endocrinol 158:145–151. [DOI] [PubMed] [Google Scholar]
- Bateman A, Bennett HPJ. 2009. The granulin gene family: From cancer to dementia. Bioessays 31:1245–1254. [DOI] [PubMed] [Google Scholar]
- Baye LM, Link BA. 2007. Interkinetic nuclear migration and the selection of neurogenic cell divisions during vertebrate retinogenesis. J Neurosci 27:10143–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill BR, Petzold AM, Clark KJ, Schimmenti LA, Ekker SC. 2009. A primer for morpholino use in zebrafish. Zebrafish 6:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieux B, Chitramuthu BP, Baranowski D, Bennett HP. 2005. The zebrafish progranulin gene family and antisense transcripts. BMC Genomics 6:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casano AM, Alberb M, Peri F. 2016. Develpmental apoptosis mediates entry and positioning of microglia in the zebrafish brain. Cell Rep 16:897–906. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. 1996. Cell fate determination in the vertebrate retina. Proc Natl Acad SciUSA 93:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig SEL, Calinescu A‐A, Hitchcock PF. 2008. Identification of the molecular signatures integral to regenerating photoreceptors in the retina of the zebra fish. J Ocul Biol Dis Infor 1:73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, et al. 2006. Null mutations in progranulin cause ubiquitin‐positive frontotemporal dementia linked to chromosome 17q21. Nature 442:920–924. [DOI] [PubMed] [Google Scholar]
- Cuadros MA, Martin C, Coltey P, Almendros A, Navascués J. 1993. First appearance, distribution, and origin of macrophages in the early development of the avian central nervous system. J Comp Neurol 330:113–129. [DOI] [PubMed] [Google Scholar]
- Cuadros MA, Navascués J. 2001. Early origin and colonization of the developing central nervous system by microglial precursors. Prog Brain Res 132:51–59. [DOI] [PubMed] [Google Scholar]
- Dai X‐M, Zong X‐H, Akhter MP, Stanley ER. 2004. Osteoclast deficiency results in disorganized matrix, reduced mineralization, and abnormal osteoblast behavior in developing bone. J Bone Miner Res 19:1441–1451. [DOI] [PubMed] [Google Scholar]
- Daniel R, He Z, Carmichael KP, Halper J, Bateman A. 2000. Cellular localization of gene expression for progranulin. J Histochem Cytochem 48:999–1009. [DOI] [PubMed] [Google Scholar]
- Das T, Payer B, Cayouette M, Harris WA. 2003. In vivo time‐lapse imaging of cell divisions during neurogenesis in the developing zebrafish retina. Neuron 37:597–609. [DOI] [PubMed] [Google Scholar]
- De Muynck L, Van Damme P. 2011. Cellular effects of progranulin in health and disease. J Mol Neurosci 45:549–560. [DOI] [PubMed] [Google Scholar]
- Dong T, Yang D, Li R, Zhang L, Zhao H, Shen Y, Zhang X, Kong B, Wang L. 2016. PGRN promotes migration and invasion of epithelial ovarian cancer cells through an epithelial mesenchymal transition program and the activation of cancer associated fibroblasts. Exp Mol Pathol 100:17–25. [DOI] [PubMed] [Google Scholar]
- Easter SS, Nicola GN. 1996. The development of vision in the zebrafish (Danio rerio). Dev Biol 180:646–663. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. 2008. Controlling morpholino experiments: Don't stop making antisense. Development 135:1735–1743. [DOI] [PubMed] [Google Scholar]
- Erblich B, Zhu L, Etgen AM, Dobrenis K, Pollard JW. 2011. Absence of colony stimulation factor‐1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One 6:e26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Joselin AP, Wang L, Kar A, Ray P, Bateman A, Goate AM, et al. 2010. Progranulin promotes neurite outgrowth and neuronal differentiation by regulating GSK‐3β. Protein Cell 1:552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, et al. 2010. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330:841–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, Prinz M. 2015. Origin of microglia: Current concepts and past controversies. Cold Spring Harb Perspect Biol 7:a020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramage E, D'Cruz T, Taylor S, Thummel R, Hitchcock PF. 2015. Midkine‐a protein localization in the developing and adult retina of the zebrafish and its function during photoreceptor regeneration. PLoS One 10:e0121789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Bateman A. 1999. Progranulin gene expression regulates epithelial cell growth and promotes tumor growth in vivo. Cancer Res 59:3222–3229. [PubMed] [Google Scholar]
- He Z, Bateman A. 2003. Progranulin (granulin‐epithelin precursor, PC‐cell‐derived growth factor, acrogranin) mediates tissue repair and tumorigenesis. J Mol Med 81:600–612. [DOI] [PubMed] [Google Scholar]
- He Z, Ismail A, Kriazhev L, Sadvakassova G, Bateman A. 2002. Progranulin (PC‐cell‐derived growth factor/acrogranin) regulates invasion and cell survival. Cancer Res 62:5590–5596. [PubMed] [Google Scholar]
- He Z, Ong CHP, Halper J, Bateman A. 2003. Progranulin is a mediator of the wound response. Nat Med 9:225–229. [DOI] [PubMed] [Google Scholar]
- Herbomel P. 2001. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M‐CSF receptor‐dependent invasive process. Dev Biol 238:274–288. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Levraud J‐P. 2005. Imaging early macrophage differentiation, migration, and behaviors in live zebrafish embryos. Methods Mol Med 105:199–214. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C. 1999. Ontogeny and behaviour of early macrophages in the zebrafish embryo. Development 126:3735–3745. [DOI] [PubMed] [Google Scholar]
- Hitchcock P, Kakuk‐Atkins L. 2004. The basic helix‐loop‐helix transcription factor neuroD is expressed in the rod lineage of the teleost retina. J Comp Neurol 477:108–117. [DOI] [PubMed] [Google Scholar]
- Hitchcock P, Ochocinska M, Sieh A, Otteson D. 2004. Persistent and injury‐induced neurogenesis in the vertebrate retina. Prog Retin Eye Res 23:183–194. [DOI] [PubMed] [Google Scholar]
- Hitchcock PF, Raymond PA. 2004. The teleost retina as a model for developmental and regeneration biology. Zebrafish 1:257–271. [DOI] [PubMed] [Google Scholar]
- Hu M, Easter SS. 1999. Retinal neurogenesis: The formation of the initial central patch of postmitotic cells. Dev Biol 207:309–321. [DOI] [PubMed] [Google Scholar]
- Kay JN. 2005. Staggered cell‐intrinsic timing of ath5 expression underlies the wave of ganglion cell neurogenesis in the zebrafish retina. Development 132:2573–2585. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni C‐W, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, et al. 2015. Reverse genetic screening revealspoor correlation between morpholino‐induced and mutant phenotypes in zebrafish. Dev Cell 32:97–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird AS, Van Hoecke A, De Muynck L, Timmers M, Van Den Bosch L, Van Damme P, Robberecht W. 2010. Progranulin is neurotrophic in vivo and protects against a mutant TDP‐43 induced axonopathy. PLoS One 5:e13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Chen MHC, Gong HY, Hu SY, Li YW, Lin GH, Lin CC, et al. 2010. Progranulin A‐mediated MET signaling is essential for liver morphogenesis in zebrafish. J Biol Chem 285:41001–41009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YH, Chen HY, Li YW, Wu SY, Wangta‐Liu, Lin GH, Hu SY, et al. 2013. Progranulin regulates zebrafish muscle growth and regeneration through maintaining the pool of myogenic progenitor cells. Sci Rep 3:1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hu M, Ochocinska MJ, Joseph NM, Easter SS. 2000. Modulation of cell proliferation in the embryonic retina of zebrafish (Danio rerio). Dev Dyn 219:391–401. [DOI] [PubMed] [Google Scholar]
- Liau LM, Lallone RL, Seitz RS, Buznikov A, Gregg JP, Kornblum HI, Nelson SF, et al. 2000. Identification of a human glioma‐associated growth factor gene, granulin, using differential immuno‐absorption. Cancer Res 60:1353–1360. [PubMed] [Google Scholar]
- Luo J, Uribe RA, Hayton S, Calinescu A‐A, Gross JM, Hitchcock PF. 2012. Midkine‐A functions upstream of Id2a to regulate cell cycle kinetics in the developing vertebrate retina. Neural Dev 7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masai I, Stemple DL, Okamoto H, Wilson SW. 2000. Midline signals regulate retinal neurogenesis in zebrafish. Neuron 27:251–263. [DOI] [PubMed] [Google Scholar]
- Meireles AM, Shiau CE, Guenther CA, Sidik H, Kingsley DM, Talbot WS. 2014. The phosphate exporter xpr1b is required for differentiation of tissue‐resident macrophages. Cell Rep 8:1659–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moisse K, Volkening K, Leystra‐Lantz C, Welch I, Hill T, Strong MJ. 2009. Divergent patterns of cytosolic TDP‐43 and neuronal progranulin expression following axotomy: Implications for TDP‐43 in the physiological response to neuronal injury. Brain Res 1249:202–211. [DOI] [PubMed] [Google Scholar]
- Monami G, Gonzalez EM, Hellman M, Gomella LG, Baffa R, Iozzo RV, Morrione A. 2006. Proepithelin promotes migration and invasion of 5637 bladder cancer cells through the activation of ERK1/2 and the formation of a paxillin/FAK/ERK complex. Cancer Res 66:7103–7110. [DOI] [PubMed] [Google Scholar]
- Nandi S, Gokhan S, Dai X‐M, Wei S, Enikolopov G, Lin H, Mehler MF, Stanley ER. 2012. The CSF‐1 receptor ligands IL‐34 and CSF‐1 exhibit distinct developmental brain expression patterns and regulate neural progenitor cell maintenance and maturation. Dev Biol 367:100–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naphade SB, Kigerl KA, Jakeman LB, Kostyk SK, Popovich PG, Kuret J. 2009. Progranulin expression is upregulated after spinal contusion in mice. Acta Neuropathol 119:123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. 2000. Effective targeted gene “knockdown” in zebrafish. Nat Genet 26:216–220. [DOI] [PubMed] [Google Scholar]
- Nayak D, Roth TL, McGavern DB. 2014. Microglia development and function. Annu Rev Immunol 32:367–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedachi T, Kawai T, Matsuwaki T, Yamanouchi K, Nishihara M. 2011. Progranulin enhances neural progenitor cell proliferation through glycogen synthase kinase 3β phosphorylation. Neuroscience 185:106–115. [DOI] [PubMed] [Google Scholar]
- Ochocinska MJ, Hitchcock PF. 2007. Dynamic expression of the basic helix‐loop‐helix transcription factor neuroD in the rod and cone photoreceptor lineages in the retina of the embryonic and larval zebrafish. J Comp Neurol 501:1–12. [DOI] [PubMed] [Google Scholar]
- Ong CH, Bateman A. 2003. Progranulin (Granulin‐epithelin precursor, PC‐cell derived growth factor, Acrogranin) in proliferation and tumorigenesis. Histol Histopathol 18:1275–1288. [DOI] [PubMed] [Google Scholar]
- Otteson DC, D'Costa AR, Hitchcock PF. 2001. Putative stem cells and the lineage of rod photoreceptors in the mature retina of the goldfish. Dev Biol 232:62–76. [DOI] [PubMed] [Google Scholar]
- Philips T, De Muynck L, Thu HNT, Weynants B, Vanacker P, Dhondt J, Sleegers K, et al. 2010. Microglial upregulation of progranulin as a marker of motor neuron degeneration. J Neuropathol Exp Neurol 69:1191–1200. [DOI] [PubMed] [Google Scholar]
- Pickford F, Marcus J, Camargo LM, Xiao Q, Graham D, Mo JR, Burkhardt M, et al. 2011. Progranulin is a chemoattractant for microglia and stimulates their endocytic activity. Am J Pathol 178:284–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polazzi E, Contestabile A. 2002. Reciprocal interactions between microglia and neurons: From survival to neuropathology. Rev Neurosci 13:221–242. [DOI] [PubMed] [Google Scholar]
- Prinz M, Priller J. 2014. Microglia and brain macrophagesin the molecular age: From origin toneuropsychiatric disease. Nature Publishing Group 15:300–312. [DOI] [PubMed] [Google Scholar]
- Quastler H, Sherman FG. 1959. Cell population kinetics in the intestinal epithelium of the mouse. Exp Cell Res 17:420–438. [DOI] [PubMed] [Google Scholar]
- Rachel RA, Dölen G, Hayes NL, Lu A, Erskine L, Nowakowski RS, Mason CA. 2002. Spatiotemporal features of early neuronogenesis differ in wild‐type and albino mouse retina. J Neurosci 22:4249–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. 2006. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol 6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. 2007. p53 activation by knockdown technologies. PLoS Genet 3:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DYR. 2015a. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524:230–233. [DOI] [PubMed] [Google Scholar]
- Rossi F, Casano AM, Henke K, Richter K, Peri F. 2015b. The SLC7A7 transporter identifies microglial precursors prior to entry into the brain. Cell Rep 11:1008–1017. [DOI] [PubMed] [Google Scholar]
- Ryan CL, Baranowski DC, Chitramuthu BP, Malik S, Li Z, Cao M, Minotti S, et al. 2009. Progranulin is expressed within motor neurons and promotes neuronal cell survival. BMC Neurosci 10:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. 1994. Early eye morphogenesis in the zebrafish, Brachydanio rerio. J Comp Neurol 344:532–542. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. 1996. Comparison of topographical patterns of ganglion and photoreceptor cell differentiation in the retina of the zebrafish, Danio rerio. J Comp Neurol 371:222–234. [DOI] [PubMed] [Google Scholar]
- Schmitt EA, Dowling JE. 1999. Early retinal development in the zebrafish, Danio rerio: Light and electron microscopic analyses. J Comp Neurol 404:515–536. [PubMed] [Google Scholar]
- Schulte‐Merker S, Stainier DYR. 2014. Out with the old, in with the new: Reassessing morpholino knockdowns in light of genome editing technology. Development 141:3103–3104. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Bilbo SD. 2014. Microglia and Neurodevelopment: Programming of Cognition throughout the Lifespan. Chapter 15. The Wiley‐Blackwell Handbook of Psychoneuroimunology. Kusnecov AW, Anisman H, Eds. John Wiley & Sons, Ltd.
- Shiau CE, Monk KR, Joo W, Talbot WS. 2013. An anti‐inflammatory NOD‐like receptor is required for microglia development. Cell Rep 5:1342–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solchenberger B, Russell C, Kremmer E, Haass C, Schmid B. 2015. Granulin knock out zebrafish lack frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis pathology. PLoS One 10:e0118956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamydas M, Nguyen D, Allen LD, Eddy J, Dréau D. 2011. Progranulin stimulated by LPA promotes the migration of aggressive breast cancer cells. Cell Commun Adhes 18:119–130. [DOI] [PubMed] [Google Scholar]
- Swinnen N, Smolders S, Avila A, Notelaers K. 2013. Complex invasion pattern of the cerebral cortex bymicroglial cells during development of the mouse embryo. Glia 61:150–163. [DOI] [PubMed] [Google Scholar]
- Tangkeangsirisin W, Serrero G. 2004. PC cell‐derived growth factor (PCDGF/GP88, progranulin) stimulates migration, invasiveness and VEGF expression in breast cancer cells. Carcinogenesis 25:1587–1592. [DOI] [PubMed] [Google Scholar]
- Taylor SM, Alvarez‐Delfin K, Saade CJ, Thomas JL, Thummel R, Fadool JM, Hitchcock PF. 2015. The bHLH transcription factor NeuroD governs photoreceptor genesis and regeneration through delta‐notch signaling. Invest Ophthalmol Vis Sci 56:7496–7515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Van Hoecke A, Lambrechts D, Vanacker P, Bogaert E, van Swieten J, Carmeliet P, et al. 2008. Progranulin functions as a neurotrophic factor to regulate neurite outgrowth and enhance neuronal survival. J Cell Biol 181:37–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verney C, Monier A, Fallet‐Bianco C, Gressens P. 2010. Early microglial colonization of the human forebrain and possible involvement in periventricular white‐matter injury of preterm infants. J Anat 217:436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Li G, Yin J, Lin T, Zhang J. 2011. Progranulin overexpression predicts overall survival in patients with glioblastoma. Med Oncol 29:2423–2431. [DOI] [PubMed] [Google Scholar]
- Xu J, Wang T, Wu Y, Jin W, Wen Z. 2015a. Microglial colonization of developing zebrafish midbrain is promoted by apoptotic neurons and lysophosphatidylcholine. Dev Cell 38:214–222. [DOI] [PubMed] [Google Scholar]
- Xu J, Zhu L, He S, Wu Y, Jin W, Yu T, Qu JY, et al. 2015b. Temporal‐spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in zebrafish. Dev Cell 34:632–641. [DOI] [PubMed] [Google Scholar]
- Youn BS, Bang SI, Kloting N, Park JW, Lee N, Oh JE, Pi KB, et al. 2009. Serum progranulin concentrations may be associated with macrophage infiltration into omental adipose tissue. Diabetes 58:627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Serrero G. 1998. Inhibition of tumorigenicity of the teratoma PC cell line by transfection with antisense cDNA for PC cell‐derived growth factor (PCDGF, epithelin/granulin precursor). Proc Natl Acad Sci USA 95:14202–14207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Gao G, Crabb JW, Serrero G. 1993. Purification of an autocrine growth factor homologous with mouse epithelin precursor from a highly tumorigenic cell line. J Biol Chem 268:10863–10869. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Figure 3.
Supporting Information Table 1.
Supporting Information Table 2.
Supporting Information Table 3.
Supporting Information Table 4.
Supporting Information Table 5.


